The small GTPase Arl13b regulates ciliary transmembrane protein localizations and anterograde IFT assembly stability.
Abstract
The small ciliary G protein Arl13b is required for cilium biogenesis and sonic hedgehog signaling and is mutated in patients with Joubert syndrome (JS). In this study, using Caenorhabditis elegans and mammalian cell culture systems, we investigated the poorly understood ciliary and molecular basis of Arl13b function. First, we show that Arl13b/ARL-13 localization is frequently restricted to a proximal ciliary compartment, where it associates with ciliary membranes via palmitoylation modification motifs. Next, we find that loss-of-function C. elegans arl-13 mutants possess defects in cilium morphology and ultrastructure, as well as defects in ciliary protein localization and transport; ciliary transmembrane proteins abnormally accumulate, PKD-2 ciliary abundance is elevated, and anterograde intraflagellar transport (IFT) is destabilized. Finally, we show that arl-13 interacts genetically with other ciliogenic and ciliary transport–associated genes in maintaining cilium structure/morphology and anterograde IFT stability. Together, these data implicate a role for JS-associated Arl13b at ciliary membranes, where it regulates ciliary transmembrane protein localizations and anterograde IFT assembly stability.
Introduction
Joubert syndrome (JS) and related cerebellar disorders (JSRDs) are rare autosomal recessive disorders characterized by multiple symptoms, including developmental delay, mental retardation, and hypotonia, as well as a defining midbrain–hindbrain junction malformation called the molar tooth sign (Millen and Gleeson, 2008). Seven JS/JSRD genes are cloned: AHI1, CEP290, NPHP1, MKS3, RPGRIP1L, ARL13B, and CC2D2A (Dixon-Salazar et al., 2004; Ferland et al., 2004; Parisi et al., 2004; Castori et al., 2005; Sayer et al., 2006; Valente et al., 2006a,b; Baala et al., 2007; Cantagrel et al., 2008; Gorden et al., 2008). Because most of these genes encode ciliary proteins, are required for cilium structure/function, and cause ciliary disease symptoms (cystic kidneys, retinitis pigmentosa, etc.), JSRDs are thought to be caused by defects in cilia.
Cilia extend from most eukaryotic cell surfaces, serving motility and sensory functions such as chemo-, photo-, and mechanotransduction, as well as facilitating developmental signaling (e.g., Shh; Christensen et al., 2007; Eggenschwiler and Anderson, 2007; Quinlan et al., 2008). Cilia are built by intraflagellar transport (IFT), a kinesin-2– and cytoplasmic dynein–driven bidirectional movement of protein complexes between the ciliary base and tip which traffics ciliary protein cargoes and turnover products (Rosenbaum, 2002; Qin et al., 2004, 2005; Blacque et al., 2008). Associated with the motor machinery and essential for IFT is the IFT particle, consisting of two subcomplexes, IFT-A (>6 proteins) and IFT-B (>12 proteins; Rosenbaum, 2002; Blacque et al., 2008). Discoveries linking IFT defects to developmental (e.g., Shh) signaling abnormalities highlight the importance of IFT for ciliary signaling and suggest that mistransport of ciliary proteins contributes to the development of ciliopathy symptoms (Blacque and Leroux, 2006; Eggenschwiler and Anderson, 2007).
Caenorhabditis elegans is an established model organism for investigating cilium biogenesis/function, IFT, and ciliopathy gene function (Inglis et al., 2007). In worms, cilia extend from the distal dendrite tips of sensory neurons, forming sensory structures that are typically environmentally exposed. IFT can be directly visualized in live worms, and mutant alleles and markers are available for most IFT proteins. In C. elegans, IFT is most extensively studied in rod-shaped amphid channel cilia, consisting of a middle segment (MS; nine outer doublet microtubules [MTs]) and distal segment (DS; nine outer singlet MTs). Two kinesin-2 motors (heterotrimeric kinesin-II and homodimeric OSM-3) act cooperatively to drive MS anterograde IFT at ∼0.7 µms−1 (Orozco et al., 1999; Snow et al., 2004; Ou et al., 2005; Pan et al., 2006). At MS tips, kinesin-II disengages, and OSM-3 completes DS IFT at ∼1.2–1.3 µms−1. Although IFT regulation is poorly understood in any organism, C. elegans BBS (Bardet-Biedl syndrome) proteins coordinate the association of kinesin-2 motors and IFT-A/B complexes (Ou et al., 2005, 2007), DYF-5 restricts kinesin-II to MSs and regulates OSM-3–kinesin translocation rates (Burghoorn et al., 2007), and NPH (nephronophthisis) proteins differentially regulate ciliary entry of IFT proteins and OSM-6/IFT52 translocation rates (Jauregui et al., 2008). Regarding IFT cargoes, the ciliary transmembrane protein OSM-9 undergoes IFT-like motility, and PKD-2 (polycystin 2) ciliary abundance is IFT regulated (Qin et al., 2005; Bae et al., 2006).
To identify new genes with ciliary functions, including ciliopathy genes, one approach has been to select candidates from published datasets enriched for cilia-related genes and proteins. These ciliomes have been identified in different systems using many experimental approaches (Inglis et al., 2006). For example, in C. elegans, bioinformatic screening for binding sites (X-boxes) of the ciliogenic transcription factor DAF-19, together with identifying genes expressed only in ciliated cells, uncovered new IFT-related components (Blacque et al., 2005; Inglis et al., 2006) and aided in cloning at least one new ciliopathy gene (Fan et al., 2004). One promising candidate ciliogenic gene to emerge from ciliomics is C. elegans arl-13 (Y37E3.5), an X-box–containing gene expressed only in ciliated cells and encoding a ciliary protein (Blacque et al., 2005). Vertebrate ARL-13 (Arl13b) also localizes to cilia and is required for ciliogenesis and Shh signaling (Sun et al., 2004; Caspary et al., 2007; Hori et al., 2008; Duldulao et al., 2009). Recently, mutations in Arl13b have been found in JS patients, thereby confirming Arl13b as a ciliopathy gene (Cantagrel et al., 2008). ARL-13/Arl13b is an Arf-like member of the Ras superfamily of small GTPases implicated in vesicle trafficking, cellular differentiation, cell movement, and cytoskeletal processes. Indeed, ciliary functions are known for other small G proteins such as ARL3, ARL6/BBS3, and RAB8, indicating that multiple GTPases serve cilia-related functions (Cuvillier et al., 2000; Fan et al., 2004; Nachury et al., 2007; Yoshimura et al., 2007).
Although Arl13b is known to function in ciliogenesis and vertebrate Shh signaling, the ciliary and molecular basis of these functions remains unclear. In this study, we used C. elegans and mammalian tissue culture models to show that ARL-13/Arl13b is a ciliary membrane–associated protein, which in certain ciliary subtypes is restricted to the proximal ciliary region. Furthermore, we find that C. elegans ARL-13 maintains cilium structure/morphology and provide evidence that ARL-13 is required for ciliary transmembrane protein localizations/abundance and the stabilization of anterograde IFT assemblies. From these data, we propose that ARL-13/Arl13b functions at ciliary membranes to stabilize ciliary protein transport processes.
Results
ARL-13 is homologous to JS-associated Arl13b
BLAST analyses identified ARL-13 (Y37E3.5) sequence homologues in mice (Arl13b), zebrafish (Scorpion), Xenopus laevis, and Chlamydomonas reinhardtii, each possessing an extended tail (100–300 residues) C-terminal to the GTPase domain (Fig. S1 A). In contrast, the top hits in Drosophila melanogaster and Tetrahymena thermophila returned lower BLAST scores and lacked extended tails, indicating that ARL-13 homologues are not present in all ciliates (Fig. S1 A). Sequence alignment showed that all ARL-13 homologues contain an N-terminal palmitoylation (Pal) modification motif (not found in most G proteins), possess the three residues (R79, W82, and R200) mutated in patients with Arl13b-associated JS (Cantagrel et al., 2008), and lack the switch two Gln residue critical for GTPase activity in most Arl/Arfs (Fig. S1 B). Together with previous findings that nematode ARL-13 and vertebrate Arl13b localize exclusively to cilia (Blacque et al., 2005; Caspary et al., 2007; Cantagrel et al., 2008; Duldulao et al., 2009), in addition to being required for proper cilium formation (see Figs. 3 and 4; Sun et al., 2004; Caspary et al., 2007; Hori et al., 2008; Duldulao et al., 2009), we conclude that ARL-13 is evolutionarily conserved and represents the orthologue of JS-associated Arl13b.
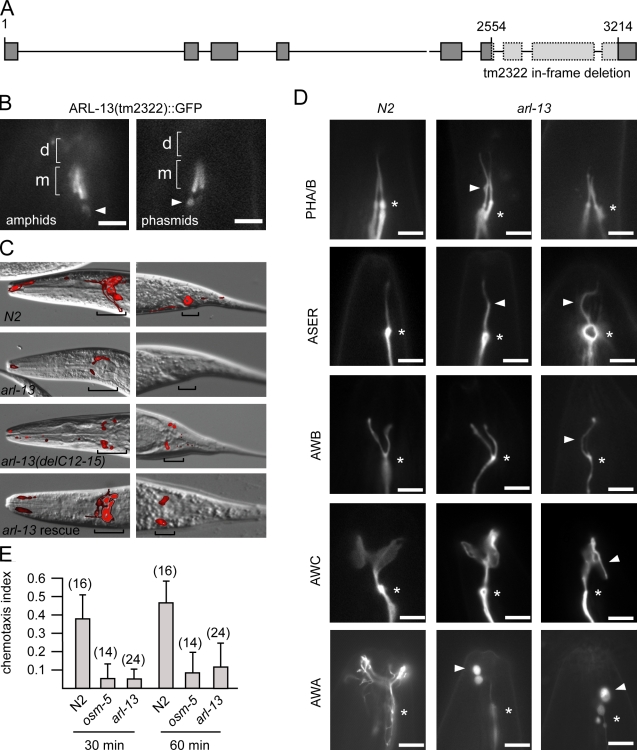
Figure 3.
C. elegans arl-13 mutants possess defective cilium structure, morphology, and functions. (A) arl-13 gene schematic, showing genomic position of in-frame tm2322 deletion. (B) tm2322 encodes a ciliary ARL-13 protein. Fluorescence images of amphid/phasmid channel cilia in worms expressing arl-13(tm2322)::gfp are shown. GFP signals are restricted to MSs (m) and excluded from DSs (d). Some abnormal accumulations are found beneath cilia (arrowheads). (C) tm2322 mutants are dye-filling (Dyf) defective. Merged differential interference contrast (DIC)–fluorescence images after a DiI uptake assay are shown. Dye uptake into amphid (head) and phasmid (tail) neuron cell bodies (denoted by brackets) is strongly reduced in tm2322 mutants and weakly reduced in WT animals expressing arl-13(delPal)::gfp but restored in tm2322 animals expressing arl-13(WT)::gfp. (D) Cilium morphologies are defective in tm2322 worms. Fluorescence images of cilia from N2 and tm2322 animals expressing ciliated cell–specific transcriptional GFP markers srb-6p::gfp (PHA/B), gcy-5p::gfp (ASER), str-1p::gfp (AWB), str-2p::gfp (AWC), and gpa-6p::gfp (AWA) are shown. All images are similarly orientated, with ciliary base denoted (asterisks). Arrowheads indicate morphology defects such as kinks (ASER), bulges (PHA/B and AWA), curls (AWB,) and ectopic projections (AWC). (E) tm2322 mutants are chemosensory defective. Indices obtained from 30- and 60-min chemoattraction assays toward isoamyl alcohol for tm2322, N2, and osm-5(p802) worms are shown. Assay numbers are shown in parentheses. Error bars indicate SEM. Bars: (B and D) 3 µm; (C) 10 µm.
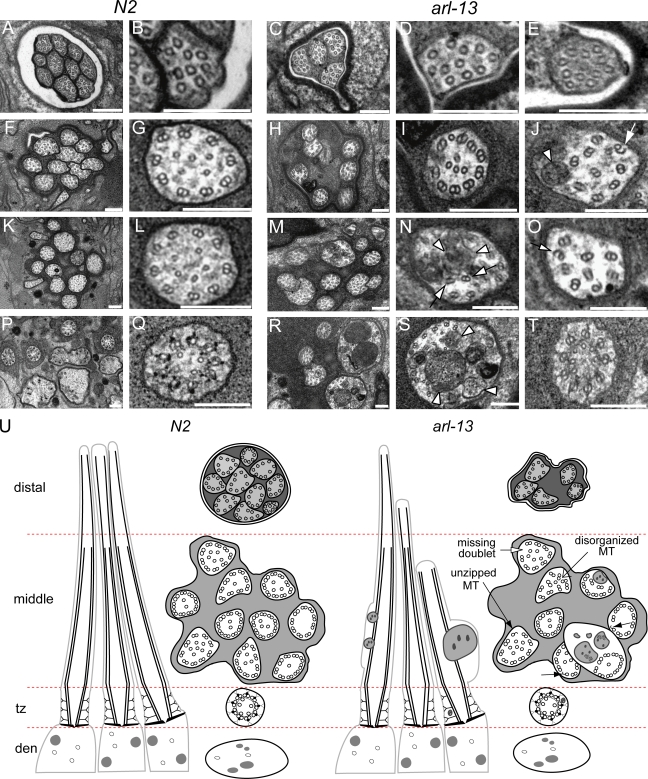
Figure 4.
arl-13(tm2322) mutant cilia possess ultrastructural defects. Transmission electron microscopy serial cross sections of amphid pore from N2 and tm2322 worms. (A–E) Distal pore showing 10 axonemes in N2 worms (A and B) but only 4 axonemes in tm2322 mutants (C–E). (F–J) 2 µm proximal to A–E (through MSs) showing axonemes still missing in tm2322 worms (H). Also, abnormal accumulation of electron-dense material (J, arrowhead) and unzipping of MTs (J, arrow) are observed. (K–O) 3 µm proximal to A–E (through MSs) showing most ciliary axonemes are present in tm2322 mutants (M). Further abnormal axonemal accumulations of amorphous electron-dense material (N, arrowheads) and missing, misplaced, and unzipped doublet MTs (N and O, arrows) are also shown. (P–T) 5 µm proximal to A and B, through MSs, transition zones, and transitional fibers. Some tm2322 cilia are abnormally enlarged (R and S), filled with electron-dense material that is amorphous, or contained within vesicle-type structures (R and S). (S) Arrowheads indicate amorphous and vesicular-like accumulations. Transition zones appear normal (T). (U) Schematics of amphid channel cilia from N2 and tm2322 mutants, showing the major ultrastructural defects observed. tz, transition zone. Bars, 200 µm.
ARL-13/Arl13b is enriched in proximal regions of C. elegans and mammalian cilia
Although C. elegans ARL-13 was previously shown to localize to cilia (Blacque et al., 2005), published images suggest that ARL-13 may not localize along the entire axoneme. To assess this further, we made new transgenic lines expressing a rescuing arl-13::gfp construct. Unlike GFP-tagged IFT proteins, which extended 6–8 µm from the ciliary base, ARL-13::GFP signals in amphid/phasmid cilia extended only ∼3–4 µm (Fig. 1 A). A similar restricted proximal ciliary localization was also observed in male tail ray neuronal cilia (Fig. 1 A). In contrast, although enriched in the proximal region of AWB cilia, ARL-13::GFP localized to the entire AWB cilium, including distal fan membranes (Fig. 1 A), indicating that ARL-13 is not excluded from the distal regions of all nematode cilia. Similarly, endogenous mammalian Arl13b localized predominantly to proximal ciliary regions of MDCKII cilia (Fig. 1 B), although in RPE1 cells, Arl13b localizes to the entire axoneme (Hori et al., 2008). Together, these findings show that in various ciliary subtypes, ARL-13/Arl13b is highly enriched in proximal ciliary regions, with little or no detectable protein in distal regions. Finally, using time-lapse videomicroscopy, no ciliary translocation was found for C. elegans ARL-13::GFP, indicating that ARL-13 does not undergo IFT (Fig. 1 A).
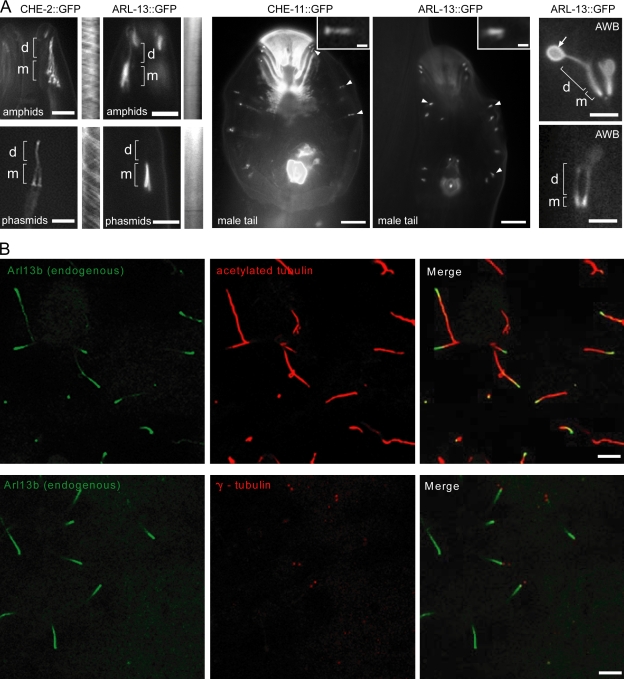
Figure 1.
ARL-13/Arl13b is enriched in proximal regions of ciliary axonemes. (A) ARL-13::GFP localizes almost exclusively to the proximal region (MSs; m) of amphid/phasmid cilia and male tail ray cilia. Although localizing to all regions of AWB cilia, ARL-13 is enriched in proximal regions and observed at the membrane of distal fans (arrow). IFT markers (CHE-2 and -11) label amphid/phasmid channel MSs and DSs (d) and RnA/B male tail ray cilia. Kymographs, from time-lapse videos, show no IFT movement for ARL-13::GFP. Arrowheads indicate RnA/B cilia, and insets show high magnification images of RnA/B cilia. (B) Arl13b is enriched in the proximal region of MDCKII cilia. Images after immunostaining for endogenous Arl13b, acetylated α-tubulin (ciliary axoneme), and γ-tubulin (basal body) are shown. Bars: (A, left and right) 3 µm; (A, middle) 10 µm; (A, insets) 1 µm; (B) 5 µm.
ARL-13/Arl13b associates with the ciliary membrane via palmitoyl anchors
CSS-Palm 2.0 software (Zhou et al., 2006) identified a putative N-terminal Pal modification motif in ARL-13 and Arl13b that may anchor this G protein to ciliary membranes (Fig. S1 B). To investigate this hypothesis, we expressed in C. elegans two Pal variants of ARL-13 (GFP tagged): delPal (deletion of C12–C15) and rPal (replacement of C12–C15 with ASAS). Both proteins were mislocalized, displaying only weak ciliary signals and abnormal strong cytosolic (diffuse) signals in cell bodies, dendrites, and axons (Fig. 2 A). Quantification of signals showed that although >90% of ARL-13(wild type [WT]) was ciliary, only 10–20% of the Pal variant signals were ciliary (Fig. 2 A). The diffuse Pal variant localizations suggest loss of membrane tethering, and in support of this notion, a C-terminal ARL-13 truncation variant (del203–370; intact Pal motif) retained membrane associations, although delocalized in cell bodies and dendrites (Fig. 2 A, arrows). Similarly, in mammalian cells, Arl13b(C8S/C9S) was also mislocalized, with little signal in cilia and diffuse extra-ciliary signals indicative of membrane dissociation (Fig. 2 B). Subcellular fractionation analysis of 293T cells transfected with HsArl13b::Flag (WT or variants) revealed that Arl13b(WT) pelleted at 20,000 g together with the Na+/K+ ATPase membrane protein (Fig. 2 C). In contrast, C8S/C9S and 20–428 Pal variants cofractionated with the cytosolic protein calpain even after 200,000 g spins (Fig. 2 C). Consistent with these data, a C-terminal truncation variant (1–357), which retains the Pal motif, fractionated in an identical manner to Arl13b(WT) (Fig. 2 C). These findings show that Pal motif disruption alters the subcellular fractionation of Arl13b, shifting it from membrane protein–associated fractions to cytosolic fractions. Finally, [3H]palmitic acid metabolic labeling of 293T cells transfected with GFP-tagged Arl13b (WT or C8S/C9S variant) revealed that Arl13b(WT)::GFP but not Arl13b(C8S/C9S)::GFP incorporated [3H]palmitate (Fig. 2 D, arrow). Together, these data indicate that ARL-13/Arl13b associates with ciliary membranes via palmitoyl anchors.
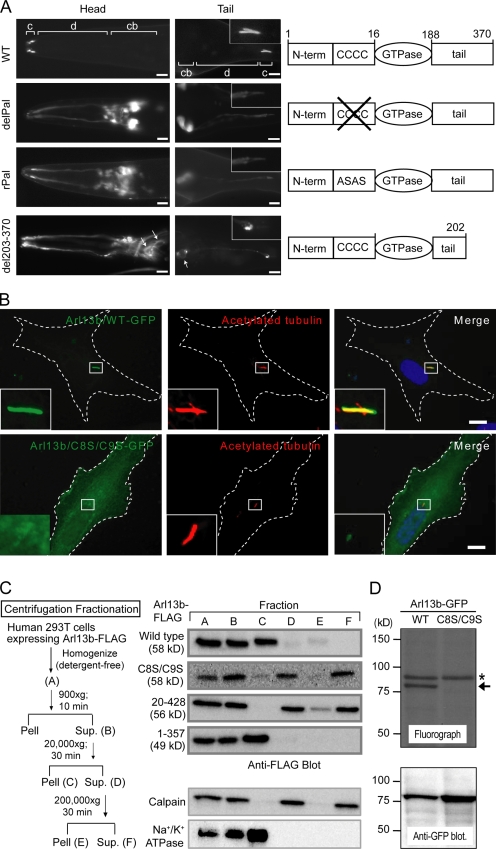
Figure 2.
ARL-13/Arl13b associates with ciliary membranes via palmitoyl anchors. (A) N-terminal Pal motif ARL-13 variants (delPal and rPal) fail to localize to amphid (head) and phasmid (tail) cilia (c) and diffusely mislocalize in cell bodies (cb) and dendrites (d). C-terminal deletion ARL-13(del203–370) is also mislocalized, but membrane associations are maintained (arrows). Insets show high magnification images of PHA/B cilia. (B) HsArl13b ciliary localization requires an N-terminal Pal motif. Ciliated RPE1 cells transfected with GFP-tagged Arl13b(WT) or a C8S/C9S variant and costained for ciliary axonemes using acetylated α-tubulin antibody are shown. Merged images show that Arl13b(C8S/C9S) is highly diffuse and nonmembrane associated, with weak signals in cilia. The boxed regions are shown at high magnification in the insets. Dashed lines outline the cell. (C) Loss of Pal motif shifts HsArl13b to cytosolic fractions. A subcellular fractionation scheme for Flag-tagged Arl13b (WT and variants) transiently expressed in 293T cells is shown. Supernatants (Sup.) and pellets (Pell) were probed for Arl13b::Flag using Western blotting and an anti-Flag antibody. Calpain and Na+/K+ ATPase proteins mark cytosol and membrane fractions, respectively. Arl13b(WT) and Arl13b(1–357) pellet with Na+/K+ ATPase membrane proteins after 20,000 g spins, whereas Arl13b(C8S/C9S) remains in calpain-enriched cytosolic supernatants even after 200,000 g spins. (D) Arl13b is palmitoylated in 293T cells. 293T cells transiently transfected with GFP-tagged Arl13b(WT) or Arl13b(C8S/C9S) were metabolically labeled with [3H]palmitate, and proteins were separated by SDS-PAGE. The top panel shows a fluorograph; the bottom panel shows a Western blot with anti-GFP antibody. The arrow indicates palmitoylated Arl13b-GFP, and the asterisk indicates an unknown endogenous palmitoylated protein. Bars: (A) 5 µm; (B) 10 µm.
arl-13 is required for normal sensory cilium morphology, ultrastructure, and function
Although Arl13b is linked to cilium biogenesis and Shh signaling, the molecular basis of these functions is poorly understood (Caspary et al., 2007; Cantagrel et al., 2008; Hori et al., 2008; Duldulao et al., 2009). In this study, we investigated C. elegans ARL-13 function using the tm2322 in-frame deletion, which is predicted to encode an ARL-13 protein lacking aa 169–342 (Fig. 3 A). Using a gfp-tagged transgene, we found that although some ARL-13(tm2322) accumulated beneath cilia and in dendrites/cell bodies, most protein still resided in cilia (Fig. 3 B), indicating that tm2322 is not likely a null mutation but instead encodes a protein with residual/altered ciliary functions.
First, we found that tm2322 mutants were partially dye-filling defective (Dyf), with 40% of phasmids failing to incorporate dye (DiI), and although amphid neurons were less affected, dye uptake was frequently reduced or absent (Fig. 3 C). Transgenic expression of arl-13(WT)::gfp in tm2322 animals rescued the Dyf phenotype (Fig. 3 C). Interestingly, expression of arl-13[delC12–C15]::gfp (delPal variant) in WT worms caused a mild Dyf phenotype, suggesting a weak dominant-negative phenotype (Fig. 3 C). Consistent with the Dyf phenotype, direct visualization of tm2322 cilia using transcriptional GFP markers revealed defective cilium shapes (Fig. 3 D). PHA/B cilia were often kinked, possessed axonemal bulges, and frequently failed to coalesce (Fig. 3 D). AWA cilia typically lacked the characteristic multibranched structure and possessed abnormal bulbous distal tips, and although less affected, ASER, AWB and AWC cilia were often kinked (ASER) or curly (ASER and AWB) or possessed ectopic projections (AWC; Fig. 3 D). Next, using transmission electron microscopy, we examined tm2322 amphid channel cilium ultrastructure. Similar to WT cilia, tm2322 cilia were segmented, possessing DSs, MSs, and transition zone and transition fiber segments (Fig. 4). However, axonemes were often missing in arl-13 animals, indicating that some cilia were truncated, missing, and/or misdirected away from the pore (Fig. 4 C). Also, arl-13 mutant cilia frequently exhibited MS defects, including enlarged axonemes, and accumulation of amorphous electron-dense matrix material, which in some cases appeared within vesicle-type structures (Fig. 4, J, M, N, R, S, and U). These MS enlargements may correspond to the axonemal bulges described in Fig. 3 D. In addition, outer doublet MTs were frequently misplaced/misaligned (Fig. 4, J, N, O, S, and U, arrows) and of incorrect number (not depicted), and there is evidence of an increased frequency of unzipped MTs (Fig. 4, J, O, and U). Consistent with cilium structure/ultrastructure defects, tm2322 mutants displayed abrogated isoamyl alcohol chemosensation, a behavior mediated by AWA/AWC cilia (Fig. 3 E).
Together, these data corroborate and extend previous vertebrate Arl13b data (Caspary et al., 2007; Cantagrel et al., 2008; Hori et al., 2008; Duldulao et al., 2009) by showing that C. elegans arl-13 is required for normal cilium morphology, ultrastructure, and function. Particularly intriguing is that arl-13 mutant cilia possessed enlarged MS diameters and axonemal accumulations of dense matrix–filled vesicle structures, as well as subtle MT abnormalities; these findings could suggest roles for ARL-13 in ciliary membrane maintenance/turnover and/or axonemal MT stability/integrity.
Ciliary transmembrane protein localizations are disrupted in arl-13 mutants
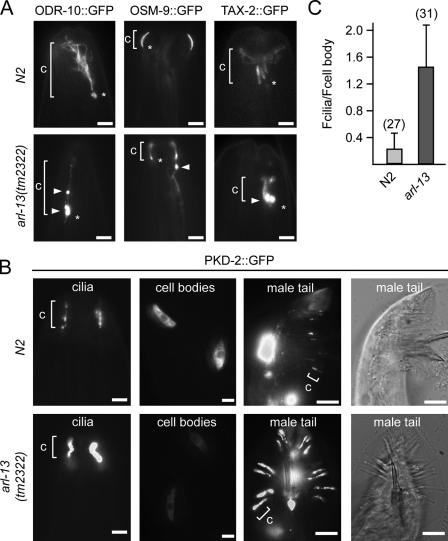
To further investigate potential ciliary membrane defects, we examined the ciliary localizations of chemo- or mechanosensory transmembrane proteins (PKD-2, ODR-10, TAX-2, and OSM-9) in tm2322 worms, and in all cases, defects were found (Fig. 5, A and B). Specifically, OSM-9::GFP accumulated in the distal dendrites of 55% of arl-13 mutants (n = 55), whereas ODR-10::GFP and TAX-2::GFP accumulated in the cilium and/or distal dendrites of 90% (n = 69) and 51% (n = 27) of animals, respectively. In WT animals, no such accumulations were observed for OSM-9::GFP, and only 9% of animals displayed weak ODR-10 ciliary accumulations. We also noticed that the ciliary abundance of PKD-2 was higher in arl-13 mutants compared with WT animals, with the former possessing five times more PKD-2 than the latter (Fig. 5, B and C). Conversely, the intensity of cell body PKD-2 signals in arl-13 worms was 50% less than that of WT worms (Fig. 5 C). This was not the result of alterations in pkd-2::gfp expression levels because the combined intensity of cell body and cilium signals was similar (unpublished data). Consistent with weak dominant-negative effects by arl-13(delPal) transgene overexpression on dye filling (Fig. 3 C), PKD-2 ciliary abundance was not increased in these animals (Fig. S2). Together, these data show that ARL-13 is required for proper targeting/retention of ciliary transmembrane proteins.
Figure 5.
Ciliary transmembrane protein localization is disrupted in arl-13(tm2322) mutants. (A and B) Representative fluorescence images of the distal head region (nose) of worms expressing gfp-tagged ODR-10, OSM-9, TAX-2, and PKD-2 are shown. In tm2322 mutants, abnormal accumulations (arrowheads) are found in ciliary axonemes (ODR-10), near the ciliary base (ODR-10 and TAX-2; asterisks), or within the distal dendrite (OSM-9; arrowhead). In tm2322 mutants, PKD-2::GFP ciliary abundance is elevated in CEM and RnB cells, with cell body levels reduced (shown for CEMs). c, cilium. (C) Analysis of PKD-2::GFP ciliary abundance in CEM cells. The ratio of PKD-2::GFP signal intensities in individual CEM cilia (Fcilia) and individual CEM cell bodies (Fcell body) is shown. All images were captured and analyzed using identical settings. The number of cilia analyzed is shown in parentheses. Error bars indicate SEM. Bars: (A and B [first and second columns]) 2 µm; (B, third and fourth columns) 10 µm.
IFT is disrupted in arl-13 mutants
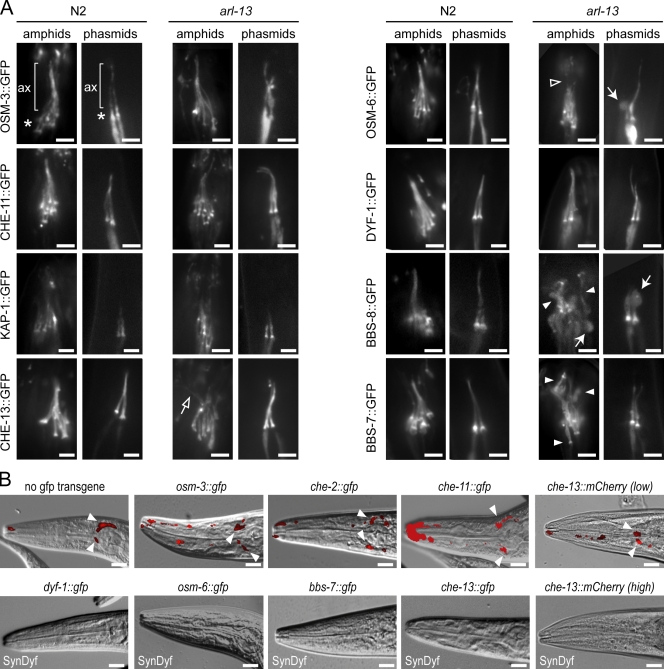
Because IFT-related gene mutants also possess defects in the ciliary localizations of transmembrane proteins (Bae et al., 2006; Tan et al., 2007), we hypothesized that IFT may be disrupted in arl-13 mutants. Using GFP markers, we found that IFT proteins localized normally at the ciliary base and axoneme of arl-13 mutants (Fig. 6 A). However, cilium structure/morphology in certain transgenic tm2322 strains appeared more disrupted than other transgenic strains; for example, in arl-13 worms overexpressing che-13::gfp, osm-6::gfp, or bbs-7/8::gfp, cilia were more highly disorganized and dispersed than other transgenic strains (Fig. 6 A), suggesting that overexpression of certain IFT transgenes enhances the cilium defects of tm2322 mutants. Using dye filling to assess this further, arl-13 mutants expressing gfp-tagged kap-1, osm-6, bbs-7, bbs-8, dyf-1, or che-13 were found to possess stronger Dyf phenotypes (SynDyf) than nontransgenic arl-13 worms or arl-13 mutants expressing gfp-tagged osm-3, che-2, or che-11 (Fig. 6 B). Together, these findings indicate that IFT transgene overexpression is toxic to tm2322 worms, causing enhanced cilium structure defects.
Figure 6.
IFT transgene overexpression enhances the structure/morphology defects of tm2322 mutant cilia. (A) Fluorescence images of amphid/phasmid cilia in N2 and arl-13(tm2322) animals expressing gfp-tagged IFT transgenes show that IFT proteins localize normally to ciliary axonemes (ax) and accumulate normally at ciliary bases (asterisks) in tm2322 worms. In tm2322 mutants overexpressing che-13::gfp, osm-6::gfp, and bbs-7/8::gfp, amphid/phasmid cilia morphologies are more severely defective than other transgenic strains. Enhanced defects include missing DS staining (open arrowhead), axonemes that are less tightly bunched and highly disorganized (closed arrowheads; open arrow), and increased frequency of large axonemal bulges (closed arrows). (B) arl-13 animals overexpressing osm-3::gfp, che-2::gfp, che-11::gfp, and che-13::mCherry(low expression level) display similar levels of DiI incorporation to nontransgenic controls (arrowheads indicate dye uptake in cell bodies). However, tm2322 mutants overexpressing che-13::gfp, osm-6::gfp, bbs-7::gfp, dyf-1::gfp, and che-13::mCherry(high expression level) are SynDyf, failing to take dye. Merged DIC–fluorescence images (head) after a DiI uptake assay are shown. Bars: (A) 2 µm; (B) 10 µm.
Next, we used IFT motility assays to measure anterograde IFT in arl-13 worms. Like previous studies (Snow et al., 2004; Burghoorn et al., 2007; Jauregui et al., 2008), IFT occurred along WT amphid channel cilium MSs at a mean speed of 0.71 µms−1 (0.67–0.75 µms−1) and along DSs at a mean speed of 1.15 µms−1 (1.11–1.19 µms−1; Fig. S3 and Table I). These rates are consistent with the cooperative model of kinesin-II (slow motor; 0.5 µms−1) and OSM-3 (fast motor; 1.3 µms−1) driving MS IFT at intermediate speeds (0.7 µms−1), with OSM-3 alone driving DS transport at fast speeds (1.3 µms−1; Snow et al., 2004). However, in tm2322 strains expressing gfp-tagged kap-1, osm-6, che-13, dyf-2, and bbs-7 (all SynDyf except the dyf-2–expressing strain), MS IFT proceeded at a mean speed of 0.51 µms−1 (0.47–0.55 µms−1), suggesting transport by kinesin-II alone (Fig. S3, Table I, and Videos 1 and 2). Surprisingly, in tm2322 worms expressing gfp-tagged osm-3, dyf-1, che-2, che-11, and ifta-1 (all non-SynDyf except the dyf-1–expressing strain), MS IFT proceeded at mean speeds of ∼0.71 µms−1 (0.66–0.76 µms−1), which is identical to WT speeds and indicates normal trafficking by OSM-3 and kinesin-II (Fig. S3 and Table I). Consistent with the latter, CHE-11::GFP slowed to ∼0.5 µms−1 in arl-13;osm-3 worms (Table I). However, although mean MS speeds were normal, OSM-3::GFP MS rate profiles showed more fast particles (>0.9 µm/s) in arl-13 mutants compared with WT worms (Fig. S3), suggesting that some OSM-3 is uncoupled from kinesin-II. Similarly, CHE-11::GFP rate profiles showed increased numbers of slow-moving particles (Fig. S3), suggesting some CHE-11 may be moving with an uncoupled kinesin-II pool.
Table I.
Anterograde IFT velocities in arl-13 mutants
| IFT Protein | Strain | MS | DS | ||||
| Mean velocity/SD | n/N | t test | Mean velocity/SD | n/N | t test | ||
| µms−1 | µms−1 | ||||||
| OSM-3::GFP | N2 | 0.70/0.12 | 258/18 | 1.17/0.21 | 63/6 | ||
| OSM-3::GFP | arl-13 | 0.76/0.29 | 210/17 | P = 0.001 | 0.98/0.26 | 199/18 | P < 0.001 |
| OSM-3::GFP | klp-11 | 1.17/0.16 | 155/12 | P < 0.001 | 1.21/0.20 | 76/8 | P = 0.09 |
| OSM-3::GFP | arl-13;klp-11 | 0.97/0.19 | 242/16 | P < 0.001 | 1.04/0.17 | 63/4 | P < 0.001 |
| KAP-1::GFP | N2 | 0.69/0.11 | 92/9 | ND | ND | ||
| KAP-1::GFP | arl-13 | 0.51/0.13 | 168/35 | P < 0.001 | ND | ND | ND |
| DYF-1::GFP | N2 | 0.67/0.29 | 189/9 | 1.12/0.22 | 71/6 | ||
| DYF-1::GFP | arl-13 | 0.68/0.24 | 107/5 | P = 0.44 | 0.98/0.25 | 60/6 | P < 0.001 |
| CHE-13::GFP | N2 | 0.72/0.14 | 114/7 | 1.11/0.24 | 93/8 | ||
| CHE-13::GFP | arl-13 | 0.54/0.18 | 262/10 | P < 0.001 | 0.92/0.30 | 62/4 | P < 0.001 |
| CHE-13::mCherry(low) | N2 | 0.73/0.10 | 73/10 | 1.17/0.22 | 54/9 | ||
| CHE-13::mCherry(low) | arl-13 | 0.72/0.22 | 106/15 | P = 0.47 | 1.02/0.21 | 58/13 | P < 0.001 |
| CHE-13::mCherry(high) | N2 | 0.73/0.16 | 79/3 | 1.21/0.23 | 33/2 | ||
| CHE-13::mCherry(high) | arl-13 | 0.55/0.14 | 69/11 | P < 0.001 | 0.91/0.15 | 20/3 | P < 0.001 |
| OSM-6::GFP | N2 | 0.75/0.14 | 137/7 | 1.15/0.21 | 59/4 | ||
| OSM-6::GFP | arl-13 | 0.53/0.11 | 291/19 | P < 0.001 | 0.94/0.29 | 55/4 | P < 0.001 |
| CHE-2::GFP | N2 | 0.71/0.19 | 156/6 | 1.17/0.24 | 68/5 | ||
| CHE-2::GFP | arl-13 | 0.68/0.15 | 120/6 | P = 0.047 | 0.93/0.21 | 48/5 | P < 0.001 |
| DYF-2::GFP | N2 | 0.75/0.14 | 173/11 | 1.08/0.19 | 62/4 | ||
| DYF-2::GFP | arl-13 | 0.53/0.15 | 90/6 | P < 0.001 | 0.83/0.15 | 66/4 | P < 0.001 |
| CHE-11::GFP | N2 | 0.69/0.19 | 116/11 | 1.19/0.28 | 67/6 | ||
| CHE-11::GFP | arl-13 | 0.66/0.28 | 120/7 | P = 0.15 | 0.86/0.39 | 63/5 | P < 0.001 |
| CHE-11::GFP | arl-13;osm-3 | 0.52/0.08 | 109/6 | P < 0.001 | ND | None | ND |
| IFTA-1::GFP | N2 | 0.69/0.15 | 94/4 | 1.14/0.25 | 73/4 | ||
| IFTA-1::GFP | arl-13 | 0.68/0.16 | 127/6 | P = 0.35 | 1.06/0.27 | 83/8 | P = 0.04 |
| BBS-7::GFP | N2 | 0.70/0.17 | 100/4 | 1.11/0.25 | 64/4 | ||
| BBS-7::GFP | arl-13 | 0.55/0.15 | 241/11 | P < 0.001 | 0.97/0.28 | 48/6 | P < 0.001 |
n, number of particles; N, number of amphids. Mean MS and DS speeds along amphid channel cilia in N2 and arl-13(tm2322) worms expressing the indicated transgene are shown. t test indicates the pairwise comparison with N2 controls. Blank cells indicate that the information is not applicable.
In DSs, all examined transgenic arl-13 strains exhibited accelerated IFT at mean speeds of 0.95 µms−1 (0.83–1.06 µms−1; Fig. S2 and Table I), indicating that although IFT protein overexpression can dislodge OSM-3 from MS assemblies, OSM-3 still docks with IFT proteins and drives DS IFT. However, the mean DS speed of 0.95 µms−1 in arl-13 worms represents a ∼17% reduction compared with WT worms (∼1.15 µms−1). As OSM-3::GFP also moved along MSs/DSs at slower rates (0.97/1.04 µms−1) in klp-11;arl-13 worms, we conclude that OSM-3 speed is slightly reduced in arl-13 worms (Table I).
Finally, we examined the IFT transgene overexpression phenotype further by expressing a che-13::mCherry transgene at two different levels in tm2322 worms. To make these strains, gonadal transformation of the che-13 construct (via microinjection) was conducted at 1× (low) and 5× (high) concentrations. che-13(high) induced a SynDyf phenotype in tm2322 worms, whereas che-13(low) did not (Fig. 6 B and Table S1). Also, CHE-13 translocated at slow MS speeds (0.55 µms−1) in che-13(high)–expressing arl-13 worms, whereas mean MS speeds were normal (0.72 µms−1) in che-13(low)–expressing arl-13 worms (Fig. S3 and Table I). However, like osm-3::gfp–expressing arl-13 worms, more fast moving MS particles were observed in che-13(low)–expressing arl-13 worms compared with WT controls, and DS speeds were reduced (Fig. S3 and Table I). From this data, we conclude that the severity of IFT defect in arl-13 worms depends on IFT transgene expression levels.
In summary, arl-13 is not required for IFT protein ciliary targeting. Instead, tm2322 worms possess modestly reduced DS rates and subtle alterations in MS rate profiles and are sensitive to overexpression of IFT transgenes (enhanced cilium morphology and MS IFT defects). Together, these data indicate that tm2322 possesses IFT defects and suggest that arl-13 stabilizes anterograde IFT.
arl-13 interacts genetically with ciliary transport–associated genes to maintain cilium structure/morphology and anterograde IFT
Destabilized IFT, cilium structure/morphology defects, and abnormalities in ciliary transmembrane protein localizations are found in other ciliopathy and ciliary transport–associated C. elegans mutants, including bbs-1/7/8, nph-4, and dyf-5, as well as klp-11 and osm-3 (Blacque et al., 2004; Snow et al., 2004; Ou et al., 2005; Burghoorn et al., 2007; Tan et al., 2007; Jauregui et al., 2008). Also, because these genes are coexpressed with arl-13 in most/all C. elegans ciliated cells and encode ciliary proteins, Arl13b function appears related to these genes. To address this notion further, double mutant phenotypic analysis was used to assess genetic relationships.
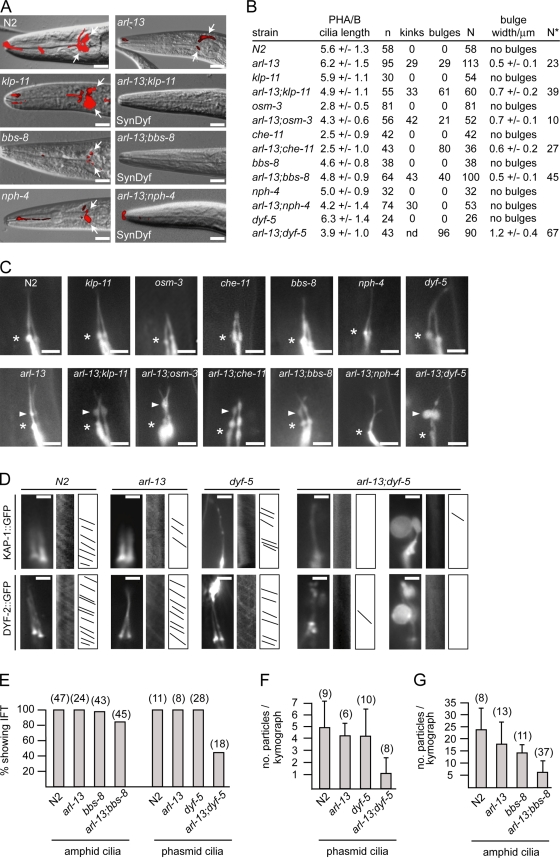
First, we used dye filling to assess cilium structure/morphology and found that compared with single mutants of arl-13 (moderate Dyf), klp-11 (not Dyf), nph-4 (weak Dyf), and bbs-8 (strong Dyf), arl-13;nph-4, arl-13;klp-11, and arl-13;bbs-8 worms were SynDyf (Fig. 7 A). In agreement with the SynDyf data, arl-13;klp-11 and arl-13;nph-4 animals possessed shorter cilia than single mutants (Fig. 7, B and C), and arl-13;klp-11 and arl-13;bbs-8 cilia exhibited specific cilium morphology defects that were worse (e.g., larger bulges) and/or observed more frequently than in single mutants (Fig. 7, B and C). Enhanced cilium morphology defects were also found in arl-13;dyf-5 (shorter cilia; very large and more frequent bulges), arl-13;osm-3 (more frequent kinks), and arl-13;che-11 (more frequent bulges) worms (Fig. 7, B and C). Together, these data show that arl-13 interacts genetically with ciliopathy/ciliary transport genes. Finally, using KAP-1::GFP and DYF-2::GFP markers, we assessed anterograde IFT in arl-13;dyf-5 and arl-13;bbs-8 animals. In arl-13, bbs-8, and dyf-5 single mutants, moving IFT particles were clearly observable, although particle frequency (DYF-2 marked) may have been slightly reduced in bbs-8 and arl-13 animals (Fig. 7, D–G; Table I; and Videos 1–5). However, in arl-13;dyf-5 double mutants, kymographs frequently failed to detect moving particles (Fig. 7, D and E; and Videos 6 and 7), and in those ciliary bunches where moving particles were detectable, particle frequently was severely reduced (Fig. 7 F and Video 8). Although not to same extent as arl-13;dyf-5 mutants, IFT particle number also appeared reduced in some arl-13;bbs-8 worms (Fig. 7, E and G; and Videos 9 and 10).
Figure 7.
arl-13 interacts synthetically with ciliopathy/ciliary transport genes to maintain cilium structure/morphology and IFT. (A) Compared with single mutants, arl-13;klp-11, arl-13;bbs-8, and arl-13;nph-4 mutants are SynDyf, failing to take up dye. Merged DIC–fluorescence head images from single and double mutants after a DiI uptake assay (arrows denote dye uptake) are shown. (B and C) PHA/B cilia morphology defects are enhanced in double mutants of arl-13 and ciliopathy/ciliary transport genes. B shows cilium morphology data from worms expressing a PHA/B cilium marker (srb-6p::gfp). Data for kinks/bulges represent the percentage of cilia with these defects. n, number of animals assayed for cilium length; N, number of cilia assessed for ciliary kinks/bulges; N*, number of cilia assessed for bulges. C shows fluorescence images of PHA/B cilia in single and double mutants. Arrowheads indicate axonemal bulges, and asterisks indicate the cilia base. (D) Anterograde IFT particle formation (marked by KAP-1::GFP and DYF-2::GFP) is defective in arl-13;dyf-5 double mutants compared with single mutants. Fluorescence images, kymographs, and kymograph schematics of single and double mutants expressing kap-1::gfp or dyf-2::gfp are shown. (E–G) Quantitative analysis of anterograde IFT kymographs (from animals expressing dyf-2::gfp). Unlike single mutants, in many arl-13;dyf-5 double mutants, moving IFT assemblies are not found (E) or are severely reduced in number (F). Fewer moving IFT particles also found in arl-13;bbs-8 double mutants compared with single mutants (G). (F and G) Error bars indicate SEM. Bars: (A) 10 µm; (C) 3 µm; (D) 2 µm.
Together, our analyses demonstrate that arl-13 interacts genetically with IFT-related genes to maintain cilium structure/morphology and, in the case of bbs-8 and dyf-5, the integrity of the IFT system. Because the ciliary localization of ARL-13 is distinct from other ciliopathy/ciliary transport–associated genes proteins and because ARL-13 does not undergo IFT-like motility, the additive/cooperative nature of these interactions may reflect a distinct function for arl-13 within a parallel/converging pathway.
Discussion
In this study, we show that JS-associated Arl13b is a ciliary membrane–associated protein required to maintain C. elegans cilium structure/morphology and ciliary protein transport processes, including anterograde IFT stability and the ciliary localizations/abundance of transmembrane proteins. In addition, we show that arl-13 functions additively with other ciliopathy/ciliary transport genes to maintain cilium structure/morphology and IFT.
ARL-13/Arl13b associates with the ciliary membrane via palmitoyl anchors
Several our findings indicate that ARL-13/Arl13b associates with ciliary membranes: (a) Arl13b is palmitoylated, (b) Pal modification motif disruption causes ARL-13/Arl13b to delocalize in a cytosolic/diffuse fashion, (c) Arl13b cofractionates with membrane proteins, whereas Arl13b(C8S/C9S) variants cofractionate with cytosolic proteins, (d) ARL-13 and Arl13b proteins reside exclusively in cilia (Blacque et al., 2005; Caspary et al., 2007; Cantagrel et al., 2008; Hori et al., 2008), and (e) ARL-13 localizes to AWB cilia membrane fans. Arl13b joins other ciliopathy/ciliogenic proteins thought to associate with membranes, including BBSome components (Nachury et al., 2007), Meckel syndrome/JS proteins with membrane-targeting C2 domains (MKS-1, MKSR-1, MKS-6, and RPGRIP1L), RAB8 (Nachury et al., 2007; Yoshimura et al., 2007), and Elipsa (Omori et al., 2008). However, Arl13b is unique as it localizes exclusively to cilia.
Localization of ARL-13/Arl13b to a distinct proximal ciliary compartment
Our finding that ARL-13/Arl13b localizes predominantly to the proximal ciliary region of most C. elegans sensory cilia and MDCKII cilia (Fig. 1, A and B) contrasts with C. elegans AWB cilia and other mammalian cell types, in which ARL-13/Arl13b localizes along the entire axoneme (Caspary et al., 2007; Cantagrel et al., 2008; Hori et al., 2008). Thus, Arl13b appears largely excluded from distal regions of some ciliary subtypes. Intriguingly, the ciliopathy-associated Inv protein (Inversin) also localizes to the proximal region or Inv compartment of mammalian renal tubule epithelial cells (Shiba et al., 2009). Conversely, Arl13b localization in mouse renal cilia was not restricted to the Inv compartment but observed along the entire cilium length (Shiba, D., and T. Yokoyama, personal communication). Although the mechanism of ARL-13/Arl13b DS exclusion in certain ciliary subtypes is not known, one possibility is that it may have an MT basis. For example, in C. elegans amphid/phasmid channel cilia, MSs contain doublet MTs, whereas DSs consist of singlet MTs (Perkins et al., 1986). However, as ARL-13 localizes to AWB cilia membrane fans, which lack MTs altogether, a correlation between MTs and restriction of ARL-13/Arl13b to discrete ciliary regions is unlikely. Instead, the answer may be related to regional differences in ciliary membrane lipid/protein compositions.
ARL-13/Arl13b and cilium formation
C. elegans ARL-13 is not required for building C. elegans sensory cilia but is required for cilium structure/morphology. This is likely true for most, if not all, nematode cilia because arl-13 is expressed in most/all ciliated cells (Fan et al., 2004) and is required for normal cilium morphology of all examined ciliary subtypes (Figs. 3 and 4). The morphological/ultrastructural phenotypes of arl-13 worms are consistent with Arl13b mutant mice (hnn), which possess truncated nodal cilia and open B-tubules (Caspary et al., 2007). However, axonemal bulges were not reported in hnn mice, indicating that this phenotype is specific to worms or restricted to cilia subtypes. Interestingly, abnormal ciliary bulges and axonemal vesicles were found in motile cilia of BBS1, -2, and -4 mice (Shah et al., 2008), and abnormal dense matrix–filled vesicles were described in C. elegans IFT mutants (che-11 and osm-5; Perkins et al., 1986). These findings suggest that arl-13 overlaps functionally with other ciliopathy/ciliary transport–associated genes and that vesicle accumulation in ciliary axonemes may be a feature of ciliopathies. Because vesicles are not found in WT cilia, what is their origin? It seems unlikely that they originate from aberrant entry into cilia because transition zones are intact in arl-13 animals (Fig. 4), indicating that the physical block to vesicles is maintained. Instead, vesicles may originate from the ciliary membrane and could reflect a specific defect in ciliary membrane processes. The MT defects also raise the possibility of MT-stabilizing functions for arl-13. Indeed, unzipped MTs are also reported in nphp-4 mutants (Jauregui et al., 2008), and one possible explanation is loss of MT posttranslational modification (e.g., polyglutamylation) such as that in zebrafish fleer mutants (Pathak et al., 2007).
ARL-13/Arl13b is required for ciliary transport processes
Several of our findings indicate that C. elegans ARL-13 stabilizes ciliary protein transport. First, ciliary transmembrane proteins abnormally distribute and accumulate within arl-13 cilia (Fig. 5). Consistent with these defects being linked to transport deficiencies, similar phenotypes are found in IFT mutants (Bae et al., 2006; Tan et al., 2007). Second, DS IFT rates are reduced in tm2322 mutants, and overexpression of IFT transgenes disrupts cilium structure/morphology and anterograde IFT, causing OSM-3 to uncouple from MS IFT assemblies (Fig. 6 and Table I). Third, arl-13 functions genetically with ciliary transport–associated genes to maintain cilium structure and IFT (Fig. 7). Interestingly, the tm2322 allele used in this study is an in-frame deletion, encoding a protein that still localizes to cilia (Fig. 3 B). Accordingly, ARL-13(tm2322) likely represents a ciliary G protein with deregulated functions.
Our observation that IFT transgene overexpression in tm2322 mutants leads to SynDyf phenotypes and IFT destabilization has also been reported in nph-4 mutants (Jauregui et al., 2008), indicating that this phenomenon is not restricted to arl-13 and extends to other ciliopathy genes. Also, although not phenotypically identical, kinesin-2 motor decoupling is a feature of other ciliary transport and ciliopathy-associated mutants such as bbs-1/7/8 and dyf-5 (Ou et al., 2005, 2007; Burghoorn et al., 2007). To explain our MS IFT rate data, we propose that endogenous MS anterograde IFT assemblies are partially destabilized (biochemically) in arl-13 worms (but still largely intact) and susceptible to further weakening (i.e., OSM-3 dislodgment) by IFT protein overexpression (Fig. 8, A and B). Consistent with this idea, when expressed at lower levels in tm2322 animals, a che-13::mCherry transgene did not cause a SynDyf phenotype nor a severe IFT defect (e.g., complete motor uncoupling). Regardless, for all IFT proteins examined, some degree of anterograde IFT defect was observed. One possible explanation is that the tm2322 allele generates a genetic background that disrupts the biochemical properties of IFT assemblies (e.g., macromolecular architecture and subunit stoichiometry), which in turn confers a reduced tolerance for changes in the stoichiometry of certain IFT protein subunits. Indeed, there is evidence in our data that enhanced destabilization correlates with overexpression of IFT-B subunits, suggesting that ARL-13 may preferentially facilitate interactions of IFT-B proteins with IFT assemblies.
Figure 8.
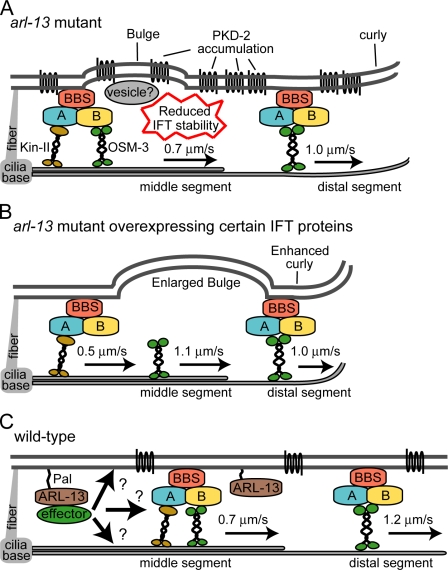
Model of ARL-13/Arl13b function. (A) Summary of arl-13(tm2322) ciliary phenotypes showing disrupted cilium ultrastructure/morphology and ciliary membrane protein localization and weakly destabilized anterograde IFT assemblies (e.g., reduced DS speeds). (B) Overexpression of IFT transgenes in tm2322 further destabilizes anterograde IFT assemblies, causing decoupling of OSM-3 and enhancement of cilium structure/morphology defects. OSM-3 retains the ability to dock with IFT/BBS assemblies in DSs. (C) Model of ARL-13/Arl13b function in WT cilia. Arl13b associates with ciliary membranes via palmitoyl anchors, where it regulates the function or functions of unknown effectors required to stabilize ciliary protein transport processes. Effectors may interact directly with transport machinery or, alternatively, regulate ciliary membrane or axonemal MT processes (e.g., membrane biogenesis/turnover or MT stabilization), which indirectly facilitate protein transport.
Model of ARL-13/Arl13b function and ciliary protein transport
We propose that ARL-13/Arl13b associates with the ciliary membrane and is required for normal ciliary transport processes (Fig. 8). Whether ARL-13 regulates transport directly or indirectly is unclear (Fig. 8 C). Our analyses reveal multiple structural and functional ciliary requirements for ARL-13 at the ciliary membrane, MTs, and protein transport. Pinpointing the primary defect is difficult because of the nonautonomous nature of these processes and structures. For example, as IFT assemblies contact MTs and membranes, defects in either could indirectly disrupt ciliary transport or vice versa (Fig. 8 C). However, an exclusive role in regulating ciliary transport seems less likely because (a) arl-13 interacts genetically with IFT genes, (b) ARL-13 does not undergo IFT, and (c) ARL-13 localizes to ciliary regions that lack MTs (e.g., AWB cilia membrane fans). In contrast, because ARL-13 associates with membranes and is required for proper cilium shape, a role for arl-13 in regulating ciliary membranes seems more likely. Therefore, the IFT connection may be indirect and simply arise because IFT assemblies contact ciliary membranes. Indeed, membrane-associated functions such as lipid metabolism, membrane curvature, and membrane remodeling are known for numerous small G proteins, and it is well established that G proteins function at membrane surfaces (D’Souza-Schorey and Chavrier, 2006; Beck et al., 2008). Future work in identifying Arl13b binding partners/effectors should provide clues to the role of Arl13b at ciliary membranes and how it affects protein transport.
Arl13b, Shh signaling, and JS
Our findings for ARL-13 suggest that mammalian Arl13b may also facilitate ciliary protein transport. Indeed, this is consistent with the role of Arl13b/hnn in Shh signaling, where it functions downstream of Smo/Ptch1 and upstream of Gli (Caspary et al., 2007), which is precisely where IFT functions (Huangfu et al., 2003; Eggenschwiler and Anderson, 2007). Indeed, Gli activity is not altogether lost in hnn mutants (unlike IFT mutants; Caspary et al., 2007; Eggenschwiler and Anderson, 2007), which is consistent with our worm data showing that IFT still operates in arl-13 animals. Because ciliary transmembrane protein localizations and levels are disrupted in arl-13 mutants, it is tempting to speculate that the ciliary localizations of Shh components are defective in hnn mice. If so, this would provide a mechanistic basis for the Shh signaling abnormalities in these animals and provide a working model showing that defects in the transport/localizations of developmental signaling proteins underlie Arl13b-associated JS.
Materials and methods
C. elegans strains
All nematode strains were maintained and cultured at 20°C using standard techniques (Brenner, 1974). Nontransgenic strains are as follows: Bristol, N2; 2322, arl-13(tm2322); PR802, osm-3(p802); VC1228, klp-11(tm324); PR813, osm-5(p813); CB1033, che-2(e1033); CB3330, che-11(e1810); MX52, bbs-8(nx77); and RB1146, dyf-5(ok1170), nph-4(tm925);him-5(e1490), and pkd-2(sy606);him-5(e1490).
Transgenic strains expressing ARL-13(WT)::GFP and ARL-13::GFP variants are as follows: OEB223, N2;oqEx58[arl-13::gfp+pRF4]; OEB161, arl-13(tm2322);oqEx58[arl-13::gfp+pRF4]; OEB97, N2;oqEx52[arl-13(delPal)::gfp+pRF4]; OEB96, N2;oqEx51[arl-13(rPal)::gfp+pRF4]; and OEB99, N2;oqEx55[arl-13(del203–370)::gfp+pRF4].
Transgenic strains expressing ciliated cell–specific transcriptional GFP markers are as follows: N2;gmIs13[srb-6p::gfp+pRF4]; CX3553, kyIs104[str-1p::gfp]; CX3695, lin-15;kyIs136[str-2p::gfp+lin-15(+)]; OH3192, kyIs164[gcy-5p::gfp] and pkIs519[gpa-6p::gfp]; OEB141, arl-13(tm2322);gmIs13[srb-6p::gfp+pRF4]; OEB49, arl-13(tm2322);klp-11(tm324);gmIs13[srb-6p::gfp+pRF4]; OEB224, arl-13(tm2322);osm-3(p802);gmIs13[srb-6p::gfp+pRF4]; OEB174, arl-13(tm2322);che-11(e1810);gmIs13[srb-6p::gfp+pRF4]; OEB51, arl-13(tm2322);bbs-8(nx77);gmIs13[srb-6p::gfp+pRF4]; OEB89, arl-13(tm2322);nph-4(tm925);gmIs13[srb-6p::gfp+pRF4]; OEB338, nph-4(tm925);gmIs13[srb-6p::gfp+pRF4]; OEB324, arl-13(tm2322);dyf-5(ok1170);gmIs13[srb-6p::gfp+pRF4]; OEB339, dyf-5(ok1170);gmIs13[srb-6p::gfp+pRF4]; OEB53, arl-13(tm2322);kyIs164[gcy-5p::gfp]; OEB180, arl-13(tm2322);kyIs104[str-1p::gfp]; OEB61, arl-13(tm2322);kyIs136[str-2p::gfp+lin-15(+)]; and OEB210, arl-13;pkIs519[gpa-6p::gfp].
Transgenic strains expressing GFP-tagged ciliary membrane protein markers are as follows: CX3260, kyIs53[odr-10::gfp]; CX3716, kyIs141[osm-9::gfp+lin-15(+)] and kyIs[tax-2::gfp+lin-15(+)]; PT572, myIs1[PKD-2::GFP+unc-122p::GFP] (gift from M. Barr, Rutgers University, Piscataway, NJ); OEB152, arl-13(tm2322);kyIs53[odr-10::gfp]; OEB175, arl-13;kyIs141[osm-9::gfp+lin-15(+)]; OEB177, arl-13(tm2322);kyIs111[tax-2::gfp]; and OEB289, arl-13;myIs1[PKD-2::GFP;unc-122p::GFP].
Transgenic strains expressing IFT::GFP markers are as follows: MX255, N2;ejEx[osm-3::gfp+pRF4] (gift from J. Scholey, University of California, Davis, Davis, CA), N2;ejEx[kap-1::gfp+pRF4] (gift from J. Scholey), xbx-1(ok279);Ex[xbx-1::gfp+pRF4], N2;yhEx[che-13::gfp+pRF4], and N2;Ex[che-12p::che-13::mcherry+unc-122p::gfp] (gift from S. Shaham, The Rockefeller University, New York, NY); PT47, N2;myEx[che-2::gfp+pRF4], N2;ejEx[dyf-1::gfp+pRF4] (gift from G. Jansen, Erasmus Medical Center, Rotterdam, Netherlands), N2;ofEx46[dyf-2::gfp+pRF4], and N2;myEx10[che-11::gfp+pRF4]; MX231, N2;nxEx[ifta-1::gfp+dpy-5(+)] (gift from M. Leroux, Simon Fraser University, Burnaby, British Columbia, Canada); MX76, dpy-5;nxEx25[bbs-7::gfp+dpy-5(+)] (gift from M. Leroux); MX68, dpy-5;nxEx[bbs-8::gfp+dpy-5(+)] (gift from M. Leroux); OEB134, arl-13(tm2322);ejEx[osm-3::gfp+pRF4]; OEB284, arl-13(tm2322);klp-11(tm324);ejEx[osm-3::gfp+pRF4]; OEB248, klp-11(tm324);ejEx[osm-3::gfp+pRF4]; OEB132, arl-13;ejEx[kap-1::gfp+pRF4]; OEB110, arl-13(tm2322);dyf-5(ok1170);ejEx[kap-1::gfp+pRF4]; OEB226, arl-13(tm2322);bbs-8(nx77);ejEx[kap-1::gfp+pRF4]; OEB133, arl-13(tm2322);nxEx[xbx-1::gfp+pRF4]; OEB163, arl-13(tm2322);mnIs17[osm-6::gfp+unc-36(+)]; OEB159, arl-13(tm2322);yhEx[che-13::gfp+pRF4]; OEB157, arl-13(tm2322);myEx[che-2::gfp+pRF4]; OEB160, arl-13(tm2322);ejEx[dyf-1::gfp+pRF4]; OEB130, arl-13(tm2322);myEx10[che-11::gfp+pRF4]; OEB117, arl-13(tm2322);osm-3(p802);myEx10[che-11::gfp+pRF4]; OEB137, arl-13(tm2322);nxEx[ifta-1::gfp+dpy-5(+)]; OEB135, arl-13(tm2322);nxEx25[bbs-7::gfp+dpy-5(+)]; OEB136, arl-13(tm2322);nxEx[bbs-8::gfp+dpy-5(+)]; OEB156, arl-13(tm2322);ofEx46[dyf-2::gfp+pRF4]; OEB95, arl-13(tm2322);klp-11(tm324);ofEx46[dyf-2::gfp+pRF4]; OEB227, arl-13(tm2322);dyf-5(ok1170);ofEx46[dyf-2::gfp+pRF4]; and OEB115, arl-13(tm2322);bbs-8(nx77);ofEx46[dyf-2::gfp+pRF4] and bbs-8(nx77);ofEx46[dyf-2::gfp+pRF4].
Genetic crossing
Standard genetic crossing techniques were used to make double mutants and to introduce transgenes into various genetic backgrounds. PCR using primers that flank deletions were used to follow the arl-13(tm2322), klp-11(tm324), bbs-8(nx77), and nph-4(tm925) deletion mutations. All other mutations were followed using the dye-filling assay.
Generation of arl-13::gfp constructs
All translational arl-13::gfp constructs (WT and variant) were generated using fusion PCR as previously described (Hobert, 2002). For the arl-13(WT) reporter, a genomic DNA fragment containing 300 bp of 5′ untranslated region (promoter) plus the entire exonic and intronic sequence of arl-13 was fused in frame with gfp. Similarly, for the C-terminal truncation variant (del203–370), gfp was fused in frame with genomic DNA fragments containing 300 bp of arl-13 5′ untranslated region plus exonic/intronic arl-13 sequence (nucleotides 1–2701 of arl-13 genomic sequence). For the Pal motif variants (delPal, deletion of C12–C15; and rPal, replacement of C12–C15 with ASAS), two fusion reactions were performed. In the first reaction, PCR fragments containing the arl-13 promotor (214 bp) plus 33 bp of exon 1 were fused to nucleotides 34–3300 of arl-13, where nucleotides 34–45 (TGCTGTTGTTGC) were altered by primer design to (TCCGCTTCTGCC; rPal) or deleted altogether (delPal). Resulting PCR products were subsequently fused in frame with gfp. All constructs were coinjected at 1–10 ng/µl with 50 ng/µl pRF4 into N2 worms to generate roller transgenic animals harboring arl-13::gfp (WT and variant) extrachromosomal arrays.
Dye-filling assay
Worms were placed in 200 µl of DiI (or DiO) solution (Invitrogen; diluted 1:200 with M9 buffer). After 1-h incubation, worms were recovered on seeded nematode growth medium plates for a further hour and then mounted on slides. Epifluorescence wide-field imaging under red (DiI) or green filters (DiO) was used to image DiI/DiO uptake into ciliated amphid/phasmid cells.
Chemosensory behavioral assays
Chemotaxis assay toward isoamyl alcohol was performed on 85-mm round plates. 1 µl of 1 M NaN3 (anesthetic) was applied to two points at opposite ends of the plate (5 mm from the edge). Then, 1 µl of attractant (diluted 1:100 in 95% ethanol) was spotted at one of these points, and 1 µl of ethanol was spotted at the other as a control. Young adult worms (∼100), washed three times with M9 buffer and once with deionized water, were then applied to plate center (∼3.5 cm from attractant). After excess liquid removal, worms were allowed to partition across the plate for 30 and 60 min. The chemotaxis index was calculated at each time point as (a − b)/n, where a equals the number of worms within 1.5 cm of the attractant, b equals the number of worms within 1.5 cm of the ethanol counterattractant, and n equals the total number of worms in the assay.
Electron microscopy
Worms were washed directly into a primary fixative of 2.5% glutaraldehyde in 0.1 M Sorensen phosphate buffer. To facilitate rapid ingress of fixative, worms were cut in half using a razor blade under a dissection microscope, transferred to Eppendorf tubes, and fixed for 1 h at room temperature. Samples were then centrifuged at 3,000 rpm for 2 min, and supernatant was removed and washed for 10 min in 0.1 M Sorensen phosphate buffer. The worms were then postfixed in 1% osmium tetroxide in 0.1 M Sorensen phosphate buffer for 1 h at room temperature. After washing in buffer, specimens were processed for electron microscopy by standard methods; in brief, they were dehydrated in ascending grades of alcohol to 100% infiltrated with epon and placed in aluminum planchettes orientated in a longitudinal aspect and polymerized at 60°C for 24 h. Using an ultramicrotome (UC6; Leica), individual worms were sectioned in cross section, from anterior tip, at 1 µm until the area of interest was located, as judged by examining the sections stained with toluidine blue by light microscopy. Thereafter, serial ultrathin sections of 80 nm were taken for electron microscopical examination. These were picked up onto 100-mesh copper grids and stained with uranyl acetate and lead citrate. Using an electron microscope (Tecnai Twin; FEI), sections were examined to locate, in the first instance, the most distal region of the ciliary region and subsequently from that point to the more proximal regions of the ciliary apparatus. At each strategic point, DSs, MSs, and transition zone/fiber regions were tilted using the Compustage of the Tecnai to ensure that the axonemal MTs were imaged in an exact geometrical normalcy to the imaging system. All images were recorded at an accelerating voltage (120 kV) and objective aperture of 10 µm using a MegaView 3 digital recording system (Olympus).
C. elegans fluorescence microscopy and IFT motility assays
Worms were anesthetized in 10 mM levamisole and mounted on 2% agarose pads for analysis on a microscope (DM5000B; Leica), fitted with epifluorescence, and a 1.4 NA 100× Plan-Apochromat lens. Images were captured using an electron-multiplying charge-coupled device camera (DV885; Andor Technology) and iQ2.0 software (Andor Technology). iQ2.0 was also used to measure signal intensities of PKD-2::GFP in the cell body and cilia of N2 and arl-13(tm2322) mutants. The region of interest was always of identical size, and background signals were subtracted to normalize values. For IFT motility assays, time-lapse videos of IFT along amphid and phasmid cilia were taken no more than 30–45 min after slide preparation, and in each case, the same exposure times (300 ms), gain, and frame rate (3 frames/s) were used. Resulting stacked multi tiff images were processed into kymographs using the multiple kymograph plug-in of ImageJ 1.38× software (National Institutes of Health; http://www.embl-heidelberg.de/eamnet/html/body_kymograph.html). Particle velocities were determined from the kymographs using the ImageJ “read velocities from tsp” macro. The frequency of IFT particles was also determined from the kymographs by counting the number of lines detected.
Mammalian cell culture and immunofluorescence microscopy
MDCKII cells were cultured on a coverglass (15-mm diameter) in DME supplemented with 10% (vol/vol) fetal bovine serum for 7 d after full confluence, with the medium changed every 3 d. RPE1 cells were transiently transfected with Arl13b-GFP expression vectors using Fugene6 (Roche) and cultured for 48 h in a starvation medium consisting of DME/F12[1:1] (Invitrogen) and 0.1% (wt/vol) BSA. For immunofluorescence staining, cells were fixed with 4% paraformaldehyde in PBS for 10 min, followed by incubation with ice-cold methanol for 5 min. After permeabilization with 0.2% Triton X-100 in PBS for 10 min, cells were treated with 5% BSA in TBS (TBS/5% BSA) and further incubated with primary antibodies in TBS/5% BSA at 37°C for 2 h. Cells were then washed three times with PBS and incubated with an Alexa Fluor 488/568–conjugated secondary antibody (Invitrogen) and DAPI for 30 min in TBS/5% BSA. After washing three times with PBS, the coverglass was mounted onto a glass slide in Permafluor mounting medium (Immunon) and viewed under a confocal microscope (LSM 510; Carl Zeiss, Inc.). The rabbit anti-Arl13b polyclonal antibody was described previously (Hori et al., 2008). Anti–acetylated tubulin, anti–γ-tubulin, and anti-Flag M2 antibodies were purchased from Sigma-Aldrich.
Subcellular fractionation and immunoblotting
293T cells cultured on a 10-cm dish (∼70% confluent) were transfected with 8 µg of expression vectors encoding WT or mutant versions of Arl13b (Flag tagged) using Lipofectamine 2000 (Invitrogen). 2 d after transfection, cells were washed twice with ice-cold wash buffer (10 mM triethanolamine–acetic acid, pH 7.6, and 250 mM sucrose) and suspended to 1 ml of ice-cold homogenization buffer (10 mM triethanolamine–acetic acid, pH 7.6, 250 mM sucrose, 5 mM MgSO4, 1 mM DTT, and 2 µg/ml aprotinin). The cell suspension was homogenized by 10 strokes in a chilled cell homogenizer with a tungsten-carbide ball (clearance of 10 µm; Isobiotec) and then subjected to standard subcellular fractionation by centrifugation (Graham and Rickman, 1997), followed by immunoblotting with anti-Flag M2, anti–µ-calpain (#MA3-940; Thermo Fisher Scientific), and anti-Na+/K+ ATPase α-1 (Millipore) antibodies.
Metabolic labeling with [3H]palmitic acid and fluorography
Palmitoylated proteins were detected as described previously (Fukata, 2006; Tsutsumi, 2009). 293T cells cultured on a 6-well dish (∼70% confluent) were transfected with 3 µg of expression vectors encoding WT or mutant versions of human GFP-tagged Arl13b using Lipofectamine 2000. 2 d after transfection, the cells were preincubated for 30 min in serum-free DME supplemented with 0.1% fatty acid–free BSA (Sigma-Aldrich). The cells were then metabolically labeled for 4 h with serum-free DME containing 0.25 mCi/ml [3H]palmitate (PerkinElmer). After washing twice with ice-cold PBS, the cells were extracted with 100 µl of ice-cold extraction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1% NP-40, 5 mM MgCl2, 1 mM DTT, 2 µg/ml aprotinin, and 2 µg/ml leupeptin). After centrifugation at 20,000 g for 20 min, the supernatants were subjected to SDS-PAGE. After fixing the gels for 30 min in a fixing solution (methanol/acetic acid/water, 3:1:6), the gels were treated with Amplify fluorographic reagent (GE Healthcare) for 30 min, dried under vacuum, and exposed to the x-ray film (RX-U; Fujifilm) without intensifying screen at −80°C for 72 h. The supernatants were also subjected to Western blot analysis with anti-GFP antibody.
Online supplemental material
Fig. S1 shows that C. elegans ARL-13 is a sequence homologue of human JS-associated Arl13b. Fig. S2 shows that overexpression of arl-13(delPal) in N2 worms does not disrupt PKD-2 ciliary abundance. Fig. S3 shows anterograde rate profiles of various IFT proteins along the MSs of arl-13(tm2322) mutants. Videos 1–10 show time-lapse recordings of KAP-1::GFP in amphid/phasmid channel cilia of arl-13 (Video 1), dyf-5 (Video 3), and arl-13;dyf-5 (Video 6) and DYF-2::GFP in amphid/phasmid channel cilia of arl-13 (Video 2), dyf-5 (Video 4), bbs-8 (Video 5), arl-13;dyf-5 (Videos 7 and 8), and arl-13;bbs-8 (Videos 9 and 10). Table S1 shows that overexpression of high levels but not low levels of a che-13::mCherry transgene enhances the Dyf defect of arl-13 mutants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908133/DC1.
Acknowledgments
We thank the Caenorhabditis Genetics Center, Japanese Bioresource Project, G. Jansen, M. Barr, J. Scholey, and M. Leroux for strains; S. Shaham for the che-12p::che-13::mCherry construct; D. Shiba and T. Yokoyama for unpublished observations; and M. Fukata for technical advice on protein Pal experiments.
This work was funded by Science Foundation Ireland’s President of Ireland Young Researcher Award (to O.E. Blacque).
Footnotes
Abbreviations used in this paper:
- BBS
- Bardet-Biedl syndrome
- DIC
- differential interference contrast
- DS
- distal segment
- IFT
- intraflagellar transport
- JS
- Joubert syndrome
- JSRD
- JS and related cerebellar disorder
- MS
- middle segment
- MT
- microtubule
- Pal
- palmitoylation
- WT
- wild type
References
- Baala L., Romano S., Khaddour R., Saunier S., Smith U.M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B., et al. 2007. The Meckel-Gruber syndrome gene, MKS3, is mutated in Joubert syndrome. Am. J. Hum. Genet. 80:186–194 10.1086/510499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y.K., Qin H., Knobel K.M., Hu J., Rosenbaum J.L., Barr M.M. 2006. General and cell-type specific mechanisms target TRPP2/PKD-2 to cilia. Development. 133:3859–3870 10.1242/dev.02555 [DOI] [PubMed] [Google Scholar]
- Beck R., Sun Z., Adolf F., Rutz C., Bassler J., Wild K., Sinning I., Hurt E., Brügger B., Béthune J., Wieland F. 2008. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc. Natl. Acad. Sci. USA. 105:11731–11736 10.1073/pnas.0805182105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Leroux M.R. 2006. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell. Mol. Life Sci. 63:2145–2161 10.1007/s00018-006-6180-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Reardon M.J., Li C., McCarthy J., Mahjoub M.R., Ansley S.J., Badano J.L., Mah A.K., Beales P.L., Davidson W.S., et al. 2004. Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 18:1630–1642 10.1101/gad.1194004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O.E., Perens E.A., Boroevich K.A., Inglis P.N., Li C., Warner A., Khattra J., Holt R.A., Ou G., Mah A.K., et al. 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15:935–941 10.1016/j.cub.2005.04.059 [DOI] [PubMed] [Google Scholar]
- Blacque O.E., Cevik S., Kaplan O.I. 2008. Intraflagellar transport: from molecular characterisation to mechanism. Front. Biosci. 13:2633–2652 10.2741/2871 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghoorn J., Dekkers M.P., Rademakers S., de Jong T., Willemsen R., Jansen G. 2007. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 104:7157–7162 10.1073/pnas.0606974104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V., Silhavy J.L., Bielas S.L., Swistun D., Marsh S.E., Bertrand J.Y., Audollent S., Attié-Bitach T., Holden K.R., Dobyns W.B., et al. 2008. Mutations in the cilia gene ARL13B lead to the classical form of Joubert syndrome. Am. J. Hum. Genet. 83:170–179 10.1016/j.ajhg.2008.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C.E., Anderson K.V. 2007. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 12:767–778 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Castori M., Valente E.M., Donati M.A., Salvi S., Fazzi E., Procopio E., Galluccio T., Emma F., Dallapiccola B., Bertini E. 2005. NPHP1 gene deletion is a rare cause of Joubert syndrome related disorders. J. Med. Genet. 42:e9 10.1136/jmg.2004.027375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.T., Pedersen L.B., Schneider L., Satir P. 2007. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 8:97–109 10.1111/j.1600-0854.2006.00516.x [DOI] [PubMed] [Google Scholar]
- Cuvillier A., Redon F., Antoine J.C., Chardin P., DeVos T., Merlin G. 2000. LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J. Cell Sci. 113:2065–2074 [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C., Chavrier P. 2006. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7:347–358 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- Dixon-Salazar T., Silhavy J.L., Marsh S.E., Louie C.M., Scott L.C., Gururaj A., Al-Gazali L., Al-Tawari A.A., Kayserili H., Sztriha L., Gleeson J.G. 2004. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 75:979–987 10.1086/425985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duldulao N.A., Lee S., Sun Z. 2009. Cilia localization is essential for in vivo functions of the Joubert syndrome protein Arl13b/Scorpion. Development. 136:4033–4042 10.1242/dev.036350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J.T., Anderson K.V. 2007. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23:345–373 10.1146/annurev.cellbio.23.090506.123249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Esmail M.A., Ansley S.J., Blacque O.E., Boroevich K., Ross A.J., Moore S.J., Badano J.L., May-Simera H., Compton D.S., et al. 2004. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 36:989–993 10.1038/ng1414 [DOI] [PubMed] [Google Scholar]
- Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., et al. 2004. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 36:1008–1013 10.1038/ng1419 [DOI] [PubMed] [Google Scholar]
- Fukata Y., Iwanaga T., Fukata M. 2006. Systematic screening for palmitoyl transferase activity of the DHHC protein family in mammalian cells. Methods. 40:177–182 10.1016/j.ymeth.2006.05.015 [DOI] [PubMed] [Google Scholar]
- Gorden N.T., Arts H.H., Parisi M.A., Coene K.L., Letteboer S.J., van Beersum S.E., Mans D.A., Hikida A., Eckert M., Knutzen D., et al. 2008. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet. 83:559–571 10.1016/j.ajhg.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.M., Rickman D., 1997. Subcellular Fractionation: A Practical Approach. Oxford University Press, Oxford: 339 pp. [Google Scholar]
- Hobert O. 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 32:728–730 [DOI] [PubMed] [Google Scholar]
- Hori Y., Kobayashi T., Kikko Y., Kontani K., Katada T. 2008. Domain architecture of the atypical Arf-family GTPase Arl13b involved in cilia formation. Biochem. Biophys. Res. Commun. 373:119–124 10.1016/j.bbrc.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 426:83–87 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Inglis P.N., Boroevich K.A., Leroux M.R. 2006. Piecing together a ciliome. Trends Genet. 22:491–500 10.1016/j.tig.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Inglis P.N., Ou G., Leroux M.R., Scholey J.M. 2007. The sensory cilia of Caenorhabditis elegans. WormBook, editor. The C. elegans Research Community, WormBook. doi/10.1895/wormbook.1.126.2. http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Jauregui A.R., Nguyen K.C., Hall D.H., Barr M.M. 2008. The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 180:973–988 10.1083/jcb.200707090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen K.J., Gleeson J.G. 2008. Cerebellar development and disease. Curr. Opin. Neurobiol. 18:12–19 10.1016/j.conb.2008.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., Jackson P.K. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Omori Y., Zhao C., Saras A., Mukhopadhyay S., Kim W., Furukawa T., Sengupta P., Veraksa A., Malicki J. 2008. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat. Cell Biol. 10:437–444 10.1038/ncb1706 [DOI] [PubMed] [Google Scholar]
- Orozco J.T., Wedaman K.P., Signor D., Brown H., Rose L., Scholey J.M. 1999. Movement of motor and cargo along cilia. Nature. 398:674 10.1038/19448 [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., Scholey J.M. 2005. Functional coordination of intraflagellar transport motors. Nature. 436:583–587 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Ou G., Koga M., Blacque O.E., Murayama T., Ohshima Y., Schafer J.C., Li C., Yoder B.K., Leroux M.R., Scholey J.M. 2007. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol. Biol. Cell. 18:1554–1569 10.1091/mbc.E06-09-0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ou G., Civelekoglu-Scholey G., Blacque O.E., Endres N.F., Tao L., Mogilner A., Leroux M.R., Vale R.D., Scholey J.M. 2006. Mechanism of transport of IFT particles in C. elegans cilia by the concerted action of kinesin-II and OSM-3 motors. J. Cell Biol. 174:1035–1045 10.1083/jcb.200606003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M.A., Bennett C.L., Eckert M.L., Dobyns W.B., Gleeson J.G., Shaw D.W., McDonald R., Eddy A., Chance P.F., Glass I.A. 2004. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am. J. Hum. Genet. 75:82–91 10.1086/421846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N., Obara T., Mangos S., Liu Y., Drummond I.A. 2007. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell. 18:4353–4364 10.1091/mbc.E07-06-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L.A., Hedgecock E.M., Thomson J.N., Culotti J.G. 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117:456–487 10.1016/0012-1606(86)90314-3 [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Burnette D.T., Bae Y.K., Forscher P., Barr M.M., Rosenbaum J.L. 2005. Intraflagellar transport is required for the vectorial movement of TRPV channels in the ciliary membrane. Curr. Biol. 15:1695–1699 10.1016/j.cub.2005.08.047 [DOI] [PubMed] [Google Scholar]
- Quinlan R.J., Tobin J.L., Beales P.L. 2008. Modeling ciliopathies: Primary cilia in development and disease. Curr. Top. Dev. Biol. 84:249–310 10.1016/S0070-2153(08)00605-4 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. 2002. Intraflagellar transport. Curr. Biol. 12:R125 10.1016/S0960-9822(02)00703-0 [DOI] [PubMed] [Google Scholar]
- Sayer J.A., Otto E.A., O’Toole J.F., Nurnberg G., Kennedy M.A., Becker C., Hennies H.C., Helou J., Attanasio M., Fausett B.V., et al. 2006. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 38:674–681 10.1038/ng1786 [DOI] [PubMed] [Google Scholar]
- Shah A.S., Farmen S.L., Moninger T.O., Businga T.R., Andrews M.P., Bugge K., Searby C.C., Nishimura D., Brogden K.A., Kline J.N., et al. 2008. Loss of Bardet-Biedl syndrome proteins alters the morphology and function of motile cilia in airway epithelia. Proc. Natl. Acad. Sci. USA. 105:3380–3385 10.1073/pnas.0712327105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba D., Yamaoka Y., Hagiwara H., Takamatsu T., Hamada H., Yokoyama T. 2009. Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J. Cell Sci. 122:44–54 10.1242/jcs.037408 [DOI] [PubMed] [Google Scholar]
- Snow J.J., Ou G., Gunnarson A.L., Walker M.R., Zhou H.M., Brust-Mascher I., Scholey J.M. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 6:1109–1113 10.1038/ncb1186 [DOI] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., Hopkins N. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 131:4085–4093 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- Tan P.L., Barr T., Inglis P.N., Mitsuma N., Huang S.M., Garcia-Gonzalez M.A., Bradley B.A., Coforio S., Albrecht P.J., Watnick T., et al. 2007. Loss of Bardet Biedl syndrome proteins causes defects in peripheral sensory innervation and function. Proc. Natl. Acad. Sci. USA. 104:17524–17529 10.1073/pnas.0706618104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., Fukata M. 2009. Identification of G protein alpha subunit-palmitoylating enzyme. Mol. Cell. Biol. 29:435–447 10.1128/MCB.01144-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente E.M., Brancati F., Silhavy J.L., Castori M., Marsh S.E., Barrano G., Bertini E., Boltshauser E., Zaki M.S., Abdel-Aleem A., et al. 2006a. AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann. Neurol. 59:527–534 10.1002/ana.20749 [DOI] [PubMed] [Google Scholar]
- Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. 2006b. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 38:623–625 10.1038/ng1805 [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. 2007. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178:363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Xue Y., Yao X., Xu Y. 2006. CSS-Palm: palmitoylation site prediction with a clustering and scoring strategy (CSS). Bioinformatics. 22:894–896 10.1093/bioinformatics/btl013 [DOI] [PubMed] [Google Scholar]