Abstract
Summary: EBImage provides general purpose functionality for reading, writing, processing and analysis of images. Furthermore, in the context of microscopy-based cellular assays, EBImage offers tools to segment cells and extract quantitative cellular descriptors. This allows the automation of such tasks using the R programming language and use of existing tools in the R environment for signal processing, statistical modeling, machine learning and data visualization.
Availability: EBImage is free and open source, released under the LGPL license and available from the Bioconductor project (http://www.bioconductor.org/packages/release/bioc/html/EBImage.html).
Contact: gregoire.pau@ebi.ac.uk
1 INTRODUCTION
Imaging cells labeled with specific markers is a powerful method to localize cellular structures and proteins, and to characterize cell morphological changes during population progression or induced by perturbing agents. Automated phenotyping from such images generates quantitative descriptors of cellular phenotypes, which are computationally analysed to infer biological roles or functional relationships. Recent examples include characterization of genes involved in cell division by analysing time-lapse image sequences of human cells (Neumann et al., 2006), estimation of drug effects based on phenotypic changes measured in human HeLa cells (Loo et al., 2007) and identification of genes involved in cell morphology in Drosophila using RNA interference (RNAi) (Kiger et al., 2003).
Cell segmentation and feature extraction are well-established steps, realized by dedicated software such as CellProfiler (Carpenter et al., 2006) or generic image processing platforms like Matlab, Labview or ImageJ. However, the analysis and interpretation of multi-parametric cellular descriptors is a more challenging task. It requires powerful statistical and machine learning methods and can be facilitated by the possibility of producing visualizations of intermediate results, by the automation of complex workflows such as cross-validation or parameter searches, and by easy access to biological metadata and genomic databases. These points motivate the use of Bioconductor (Gentleman et al., 2004), a software project based on the feature-rich R programming language, providing tools for the analysis and comprehension of genomic data.
Several R packages provide some level of functionality for processing and analysing images. Rimage offers diverse filtering functions but supports only the JPEG format and cannot save images. The package ripa is dedicated to the analysis of hyperspectral images but does not provide for image segmentation. biOps offers a wide range of image filters, but only supports the JPEG and TIFF formats and lacks a fast interactive display interface. The recent package RImageJ provides R bindings to ImageJ, but does not allow easy access to the image data by R.
EBImage is an image processing toolbox for R, which has been developed over the past 4 years (Sklyar and Huber, 2006). The current release 3.0 is a major redesign whose features include multi-dimensional image processing, a range of fast image processing functions, support of more than 80 image formats, fast interactive image display, seamless integration with R's native array data structures and coherence of the user interface.
2 DESCRIPTION
Images are represented in EBImage as multi-dimensional arrays containing pixel intensity values. The two first dimensions are typically meant to be spatial, while the other ones are unspecified and can contain, e.g. colour channels, z-slices, replicates, time points or combinations of different conditions. Image representation is dissociated from rendering, and multi-dimensional arrays can be displayed as animated sequences of images in greyscale or colour mode. The interactive display interface is powered by GTK+ and supports animation, zoom and pan.
As matrices, images can be manipulated in R with algebraic operators such as sum, product, comparison or convolution. These elementary operators allow a broad range of image transformations. For example, if we denote by x an image, α+βxγ is an enhanced image where the parameter α controls the brightness, β the contrast and γ the γ-factor of the transformed image. Another example includes adaptive thresholding, performed by x>x⋆m+μ, where ⋆ is the fast convolution product, m a neighbourhood mask and μ an offset parameter. R also offers statistical tools to model images with natural 2D splines and provides Fourier analysis tools to detect regular patterns and deconvolute noisy images using Wiener filters.
EBImage uses ImageMagick to read and save images, and supports more than 80 image formats, including JPEG, TIFF, TGA, GIF and PNG. The package also supports standard geometric transformations such as rotation, reflection, cropping, translation and resizing. Classical image processing tools are available: linear filtering, morphological erosion and dilation, fast distance map computation, contour delineation and area filling.
Object segmentation can be performed with global or adaptive thresholding followed by connected set labeling. Specific algorithms such as watershed transform or Voronoi segmentation (Jones et al., 2005) are provided to segment touching objects. Computation of geometric and texture features (image moments, Haralick features, Zernike moments) from segmented objects is supported.
3 ANALYSIS OF CELLULAR PHENOTYPES
RNAi is a powerful method to study the role of genes in loss-of-function phenotypes. We measured the effects of two RNAi reagents on human HeLa cells by fluorescence microscopy. One cell population was transfected by a negative control, siRluc, a small interfering RNA (siRNA) targeting the Renilla firefly luciferase gene that is not present in the HeLa genome. The other population was treated with siCLSPN, an siRNA targeting the CLSPN mRNA, whose protein is involved in DNA damage response mediation. Cells were grown for 48 h, stained with immunofluorescent markers and imaged (Fig. 1).
Fig. 1.
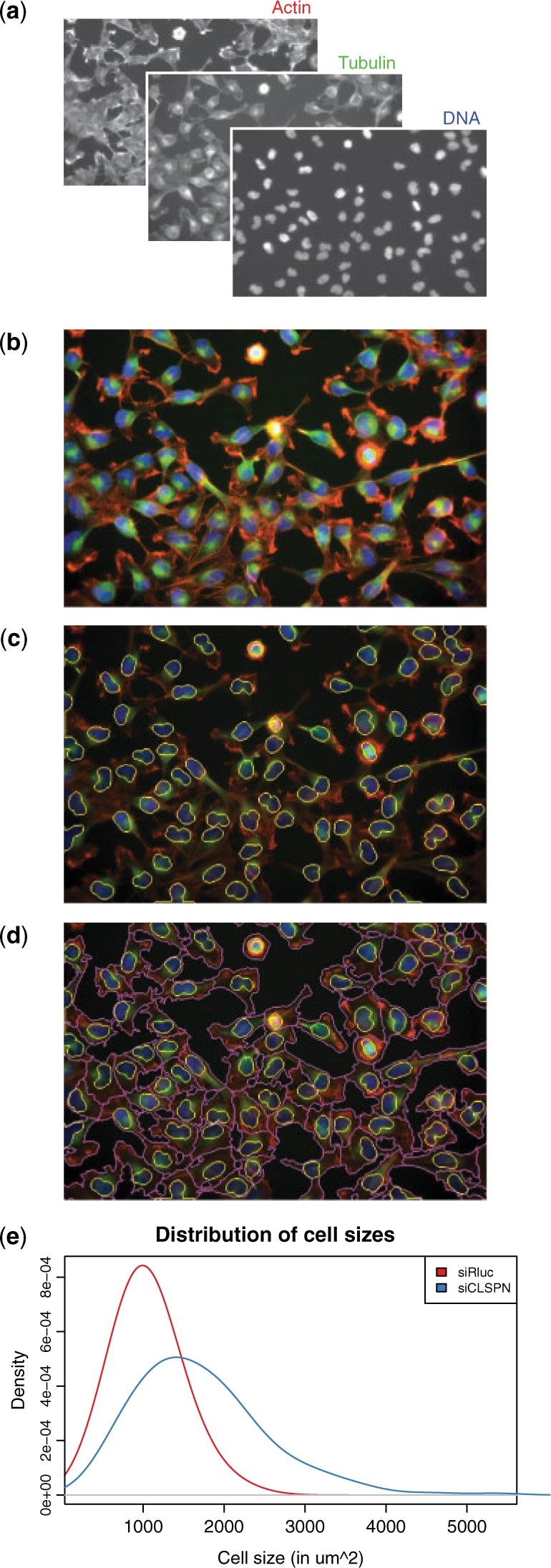
From microscope images to cellular phenotypes. (a) Fluorescent microscopy images from three channels of the same population of HeLa cells perturbed by siRluc. (b) A false colour image combining the actin (red), the tubulin (green) and the DNA (blue) channels. (c) Nuclei boundaries (yellow) were segmented with adaptive thresholding followed by connected set labeling. (d) Cell membranes (magenta) were determined by Voronoi segmentation. (e) Distribution of the cell sizes compared to a population of HeLa cells perturbed by siCLSPN. Cells treated with siCLSPN were significantly enlarged compared to those perturbed with siRluc (Wilcoxon rank sum test, P<10−15).
For visualization, the three channels were combined (Fig. 1a) into a colour image (Fig. 1b). Nuclei were segmented by adaptive thresholding, morphological opening and connected set labeling (Fig. 1c). Cell boundaries were determined by Voronoi segmentation, using nuclei as seeds and propagating the boundaries using a Riemann metric based on the image gradient (Fig. 1d; Jones et al., 2005). Quantitative descriptors were extracted from the cell shapes and fluorescence distributions. Negative control cells treated with siRluc showed a median cell size of 1024 μm2, while targeting CLSPN led to a population of significantly enlarged cells with a median cell size of 1577 μm2 (Wilcoxon rank sum test, P<10−15) (Fig. 1e). The median Zernike (4,4) actin moment descriptor, capturing high-frequency radial structures, was also strongly discriminating between the two cell populations and can serve to characterize the actin stress fibers displayed by the siCLSPN perturbed cells.
4 CONCLUSION
Automated phenotyping of cells promises to be useful in many fields of biology. It relies on imaging, cell segmentation, feature extraction and statistical analysis. EBImage offers the essential functionality for performing these tasks. Future improvements are expected to include handling large multi-dimensional objects using NetCDF format, 3D objects (segmentation and reconstruction) and time-lapse image sequences (cell tracking and extraction of spatio-temporal features).
ACKNOWLEDGEMENTS
We thank Rémy Clément and Mike Smith for their helpful comments and proposals.
Funding: CancerPathways project (EU FP7, grant HEALTH-F2-2008-201666); Research Grant, Human Frontier Sciences Program.
Conflict of Interest: none declared.
REFERENCES
- Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, et al. Voronoi-based segmentation of cells on image manifolds. Computer Vision for Biomedical Image Applications. 2005;3765:535–543. doi: 10.1007/11569541_49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, et al. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LH, et al. Image-based multivariate profiling of drug responses from single cells. Nat. Methods. 2007;4:445–453. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- Neumann B, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Sklyar O, Huber W. Image analysis for microscopy screens. R News. 2006;6:12–16. [Google Scholar]


