Abstract
OBJECTIVE
Genetic factors have been considered to contribute to the development and progression of diabetic nephropathy. The KCNQ1 gene (potassium voltage-gated channel, KQT-like subfamily, member 1) was originally identified as a strong susceptibility gene for type 2 diabetes in two Japanese genome-wide association studies. In this study, we examined the association of single nucleotide polymorphisms (SNPs) within KCNQ1 with diabetic nephropathy in Japanese subjects with type 2 diabetes.
RESEARCH DESIGN AND METHODS
We genotyped 33 SNPs in KCNQ1 using 754 type 2 diabetic patients with overt nephropathy and 558 control subjects (an initial study), and we further examined the association of a candidate SNP using three other independent Japanese populations (replications 1–3).
RESULTS
We found that five SNPs were nominally associated with diabetic nephropathy, and the association of rs2237897 was the strongest. We also found that the T allele frequencies of rs2237897 were consistently higher in the nephropathy groups than in the control groups for all study populations (initial study: 0.33 vs. 0.27; replication 1: 0.32 vs. 0.30; replication 2: 0.33 vs. 0.28; and replication 3: 0.32 vs. 0.28), although the individual associations did not reach statistically significant levels. Combined analysis by a meta-analysis revealed that the T allele of rs2237897 was significantly associated with susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes (odds ratio 1.22 [95% CI 1.10–1.34], P = 3.1 × 10–4, corrected P = 0.01).
CONCLUSIONS
These results suggest that KCNQ1 is a new candidate gene for conferring susceptibility to diabetic nephropathy.
Diabetic nephropathy is a serious microvascular complication of diabetes, and it is a leading cause of end-stage renal disease in western countries (1) and in Japan (2). Several genetic and environmental factors are likely to contribute to its development and progression (3,4), but the precise mechanism for this contribution is unknown.
The recent genome-wide association studies using Japanese populations led to the identification of the KCNQ1 (potassium voltage-gated channel, KQT-like subfamily, member 1) gene as a novel gene for susceptibility to type 2 diabetes (5,6). KCNQ1 encodes the pore-forming α subunit of the voltage-gated potassium channel expressed mainly in the heart (7). The mutations of KCNQ1 have been known to cause hereditary long QT syndromes (Romano-Ward syndrome [8] and Jervell and Lange-Nielsen syndrome [9]), which are characterized by the prolongation of QT intervals on the electrocardiogram; in some instances, these syndromes cause sudden cardiac death in the young (10) through the loss of function of the potassium channel in the heart. The expression of KCNQ1 could also be observed in the human kidney. In the kidney, KCNQ1 has been shown to assemble with KCNE1, the β subunit of the potassium channel, forming a potassium channel complex localized to the brush border of the mid to late proximal tubule (11,12); moreover, it has been shown to play a role in the Na+ secretion at the proximal tubule by maintaining a driving force for Na+ transport across the membrane (13). In previous studies, kcnq1 knockout mice were reported to show lower blood pressure (14). These observations suggest the possibility that KCNQ1 is a candidate for conferring susceptibility to diabetic nephropathy. To test this hypothesis, we focused on KCNQ1 as a candidate gene for diabetic nephropathy and investigated the association between the single nucleotide polymorphisms (SNPs) within KCNQ1 and diabetic nephropathy in Japanese subjects with type 2 diabetes.
RESEARCH DESIGN AND METHODS
DNA preparation and SNP genotyping
For the initial study, DNA samples were obtained from the peripheral blood of patients with type 2 diabetes who regularly visited the outpatient clinic at Shiga University of Medical Science, Tokyo Women's Medical University, Juntendo University, Kawasaki Medical School, Iwate Medical University, Toride Kyodo Hospital, Kawai Clinic, Osaka City General Hospital, Chiba Tokushukai Hospital, or Osaka Rosai Hospital. All subjects provided informed consent before enrolling in this study. Diabetes was diagnosed according to the World Health Organization criteria. Type 2 diabetes was clinically defined as a disease with gradual adult onset. Subjects who tested positive for anti-GAD antibodies and those with mitochondrial disease (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes) or maturity-onset diabetes of the young (MODY) were not included. DNA was extracted by a standard phenol-chloroform method. Diabetic patients were divided into two groups according to the following diagnostic criteria: 1) the nephropathy group (n = 754) comprised patients with diabetic retinopathy and overt nephropathy indicated by a urinary albumin excretion rate (AER) ≥200 μg/min or a urinary albumin-to-creatinine ratio (ACR) ≥300 mg/g creatinine; and 2) the control group (n = 558) comprised patients with diabetic retinopathy but with no evidence of renal dysfunction (i.e., AER <20 μg/min or ACR <30 mg/g creatinine). Measurements of AER or ACR were performed at least twice for each patient. The genotype of 33 SNPs within KCNQ1 was analyzed with multiplex PCR-Invader assays as described previously (15–17). The statistical power of the initial study was estimated to be 0.35 for SNPs with minor allele frequency of 0.3, if we set a cutoff value at the P = 0.0015 level (significant level after Bonferroni correction) and a genotypic relative risk (γ) of 1.2 and the prevalence of diabetic nephropathy is assumed to be 10%. All patients participating in this study provided written informed consent, and the study protocol was approved by the ethics committees of RIKEN Yokohama Institute and each participating institution.
Subjects in replication studies
Replication 1, BioBank Japan case-control study.
We selected diabetic nephropathy case and control samples from the subjects enrolled in BioBank Japan. Nephropathy was defined as patients with type 2 diabetes having both overt diabetic nephropathy and diabetic retinopathy (n = 449). The control subjects included patients with type 2 diabetes having diabetic retinopathy and normoalbuminuria (n = 965). All patients participating in this study provided written informed consent, and the study protocol was approved by the ethics committees of RIKEN Yokohama Institute.
Replication 2, Tokai University case-control study.
Patients with type 2 diabetes regularly visiting Tokai University Hospital or its affiliated hospitals were enrolled. All nephropathy patients were receiving chronic hemodialysis therapy, and control patients included those with normoalbuminuria determined by at least two measurements of the urinary ACR and with diabetes for >10 years (case subjects, n = 310; control subjects, n = 224). All patients provided written informed consent, and the protocol was approved by the ethics committees of Tokai University School of Medicine and RIKEN Yokohama Institute.
Replication 3, Shiga prospective study.
Patients with type 2 diabetes were recruited from among the participants of the Shiga Prospective Observational Follow-Up Study for Diabetic Complications (18). Patients classified as having microalbuminuria (200 μg/ml > AER ≥20 μg/min) on the basis of at least two measurements of AER in 24-h urine collections were followed for up to 6 years. The progressors (case subjects, n = 32) were defined as those who had progressed to overt proteinuria (AER ≥200 μg/min), and the remaining patients were defined as nonprogressors (control subjects, n = 168). The study protocol and informed consent procedure were approved by the ethics committees of Shiga University of Medical Science and RIKEN Yokohama Institute.
The clinical characteristics of subjects are shown in supplementary Table A1 (available in an online appendix at http://care.diabetesjournals.org/cgi/content/full/dc09-1933/DC1).
Statistical analyses
We tested the genotype and allele frequencies for Hardy-Weinberg equilibrium proportions by the χ2 test (19). We calculated the linkage-disequilibrium index, D′, and r2 as described elsewhere (20). We analyzed the differences between the case and control groups in terms of the distribution of genotypes scored with an additive model by using a logistic regression analysis with or without adjustment for age, sex, BMI, and duration of type 2 diabetes. A haplotype structure within the KCNQ1 locus was analyzed by using Haploview software, version 4.1 (21). Combined meta-analysis was performed by using the Mantel-Haenszel procedure with a fixed-effects model after testing for heterogeneity.
RESULTS
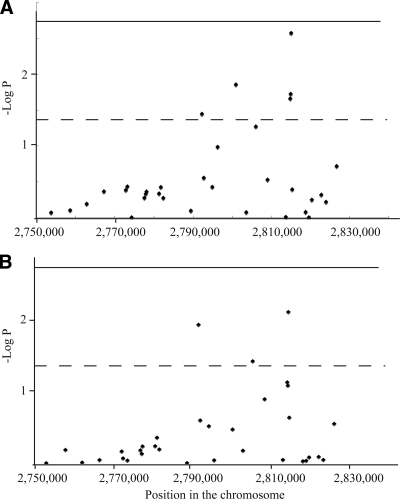
We first examined the association of 33 SNPs within the KCNQ1 susceptibility locus for type 2 diabetes with diabetic nephropathy in Japanese subjects with type 2 diabetes (initial study: case subjects, n = 754; control subjects, n = 558). The genotype distributions for all except two SNPs (rs234881 [P = 0.046] in case subjects and rs2283228 [P = 0.029] in control subjects) (supplementary Table A2, available in an online appendix) did not deviate from the Hardy-Weinberg equilibrium. We identified five SNPs that were significantly associated with diabetic nephropathy (nominal P < 0.05). Among them, the T allele of rs2237897 was the most strongly associated with susceptibility to the disease (unadjusted odds ratio [OR] 1.30 [95% CI 1.09–1.54], P = 0.0027; OR adjusted for age, sex, BMI, and duration of diabetes 1.33 [1.08–1.63], P = 0.008) (Table 1, Fig. 1, supplementary Table A3, available in an online appendix). We also found that age, sex (male), and duration of type 2 diabetes were independent risk factors for diabetic nephropathy in this analysis (data not shown). Subsequent analysis for the haplotype structure within this locus revealed that none of the haplotypes were more strongly associated with diabetic nephropathy than the single SNP alone (rs2237897) (supplementary Table A4, available in an online appendix).
Table 1.
SNPs nominally associated with diabetic nephropathy in the initial study
| SNP | Risk allele | Unadjusted |
Adjusted* |
||
|---|---|---|---|---|---|
| P | OR (95% CI) | P | OR (95% CI) | ||
| rs123381 | G | 0.036 | 1.24 (1.02–1.52) | 0.012 | 1.39 (1.08–1.79) |
| rs163183 | C | 0.014 | 1.78 (1.13–2.81) | 0.328 | 1.32 (0.76–2.11) |
| rs2299620 | T | 0.022 | 1.21 (1.03–1.42) | 0.074 | 1.20 (0.98–1.46) |
| rs2237896 | A | 0.019 | 1.22 (1.03–1.44) | 0.082 | 1.20 (0.98–1.47) |
| rs2237897 | T | 0.0027 | 1.30 (1.09–1.54) | 0.008 | 1.33 (1.08–1.63) |
*Adjusted for age, sex, BMI, and duration of type 2 diabetes.
Figure 1.
Association of the SNPs within KCNQ1 in the initial study. P values for each SNP, calculated by a logistic regression analysis, are plotted. A: Unadjusted data. B: Data adjusted for age, sex, BMI, and duration of type 2 diabetes. – – –, Nominally significant level (P = 0.05); ——, statistically significant level after Bonferroni adjustment (P = 0.0015).
We also examined the association of rs2237897 with hypertension. In this analysis, we divided subjects with type 2 diabetes into two groups: 1) hypertensive group, comprising subjects with systolic blood pressure ≥140 mmHg and/or diastolic blood pressure ≥90 mmHg or subjects taking antihypertensive agents and 2) the normotensive group, comprising subjects with systolic blood pressure <140 mmHg and diastolic blood pressure <90 mmHg without taking any antihypertensive agent. The result indicated that rs2237897 was not significantly associated with hypertension (OR 1.19 [95% CI 0.90–1.58], P = 0.23). rs2237897 was also not associated with plasma glucose or A1C in subjects with type 2 diabetes (data not shown).
The association of rs2237897 with diabetic nephropathy did not achieve statistically significant levels after Bonferroni correction, probably due to insufficient study power to detect a true association in the initial study (see research design and methods). Therefore, to validate the association of the T allele of rs2237897, we further examined the association of rs2237897 with diabetic nephropathy in three other independent studies (BioBank Japan case-control study, replication 1: case subjects, n = 449; control subjects, n = 965; Tokai University case-control study, replication 2: case subject, n = 310; control subjects, n = 224; Shiga prospective study, replication 3: progression case subjects, n = 32; control subjects, n = 168). The results revealed that the T allele frequencies of rs2237897 were consistently higher in the case groups than in the control groups (replication 1: 0.32 vs. 0.30; replication 2: 0.33 vs. 0.28; and replication 3: 0.32 vs. 0.28), although the individual associations did not reach statistically significant levels (Table 2). Combined analysis by a meta-analysis with the Mantel-Haenszel test revealed that the T allele of rs2237897 was significantly associated with susceptibility to diabetic nephropathy (proteinuria: OR 1.21 [95% CI 1.08–1.36], P = 1.4 × 10–3, corrected P = 0.046; proteinuria plus end-stage renal disease: 1.22 [1.10–1.34], P = 3.1 × 10–4, corrected P = 0.01) (Table 2).
Table 2.
Association of rs2237897 with diabetic nephropathy in four independent Japanese studies
| Population | Case |
Control |
P * | OR (95% CI)* | |||
|---|---|---|---|---|---|---|---|
| Phenotype | RAF | 11/12/22 | RAF | 11/12/22 | |||
| Initial study | Overt proteinuria | 0.33 | 331/336/81 | 0.27 | 294/210/48 | 0.0027 | 1.30 (1.09–1.54) |
| Replication 1 | Overt proteinuria | 0.32 | 209/188/50 | 0.30 | 473/404/82 | 0.156 | 1.13 (0.95–1.34) |
| Replication 3 | Overt proteinuria | 0.32 | 14/13/3 | 0.28 | 79/61/13 | 0.617 | 1.16 (0.64–2.11) |
| Meta-analysis† | 1.4 × 10−3 | 1.21 (1.08–1.36) | |||||
| Replication 2 | End-stage renal disease | 0.33 | 133/132/33 | 0.28 | 111/98/14 | 0.080 | 1.28 (0.97–1.68) |
| Meta-analysis (all studies)‡ | 3.08 × 10−4 | 1.22 (1.10–1.34) | |||||
| Meta-analysis (replication 1–3)§ | 0.03 | 1.16 (1.01–1.34) | |||||
*Unadjusted data.
†Meta-analysis was performed by Mantel-Haenszel test (heterogeneity P = 0.51).
‡Meta-analysis was performed by Mantel-Haenszel test (heterogeneity P = 0.70).
§Meta-analysis was performed by Mantel-Haenszel test (heterogeneity P = 0.79). RAF, risk allele frequency; 11, homozygous of major allele; 12, heterozygous; 22, homozygous of minor allele.
CONCLUSIONS
In the present study, we found that an SNP (rs2237897) within KCNQ1 was associated with susceptibility to diabetic nephropathy in a Japanese population. The same trend of the association of rs2237897 was consistently observed in three additional replication studies, and the association was found to be statistically significant by a meta-analysis (corrected P = 0.01).
KCNQ1, located in 11p15.5, encodes the α subunit of the voltage-gated potassium channel that is mainly expressed in the heart, inner ear, and to a lesser extent in the stomach, intestine, liver, and kidney (7). In the kidney, KCNQ1 assembles with KCNE1 to form a potassium channel complex that is localized to the brush border of the mid to late proximal tubule (11,12). This potassium channel has been shown to be responsible for maintaining a driving force for Na+ reabsorption by the repolarization of the membrane and to affect Na+ secretion at the proximal tubule (13). Therefore, KCNQ1 may be considered as a plausible candidate for conferring susceptibility to diabetic nephropathy.
Our present study revealed that the T allele of rs2237897 in KCNQ1 was the most strongly associated with susceptibility to diabetic nephropathy, and no other SNP or haplotype was more strongly associated with the disease than rs2237897 alone in our Japanese populations. Furthermore, in our previous genome-wide association study for type 2 diabetes, we demonstrated that rs2237897 was also the most strongly associated with type 2 diabetes (5); therefore, it is likely that rs2237897 could directly contribute to the susceptibility to diabetic nephropathy, although the mechanism underlying the contribution of this variation is unknown.
From cumulative evidence, it is well known that poor glycemic control and/or high blood pressure contribute to the progression of nephropathy in patients with diabetes. Because rs2237897 was originally shown to be associated with type 2 diabetes as well as with insulin secretion or fasting plasma glucose in subjects with normal glucose tolerance (22,23), the effects of rs2237897 on conferring susceptibility to diabetic nephropathy might be mediated by a difference in glycemic control according to the genotype. In our present study, however, rs2237897 was not associated with plasma glucose or A1C in subjects with type 2 diabetes, and the association between rs2237897 and diabetic nephropathy was not affected by adjusting A1C in the logistic regression analysis. Moreover, the T allele of rs2237897, a risk allele for diabetic nephropathy in the present study, was shown to be a nonrisk allele for type 2 diabetes, reduction of insulin secretion, or elevation of blood glucose level. Therefore, it is suggested that rs2237897 contributes to conferring susceptibility to diabetic nephropathy through a mechanism independent of glycemic control.
There may be a possibility that rs2237897 affects systemic blood pressure because KCNQ1 has been shown to play some roles in the regulation of renal Na+ reabsorption as described above. To evaluate this possibility, we also examined the association of rs2237897 with hypertension using 1,312 subjects with type 2 diabetes (initial study). The result indicated that rs2237897 was not significantly associated with hypertension (OR 1.19 [95% CI 0.90–1.58], P = 0.23). In addition, a nominal association of rs2237897 with diabetic nephropathy could also be observed by a logistic regression analysis using the presence of hypertension or systolic and/or diastolic blood pressures as covariables (data not shown). Therefore, it is likely that the effect of rs2237897 is also independent of systemic blood pressure.
In previous studies, subjects with a loss-of-function mutation of KCNQ1 and kcnq1 knockout mice did not exhibit glucose intolerance (14). In contrast, it was reported that an inhibitor of KCNQ1 (chromanol 293B) significantly increased insulin secretion in the presence of sulfonylurea (24); this finding suggested that the activity of KCNQ1 in subjects with the C allele of rs2237897, a risk allele for type 2 diabetes, might be higher than that in subjects with the T allele, a risk allele for diabetic nephropathy in the present study. However, further studies will be required to elucidate the precise molecular mechanism by which KCNQ1 and its polymorphism contribute to susceptibility to diabetic nephropathy.
In summary, we found that the T allele of rs2237897 within KCNQ1 was significantly associated with susceptibility to diabetic nephropathy in Japanese populations. The present data suggest that KCNQ1 may be a good candidate for diabetic nephropathy and a target to develop new drugs for treatment of the disease.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
No potential conflicts of interest relevant to this article were reported.
We thank the technical staff of the Laboratory for Endocrinology and Metabolism at the Center for Genomic Medicine for providing technical assistance.
APPENDIX
Participating investigators were as follows: Dr. Koichi Kawai, MD, PhD, Kawai Clinic, Tsukuba, Japan; Dr. Masahito Imanishi, MD, PhD, Osaka City General Hospital, Osaka, Japan; Dr. Takashi Uzu, MD, PhD, and Dr. Atsunori Kashiwagi, MD, PhD, Shiga University of Medical Science, Otsu, Japan; and Dr. Kohei Kaku, MD, PhD, Kawasaki Medical School, Kurashiki, Japan.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1. U.S. Renal Data System. USRDS 2008 Annual Data Report. Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 2008. [Google Scholar]
- 2. Nakai S, Masakane I, Akiba T, Shigematsu T, Yamagata K, Watanabe Y, Iseki K, Itami N, Shinoda T, Morozumi K, Shoji T, Marubayashi S, Morita O, Kimata N, Shoji T, Suzuki K, Tsuchida K, Nakamoto H, Hamano T, Yamashita A, Wakai K, Wada A, Tsubakihara Y. Overview of regular dialysis treatment in Japan as of 31 December 2006. Ther Apher Dial 2008;12:428–456 [DOI] [PubMed] [Google Scholar]
- 3. Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 1989;320:1161–1165 [DOI] [PubMed] [Google Scholar]
- 4. Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia 1996;39:940–945 [DOI] [PubMed] [Google Scholar]
- 5. Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jørgensen T, Sandbaek A, Lauritzen T, Hansen T, Nurbaya S, Tsunoda T, Kubo M, Babazono T, Hirose H, Hayashi M, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Tai ES, Pedersen O, Kamatani N, Kadowaki T, Kikkawa R, Nakamura Y, Maeda S. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008;40:1098–1102 [DOI] [PubMed] [Google Scholar]
- 6. Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, Yamagata K, Hinokio Y, Wang HY, Tanahashi T, Nakamura N, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Takeda J, Maeda E, Shin HD, Cho YM, Park KS, Lee HK, Ng MC, Ma RC, So WY, Chan JC, Lyssenko V, Tuomi T, Nilsson P, Groop L, Kamatani N, Sekine A, Nakamura Y, Yamamoto K, Yoshida T, Tokunaga K, Itakura M, Makino H, Nanjo K, Kadowaki T, Kasuga M. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008;40:1092–1097 [DOI] [PubMed] [Google Scholar]
- 7. Lee MP, Ravenel JD, Hu RJ, Lustig LR, Tomaselli G, Berger RD, Brandenburg SA, Litzi TJ, Bunton TE, Limb C, Francis H, Gorelikow M, Gu H, Washington K, Argani P, Goldenring JR, Coffey RJ, Feinberg AP. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J Clin Invest 2000;106:1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet 1996;12:17–23 [DOI] [PubMed] [Google Scholar]
- 9. Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Fauré S, Gary F, Coumel P, Petit C, Schwartz K, Guicheney P. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet 1997;15:186–189 [DOI] [PubMed] [Google Scholar]
- 10. Schwartz PJ, Moss AJ, Vincent GM, Crampton RS. Diagnostic criteria for the long QT syndrome. An update. Circulation 1993;88:782–784 [DOI] [PubMed] [Google Scholar]
- 11. Sugimoto T, Tanabe Y, Shigemoto R, Iwai M, Takumi T, Ohkubo H, Nakanishi S. Immunohistochemical study of a rat membrane protein which induces a selective potassium permeation: its localization in the apical membrane portion of epithelial cells. J Membr Biol 1990;113:39–47 [DOI] [PubMed] [Google Scholar]
- 12. Vallon V, Grahammer F, Richter K, Bleich M, Lang F, Barhanin J, Völkl H, Warth R. Role of KCNE1-dependent K+ fluxes in mouse proximal tubule. J Am Soc Nephrol 2001;12:2003–2011 [DOI] [PubMed] [Google Scholar]
- 13. Lang F, Rehwald W. Potassium channels in renal epithelial transport regulation. Physiol Rev 1992;72:1–32 [DOI] [PubMed] [Google Scholar]
- 14. Vallon V, Grahammer F, Volkl H, Sandu CD, Richter K, Rexhepaj R, Gerlach U, Rong Q, Pfeifer K, Lang F. KCNQ1-dependent transport in renal and gastrointestinal epithelia. Proc Natl Acad Sci U S A 2005;102:17864–17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tanaka N, Babazono T, Saito S, Sekine A, Tsunoda T, Haneda M, Tanaka Y, Fujioka T, Kaku K, Kawamori R, Kikkawa R, Iwamoto Y, Nakamura Y, Maeda S. Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes 2003;52:2848–2853 [DOI] [PubMed] [Google Scholar]
- 16. Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes 2005;54:1171–1178 [DOI] [PubMed] [Google Scholar]
- 17. Kamiyama M, Kobayashi M, Araki S, Iida A, Tsunoda T, Kawai K, Imanishi M, Nomura M, Babazono T, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Ng DP, Hansen T, Gaede P, Pedersen O, Nakamura Y, Maeda S. Polymorphisms in the 3′ UTR in the neurocalcin δ gene affect mRNA stability, and confer susceptibility to diabetic nephropathy. Hum Genet 2007;122:397–407 [DOI] [PubMed] [Google Scholar]
- 18. Araki S, Haneda M, Sugimoto T, Isono M, Isshiki K, Kashiwagi A, Koya D. Factors associated with frequent remission of microalbuminuria in patients with type 2 diabetes. Diabetes 2005;54:2983–2987 [DOI] [PubMed] [Google Scholar]
- 19. Nielsen DM, Ehm MG, Weir BS. Detecting marker-disease association by testing for Hardy-Weinberg disequilibrium at a marker locus. Am J Hum Genet 1998;63:1531–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 1995;29:311–322 [DOI] [PubMed] [Google Scholar]
- 21. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265 [DOI] [PubMed] [Google Scholar]
- 22. Tan JT, Nurbaya S, Gardner D, Ye S, Tai ES, Ng DP. Genetic variation in KCNQ1 associates with fasting glucose and β-cell function: a study of 3,734 subjects comprising three ethnicities living in Singapore. Diabetes 2009;58:1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Müssig K, Staiger H, Machicao F, Kirchhoff K, Guthoff M, Schäfer SA, Kantartzis K, Silbernagel G, Stefan N, Holst JJ, Gallwitz B, Häring HU, Fritsche A. Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes 2009;58:1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ullrich S, Su J, Ranta F, Wittekindt OH, Ris F, Rösler M, Gerlach U, Heitzmann D, Warth R, Lang F. Effects of IKs channel inhibitors in insulin-secreting INS-1 cells. Pflugers Arch 2005;451:428–436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.