Abstract
Summary: Metscape is a plug-in for Cytoscape, used to visualize and interpret metabolomic data in the context of human metabolic networks. We have developed a metabolite database by extracting and integrating information from several public sources. By querying this database, Metscape allows users to trace the connections between metabolites and genes, visualize compound networks and display compound structures as well as information for reactions, enzymes, genes and pathways. Applying the pathway filter, users can create subnetworks that consist of compounds and reactions from a given pathway. Metscape allows users to upload experimental data, and visualize and explore compound networks over time, or experimental conditions. Color and size of the nodes are used to visualize these dynamic changes. Metscape can display the entire metabolic network or any of the pathway-specific networks that exist in the database.
Availability: Metscape can be installed from within Cytoscape 2.6.x under ‘Network and Attribute I/O’ category. For more information, please visit http://metscape.ncibi.org/tryplugin.html
Contact: jinggao@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Metabolomics is a rapidly emerging field that is joining other high-throughput omics, such as proteomics and transcriptional profiling. It promises to be a powerful systems approach for studying metabolic profiles pertinent to a variety of normal and disease states. Transcriptional profiling and proteomics have established data analysis tools; a metabolomics analytical toolkit is yet to be developed. An important step in metabolomic data analysis is examining experimental data in the context of metabolic networks. A number of tools have been developed for the interactive exploration of metabolic pathways (Okuda et al., 2008; Paley and Karp, 2006). Several databases describing human metabolic networks have been published (KEGG, EHMN, HMDB).
Visual analysis has been shown to be effective in facilitating human cognition in exploratory analysis with bioinformatics tools (Mirel, 2009). Cytoscape is a highly popular Java-based open source software tool that is used by biomedical researchers to visualize and dynamically analyze gene/protein interaction networks (Shannon et al., 2003). Cytoscape is a powerful stand-alone tool, but its strength is even more apparent when it is coupled with databases. A number of plug-ins (MiMI plug-in, Pathway Commons, etc.) have been developed to interact with various databases.
Here we describe Metscape, a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks.
2 OVERVIEW
2.1 The human metabolomic database
A number of databases contain information about human metabolic networks consisting of compounds, chemical reactions, pathways, enzymes and genes. In Metscape, we used data from HUMDB (Beecher, 2003; C.Beecher, unpublished data), KEGG (Kanehisa et al., 2006) and EHMN (Ma et al., 2007) to trace the connections between metabolites and genes. We created a Microsoft SQL Server database to store these data. KEGG IDs are used as the primary compound identifier. Our database contains nearly 2700 compounds, 870 enzymes, 1400 genes and 3000 metabolic reactions involved in over 70 human-specific metabolic pathways defined in EHMN. We decided to use EHMN as a starting point because it appeared to be very well cross-referenced with the KEGG database. KEGG is widely known and many biologists and potential users of our tools use it as a reference. We fully intend to merge additional databases—e.g. we are currently in the process of merging in BIGG. We expect HumanCyc will be next.
Compounds, reactions, enzymes, genes, pathways and the relationships between them provide the initial framework for the analysis of metabolomic data.
2.2 Visualization of metabolomic network
Visualization of metabolomic networks poses a number of challenges. Typically, metabolic networks possess a high degree of connectivity (Bourqui et al., 2007; Wegner and Kummer, 2005). A number of different layout algorithms had been developed for Cytoscape that attempt to minimize the crossings between the edges, the distances between nodes and the bending of the edges. Users can easily apply Cytoscape layouts through the Cytoscape layout menu to the compound network generated by Metscape. To start-up Metscape, users select Metscape from the plug-ins menu in Cytoscape. The Metscape main window has three tabs that provide users with the following options: (i) load a list of compounds (compound names or KEGG IDs), (ii) load a file containing normalized experimental metabolite data with metabolite KEGG IDs and corresponding values at given time points or under specific experimental conditions and (iii) display pathway-specific networks by choosing a pathway from the drop-down list.
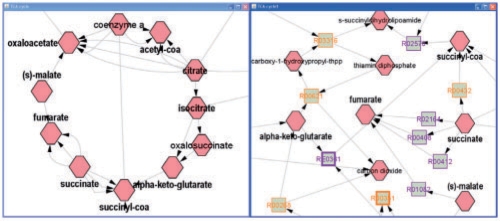
Metscape can generate two types of networks—a compound network, where compounds are nodes and reactions are edges, and a compound reaction network represented by a bipartite graph (Lacroix et al., 2008) (Fig. 1). The latter consists of two types of nodes, compounds (hexagons) and reactions (rectangles). In both network styles, edges are directional. A compound node with an outgoing edge is a substrate, while a compound with an incoming edge is the product of a specific reaction. In compound-centric networks, the reversible reactions are shown by bidirectional arrows, while in reaction-centric networks reversible (purple) and irreversible (maize) reactions can be distinguished by the node edge color.
Fig. 1.
Part of TCA (citric acid) cycle is shown as a compound network with compounds as nodes and reactions as edges (left) and as a compound reaction network with compounds (hexagons) and reactions (rectangles) as nodes (right).
Metscape provides two network styles for users to choose before creating a network and allows users to switch between two network views at any time after the network is created.
Metscape takes advantage of Cytoscape's VizMapper to create the Metscape visual style. User-inputted compounds are shown in blue, while the rest of the compound nodes are shown in pink. Metscape also uses a thicker node border to denote reactions with multiple enzymes. Detailed information can be found in the Metscape legend that is displayed at the beginning of each Metscape session.
When a user hovers the cursor over a reaction node, all compounds that participate in the reaction are highlighted. Metscape allows users to create a reaction-specific subnetwork from the highlighted compounds. Users can expand a compound network by adding reactions and compounds from the database; expanding the network will keep the current view that the original network is using. When done, users can collapse the expanded network or restore the original network.
2.3 Interpretation of metabolomic data
Once the network has been displayed in Metscape, users can import normalized experimental metabolite data from a tab-delimited file, where the first column always contains compound IDs. The detailed description of the format for experimental data file is contained in the online Metscape documentation. Node color and size can be used to represent two different parameters simultaneously, e.g. concentration and fold change. In order to visualize the experimentally measured changes in metabolite concentrations over time or under specific conditions, we adopted and modified plug-in—dynamicXpr (http://chianti.ucsd.edu/cyto_web/plugins/). Metscape can handle up to 80 data points (conditions or time points). Users can cycle through the data points sequentially. If the experimental values have wide range and/or the distribution of the data values is skewed, users can choose to represent the data on a log scale or rescale input data to a range from 0 to 10. This allows for better and more obvious visualized results.
Users can obtain detailed information about compounds and reactions by double clicking a node. For compounds, Metscape will display compound name, synonyms, formula, 2D structure, molecular weight, CAS number and smiles string. Metscape also provides links to MiMI (Tarcea et al., 2009), HMDB (Wishart et al., 2009), KEGG, PubChem, ChEBI and BioCyc. For reactions, Metscape shows the equation, pathway and reversibility. In addition, enzymes and genes involved in the reaction are shown.
Besides selecting a pathway from a drop-down list as described above, Metscape provides a pathway filter that can highlight pathway-specific reactions, which then can be used to create a pathway-specific subnetwork in a new Cytoscape window.
A Metscape tutorial is available from http://metscape.ncibi.org.
3 CONCLUSION
Metscape (See Supplementary material for the comparison of Metscape and other tools) provides a bioinformatics framework for the analysis and interpretation of metabolomic data. Metabolomic data from HUMDB, EHMN and KEGG are integrated and stored in a database at NCIBI. Metscape provides users an interface to query the database. Visualizing a compound network helps users to trace connections between metabolites and genes, and understand the relationships between compounds, reactions, enzymes, genes and pathways. Visualizing metabolites with node color and size changes in a network at a series of time points can help users to predict functions of metabolites. Linking compounds and reactions to well-known metabolomic data sources helps researchers to verify and better understand the presented metabolites.
Since March 2009 when the first version of Metscape was released, we have had >200 unique IPs (users/user groups), and the number is still growing. For future work, we plan to integrate more data from additional databases and provide more information like enzyme activators and inhibitors from databases such as BRENDA (Chang et al., 2009).
Supplementary Material
ACKNOWLEDGEMENTS
We thank David States, Carlo Laudanna, Allan Kuchinsky and John ‘Scooter’ Morris for helpful comments and support.
Funding: National Institutes of Health (U54 DA021519 to J.G., V.G.T., A.K., B.R.M., T.E.W., J.D.C., B.D.A., G.S.O., C.F.B., H.V.J. and R01DK079084 to B.D.A., C.W.B. and C.F.B.).
Conflict of Interest: none declared.
REFERENCES
- Beecher C. The Human Metabolome. In: Harrigan G, Goodacre R, editors. Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis. Boston: Kluwer Academic Publishers; 2003. pp. 311–319. [Google Scholar]
- Bourqui R, et al. Metabolic network visualization eliminating node redundance and preserving metabolic pathways. BMC Syst. Biol. 2007;1:29. doi: 10.1186/1752-0509-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, et al. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix V, et al. An introduction to metabolic networks and their structural analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2008;5:594–617. doi: 10.1109/TCBB.2008.79. [DOI] [PubMed] [Google Scholar]
- Ma H, et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirel B. Supporting cognition in systems biology analysis: findings on users' processes and design implications. J. Biomed. Discov. Collab. 2009;4:2. doi: 10.1186/1747-5333-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, et al. Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley SM, Karp PD. The Pathway Tools cellular overview diagram and Omics Viewer. Nucleic Acids Res. 2006;34:3771–3778. doi: 10.1093/nar/gkl334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarcea VG, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res. 2009;37:D642–D646. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner K, Kummer U. A new dynamical layout algorithm for complex biochemical reaction networks. BMC Bioinformatics. 2005;6:212. doi: 10.1186/1471-2105-6-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.