Arf assembles a complex containing Miz1, heterochromatin, and histone H3K3 to block expression of genes involved in cell adhesion and signal transduction. The resulting blockade of cell–cell and cell–matrix interactions facilitates elimination of cells carrying oncogenic mutations.
Abstract
Oncogenic stress induces expression of the alternate reading frame (Arf) tumor suppressor protein. Arf then stabilizes p53, which leads to cell cycle arrest or apoptosis. The mechanisms that distinguish both outcomes are incompletely understood. In this study, we show that Arf interacts with the Myc-associated zinc finger protein Miz1. Binding of Arf disrupts the interaction of Miz1 with its coactivator, nucleophosmin, induces the sumoylation of Miz1, and facilitates the assembly of a heterochromatic complex that contains Myc and trimethylated H3K9 in addition to Miz1. Arf-dependent assembly of this complex leads to the repression of multiple genes involved in cell adhesion and signal transduction and induces apoptosis. Our data point to a tumor-suppressive pathway that weakens cell–cell and cell–matrix interactions in response to expression of Arf and that may thereby facilitate the elimination of cells harboring an oncogenic mutation.
Introduction
Enhanced expression of the c-myc oncogene is a hallmark of multiple human tumors, and many experiments using transgenic animals document the oncogenic potential of deregulated c-myc expression (Oster et al., 2002). c-myc encodes a nuclear protein (Myc) that forms several distinct chromatin-bound complexes (Eilers and Eisenman, 2008). As part of a binary complex with a partner protein, Max, Myc binds to specific DNA sequences termed E-boxes and activates transcription of RNA polymerase II–dependent genes. Myc represses transcription when the Myc/Max heterodimer is recruited to core promoter sequences by the zinc finger transcription factor Miz1.
Several genome-wide expression and DNA-binding studies show that Myc has an extraordinary large number of binding sites and target genes and can enhance the expression of large groups of genes. In contrast, the spectrum of target genes of the Myc–Miz1 complex is more limited; among its best characterized targets are those encoding the cyclin-dependent kinase inhibitors p15Ink4b, p21Cip1, and p57Kip2 and a group of genes encoding proteins involved in cell–cell and cell–matrix adhesion (Staller et al., 2001; Seoane et al., 2002; Gebhardt et al., 2006). In the absence of Myc, Miz1 binds to the core promoter of these genes and activates their expression in response to antimitogenic signals; for example, addition of TGF-β (Seoane et al., 2001, 2002; Staller et al., 2001), exposure to DNA damage (Seoane et al., 2002), and disturbance of protein translation (Wanzel et al., 2008) can all activate Miz1 function. To activate its target genes, Miz1 needs to bind to nucleophosmin (NPM; Wanzel et al., 2008). In unstressed cells, NPM shuttles between cytosol and nucleolus and acts as a chaperone for the nuclear export of ribosomal subunits; at steady state, the majority of NPM resides in the nucleolus (Maggi et al., 2008). Exposure of cells to stress such as DNA damage leads to accumulation of a fraction of NPM in the nucleus, where it interacts with Miz1 to activate its target genes.
Both NPM and Myc also interact with the alternate reading frame (Arf) tumor suppressor protein (Bertwistle et al., 2004; Qi et al., 2004; Korgaonkar et al., 2005). Arf is not expressed under physiological conditions, but its expression is induced in response to oncogenic stress signals (Zindy et al., 2003). Arf stabilizes p53 because it inhibits the Mdm2 and HectH9 (Arf-Bp1) ubiquitin ligases that degrade p53 in unstressed cells (Stott et al., 1998; Zhang et al., 1998; Chen et al., 2005). Arf also contributes to the cellular stress response by inhibiting the functions of NPM in ribosome assembly (Itahana et al., 2003; Bertwistle et al., 2004). Finally, Arf induces the sumoylation of proteins to which it binds, including NPM; this may be mediated by its ability to inhibit the Sumo protease Senp3 and trigger its degradation via the proteasome (Tago et al., 2005; Haindl et al., 2008; Kuo et al., 2008).
Ectopic expression of Arf induces G1 arrest, but it is also required for oncogene-induced senescence and apoptosis, arguing that mechanisms must exist that regulate these cellular responses to induction of Arf (Kamijo et al., 1997; Zindy et al., 1998). How a cell chooses between G1 arrest and apoptosis in response to expression of Arf is unclear. One factor that favors apoptosis is enhanced expression of Myc, and Arf-dependent apoptosis limits the oncogenic potential of Myc (Zindy et al., 1998). In this study, we show that Arf facilitates the assembly of a heterochromatic Myc–Miz1 complex and that this event provides a critical switch from G1 arrest to apoptosis in response to Arf expression.
Results
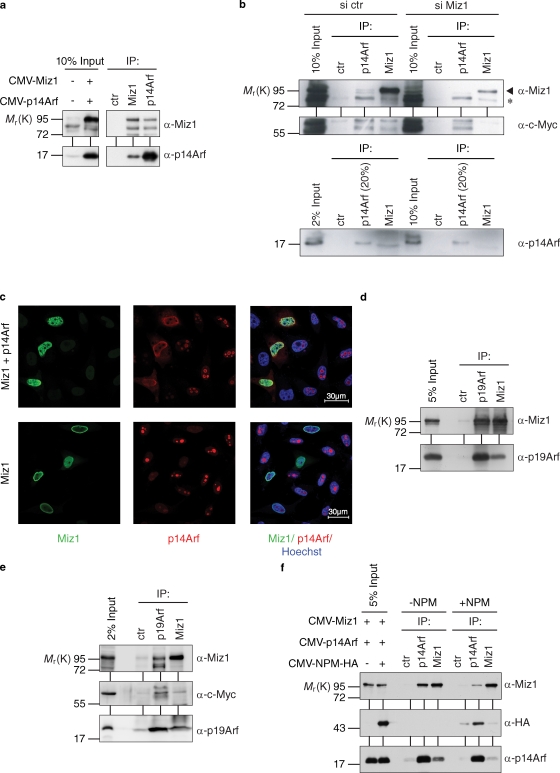
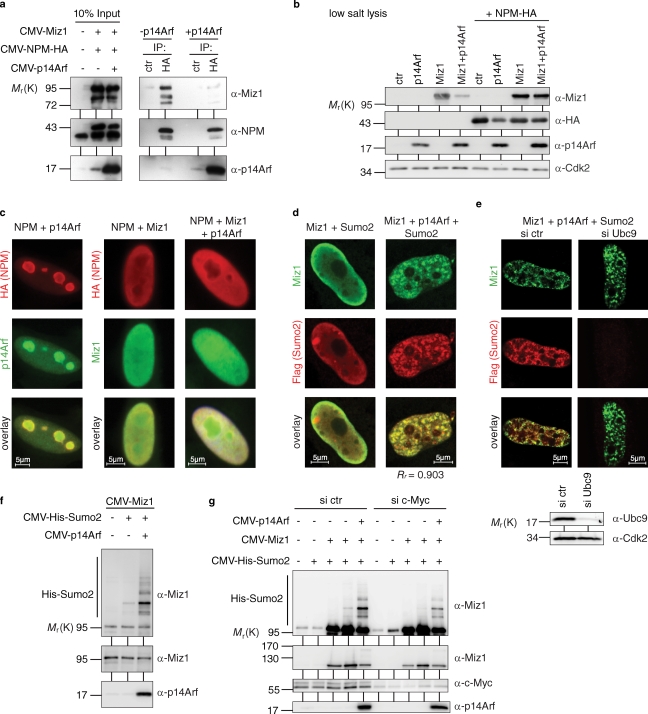
To test whether human p14Arf associates with Miz1, HeLa cells were transfected with cytomegalovirus (CMV)-driven expression vectors encoding both proteins; lysates were prepared and immunoprecipitated with antibodies directed against either protein. Immunoblots revealed the presence of p14Arf in α-Miz1 immunoprecipitates; conversely, Miz1 was present in α-p14Arf precipitates (Fig. 1 a). Neither protein was present in control immunoprecipitates, suggesting that both proteins associate in vivo. To determine whether the endogenous Miz1 and p14Arf proteins form a complex, HeLa cell lysates were immunoprecipitated using specific antibodies. Immunoblotting showed that Miz1 was present in α-p14Arf immunoprecipitates and p14Arf in α-Miz1 immunoprecipitates; neither protein was present in control immunoprecipitates (Fig. 1 b). Furthermore, Myc was associated with both endogenous p14Arf and Miz1, confirming previous results (Staller et al., 2001; Qi et al., 2004). To rule out the idea that the reciprocal coimmunoprecipitation of p14Arf and Miz1 was caused by a cross reactivity of the Miz1 antibody, we used siRNA to deplete Miz1. Depletion of Miz1 strongly reduced the amount of Miz1, Myc, and p14Arf present in α-Miz1 immunoprecipitates. Depletion of Miz1 also eliminated the Miz1 signal present in p14Arf immunoprecipitates but had no effect on the amount of p14Arf and Myc present in α-p14Arf immunoprecipitates. We concluded that endogenous Miz1 and human p14Arf associate with each other. p14Arf is localized in the nucleolus, whereas Miz1 is localized in the nucleoplasm, raising the question of where the Miz1–Arf complex might reside. Ectopic expression of Miz1 did not affect levels of Arf protein but recruited both ectopically expressed and endogenous p14Arf protein from the nucleolus to the nucleoplasm (Fig. 1 c). Similar to human p14Arf, murine p19Arf associated with Miz1 when both proteins were ectopically expressed (Fig. 1 d). Furthermore, immunoprecipitations from mouse embryonic fibroblasts (MEFs) that lack p53 and NPM and express elevated levels of endogenous p19Arf showed that a significant fraction of endogenous Miz1 and a smaller fraction of endogenous Arf were associated with each other in these cells (Fig. 1 e). Collectively, the data show that Miz1 and Arf are associated with each other in human and mouse cells.
Figure 1.
In vivo interaction of Arf with Miz1. (a) p14Arf and Miz1 form a complex after ectopic expression in HeLa cells. Where indicated (+), cells were transfected with CMV-driven expression plasmids encoding Miz1 or p14Arf. 48 h later, lysates were immunoprecipitated with the indicated antibodies. (left) Input blots are shown. (right) The results of the coimmunoprecipitation experiments are shown. IP, immunoprecipitate. (b) Interaction of endogenous Miz1 and p14Arf. (left) HeLa cells were lysed, and lysates were immunoprecipitated with control (ctr), α-p14Arf, or α-Miz1 antibodies; immunoblots of the precipitates were probed with the indicated antibodies. (right) To confirm that coprecipitation of p14Arf with α-Miz1 antibodies is not caused by antibody cross reactivity, cells were transfected with siRNA targeting Miz1 72 h before harvesting. Immunoprecipitation was performed as described in a. The arrowhead points to Miz1, and the asterisk denotes a nonspecific band. (c) Miz1 recruits p14Arf from the nucleolus to the nucleoplasm. HeLa cells were transfected with the indicated expression plasmids and fixed 48 h later. Indirect immunofluorescence was performed using the indicated antibodies. The localization of ectopically expressed p14Arf (top) and the localization of endogenous p14Arf (bottom) are shown. Note that Miz1 does not affect total levels of either ectopically expressed or endogenous Arf proteins. (d) Murine p19Arf interacts with Miz1. NPM−/−/p53−/− MEFs were transfected with expression plasmids encoding p19Arf and Miz1. Cell lysates were prepared 48 h after transfection, immunoprecipitated, and immunoblots were probed with p19Arf and Miz1 antibodies. (e) Endogenous p19Arf and Miz1 form a complex in NPM−/−/p53−/− MEFs. Cell lysates were immunoprecipitated with control, p19Arf, or Miz1 antibodies; immunoblots of the precipitates and input material (2%) were probed with the indicated antibodies. (f) Miz1 interacts with p14Arf independently of NPM. NPM−/−/p53−/− MEFs were transfected with expression plasmids encoding p14Arf, NPM, and Miz1 as indicated. Cell lysates were prepared 48 h after transfection, immunoprecipitated with the indicated antibodies, and immunoblots were probed as described in a.
Miz1 requires binding to NPM for transcriptional activation of its target genes (Wanzel et al., 2008). Because NPM forms a stoichiometric complex with p14Arf, this raises the possibility that binding of Arf to Miz1 occurs indirectly via NPM (Itahana et al., 2003; Bertwistle et al., 2004; Korgaonkar et al., 2005). To determine whether this is the case, we expressed Miz1 and p14Arf in MEFs lacking both NPM and p53 (Fig. 1 f; Colombo et al., 2005). Miz1 and p14Arf efficiently bound to each other in such cells, and restoration of NPM reduced rather than enhanced the amount of Arf that was bound to Miz1. Furthermore, interaction of Miz1 with Arf did not require the amino-terminal POZ domain of Miz1, which is required for transcriptional activation and interaction with NPM (Fig. S1 a). Similarly, both Miz1 and Myc interact with p14Arf, suggesting that Myc might be required for Arf to interact with Miz1 (Qi et al., 2004); however, Myc and Miz1 bound to different domains of Arf (see Fig. 3 a). We concluded that Miz1 binds to p14Arf independently of its interactions with Myc and NPM.
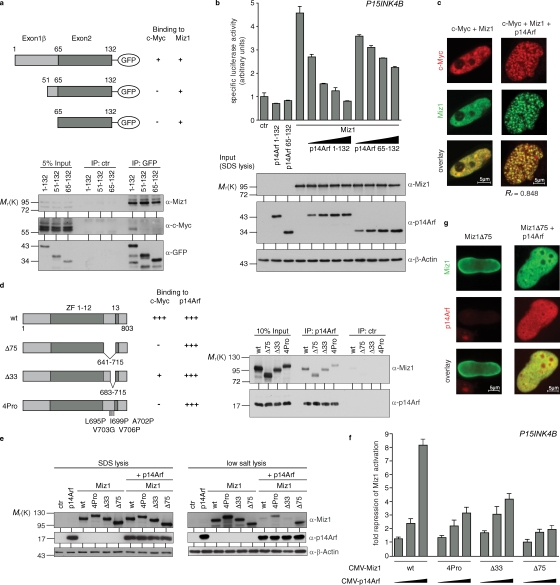
Figure 3.
Inhibition of Miz1 by p14Arf requires assembly of the Myc–Miz1 complex. (a) Miz1 and Myc interact with different domains of p14Arf. U2OS cells were transfected with expression vectors encoding Miz1 and the indicated p14Arf-GFP fusion proteins as indicated. Lysates were precipitated with α-GFP antibodies and immunoblots of the precipitates probed with the indicated antibodies. The top diagram illustrates the amino acids encoded by exons 1β and 2 and gives a summary of the interaction assays. The blots below document the results of the coimmunoprecipitation experiments. (b) Binding of p14Arf to Miz1 is not sufficient to inhibit transactivation. (top) The result of transient reporter assays using the indicated expression plasmids and a P15INK4B reporter plasmid is shown. (bottom) Immunoblots documenting expression of the indicated proteins are shown. (c) Expression of p14Arf recruits Myc to Miz1 foci. Confocal microscopy pictures of HeLa cells transfected with the indicated expression vectors are shown. Cells were stained with the antibodies shown. Pictures of individual cells were used to calculate correlation coefficients indicating colocalization. The numbers shown are mean values for 10 cells each. (d) Mutants of Miz1 that are impaired in binding to Myc bind to Arf. (left) The mutants used are shown and their strength of interaction with Myc is indicated (data adapted from Peukert et al., 1997). (right) Immunoblots documenting that all three mutant alleles bind to Arf with similar efficiency are shown. The experiment was performed as in Fig. 1 a. wt, wild type. (e) Miz1 mutants that are deficient in Myc binding are poorly sequestered by Arf. HeLa cells were transfected with the indicated expression vectors and extracted with either a buffer containing low salt and nonionic detergent (low salt lysis; see Materials and methods) or with buffer containing 4% SDS at 95°C (SDS lysis). Immunoblots of the soluble material are shown. (f) Mutants of Miz1 that are impaired in binding to Myc are resistant to inhibition by Arf. The degree by which increasing amounts of Arf inhibit transactivation by each mutant (calculated from P15INK4B reporter assays) is indicated. (g) Miz1Δ75 does not form foci in response to expression of Arf. The experiment was performed as described in Fig. 2 e for wild-type Miz1. IP, immunoprecipitate; ctr, control. Error bars represent standard deviation of biological triplicates.
To determine the functional consequences of the interaction of p14Arf with Miz1, we performed reporter assays using a P15INK4B promoter plasmid (Fig. 2 a). Consistent with previous results, Miz1 enhanced P15INK4B promoter activity (Wanzel et al., 2008). Expression of p14Arf had no effect on the basal activity of the promoter but abrogated Miz1-dependent transactivation. Identical results were obtained using a P21CIP1 reporter plasmid (unpublished data).
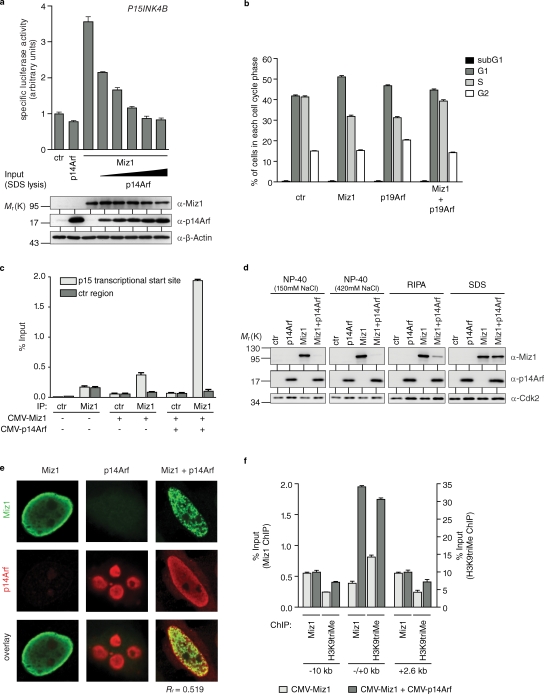
Figure 2.
p14Arf inhibits Miz1-dependent transactivation and G1 arrest. (a) p14Arf inhibits transactivation of the P15INK4B promoter by Miz1. The result of reporter assays using a luciferase reporter plasmid that contains the transcription start site of the P15INK4B promoter is shown. HeLa cells were transfected with the indicated expression plasmids, and the specific luciferase activity was determined 48 h after transfection. (b) p19Arf inhibits the Miz1-induced arrest in the G1 phase of the cell cycle. MEFs that are deficient for p53, Mdm2, and p19Arf were infected with retroviruses encoding Miz1, p19Arf, or both, and resistant cells were selected. FACS analysis was performed immediately after selection. (a and b) Error bars show the standard deviation obtained from three independent samples per experimental condition. (c) p14Arf does not interfere with DNA binding of Miz1. HeLa cells were transfected with expression vectors encoding Miz1, p14Arf, or both. Results of chromatin immunoprecipitation assays documenting the percentage of Miz1 bound to the P15INK4B promoter and a control (ctr) region 10 kb upstream under these conditions are shown. (d) Expression of p14Arf renders Miz1 insoluble. HeLa cells were transfected with expression plasmids encoding Miz1 and p14Arf as shown; in these experiments, the relative amount of CMV-p14Arf expression plasmid was higher than used before. Lysates were prepared using the indicated buffer conditions, cleared by centrifugation, and immunoblots of soluble extracts were probed with the indicated antibodies. (e) Expression of p14Arf induces relocalization of Miz1 within the nucleus. Confocal microscopy images of individual HeLa cell nuclei transfected with the indicated expression plasmids are shown. Rr indicates the Pearson’s coefficient for the correlation of the localization of both proteins. (f) Arf induces formation of heterochromatin around Miz1-binding sites. A summary of the results of chromatin immunoprecipitation (ChIP) assays using antibodies directed against Miz1 and trimethylated H3K9 is shown. The y axis shows the amount of chromatin recovered in α-Miz1 and in α-trimethylated H3K9 immunoprecipitates plotted as a percentage of input chromatin. For each region of the P15INK4B gene, the data are shown for cells transfected with expression vectors encoding Miz1 or Miz1 together with Arf (c and f). Errors bars show the standard deviation of triplicate PCR reactions.
Ectopic expression of Miz1 inhibits cell proliferation in a p21-dependent manner in the G1 phase of the cell cycle and in a p21-independent manner in the S and G2 phases of the cell cycle (Herold et al., 2008). To ascertain whether Arf affects Miz1-dependent cell cycle arrest, we expressed Miz1 alone or together with (murine) p19Arf by retroviral infection in triple-knockout MEFs that lack p53, mdm2, and arf (Weber et al., 2000). Expression of Miz1 by itself induced an accumulation of cells in the G1 phase of the cell cycle (Fig. 2 b). Coexpression of p19Arf abrogated the Miz1-induced G1 arrest and shifted the arrest to the S phase of the cell cycle, which is consistent with its ability to abrogate transcriptional activation by Miz1.
To understand how p14Arf inhibits transactivation by Miz1, we initially used chromatin immunoprecipitation assays to measure DNA binding of Miz1 in the absence and presence of p14Arf. These assays showed that expression of Arf did not interfere with DNA binding of Miz1 to the P15INK4B (Fig. 2 c) and P21CIP1 (not depicted) promoters; instead, Arf appeared to enhance DNA binding by Miz1 (see Fig. 6 d).
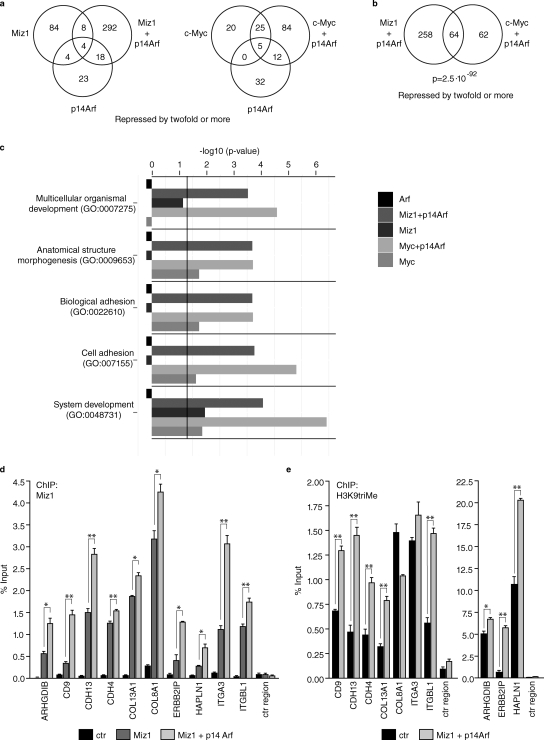
Figure 6.
Arf-mediated assembly of the Myc–Miz1 complex represses genes involved in cell adhesion. (a) Expression of Arf regulates gene repression by Myc and Miz1. A summary of several microarray experiments is shown. U2OS cells were infected with either control retroviruses or with retroviruses expressing the indicated proteins. After selection of resistant cell pools, RNA was extracted and used for microarray experiments. Two Venn diagrams describing the overlap in genes down-regulated more than twofold in each experimental condition are shown. (b) Myc and Miz1 repress an overlapping set of genes in the presence of Arf. A Venn diagram illustrating the overlap in genes repressed by twofold or more in cells expressing Myc plus Arf or Miz1 plus Arf relative to control cells is shown. The p-value was calculated with a hypergeometric test against a null hypothesis of random selection. (c) Myc and Miz1 jointly inhibit cell adhesion genes in the presence of Arf. Results of a GO term analysis using DAVID are shown. The five most significant GO terms describing biological processes affected by genes down-regulated by Miz1 + Arf and Myc + Arf are shown. For each term, the p-value is given for genes down-regulated in cells infected with the indicated expression vectors. Whenever a GO term was not significant for a gene set, the value was arbitrarily set to −1. (d) Miz1 binds to the start sites of multiple cell adhesion genes and promotes H3K9 trimethylation. The results of chromatin immunoprecipitation (ChIP) experiments from U20S cells expressing the indicated proteins are shown. (e) Expression of Miz1 and Arf promotes H3K9 trimethylation at promotors of multiple cell adhesion genes. The result of chromatin immunoprecipitation experiments performed as described in d is shown. ctr, control. Asterisks indicate statistical significance: *, P < 0.05; **, P < 0.01. Error bars represent standard deviation of triplicate PCR reactions.
In these experiments, we noted that the amount of Miz1 present in detergent lysates was decreased upon cotransfection of p14Arf; this effect was particularly apparent when high levels of Arf were expressed (Fig. 2 d). Arf had no effect on the expression of the MIZ1 mRNA levels; therefore, the result is not caused by Arf-dependent inhibition of the CMV promoter (unpublished data). Furthermore, treatment of cells with MG132, an inhibitor of the proteasome, did not enhance recovery of Miz1 (unpublished data). In contrast, extracting cells with 4% SDS at 95°C led to recovery of Miz1, demonstrating that p14Arf renders Miz1 insoluble (Fig. 2 d). Notably, this change in solubility correlated with a striking intranuclear relocalization of Miz1 from a homogeneous distribution throughout the nucleoplasm to a localization in distinct subnuclear foci in response to Arf (Fig. 2 e and Fig. S1 b).
Because resistance to detergent extraction often correlates with formation of heterochromatin, we used chromatin immunoprecipitation with antibodies directed against several histone modifications to analyze how repression of Miz1 by Arf affects the chromatin structure of Miz1 target genes. No change was observed using antibodies directed against trimethylated K27 of histone H3 (unpublished data). In contrast, expression of Arf led to a strong increase in the amount of histone H3 trimethylated at lysine 9 (H3K9trime), a defining feature of heterochromatin, at the Miz1-binding site of the P15INK4B and P21CIP1 genes (Fig. 2 f and not depicted). Collectively, the data argue that Arf induces the local formation of heterochromatin at Miz1-binding sites.
p14Arf is encoded by two exons: amino acids 1–64 are encoded by exon 1β of the CDKN2A locus and are sufficient to inhibit Mdm2 and cause p53-dependent cell cycle arrest (Kamijo et al., 1998; Clark et al., 2002). Amino acids 65–132 are encoded by exon 2, which is shared with p16Ink4a; these amino acids contribute to induction of apoptosis by Arf (Itahana and Zhang, 2008). To determine which of the two domains binds to Miz1, we coexpressed Miz1 with a full-length Arf-GFP fusion protein and two deletion mutants lacking the first 50 and 64 amino acids, respectively (Fig. 3 a). All three proteins efficiently coimmunoprecipitated Miz1, demonstrating that amino acids 65–132 of the human Arf protein are sufficient for the interaction.
Reporter assays revealed that binding of Arf to Miz1 is not sufficient to inhibit Miz1 function because amino acids 65–132 of Arf were inefficient in inhibiting Miz1-dependent transactivation and sequestering of Miz1 (Fig. 3 b and Fig. S2). The amino terminus of p14Arf is required for interaction with Myc (Fig. 3 a; Datta et al., 2004). Because Myc can repress transactivation by Miz1, we speculated that recruitment of Myc might be required for p14Arf to repress Miz1 function. Consistent with such a model, confocal microscopy revealed that both transfected and endogenous Myc colocalized with Miz1 foci after expression of Arf (Fig. 3 c and not depicted).
To test whether recruitment of Myc is required for Arf to inhibit Miz1 function, we used three previously characterized mutations of Miz1 that disrupt the Myc-binding domain localized between the 12th and 13th zinc finger: one deletion (Miz1 Δ33) partially impairs Myc binding, whereas a larger deletion (Miz1 Δ75) and a quadruple point mutant (Miz1 4Pro) abrogate binding to Myc (Fig. 3 d; Peukert et al., 1997). Although all three mutant proteins interacted equally with Arf (Fig. 3 d), inhibition of Miz1 transactivation as well as sequestration (as indicated by their increased solubility when coexpressed with Arf) was moderately (Δ33) or strongly (Δ75 and 4Pro) impaired relative to wild-type Miz1 (Fig. 3, e and f). Confocal microscopy confirmed that Miz1Δ75 was not sequestered upon coexpression of Arf (Fig. 3 g). We concluded that recruitment of Myc is essential for Arf to inhibit transactivation by Miz1.
Miz1 forms a soluble complex with NPM to activate transcription. Myc competes with NPM for binding to Miz1 and forms a complex that is resistant to mild extraction; therefore, the altered properties of Miz1 in response to expression of Arf might reflect an Arf-induced assembly of the Myc–Miz1 complex (Peukert et al., 1997). To analyze how expression of Arf affects complex formation of Miz1 with NPM, we performed immunoprecipitation assays and found that expression of Arf led to a dissociation of Miz1 from NPM (Fig. 4 a). Strikingly, the converse was also true: elevated expression of NPM prevented the Arf-induced decrease in solubility and the change in subnuclear localization of Miz1 (Fig. 1 f; and Fig. 4, b and c). Collectively, the data strongly suggest a model in which Arf and NPM antagonize each other’s function in the assembly of the Myc–Miz1 complex (see Fig. 8).
Figure 4.
Arf antagonizes NPM in the assembly of a repressive Myc–Miz1 complex and promotes sumoylation of Miz1. (a) Expression of p14Arf disrupts the NPM–Miz1 complex. HeLa cells were transfected with expression plasmids encoding Miz1, p14Arf, and HA-tagged NPM as shown. 48 h after transfection, cells were lysed and lysates were immunoprecipitated with either control (ctr) or α-HA antibodies. Immunoblots of the precipitates were probed with the indicated antibodies. (left) Input blots are shown. IP, immunoprecipitate. (b) Expression of NPM inhibits sequestration of Miz1 by Arf. HeLa cells were transfected with expression vectors for Miz1, Arf, and NPM as shown. 48 h after transfection, cells were lysed in low salt buffer. Immunoblots of the soluble fraction were probed with the indicated antibodies. (c) Expression of NPM antagonizes Arf-induced foci formation. HeLa cells were transfected with expression vectors for NPM and either Miz1, Arf, or both and processed for immunofluorescence. Note that the relocalization of NPM and Arf to the nucleus is almost certainly caused by residual binary Miz1–NPM and Miz1–Arf complexes in the transfected cells. (d) p14Arf induces colocalization of Miz1 into foci with Sumo2. Immunofluorescence pictures of HeLa cells transfected with expression plasmids encoding Miz1, p14Arf, and Sumo2 as shown; transfected cells were stained with the indicated antibodies. (e) Recruitment of Sumo2 to Myc–Miz1 foci requires Ubc9. Cells were transfected with either control siRNA or siRNA targeting Ubc9 and subsequently transfected and processed for immunofluorescence as described in b. (bottom) The Ubc9 knockdown is documented. (f) Arf induces sumoylation of Miz1. HeLa cells were transfected with the indicated expression plasmids; 48 h later, cells were lysed in denaturing buffer, and sumoylated proteins were recovered on Ni-NTA agarose. (g) Depletion of Myc reduces Arf-induced sumoylation of Miz1. Sumoylation assays were performed as described in e except that HeLa cells were transfected with either control siRNA or siRNA targeting Myc. Rr indicates the Pearson’s coefficient for the correlation of the localization of both proteins.
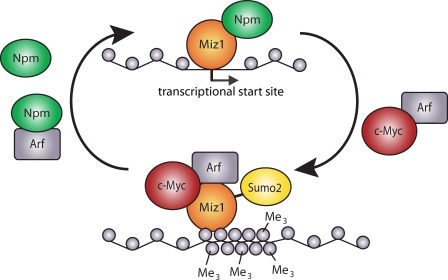
Figure 8.
Model summarizing our findings. We propose that NPM and Arf have antagonistic roles in Miz1-dependent transactivation and Myc–Miz1-mediated repression of genes. Arf also stimulates the sumoylation of Miz1 potentially by antagonizing the Sumo-specific protease Senp3.
Expression of Arf induces sumoylation of several of its substrate proteins (Tago et al., 2005). Consistent with these observations, Miz1 colocalized with Flag-tagged Sumo2 upon expression of Arf but not in its absence (Fig. 4 d); this was not observed for Sumo1, suggesting that it is specific for Sumo2 (not depicted). siRNA-mediated depletion showed that Ubc9, the E2 enzyme responsible for conjugation of Sumo moieties, was not required for Arf-induced sequestration of Miz1 and focus formation but was strictly required for recruitment of Sumo2 to these foci (Fig. 4 e and not depicted). In vitro sumoylation assays revealed that Miz1 is sumoylated in vitro (unpublished data). Transient transfection assays showed that expression of Arf strongly stimulated sumoylation of Miz1 in vivo (Fig. 4 f). In contrast, we did not detect sumoylation of Myc (unpublished data). Depletion of Myc using specific siRNA reduced the Arf-dependent sumoylation of Miz1, suggesting that assembly of the Myc–Miz1 complex enhances sumoylation of Miz1; consistent with such a model, sumoylation of Miz1Δ75 was reduced relative to wild-type Miz1 (Fig. 4 g and not depicted).
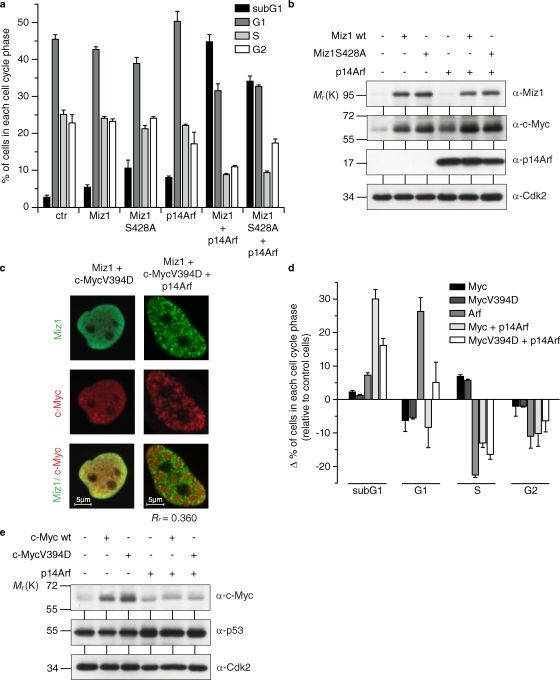
As shown in Fig. 2 b, Arf negates the ability of Miz1 to arrest proliferation of p53-deficient cells in the G1 phase of the cell cycle. To determine how Arf affects Miz1 function in a p53-proficient cell, we expressed p14Arf and Miz1 either alone or together via retroviral infection in U2OS cells in which the endogenous ARF gene is silenced but that have wild-type p53 (Park et al., 2002). Expression of Arf inhibited cell proliferation and led to an increase in the percentage of cells in the G1 phase of the cell cycle, which is consistent with published data (Fig. 5 a; Quelle et al., 1995). Expression of either wild-type Miz1 or Miz1 S428A, a point mutant that cannot be phosphorylated by Akt, had only moderate effects on cell cycle progression by itself, potentially because U2OS cells express significant levels of endogenous Myc when grown exponentially in culture (Fig. 5 b). In clear contrast to cells expressing either Miz1 or Arf alone, cells expressing both proteins underwent rampant apoptosis as demonstrated by the accumulation of cells with a sub-G1 DNA content. Virtually identical results were obtained when activation of caspase 3 was used as a marker for apoptosis (Fig. S3 a). Immunoblotting revealed that this change in cellular behavior occurred without any detectable changes in the expression of either Miz1 or Arf (Fig. 5 b). We noted that levels of endogenous Myc were elevated in cells expressing Miz1, reflecting the ability of Miz1 to stabilize Myc (Salghetti et al., 1999).
Figure 5.
Binding of Myc to Miz1 switches the cellular response to Arf from cell cycle arrest to apoptosis. (a) Coexpression of Miz1 and Arf induces apoptosis in p53-proficient cells. U2OS cells were sequentially infected with either control retroviruses or viruses expressing the indicated proteins. Pools of resistant cells were selected and harvested for FACS analysis immediately after selection. (b) Immunoblots documenting expression of the indicated proteins in the U2OS cells used for the analysis shown in a. Note the enhanced levels of endogenous Myc expression in cells expressing ectopic Miz1 or Miz1S428A. (c) Arf does not recruit MycV394D into nuclear foci. Confocal microscopy pictures documenting the distribution of MycV394D and of Miz1 in HeLa cells expressing both proteins and p14Arf are shown. (d) Binding of Myc to Miz1 stimulates induction of apoptosis by Myc and Arf. U2OS cells were infected and harvested as described in a. The plot shows the difference in the percentage of apoptotic cells and of cells in the indicated phase of the cell cycle observed in cells expressing the indicated proteins relative to control-infected cells. (e) Immunoblots documenting expression of the indicated proteins in the cells used for the analysis shown in d. wt, wild type. Rr indicates the Pearson’s coefficient for the correlation of the localization of both proteins. Error bars represent the standard deviation of biological triplicates.
Levels of endogenous Myc were even further elevated in cells expressing Miz1 and Arf, suggesting that the switch from cell cycle arrest to apoptosis requires formation of the Myc–Miz1 complex. Consistent with this idea, coexpression of Arf with Miz1Δ75 induced significantly less apoptosis relative to coexpression of Arf with wild-type Miz1 (Fig. S3 c). As a further test of this idea, we coexpressed Arf with either wild-type Myc or MycV394D, a point mutant of Myc that does not associate with Miz1 (Herold et al., 2002). Confocal immunofluorescence confirmed that Arf does not recruit MycV394D into Miz1-containing nuclear foci (Fig. 5 c). Expression of wild-type Myc abrogated Arf-induced G1 arrest and induced apoptosis, similar to what was observed in the presence of Miz1 and Arf (Fig. 5, d and e; and Fig. S3 b). In contrast, MycV394D was significantly compromised in its ability to overcome Arf-induced G1 arrest and induce apoptosis when coexpressed with Arf, demonstrating that induction of apoptosis by Arf requires formation of the Myc–Miz1 complex (Fig. 5, d and e; and Fig. S3 b).
To understand how Arf affects Miz1-dependent gene expression and induces apoptosis, we performed genome-wide microarray analysis on RNA extracted from U2OS cells infected with retroviruses expressing either Miz1, Myc, or Arf either by themselves or in combination to identify the target genes jointly regulated by Myc and Miz1 in the presence of Arf. This analysis showed that expression of Arf enhanced the ability of Myc and Miz1 to repress transcription. Specifically, Miz1 repressed 322 genes more than twofold in the presence of Arf but only 100 genes in its absence; similarly, expression of Arf enhanced the number of Myc-repressed genes from 50 to 126 (Fig. 6 a). Importantly, 64 of the 126 genes were also repressed when Miz1 and Arf were coexpressed, identifying a highly significant overlap in target genes (P = 2.5 × 10−92) and defining a core program of genes that are jointly repressed by Myc and Miz1 in the presence of Arf (Fig. 6 b). Arf also induced qualitative differences in gene repression by Myc and Miz1; notably, only 30 of the 126 genes that were repressed by Myc in the presence of Arf and 12 of the 322 genes repressed by Miz1 in the presence of Arf were also repressed in its absence.
We considered several hypotheses of how repression of the target genes of the Myc–Miz1 complex might shift the cellular response to expression of Arf toward apoptosis. First, none of the 64 genes that were repressed by either Myc or Miz1 in the presence of Arf were a target gene of p53 as judged by comparison with a database of genomic binding sites for p53 (Wei et al., 2006). Therefore, we ruled out a model in which Arf induces apoptosis by shifting the p53-dependent transcriptional response toward proapoptotic genes via its effects on the Myc–Miz1 complex. Second, none of the genes encoded a mitochondrial protein, suggesting that direct alterations of the apoptotic machinery are also not responsible for the shift in the cellular response. Instead, Gene Ontology (GO) term analysis (Fig. 6 c) showed that coexpression of either Myc or Miz1 with Arf repressed a set of genes that is highly enriched for genes encoding proteins involved in cell adhesion and as coreceptors in signal transduction. This group of genes was not repressed by either Miz1 or Arf alone and only weakly by expression of Myc alone (Fig. 6 c). Arf-enhanced repression was also apparent when the expression of each member of this group was analyzed individually (Fig. S4 a). We analyzed 10 of these genes by chromatin immunoprecipitation and found that Miz1 bound to the start site of all of them (Fig. 6 d). Coexpression of Arf and Miz1 promoted the accumulation of trimethylated H3K9 around the Miz1-binding sites on 8/10 genes (Fig. 6 e). Furthermore, real-time quantitative PCR (RQ-PCR) experiments from independent experiments showed that expression of these genes was repressed upon coexpression of Miz1 and Arf but much less upon coexpression of Miz1Δ75 and Arf, paralleling the effect of adhesion, and arguing that repression by Miz1 and Arf requires recruitment of Myc (Fig. S4 b and Table S1). We used chromatin immunoprecipitations to analyze a subset of these genes and confirmed that endogenous Myc was bound to the Miz1-binding site in vivo (Fig. S4 c).
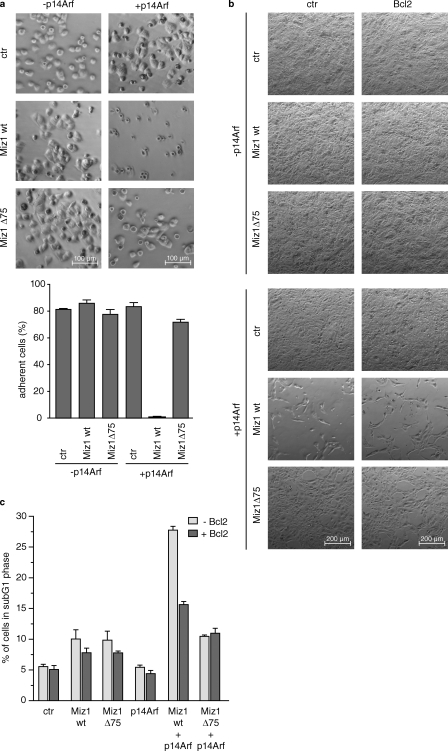
To determine how this change in gene expression translates into alterations in cellular behavior, we infected U2OS cells as before and trypsinized the cells 2 d after infection; at this time point, only a minority of cells had already undergone apoptosis. Upon replating, cells expressing either Miz1 or Arf by itself rapidly reattached; in contrast, cells coexpressing Miz1 and Arf failed to attach to the dish (Fig. 7 a). Loss of adhesion was not observed in cells coexpressing Arf and Miz1Δ75, arguing that it was depending on binding of Myc to Miz1.
Figure 7.
Expression of Miz1 and Arf results in reduced cell adhesion and subsequent apoptosis. (a) U2OS cells expressing Miz1 and Arf show strongly reduced cell adhesion. Morphology of U2OS cells expressing the indicated proteins. Pools of resistant cells expressing the indicated protein were selected, trypsinized, and replated. (top) Photographs taken 2 h after replating are shown. (bottom) A quantitation of the experiment is shown. Similar results were obtained after 4 h and also in cells expressing Myc plus Arf (not depicted). (b) Loss of cell adhesion upon coexpression of Miz1 and Arf is not affected by expression of Bcl2. U2OS cells were infected with either control (ctr) retroviruses or retroviruses expressing the indicated proteins. Pools of resistant cells were selected without trypsinization. Photographs were taken 4 d after infection. (c) Expression of Bcl2 reduces apoptosis induced by coexpression of Miz1 and Arf. U2OS cells described in b were harvested for FACS analysis. The graph shows the percentage of apoptotic cells. wt, wild type. Error bars represent the standard deviation of biological triplicates.
We repeated the experiment by infecting adherent cells that express either Miz1 or Miz1Δ75 with a retrovirus expressing Arf and selecting pools of infected cells without trypsinization. In this experimental setting, expression of Arf led to rapid loss of adhesion of cells expressing Miz1 but not of control cells or cells expressing Miz1Δ75 (Fig. 7 b). Similarly, cells expressing Myc + Arf but not MycV394D + Arf rapidly detached from the plate (Fig. S5 a). Furthermore, identical results were obtained when adherent cells were infected with a high titer lentivirus expressing Arf and subsequently cultured without drug selection (unpublished data). Under these experimental conditions, cells that detached from the plate underwent apoptosis, whereas the remaining adherent cells did not, showing that loss of adhesion is tightly linked to induction of apoptosis (Fig. S5 b). Importantly, retroviral expression of Bcl2 suppressed the apoptosis induced by coexpression of Miz1 + Arf or Myc + Arf but had no discernible effect on the loss of adhesion (Fig. 7, b and c; and Fig. S5 a). Together, the data strongly argue that the repression of cell adhesion genes and the resultant altered adhesion properties are the cause, not the consequence, of induction of apoptosis.
Discussion
In this study, we show that the Arf tumor suppressor protein facilitates two distinct steps in the assembly of a heterochromatic complex that contains Myc, Miz1, Sumo2, and trimethylated H3K9 (Fig. 8). One of these steps occurs independently of Ubc9: this step requires binding of Myc to Miz1 and leads to an altered intranuclear localization and solubility of Miz1 that is characteristic of Myc–Miz1 complexes. Previous work has shown that NPM is a coactivator of Miz1 and that Myc inhibits binding of NPM to Miz1 (Wanzel et al., 2008). Also, direct binding of Arf to Myc has been demonstrated previously (Qi et al., 2004). Therefore, the simplest model that explains our data is that Arf facilitates the recruitment of Myc to Miz1, thereby displacing NPM. This model can explain our observation that elevated levels of NPM disrupt foci formation because free NPM would compete with Miz1–NPM complexes for binding to Arf and Myc. Next, Arf promotes sumoylation of Miz1, similar to other proteins to which it binds (Tago et al., 2005). Depletion of Myc attenuates Arf-dependent sumoylation of Miz1, suggesting that formation of the Myc–Miz1 complex facilitates sumoylation. Because sumoylation of transcription factors can mediate the recruitment of corepressors, it is tempting to speculate that sumoylation of Miz1 enhances repression by the Myc–Miz1 complex (Stielow et al., 2008; Ouyang et al., 2009). It remains to be determined whether Arf-induced sumoylation of Miz1 is mediated via inhibition of the Senp3 Sumo-specific protease (Haindl et al., 2008; Kuo et al., 2008).
Miz1 arrests cell proliferation in a p21-dependent manner in the G1 phase of the cell cycle and in a p21-independent, Atr-dependent manner in the S and G2 phases of the cycle (Herold et al., 2008). Expression of Arf has no effect on Miz1-mediated G2 arrest but abrogates the G1 arrest, which is consistent with the ability of Arf to inhibit Miz1-dependent transcriptional activation. Furthermore, the interaction of Arf with Miz1 leads to repression of a large group of genes that encode proteins involved in cell adhesion and cell signaling and closely correlates with induction of apoptosis by Miz1 and Arf. Two previous studies have linked Myc’s apoptotic function to Miz1-dependent repression of Bcl-2 and p21Cip1 (Seoane et al., 2002; Patel and McMahon, 2007), respectively. Furthermore, a recent study suggests that Miz1 might inhibit expression of Bax and Puma and that Arf might relieve this inhibition (Miao et al., 2010). However, we did not observe regulation of any of these genes in our experimental settings, arguing that additional targets of this pathway must exist. Indeed, expression of either Miz1 or Myc together with Arf led to repression of a group of genes encoding proteins involved in both cell–cell and cell–matrix adhesion. Previous studies have shown that Myc can inhibit cell adhesion in hematopoietic and epithelial cells and have implicated Miz1-mediated repression in this process (Frye et al., 2003; Wilson et al., 2004; Gebhardt et al., 2006).
The findings suggest a model in which induction of Arf and subsequent repression via the Myc–Miz1 complex reduces or disrupts the interaction of a cell that has sustained an oncogenic mutation with its environment. This mechanism would be tumor suppressive because many epithelial cells depend on continuing contact with a substratum to escape anoikis (Reginato et al., 2003). Indeed, several of the genes repressed by Myc–Miz1 in the presence of Arf encode coreceptors of growth factor receptors, arguing that their repression may lead to loss of critical survival signals (Orian-Rousseau et al., 2002). Second, most epithelial cells undergo rapid turnover and do not reside long enough in a tissue to acquire the multiple mutations required for tumorigenesis. The exceptions are stem cells, which depend on specific cell adhesion molecules for retention in the niche. Several of the genes encoding these molecules are targets of Myc–Miz1 complexes and, therefore, would be repressed in cells expressing Arf (Frye et al., 2003; Gebhardt et al., 2006). This second mechanism could explain the p53-independent tumor-suppressive role of Arf that has been demonstrated during skin carcinogenesis (Kelly-Spratt et al., 2004). Finally, we note that the enhanced levels of NPM that are found in many tumor cells would antagonize repression of cell adhesion genes and subsequent apoptosis mediated by Myc–Miz1 complexes, providing a potential mechanistic explanation for the potent ability of NPM to enhance transformation by Myc (Li et al., 2008).
Materials and methods
Cell culture
All cell lines were grown in DME. Media were supplemented with 10% heat-inactivated fetal calf serum (Sigma-Aldrich), 2 mM L-glutamine, 100 U ml−1 penicillin, and 100 µg ml−1 streptomycin. mdm2/p53/arf- and NPM/p53-deficient MEFs were provided by C. Sherr and M. Roussel (St. Jude Children’s Research Hospital, Memphis, TN) and P.-G. Pelicci (Istituto Europeo di Oncologia, Milan, Italy), respectively. Recombinant retroviruses were generated and used as described previously (Pear et al., 1993).
U2OS cells were stably transfected with an ecotropic receptor expression plasmid before infection. Infected cells were selected and analyzed within one to two passages after selection. Cells used for double infection with two different viruses were split 1 d after infection and superinfected 24 h later. Selection was performed 2 d after the second infection.
For lentiviral infections, the pLV-tTRKRAB-red vector with p14Arf replacing the tTRKRAB element, the packaging plasmid psPAX.2, and the envelope plasmid pMD2.G were used. 293T producer cells were triple transfected with these vectors, supernatant containing lentiviral particles was harvested 36 h after transfection, and U2OS cells were infected in the presence of 4 µg/ml protamine sulfate (Sigma-Aldrich).
Transient transfection and siRNA transfection
Transient transfection of plasmid DNA was performed using a standard CaPO4 protocol. The expression and reporter plasmids were described previously (Wanzel et al., 2008). siRNAs targeting Ubc9 and c-Myc were purchased from Thermo Fisher Scientific, and transfection was performed using reagent (Lipofectamine RNAiMAX; Invitrogen) according to the manufacturer’s instructions. Cells were harvested 48 h after transfection and lysed directly in buffer containing 190 mM Tris, pH 6.8, 30% glycerin, 2 M β-mercaptoethanol, and 4% SDS at 95°C (SDS lysis). Low salt lysis was performed with a buffer containing 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, and 1% (vol/vol) Triton X-100.
Antibodies, chromatin immunoprecipitation, and immunofluorescence
A rabbit polyclonal antibody was used to precipitate Miz1 (H-190; Santa Cruz Biotechnology, Inc.), mouse monoclonal antibody 10E2 was used for immunoblotting, and both antibodies were used for immunofluorescence. The following antibodies were used for immunofluorescence and immunoblotting: p19Arf (Abcam), p14Arf (Novus Biologicals), mouse monoclonal anti-Myc antibody (9E10), anti-NPM (FC82291; Abcam), anti-HA (16B12; Covance), anti-Cdk2 (Santa Cruz Biotechnology, Inc.), anti-Flag (F3165; Sigma-Aldrich), anti–β-actin (clone AC-15; Sigma-Aldrich), anti-Ubc9 (Abcam), anti-GFP (Sigma-Aldrich), anti-H3K9trime (Abcam), and anti-p53 (DO-1; Santa Cruz Biotechnology, Inc.).
Fluorescence images were recorded on a confocal microscope (SP5; Leica) with a 63× NA 1.4 oil immersion objective. The fluorescence emission resulted from excitation with 405- (for Hoechst), 488- (for Alexa Fluor 488, fluorescein, and GFP), and 633 (for Alexa Fluor 647)-nm laser lines with a confocal microscope (SP5 LSM; Leica) using internal spectral parameter settings. The fluorescence was detected using a 430–470-nm spectral bandwidth setting for Hoechst, a 505–570-nm spectral bandwidth setting for Alexa Fluor 488, and a 650–720-nm spectral bandwidth setting for Alexa Fluor 647. To avoid bleeding between channels, each excitation and emission was recorded individually and sequentially in each focal plane. Each image was line and frame averaged twice. For consistency, the internal settings were kept constant for all samples. The correlation of fluorescence intensities was evaluated with LAS AF software (Leica) to determine the Pearson’s coefficient.
FACS analysis
For analysis of the cell cycle profile/distribution, cells were fixed in ethanol and stained with propidium iodide. To determine the level of apoptosis, cells were labeled using the CaspGlow Fluorescein Active Caspase-3 Staining kit (BioVision) according to the manufacturer’s instructions.
In vivo sumoylation assays
HeLa cells were transfected with combinations of CMV-driven expression vectors for Miz1, Arf (provided by G. Peters, London Research Institute, London, England, UK), and a His-Sumo2 expression plasmid (provided by G. Suske, University of Marburg, Marburg, Germany). 48 h after transfection, cells were harvested, lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, 250 mM sodium chloride, 10 mM imidazole, and 0.1 mM N-ethyl maleimide), sonicated, and cellular debris was removed by centrifugation. The lysate was incubated with Ni-NTA beads (QIAGEN) for 4 h at 4°C to purify His-tagged proteins. The beads were washed two times with buffer A, two times with a 1:4 mixture of buffer A and buffer B (25 mM Tris–HCl, pH 6.8, and 20 mM imidazole), and two times with buffer B. Bound proteins were eluted by boiling in SDS sample buffer containing 200 mM imidazole, and eluted proteins were analyzed by Western blotting.
Microarray procedures
We used a whole human genome microarray kit (G4112F; Agilent Technologies), and procedures were performed according to the manufacturer’s instructions. Samples were prepared from 100 ng total RNA. The Cy3- and Cy5-labeled probes were hybridized to microarrays for 16 h at 65°C and according to the manufacturer’s instructions. Data analysis was performed using the limma package (Wettenhall and Smyth, 2004) and R (http://www.R-project.org). Genes were selected as regulated if they had minimum mean log2 expression of 5 and a minimum fold change of 2. Multiple probe sets for one gene were averaged before selection. The experiments were performed in duplicates, and independent arrays were performed from each sample. Genes were filtered by reproducibility, and a set of genes was validated by RQ-PCR (Absolute QPCR SYBR green mix; Thermo Fisher Scientific). (max (1, (array_1 + array_2)/2) > |array_1 − array_2|). GO term group analysis was performed using DAVID (Database for Annotation, Visualization, and Integrated Discovery; Dennis et al., 2003).
Primers for RQ-PCR analysis
S14 (forward), 5′-GGCAGACCGAGATGAATCCTCA-3′ and (reverse) 5′-CAGGTCCAGGGGTCTTGGTCC-3′; CDH13 (forward), 5′-GAGAACTCCGCTCGTTCTGT-3′ and (reverse) 5′-AGTGCAGTCCAAATCTTCTGC-3′; GSG1 (forward), 5′-GCTGCTGTTTCCTCTGTCCT-3′ and (reverse) 5′-TCGCTTGGAAGACTTGTGAAT-3′; ARHGDIB (forward), 5′-AAATGGACAAAGATGATGAGAGTCTA-3′ and (reverse) 5′-ACGACATTGGGGGCTTTC-3′; CD9 (forward), 5′-CTGCTGTTCGGATTTAACTTCA-3′ and (reverse) 5′-GTCGAATCGGAGCCATAGTC-3′; HAPLN1 (forward), 5′-GACAGAGCTATTCACATCCAAGC-3′ and (reverse) 5′-TGCCACCTCTGTGTGAAAAC-3′; LOXL2 (forward), 5′-ACTGCCAGCTCCTCCTACG-3′ and (reverse) 5′-TCGTTGCCAGTACAGTGGAG-3′; CDH4 (forward), 5′-CAGACCCCGTAACCAACG-3′ and (reverse) 5′-TGAAAGCTCTGTTGAGCTCGT-3′; ITGA6 (forward), 5′-AGCCTCTTCGGCTTCTCG-3′ and (reverse) 5′-CTCCCGTTCTGTTGGCTCT-3′; COL8A1 (forward), 5′-CATGGACTTCCTGGCATTG-3′ and (reverse) 5′-TCGATCACCCTTTGGTCCT-3′; COL13A1 (forward), 5′-GGGAGAAGCAGGTGTCGAT-3′ and (reverse) 5′-GGCCATCTGGTCCCTGTT-3′; and PTX3 (forward), 5′-TGTATGTGAATTTGGACAACGAA-3′ and (reverse) 5′-CATTCCGAGTGCTCCTGAC-3′.
Online supplemental material
Fig. S1 shows cellular Interactions of Miz1 and Arf. Fig. S2 shows that p14Arf(65–132) fails to sequester Miz1. Fig. S3 shows that Arf-induced assembly of the Myc–Miz1 complex promotes apoptosis. Fig. S4 shows expression data for individual genes in the adhesion cluster. Fig. S5 shows loss of adhesion in cells expressing Myc and Arf. Table S1 shows p-values for the differences in expression of the adhesion genes shown in Fig. S4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908103/DC1.
Acknowledgments
We thank Charles Sherr and Martine Roussel for mdm2/p53/arf-deficient cells, Pier-Guiseppe Pelicci for NPM/p53-deficient cells, Guntram Suske and Ivan Dikic for Sumo expression plasmids, and Gordon Peters for expression plasmids encoding p14Arf.
This study was supported by grants from the Deutsche Forschungsgemeinschaft (Transregio 17 to M. Eilers), the Rudolf-Virchow Center (grant FZ-82 to M. Eilers and G.S. Harms), and the German-Israeli Research Foundation (to A. Dwertmann and M. Abed).
Footnotes
Abbreviations used in this paper:
- Arf
- alternate reading frame
- CMV
- cytomegalovirus
- GO
- Gene Ontology
- MEF
- mouse embryonic fibroblast
- NPM
- nucleophosmin
- RQ-PCR
- real-time quantitative PCR
References
- Bertwistle D., Sugimoto M., Sherr C.J. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24:985–996 10.1128/MCB.24.3.985-996.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Kon N., Li M., Zhang W., Qin J., Gu W. 2005. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 121:1071–1083 10.1016/j.cell.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Clark P.A., Llanos S., Peters G. 2002. Multiple interacting domains contribute to p14ARF mediated inhibition of MDM2. Oncogene. 21:4498–4507 10.1038/sj.onc.1205558 [DOI] [PubMed] [Google Scholar]
- Colombo E., Bonetti P., Lazzerini Denchi E., Martinelli P., Zamponi R., Marine J.C., Helin K., Falini B., Pelicci P.G. 2005. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 25:8874–8886 10.1128/MCB.25.20.8874-8886.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Nag A., Pan W., Hay N., Gartel A.L., Colamonici O., Mori Y., Raychaudhuri P. 2004. Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J. Biol. Chem. 279:36698–36707 10.1074/jbc.M312305200 [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Eilers M., Eisenman R.N. 2008. Myc’s broad reach. Genes Dev. 22:2755–2766 10.1101/gad.1712408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., Gardner C., Li E.R., Arnold I., Watt F.M. 2003. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 130:2793–2808 10.1242/dev.00462 [DOI] [PubMed] [Google Scholar]
- Gebhardt A., Frye M., Herold S., Benitah S.A., Braun K., Samans B., Watt F.M., Elsässer H.P., Eilers M. 2006. Myc regulates keratinocyte adhesion and differentiation via complex formation with Miz1. J. Cell Biol. 172:139–149 10.1083/jcb.200506057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haindl M., Harasim T., Eick D., Muller S. 2008. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9:273–279 10.1038/embor.2008.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S., Wanzel M., Beuger V., Frohme C., Beul D., Hillukkala T., Syvaoja J., Saluz H.P., Haenel F., Eilers M. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 10:509–521 10.1016/S1097-2765(02)00633-0 [DOI] [PubMed] [Google Scholar]
- Herold S., Hock A., Herkert B., Berns K., Mullenders J., Beijersbergen R., Bernards R., Eilers M. 2008. Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1. EMBO J. 27:2851–2861 10.1038/emboj.2008.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K., Zhang Y. 2008. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 13:542–553 10.1016/j.ccr.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K., Bhat K.P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 12:1151–1164 10.1016/S1097-2765(03)00431-3 [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy F., Roussel M.F., Quelle D.E., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 91:649–659 10.1016/S0092-8674(00)80452-3 [DOI] [PubMed] [Google Scholar]
- Kamijo T., Weber J.D., Zambetti G., Zindy F., Roussel M.F., Sherr C.J. 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA. 95:8292–8297 10.1073/pnas.95.14.8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Spratt K.S., Gurley K.E., Yasui Y., Kemp C.J. 2004. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2:E242 10.1371/journal.pbio.0020242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar C., Hagen J., Tompkins V., Frazier A.A., Allamargot C., Quelle F.W., Quelle D.E. 2005. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 25:1258–1271 10.1128/MCB.25.4.1258-1271.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M.L., den Besten W., Thomas M.C., Sherr C.J. 2008. Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle. 7:3378–3387 [DOI] [PubMed] [Google Scholar]
- Li Z., Boone D., Hann S.R. 2008. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc. Natl. Acad. Sci. USA. 105:18794–18799 10.1073/pnas.0806879105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi L.B., Jr., Kuchenruether M., Dadey D.Y., Schwope R.M., Grisendi S., Townsend R.R., Pandolfi P.P., Weber J.D. 2008. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the Mammalian ribosome. Mol. Cell. Biol. 28:7050–7065 10.1128/MCB.01548-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L., Song Z., Jin L., Zhu Y.M., Wen L.P., Wu M. 2010. ARF antagonizes the ability of Miz-1 to inhibit p53-mediated transactivation. Oncogene. 29:711–722 10.1038/onc.2009.372 [DOI] [PubMed] [Google Scholar]
- Orian-Rousseau V., Chen L., Sleeman J.P., Herrlich P., Ponta H. 2002. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev. 16:3074–3086 10.1101/gad.242602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster S.K., Ho C.S., Soucie E.L., Penn L.Z. 2002. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84:81–154 10.1016/S0065-230X(02)84004-0 [DOI] [PubMed] [Google Scholar]
- Ouyang J., Shi Y., Valin A., Xuan Y., Gill G. 2009. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol. Cell. 34:145–154 10.1016/j.molcel.2009.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.B., Park M.J., Kimura K., Shimizu K., Lee S.H., Yokota J. 2002. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet. Cytogenet. 133:105–111 10.1016/S0165-4608(01)00575-1 [DOI] [PubMed] [Google Scholar]
- Patel J.H., McMahon S.B. 2007. BCL2 is a downstream effector of MIZ-1 essential for blocking c-MYC-induced apoptosis. J. Biol. Chem. 282:5–13 10.1074/jbc.M609138200 [DOI] [PubMed] [Google Scholar]
- Pear W.S., Nolan G.P., Scott M.L., Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 90:8392–8396 10.1073/pnas.90.18.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peukert K., Staller P., Schneider A., Carmichael G., Hänel F., Eilers M. 1997. An alternative pathway for gene regulation by Myc. EMBO J. 16:5672–5686 10.1093/emboj/16.18.5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Gregory M.A., Li Z., Brousal J.P., West K., Hann S.R. 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 431:712–717 10.1038/nature02958 [DOI] [PubMed] [Google Scholar]
- Quelle D.E., Zindy F., Ashmun R.A., Sherr C.J. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 83:993–1000 10.1016/0092-8674(95)90214-7 [DOI] [PubMed] [Google Scholar]
- Reginato M.J., Mills K.R., Paulus J.K., Lynch D.K., Sgroi D.C., Debnath J., Muthuswamy S.K., Brugge J.S. 2003. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat. Cell Biol. 5:733–740 10.1038/ncb1026 [DOI] [PubMed] [Google Scholar]
- Salghetti S.E., Kim S.Y., Tansey W.P. 1999. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18:717–726 10.1093/emboj/18.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J., Pouponnot C., Staller P., Schader M., Eilers M., Massagué J. 2001. TGFbeta influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400–408 10.1038/35070086 [DOI] [PubMed] [Google Scholar]
- Seoane J., Le H.V., Massagué J. 2002. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 419:729–734 10.1038/nature01119 [DOI] [PubMed] [Google Scholar]
- Staller P., Peukert K., Kiermaier A., Seoane J., Lukas J., Karsunky H., Möröy T., Bartek J., Massagué J., Hänel F., Eilers M. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 3:392–399 10.1038/35070076 [DOI] [PubMed] [Google Scholar]
- Stielow B., Sapetschnig A., Krüger I., Kunert N., Brehm A., Boutros M., Suske G. 2008. Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell. 29:742–754 10.1016/j.molcel.2007.12.032 [DOI] [PubMed] [Google Scholar]
- Stott F.J., Bates S., James M.C., McConnell B.B., Starborg M., Brookes S., Palmero I., Ryan K., Hara E., Vousden K.H., Peters G. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001–5014 10.1093/emboj/17.17.5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K., Chiocca S., Sherr C.J. 2005. Sumoylation induced by the Arf tumor suppressor: a p53-independent function. Proc. Natl. Acad. Sci. USA. 102:7689–7694 10.1073/pnas.0502978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzel M., Russ A.C., Kleine-Kohlbrecher D., Colombo E., Pelicci P.G., Eilers M. 2008. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nat. Cell Biol. 10:1051–1061 10.1038/ncb1764 [DOI] [PubMed] [Google Scholar]
- Weber J.D., Jeffers J.R., Rehg J.E., Randle D.H., Lozano G., Roussel M.F., Sherr C.J., Zambetti G.P. 2000. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 14:2358–2365 10.1101/gad.827300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.L., Wu Q., Vega V.B., Chiu K.P., Ng P., Zhang T., Shahab A., Yong H.C., Fu Y., Weng Z., et al. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell. 124:207–219 10.1016/j.cell.2005.10.043 [DOI] [PubMed] [Google Scholar]
- Wettenhall J.M., Smyth G.K. 2004. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 20:3705–3706 10.1093/bioinformatics/bth449 [DOI] [PubMed] [Google Scholar]
- Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.C., Knabenhans C., Macdonald H.R., Trumpp A. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18:2747–2763 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong Y., Yarbrough W.G. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 92:725–734 10.1016/S0092-8674(00)81401-4 [DOI] [PubMed] [Google Scholar]
- Zindy F., Eischen C.M., Randle D.H., Kamijo T., Cleveland J.L., Sherr C.J., Roussel M.F. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424–2433 10.1101/gad.12.15.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Williams R.T., Baudino T.A., Rehg J.E., Skapek S.X., Cleveland J.L., Roussel M.F., Sherr C.J. 2003. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc. Natl. Acad. Sci. USA. 100:15930–15935 10.1073/pnas.2536808100 [DOI] [PMC free article] [PubMed] [Google Scholar]