Loss of FGFRs results in skin abnormalities due to activation of keratinocytes and epidermal T cells.
Abstract
Fibroblast growth factors (FGFs) are master regulators of organogenesis and tissue homeostasis. In this study, we used different combinations of FGF receptor (FGFR)-deficient mice to unravel their functions in the skin. Loss of the IIIb splice variants of FGFR1 and FGFR2 in keratinocytes caused progressive loss of skin appendages, cutaneous inflammation, keratinocyte hyperproliferation, and acanthosis. We identified loss of FGF-induced expression of tight junction components with subsequent deficits in epidermal barrier function as the mechanism underlying the progressive inflammatory skin disease. The defective barrier causes activation of keratinocytes and epidermal γδ T cells, which produce interleukin-1 family member 8 and S100A8/A9 proteins. These cytokines initiate an inflammatory response and induce a double paracrine loop through production of keratinocyte mitogens by dermal cells. Our results identify essential roles for FGFs in the regulation of the epidermal barrier and in the prevention of cutaneous inflammation, and highlight the importance of stromal–epithelial interactions in skin homeostasis and disease.
Introduction
FGFs comprise a family of 22 polypeptides that regulate migration, proliferation, differentiation, and survival of different cell types. They exert these functions through activation of four transmembrane tyrosine kinase receptors, designated FGF receptor (FGFR) 1–4 (Ornitz and Itoh, 2001; Beenken and Mohammadi, 2009). Further complexity is achieved by alternative splicing in the FGFR genes. Of particular importance is alternative splicing in the third immunoglobulin-like domain of FGFR1-3, which generates IIIb and IIIc variants of these receptors that are characterized by different ligand-binding specificities (Ornitz and Itoh, 2001). For example, the IIIb splice variant of FGFR2 (FGFR2IIIb) is a high-affinity receptor for FGF7, FGF10, and FGF22, whereas the IIIc variant (FGFR2IIIc) binds a variety of other FGF ligands (Zhang et al., 2006).
Previous studies revealed important roles of FGFs in development, homeostasis, and repair of the skin (Steiling and Werner, 2003). Several FGFs are expressed in this tissue, and most of them are up-regulated upon injury (Werner et al., 1992, 1993; Komi-Kuramochi et al., 2005). Of particular interest are ligands of FGFR2IIIb because transgenic mice expressing a dominant-negative mutant of this receptor in keratinocytes showed epidermal atrophy, hair follicle abnormalities, and impaired wound reepithelialization (Werner et al., 1994). However, the responsible receptors remain to be identified, as the dominant-negative mutant blocks the action of all FGF receptors in response to common FGF ligands (Ueno et al., 1992). The abnormalities seen in these animals were not observed in FGF7 knockout mice (Guo et al., 1996), which suggests functional redundancy among different FGFs and possibly FGF receptors. Indeed, expression studies revealed that FGF10 and FGF22 are also expressed in normal and wounded skin (Beer et al., 1997; Nakatake et al., 2001; Beyer et al., 2003). Together with FGF7, they can activate the “b” splice variants of FGFRs 1 and 2 (Zhang et al., 2006) that are expressed in keratinocytes (Beer et al., 2000; Zhang et al., 2004). In the case of FGF7 and FGF10, the activation occurs in a paracrine manner because both ligands are produced by fibroblasts in the dermal papilla and in the interfollicular dermis as well as by epidermal γδ T cells (Werner et al., 1993; Rosenquist and Martin, 1996; Jameson and Havran, 2007). In contrast, FGF22 is mainly expressed in the inner root sheath of the hair follicle (FGF22; Nakatake et al., 2001) and most likely acts in an autocrine manner (Fig.1 A).
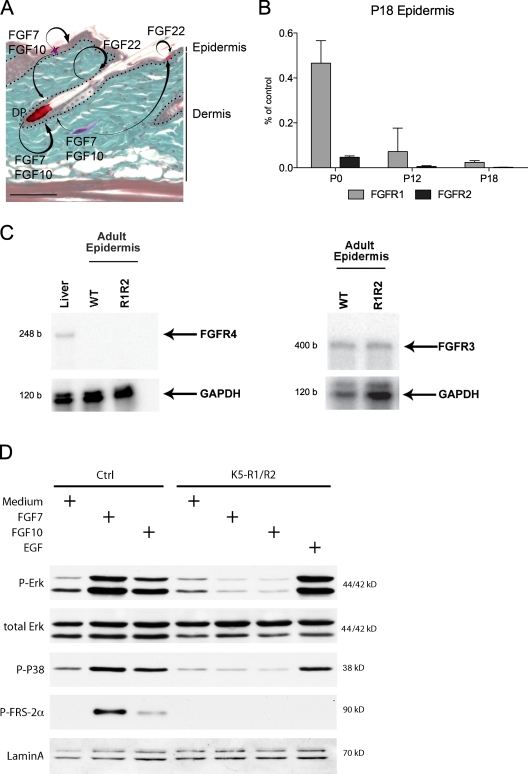
Figure 1.
Expression and activation of FGFR1IIIb and FGFR2IIIb in the skin of control and K5-R1/R2 mice. (A) The expression pattern of FGF7, FGF10, and FGF22 in the skin is shown schematically. FGF7 and FGF10 are expressed by fibroblasts of the dermis and the dermal papilla (DP) of the hair follicles and by epidermal γδ T cells. FGF22 is expressed by keratinocytes. These FGFs activate FGFR1IIIb and FGFR2IIIb on keratinocytes. Bar, 50 µm. (B) RNA from P0, P12, and P18 back skin epidermis of control and K5-R1/R2 mice was analyzed by real-time RT-PCR for the levels of Fgfr1 and Fgfr2 mRNAs. Error bars indicate mean ± SD. n = 3 K5-R1/R2 mice and 2 control mice at P0, n = 5 mice per genotype for P12 and P18. Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) mRNA was used for normalization. Data are indicated as the percentage of control. (C) RNA was isolated from the epidermis of adult K5-R1/R2 mice and age-matched control mice. RNA from mouse liver was used as a positive control for FGFR4. Samples of 20 µg of RNA were analyzed by an RNase protection assay for expression of FGFR3, FGFR4, or GAPDH. b, bases. (D) Primary keratinocytes from control and K5-R1/R2 mice were grown to confluency, serum-starved, and treated for 10 min with FGF7, FGF10, EGF, or medium without growth factors (medium). Lysates were analyzed by Western blotting using antibodies against total and phosphorylated signaling proteins or lamin A (loading control).
To determine the function of these FGFs and their receptors in the skin, we generated mice lacking FGFR1, FGFR2, or both receptors in keratinocytes. Our results revealed that these receptors cooperate to maintain the epidermal barrier and cutaneous homeostasis.
Results
Generation of mice lacking FGFR1, FGFR2, or both receptors in keratinocytes
Mice with floxed Fgfr1 (Pirvola et al., 2002) and Fgfr2 alleles (Yu et al., 2003) were mated with transgenic mice expressing Cre recombinase under the control of the keratin 5 (K5) promoter. This promoter allows excision of floxed alleles in basal cells of stratified epithelia after embryonic day 15.5 (Ramirez et al., 2004).
The progeny of our breeding included mice lacking FGFR1, FGFR2, or both receptors in keratinocytes (designated K5-R1, K5-R2, and K5-R1/R2 mice). Mice with floxed Fgfr alleles but without the Cre transgene were used as controls. Mice heterozygous for the floxed alleles that express Cre were used as an additional control in some experiments, and they never revealed phenotypic abnormalities (unpublished data).
Real-time RT-PCR using RNA from isolated epidermis of control and mutant mice demonstrated a strong reduction of Fgfr1 and Fgfr2 expression (Fig. 1 B) in newborn single and double knockout mice, which further declined until postnatal day 18 (P18; Fig. 1 B and not depicted). There was no compensatory up-regulation of Fgfr3 expression, and Fgfr4 mRNA could not be detected in mice of all genotypes using an RNase protection assay (Fig. 1 C).
When primary keratinocytes from P3 mice were stimulated with FGF7 or FGF10, efficient phosphorylation of FGFR substrate 2α (FRS2α), extracellular signal-regulated kinase 1/2 (Erk1/2), and p38 was observed in cells from control but not from K5-R1/R2 mice, which demonstrates efficient inhibition of FGFR signaling in the latter. EGF activated these signaling pathways in cells of both genotypes (Fig. 1 D).
Overlapping functions of FGFR1 and FGFR2 in the skin
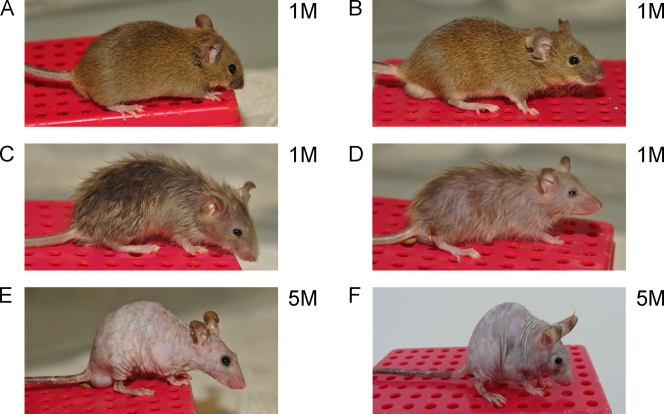
Macroscopically, no obvious abnormalities were observed in K5-R1 mice at any stage of postnatal development (Fig. 2 B). In contrast, K5-R2 mice revealed a phenotype (Fig. 2 C) that fully matches the abnormalities seen in mice lacking FGFR2IIIb in keratinocytes (K5-R2IIIb mice; Grose et al., 2007). The latter show hair abnormalities, a reduction in the number of hairs, and loss of sebaceous glands. The phenotype was much more severe in the double knockout mice (Fig. 2 D), whereas the loss of only one Fgfr2 allele in addition to both Fgfr1 alleles did not cause obvious phenotypic abnormalities (not depicted). All females and ∼60% of the males were infertile, and both were much smaller than control littermates. They also progressively lost their hair and were hairless by the age of 2–4 mo (Fig. 2 E). However, we did not observe an increased mortality rate, and we were able to maintain the animals for up to 2 yr.
Figure 2.
Macroscopic abnormalities in FGFR mutant mice. Photographs were taken from control (A), K5-R1 (B), K5-R2 (C), K5-R1/R2 (D and E), and K5-R2/R1IIIb (F) mice at the age of 1 mo (1M; A–D) or 5 mo (5M; E and F).
FGFR1IIIb cooperates with FGFR2IIIb in the regulation of epidermal homeostasis
To identify the splice variant that cooperates with FGFR2 in keratinocytes, we generated mice lacking FGFR1IIIb in all cells and FGFR2 in keratinocytes (K5-R2/R1IIIb mice). FGFR1IIIb-deficient mice are phenotypically normal and do not display an obvious skin phenotype because only the IIIb exon of the Fgfr1 gene was deleted in these mice, whereas all other FGFR1 splice variants are normally expressed (Zhang et al., 2004). The phenotype of K5-R2/R1IIIb mice was identical to the phenotype observed in K5-R1/R2 mice at the macroscopic (Fig. 2, E and F) and histological level (Fig. 3 E and not depicted). Therefore, we conclude that FGFR1IIIb and FGFR2IIIb cooperate in the regulation of epidermal homeostasis.
Figure 3.
Progressive loss of skin appendages in K5-R1/R2 mice. (A–D) Longitudinal paraffin sections from back skin of K5-R1/R2 female mice and control (ctrl) female littermates at P5, P18, 1 mo (1M), or 6 mo (6M) were stained with H/E. Bars, 100 µm. (E) Paraffin sections from the tail skin of control, K5-R1/R2, and K5-R2/FGFR1IIIb animals were stained with H/E. D, dermis; E, epidermis; HF, hair follicles. Bars, 22 µm.
Loss of skin appendages and progressive acanthosis in K5-R1/R2 mice
We next focused on the phenotype of the double mutant mice. At P5 (first anagen), a mild hypotrophy of the epidermis was observed, but the dermis and appendages appeared normal (Fig. 3 A). Although smaller and abnormally shaped, hair follicles were still present at P18 (first telogen), and their number was similar to control mice (24 follicles/mm in control mice vs. 25 follicles/mm in K5-R1/R2 mice; n = 6 control and 5 K5-R1/R2 mice). At this stage, the epidermis had a normal thickness, and no dermal abnormalities were observed (Fig. 3 B). By P30, control mice had entered the second anagen. However, most follicles from K5-R1/R2 mice were in telogen, although their number was still similar to the number in control mice (∼5 follicles/mm in mice of both genotypes; n = 4 control and 3 K5-R1/R2 mice; Fig. 3 C). Hair follicles and sebaceous glands were virtually absent in the back skin of older mice, and only a few cysts were present in the dermis (unpublished data), which demonstrates a progressive loss of appendages. Concomitantly, fibrosis developed in the dermis (Fig. 3 D). These abnormalities were reminiscent of the phenotype seen in K5-R2IIIb mice (Grose et al., 2007), but they were more severe in double knockout mice. In contrast to all single knockout mice, however, K5-R1/R2 as well as K5-R2/R1IIIb mice developed epidermal hyperthickening (acanthosis) combined with disorganization of the keratinocytes at the age of 2–3 mo, and this phenotype further progressed upon aging (Fig. 3, D and E). Acanthosis was seen in all areas of the skin and was particular severe in the tail skin (Fig. 3 E). This phenotype was unexpected because FGFs are potent mitogens for keratinocytes (Steiling and Werner, 2003). Therefore, we next focused on the mechanisms underlying the epidermal abnormalities and the progressive skin disease that developed in these mice.
Loss of FGFR1 and FGFR2 in keratinocytes causes hyperproliferation in vivo but not in vitro
Acanthosis may result from reduced apoptosis, impaired differentiation, or enhanced proliferation of keratinocytes, and these possibilities were explored. Apoptotic cells were extremely rare in the epidermis of control or K5-R1/R2 mice (unpublished data). Immunofluorescence analysis of epidermal differentiation markers revealed appropriate expression of keratin 14 (K14) in the basal layer. K10 expression started in the first suprabasal layer in mice of both genotypes, but the number of K10-positive layers was increased in K5-R1/R2 mice. Loricrin was expressed in the granular and cornified layers in mice of all genotypes, although the loricrin-positive part of the epidermis was thicker in K5-R1/R2 animals. K6, which is restricted to hair follicle keratinocytes in normal skin, was abnormally expressed in the interfollicular epidermis of adult K5-R1/R2 mice (Fig. S1). Interfollicular expression of this keratin is characteristic for hyperplastic and hyperproliferative skin. Overexpression of K6 was already seen at the RNA level at P18, but only a weak immunoreactivity was observed in the interfollicular epidermis at this time point (unpublished data).
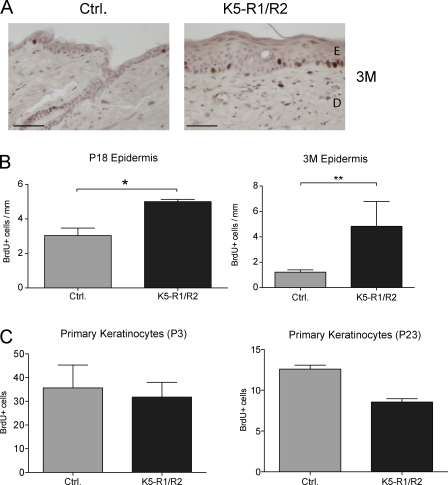
Cell proliferation was assessed by in vivo labeling with BrdU. At P18, keratinocyte proliferation was only mildly increased in K5-R1/R2 mice (Fig. 4 B, left). At the age of 3 mo, however, keratinocyte proliferation was strongly increased in the back and tail skin of K5-R1/R2 mice but not of K5-R1 or K5-R2 mice (Fig. 4, A and B, right; and not depicted). The increase in keratinocyte proliferation upon aging suggests that the hyperproliferation is not a cell-autonomous effect but results from the progressive inflammation. This hypothesis is supported by the normal in vitro proliferation rate of primary keratinocytes isolated from K5-R1/R2 mice at P3 (Fig. 4 C, left). Proliferation was even reduced in cells from P23 K5-R1/R2 mice compared with cells from control mice of the same age (Fig. 4 C, right).
Figure 4.
Enhanced keratinocyte proliferation in aged K5-R1/R2 mice in vivo but not in vitro. K5-R1/R2 mice and control littermates were injected with BrdU at P18 or 3 mo of age. Tail skin sections were stained with a peroxidase-conjugated antibody against BrdU. (A) Representative sections from 3-mo-old mice are shown. Bars, 100 µm. (B) The number of BrdU-positive cells/mm of basement membrane was counted using at least 10 sections per mouse. Error bars indicate mean ± SD. n = 8 control and 6 K5-R1/R2 mice for P18; n = 4 control and 6 K5-R1/R2 mice for 3 mo (3M). *, P ≤ 0.05; **, P ≤ 0.005. (C) Primary keratinocytes of control and K5-R1/R2 mice were isolated at P3 or P23, seeded at equal density, and labeled with BrdU. The percentage of BrdU positive cells was determined. Error bars indicate mean ± SD. n = 4 per genotype for P3 and n = 3 for P23. At least two microscopic areas were counted per dish.
Progressive skin inflammation in K5-R1/R2 mice
To test the possible role of inflammation in the hyperthickening of the epidermis, we analyzed the immune cells. The most obvious difference that we observed by immunofluorescence was the strong (60%) increase in epidermal γδ T cells (Fig. 5 A). This was verified by FACS analysis of cells from isolated epidermis (unpublished data). Interestingly, the number of γδ T cells was already significantly increased at P18, whereas no difference was observed at P12 (Fig. S2 A). The number of Langerhans cells was similar in control and K5-R1/R2 mice (unpublished data). Toluidine blue staining revealed significantly more mast cells in the dermis of K5-R1/R2 mice at the age of 6 mo and P18, but no differences at P12 (Fig. 5 A and Fig. S2 B).
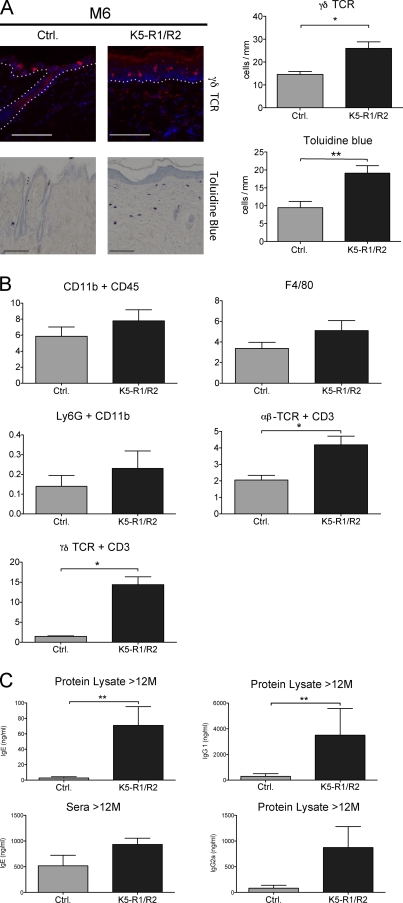
Figure 5.
Immune cell infiltrate in the skin of K5-R1/R2 mice. (A) Back skin sections from 6-mo-old (M6) control and K5-R1/R2 mice were stained with antibodies against the γδ T cell receptor. The white dotted line indicates the basement membrane. In addition, sections were stained with toluidine blue to identify mast cells. The number of γδ T cells and mast cells/mm of basement membrane was counted using at least five sections per mouse. n = 5 mice per genotype for γδ T cells and 7 mice per genotype for mast cells. Bar, 33 µm. (B) Cells from the dermis of 12-mo-old mice were analyzed by FACS using antibodies against different inflammatory cell markers. n = 4 mice per genotype. The frequencies of the individual inflammatory cells are shown. Original FACS data are shown in Fig. S3 A. (C) Dermal protein lysates (n = 3 control and 4 K5-R1/R2 mice) or serum (n = 3 per genotype) of mice at the age of 12 mo were analyzed for the levels of IgG1, IgG2a, and IgE by ELISA. Error bars indicate mean ± SD. *, P ≤ 0.05; **, P ≤ 0.005.
FACS analysis of dermal cells showed a significant increase in the number of CD45-positive immune cells, particularly αβ and γδ T cells, in adult mice (Figs. 5 B and Fig. S3 A). This was confirmed by immunofluorescence, and an increased number of CD45-positive cells was already seen at P36 (Fig. S2 C). In contrast, no significant difference in the number of macrophages and neutrophils was detected using antibodies against CD11b and F4/80 or Ly-6G, respectively (Fig. 5 B and Fig. S3 A). The lack of a macrophage or neutrophil infiltrate was also confirmed by immunohistochemistry (unpublished data).
B cells do not accumulate in the skin, but their activation in adult K5-R1/R2 mice was demonstrated by the presence of enhanced levels of Igs G1, G2a, and E in the dermis and of IgE in the serum (Fig. 5 C).
These results demonstrate that the loss of FGFR1 and FGFR2 in keratinocytes initiates an inflammatory response. This is also reflected by enhanced levels of phosphorylated (activated) and total nuclear factor κB (NF-κB) in the epidermis of aged mutant mice. Levels of total and phosphorylated STAT3 were also higher compared with controls, whereas expression and activation of p38 were not affected (Fig. 6 A).
Figure 6.
Progressive inflammation in the skin of K5-R1/R2 mice through production of cytokines by activated keratinocytes and γδ T cells. (A) Epidermal lysates from control and K5-R1/R2 mice at the age of P18 and 5 mo (5M) were analyzed by Western blotting for the levels of phosphorylated and total NF-κB (p65), Stat3, Erk1/2, and p38. (B) RNAs from isolated dermis and epidermis of K5-R1/R2 mice and littermate controls (8M) were analyzed for expression of inflammatory markers using real-time RT-PCR. Gapdh mRNA was used for normalization. Results are shown in the table. −, not detectable; +, weak expression; ++, moderate expression; +++, strong expression. (C) RNAs from the epidermis of K5-R1/R2 mice and littermate controls at P18 were analyzed for expression of inflammatory markers using real-time RT-PCR. Gapdh mRNA was used for normalization. Error bars represent mean ± SD. n = 3 per genotype. *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.001. (D) Keratinocytes and γδ T cells were purified from epidermal sheets by preparative FACS. Original FACS data are shown in Fig. S3 (A and B). RNAs from the purified cell populations were analyzed by RT-PCR for the expression of S100A8, S100A9, and IL-1F8. Ribosomal protein S29 (RPS29) mRNA was used for normalization; expression of keratin 14 and of the γδ T cell receptor was analyzed to verify the enrichment of the two cell populations.
Real-time RT-PCR analysis of RNAs from dermis and epidermis of aged mice revealed enhanced expression of the pro-inflammatory cytokines TNF, interleukin-1β (IL-1β), and of IL-1 family member 8 (IL-1F8), a new member of the IL-1 family (Fig. 6 B; Barksby et al., 2007). We also found up-regulation of S100A8 and S100A9, which are expressed by activated keratinocytes in hyperproliferative epidermis of psoriatic patients and in epidermal skin cancers and which act as chemoattractants for inflammatory cells (Gebhardt et al., 2006). In addition, intercellular adhesion molecule 1 (ICAM-1) was strongly expressed in the dermis (Fig. 6 B). This reflects the progressive skin inflammation in K5-R1/R2 mice.
Role of keratinocyte- and γδ T cell–derived cytokines in the initiation of the inflammatory response
To identify the factors that initiate the inflammation in K5-R1/R2 mice, we determined the expression of the above-mentioned cytokines at P18 using RNA from isolated epidermis. The efficient separation of epidermis from dermis was verified by RT-PCR for K14 (epidermal marker) and vimentin (dermal marker; unpublished data). At P18, the loss of FGFR expression was almost complete (Fig. 1 B), but the epidermis was not yet hyperthickened (Fig. 3 B). We found a strongly increased expression of the genes encoding S100A8, S100A9, and IL-1F8 in the epidermis of K5-R1/R2 mice, whereas TNF and IL-1β expression was unaltered (Fig. 6 C and not depicted). However, S100A8, S100A9, and IL-1F8 were not up-regulated in cultured keratinocytes from K5-R1/R2 mice, neither in exponentially growing cells nor in quiescent cells that had undergone in vitro differentiation (unpublished data). This finding suggests that these cytokines are not directly regulated by the loss of FGFR1 and FGFR2, but rather through a pro-inflammatory stimulus that is only present in vivo. Alternatively, they may be produced by other cell types in the epidermis, e.g., by γδ T cells. To distinguish between these possibilities, we separated γδ T cells from other epidermal cells (predominantly keratinocytes) by FACS using dissociated cells from epidermal sheets (Fig. S3, B and C). The efficient enrichment of keratinocytes and γδ T cells was verified by semiquantitative RT-PCR analysis of mRNAs encoding K14 or the γδ T cell receptor, respectively (Fig. 6 D). Using RNAs from the purified cell populations, we found that S100A8/A9 are mainly produced by keratinocytes and up-regulated in this cell type in K5-R1/R2 mice, whereas IL-1F8 was predominantly expressed by γδ T cells, particularly in the knockout mice (Fig. 6 D). Therefore, both keratinocytes and γδ T cells appear to contribute to the inflammatory phenotype.
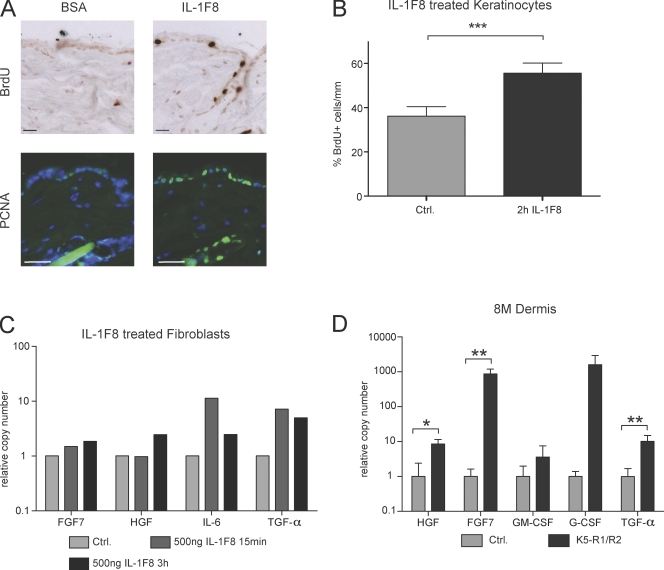
IL-1F8 stimulates keratinocyte proliferation and production of keratinocyte mitogens by stromal cells
IL-1 released from keratinocytes is a potent inducer of keratinocyte mitogens in fibroblasts, resulting in keratinocyte proliferation through a double paracrine loop (Szabowski et al., 2000). To determine if IL-1F8 plays a similar role, we injected IL-1F8 or BSA as a control intradermally into the skin of wild-type mice. 24 h later, increased keratinocyte proliferation was observed in IL-1F8–injected mice as determined by BrdU labeling as well as by staining of skin sections with an antibody against proliferating cell nuclear antigen (PCNA; Fig. 7 A). To determine if this is a direct effect of IL-1F8 or mediated via stromal cells, we treated murine keratinocytes with recombinant IL-1F8 at concentrations used in previous studies (Magne et al., 2006) and found a mild pro-mitogenic effect of this cytokine (60% increase; Fig. 7 B). Treatment of serum-starved murine fibroblasts with IL-1F8 induced the expression of IL-6 and IL-8, as described previously (Fig. 7 C and not depicted; Magne et al., 2006). In addition, increased mRNA levels of TGF-α, hepatocyte growth factor (HGF), and FGF7 were observed in response to IL-1F8 treatment (Fig. 7 C). Most importantly, these mitogens were also overexpressed in the dermal compartment of K5-R1/R2 skin, particularly in aged mice, together with granulocyte colony-stimulating factor (G-CSF; Fig. 7 D). Although the FGFR1/2-deficient keratinocytes can no longer respond to FGF7, the other growth factors are likely to contribute strongly to the hyperproliferative phenotype.
Figure 7.
IL-1F8 stimulates keratinocyte proliferation and production of keratinocyte mitogens by fibroblasts. (A) IL-1F8 or BSA were intradermally injected into wild-type mice. 24 h later, proliferating cells were identified by BrdU labeling or by immunostaining with an antibody against PCNA. Bars, 33 µm. (B) Immortalized keratinocytes from wild-type mice were treated with 500 ng/ml IL-1F8 for 2 h and labeled with BrdU. The percentage of BrdU-positive cells was determined. Error bars indicate mean ± SD; n = 3. (C) Immortalized embryonic fibroblasts were starved overnight in medium with 0.1% FCS and subsequently treated for 15 min or 3 h with 500 ng/ml IL-1F8. RNA was isolated from these cells before and after IL-1F8 treatment and analyzed by real-time RT-PCR for the mRNA levels of different keratinocyte mitogens as indicated. Gapdh mRNA was used for normalization. Bars represent means from duplicate determinations. The IL-1F8–induced expression of keratinocyte mitogens was reproduced with an independent fibroblast cell line. (D) RNA from the dermis of 8-mo-old (8M) control and K5-R1/R2 mice was analyzed by real-time RT-PCR for the mRNA levels of different keratinocyte mitogens as indicated. RPS29 mRNA was used for normalization. Error bars represent mean ± SD. n = 3 per genotype (5 for TGF-α). *, P ≤ 0.05; **, P ≤ 0.005; ***, P ≤ 0.001.
Epidermal barrier function is disturbed in K5-R1/R2 mice
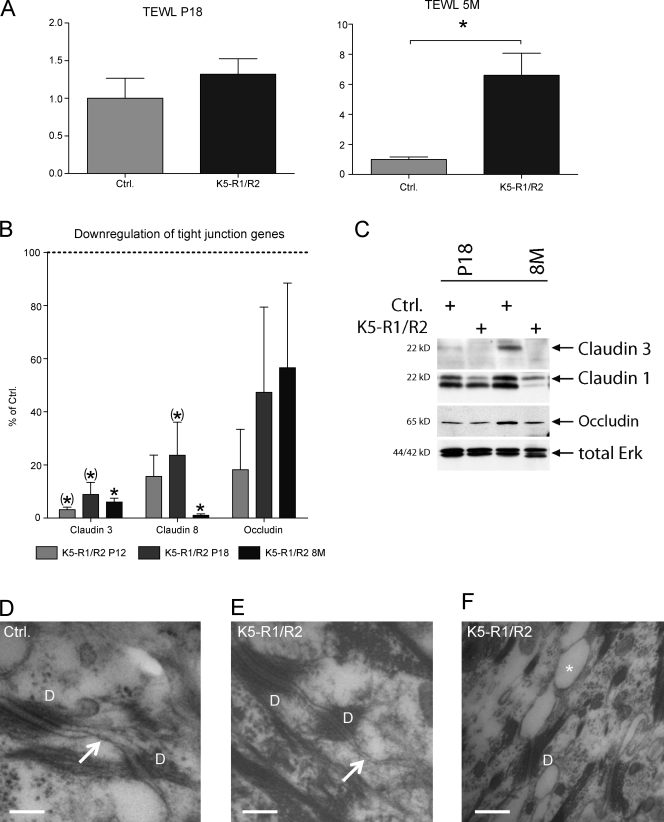
Because cutaneous inflammation frequently results from a defect in epidermal barrier function (Segre, 2006), we analyzed the transepidermal water loss (TEWL) that reflects the status of the permeability barrier (Fluhr et al., 2006). At P18, TEWL was slightly increased in K5-R1/R2 mice. This phenotype strongly increased with age, and the TEWL was significantly higher in the double knockout mice compared with control animals at the age of 6 mo (Fig. 8 A). This is consistent with their high consumption of drinking water (unpublished data) as well as with the dry and fragile appearance of the skin.
Figure 8.
Impaired barrier function in K5-R1/R2 mice. (A) TEWL was determined in control and K5-R1/R2 mice at the age of P18 or 5 mo (5M). Error bars indicate mean ± SD. n = 6 control mice at P18 and 9 K5-R1/R2 mice at P18; n = 4 mice per genotype at 5M. *, P ≤ 0.05. (B) RNA from isolated epidermis of mice at P12, P18, or 8 mo was analyzed by real-time RT-PCR for expression of different claudins and occludin. RPS29 mRNA was used for normalization. n = 3–5 per time point and genotype. Expression in control mice was arbitrarily set as 100%. Error bars represent mean ± SD. (*), 0.05 < P < 0.06; *, P ≤ 0.05. (C) Epidermal lysates of control and K5-R1/R2 mice (P18 or 8 mo old [8M]) were analyzed by Western blotting for the levels of claudin 1, claudin 3, occludin, and total ERK (loading control). (D–F) High-magnification ultrastructure revealed a typical desmosome–tight junction (arrow) complex between keratinocytes of the stratum granulosum in a 4-mo-old control mouse (D), whereas only a rudimentary cell–cell contact is formed adjacent to a desmosome of a 4-mo-old K5-R1/R2 mouse (E). In these animals, bubble-like clefts (asterisk) were frequently seen between the keratinocytes in the stratum granulosum, particularly in aged mice (F, 6-mo old mouse). D, desmosome. Bars: (D and E) 150 nm; (F) 250 nm.
Epidermal barrier function is conferred by the cornified envelope (Segre, 2006) and by tight junctions (Pummi et al., 2001; Brandner et al., 2002; Furuse et al., 2002; Langbein et al., 2002). Expression of loricrin was enhanced in the knockout mice (Fig. S1), and the mRNA levels of SPRR2A, another component of the cornified layer, were up-regulated >30-fold (unpublished data). In contrast, expression of tight junction components was strongly reduced. The mRNA levels of claudin 3, claudin 8, and occludin were much lower in K5-R1/R2 mice, as determined by real-time RT-PCR analysis of epidermal RNAs (Fig. 8 B). A strong down-regulation was already seen at P12 (2–18% of control). Western blot analysis of epidermal lysates confirmed the down-regulation of claudin 3 and occludin at the protein level and also revealed reduced claudin-1 expression in K5-R1/R2 mice (Fig. 8 C). Importantly, the down-regulation of tight junction gene expression preceded the onset of inflammation and hair loss, which strongly suggests that this is a direct consequence of the loss of FGFR1 and -2.
As a consequence of the down-regulation of tight junction gene expression, only a rudimentary development of tight junctions was observed in the epidermis of K5-R1/R2 mice using transmission electron microscopy (Fig. 8 E), whereas tight junctions were well developed in control mice (Fig. 8 D). In most of the sections from adult K5-R1/R2 animals, we also found bubble-like intercellular clefts between the keratinocytes of the stratum granulosum, which most likely result from water-filled cavities (Fig. 8 F). Enlargement of the intercellular gaps was already seen occasionally in mutant mice at P18, and the phenotype progressed upon aging (unpublished data).
To determine if the down-regulation of claudin/occludin expression is a cell-autonomous effect, we analyzed the expression of these tight junction components in immortalized keratinocytes. Indeed, a strong down-regulation was observed in three independent cell lines from K5-R1/R2 mice compared with controls (Fig. 9, A–C). When confluent keratinocytes from wild-type mice were stimulated with FGF7, a slight increase in the levels of occludin and a stronger increase in claudin 1 and claudin 3 were observed, which indicates that these genes are targets of FGFs in keratinocytes (Fig. 9 B). Their regulation occurs at the RNA level because the mRNA levels of claudin 1 and claudin 3 were 5- or 30-fold elevated in FGF7-treated keratinocytes compared with nontreated cells (unpublished data). EGF also induced their expression under these conditions (unpublished data), but it can obviously not compensate for the loss of FGF receptors because expression of claudin 1, claudin 3, and occludin was reduced in keratinocytes of K5-R1/R2 mice, which were cultured in the presence of EGF (Fig. 9 B).
Figure 9.
Reduced expression of tight junction proteins in cultured keratinocytes from K5-R1/R2 mice reduces the TER. (A) RNAs from cultured, immortalized keratinocytes of control and K5-R1/R2 mice were analyzed by real-time RT-PCR for expression of different claudins and occludin. RPS29 mRNA was used for normalization. Bars represent means from duplicate determinations. The down-regulation of these tight junction components in K5-R1/R2 cells was reproduced with two independent cell lines from control and mutant mice. (B) Cultured keratinocytes of control and K5-R1/R2 mice were starved for 24 h and incubated for 96 h in starvation medium with or without FGF7. Lysates were analyzed by Western blotting for expression of claudin 1, claudin 3, occludin, or GAPDH (loading control). (C) Cultured primary keratinocytes were grown to confluency and analyzed for the expression of claudin 3 by immunofluorescence (green). Bars, 33 µm. (D) TER was measured using immortalized keratinocytes from control and K5-R1/R2 mice that had been grown to confluency and incubated in differentiation medium until stable values were obtained. A representative measurement is shown using six independent filters with cells from each genotype. Error bars represent mean ± SD. The result was reproduced at different days and with two independent cell lines from control and K5-R1/R2 mice. **, P ≤ 0.005.
Finally, we determined if the reduction in claudin/occludin expression is functionally important. Because of the lack of a functional assay to determine tight junction permeability in adult mice, we addressed this question in vitro. For this purpose, we measured the transepithelial electrical resistance (TER) of confluent immortalized keratinocytes from control and K5-R1/R2 mice because tight junctions restrict paracellular diffusion of ions. Indeed, the TER was reduced by >50% in three independent cell lines from FGFR-deficient mice compared with control mice (Fig. 9 D and not depicted). The same difference was observed on several consecutive days (unpublished data).
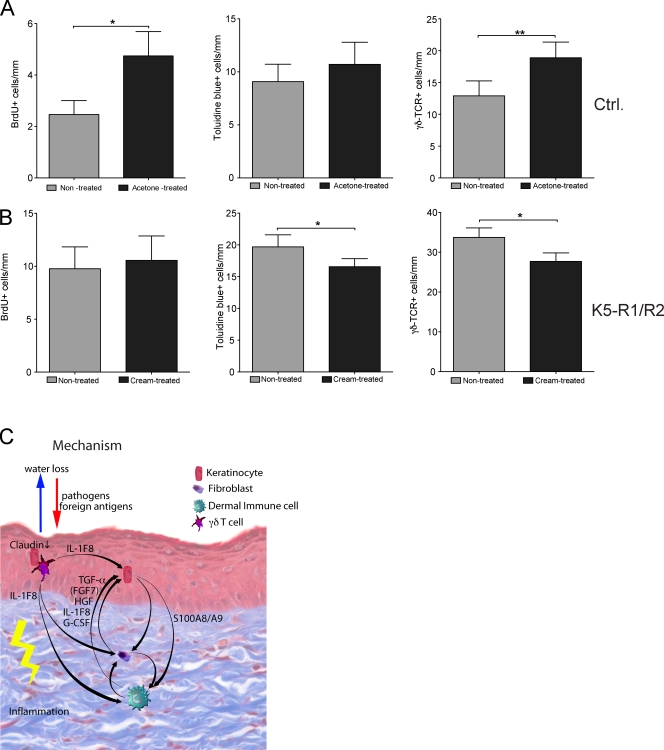
Taken together, our results suggest that reduced expression of several tight junction components disrupts the epidermal barrier, resulting in activation of keratinocytes and γδ T cells. To further test this hypothesis, we determined if disruption of the epidermal barrier causes a similar phenotype. Treatment of wild-type mice with acetone, which damages the epidermal barrier, caused the expected hyperproliferation of keratinocytes (Fig. 10 A; Proksch et al., 1991). Similar to the case of K5-R1/R2 mice, this was associated with a minor increase in the number of mast cells and with a significant increase in epidermal γδ T cells (Fig. 10 A). In a complementary experiment, we tested if treatment of the skin of K5-R1/R2 mice with a moisturizing cream ameliorates the phenotype. Although we could not observe a reduction in keratinocyte proliferation, the number of mast cells and of γδ T cells was significantly reduced (Fig. 10 B). Concomitantly, expression of IL-1F8 was reduced by 30%, whereas no obvious change in the expression of claudin 1, claudin 3, or occludin was observed (unpublished data).
Figure 10.
The epidermal barrier controls the number of mast cells and γδ T cells. (A) Topical treatment of wild-type mice with acetone enhanced keratinocyte proliferation and increased the number of mast cells and γδ T cells. (B) Topical treatment of K5-R1/R2 with moisturizing cream did not affect keratinocyte proliferation, but reduced the number of mast cells and of γδ T cells. Error bars indicate mean ± SD. n = 7 mice for acetone and 5 mice for moisturizing cream treatment. (C) Model describing the pathogenic mechanisms in the skin of K5-R1/R2 mice. Loss of FGFR signaling in keratinocytes results in reduced expression of tight junction components. The resulting defective epidermal barrier, together with the loss of sebaceous glands, causes skin dryness. This activates a stress response in keratinocytes, resulting in activation of γδ T cells and up-regulation of cytokine expression in activated γδ T cells and keratinocytes. The latter stimulate keratinocyte proliferation directly (IL-1F8) and/or indirectly (IL-1F8 and S100A8/A9) through induction of a double paracrine loop involving several keratinocyte mitogens that are produced by stromal cells. In addition, invasion of foreign antigens and pathogens may further activate immune cells and accelerate the inflammatory response. *, P ≤ 0.05; **, P ≤ 0.005.
Together, these findings support the hypothesis that a defect in the epidermal barrier initiates and maintains the cutaneous inflammatory response in K5-R1/R2 mice. This induces keratinocyte hyperproliferation through a double paracrine loop involving γδ T cell–derived IL-1F8, keratinocyte-derived S100A8 and A9, and several keratinocyte mitogens produced by dermal cells (summarized in Fig. 10 C).
Discussion
Cooperative functions of FGFR1 and FGFR2 in keratinocytes
We identified essential roles of FGFR1IIIb and FGFR2IIIb in the maintenance of skin appendages and epidermal barrier function. The responsible ligands are most likely FGF1, FGF7, FGF10, and FGF22, which are expressed in different compartments of the skin (Steiling and Werner, 2003). Our results identified FGFR2IIIb as the most important receptor for these ligands in keratinocytes, whereas FGFR1IIIb provides a back-up function. FGFR3 is also expressed in keratinocytes, particularly in the suprabasal layers (Logié et al., 2005). However, it binds a different set of FGFs than FGFR1IIIb and FGFR2IIIb (Zhang et al., 2006), which suggests distinct functions. Although mice lacking FGFR3 have no obvious skin abnormalities (unpublished data), it will be interesting to determine the consequences of the loss of all FGF receptors in keratinocytes.
Loss of FGFR1 and 2 in keratinocytes impairs hair regeneration
The loss of skin appendages that we observed in K5-R1/R2 mice is consistent with the stimulatory effect of FGF7 on hair follicle growth, development, and differentiation (Danilenko et al., 1995). However, only a rough hair coat but no hair loss was seen in FGF7-deficient animals (Guo et al., 1996), which indicates functional redundancy among FGF family members. It seems likely that FGF7 together with FGF10 and FGF22 cooperatively orchestrate hair regeneration via activation of FGFR1IIIb and FGFR2IIIb on hair follicle keratinocytes. This hypothesis is further supported by the up-regulation of these FGFs in the anagen phase of the hair cycle (Kawano et al., 2005). It was recently reported that expression of FGF7 and FGF10 increases in the dermal papilla during the transition from early to late telogen. Functional in vitro studies suggested that this results in stimulation of hair germ cells adjacent to the papilla and subsequent hair cycle activation (Greco et al., 2009). The failure of K5-R1/R2 mice to regenerate hair follicles is fully consistent with this predicted function. The reason for the subsequent hair loss is presently unclear. It may well be that the progressive inflammation contributes to this phenotype, but this needs to be further studied in the future.
Keratinocyte hyperproliferation in K5-R1/R2 mice is mediated via the stroma
A particularly striking and unexpected phenotype of the K5-R1/R2 mice is the progressive acanthosis, which results from enhanced proliferation of keratinocytes. This was surprising because FGFs are potent mitogens for keratinocytes, and because epidermal hypotrophy was observed in the skin of newborn mice lacking FGFR2IIIb in all cells (Petiot et al., 2003) as well as in the skin of adult mice expressing a dominant-negative FGFR2IIIb mutant in keratinocytes (Werner et al., 1994). In fact, we also observed a slight epidermal hypotrophy in very young K5-R1/R2 mice (Fig. 3 A), but this reversed upon aging, and the reversal correlated with the onset of inflammation. Therefore, we propose that loss of FGFRs in keratinocytes results in activation of immune cells, which subsequently disrupts cutaneous homeostasis.
FGFs regulate tight junction components
Several mechanisms may be responsible for the activation of immune cells, including direct FGF-mediated suppression of pro-inflammatory cytokine expression, inflammation mediated by degenerating hair follicles, or a defect in barrier function that results in dry skin and possibly invasion of irritants/allergens and bacteria. The first possibility seems unlikely because expression levels of S100A8/A9 and IL-1F8 were similar in cultured keratinocytes from mice of both genotypes. Inflammation as a result of hair follicle degeneration has been postulated for mice lacking β1 integrins in keratinocytes (Brakebusch et al., 2000). However, the macrophage infiltrate around the hair follicles that was seen in β1 integrin-deficient mice was not observed in K5-R1/R2 animals. Furthermore, the phenotype progressed upon loss of all follicles, which indicates that degenerating follicles are not or at least not exclusively responsible for the development of skin inflammation. Therefore, the most likely explanation for the skin inflammation is the defect in the epidermal barrier. Surprisingly, expression of several genes involved in the formation of the cornified envelope was even increased (unpublished data). This was also the case for filaggrin, a gene that is mutated in a large percentage of patients with atopic dermatitis (Sandilands et al., 2009). In addition, the filaggrin protein was normally processed in K5-R1/R2 mice (unpublished data). In contrast, expression of several claudins and of occludin was much lower in the epidermis and in cultured keratinocytes of K5-R1/R2 mice. The reduced TER in vitro as well as the increased TEWL in vivo strongly suggest that the down-regulation of these tight junction components is functionally important. Consistent with this hypothesis, changes in the tight junction composition affected their permeability (Inai et al., 1999; Furuse et al., 2002; Tunggal et al., 2005). Unfortunately, functional in vivo assays, such as biotin penetration, can only be performed in newborn mice, where the phenotype was not sufficiently developed. Therefore, a final proof for an in vivo deficit in tight junction permeability will await improved in vivo assays. Nevertheless, the rudimentary development of the tight junctions and the adjacent large gaps between the keratinocytes of the granular layer that we observed by electron microscopy strongly support our hypothesis of an increased tight junctional permeability that results in severe water loss. Down-regulation of tight junction proteins was already observed at P12, whereas enhanced water loss was only seen after P18. It may well be that a certain threshold of tight junction protein down-regulation must be reached to allow significant water loss. In addition, we found that expression levels of claudins and occludin strongly declined in control mice after P18 (Fig. S4). Therefore, a further loss in K5-R1/R2 mice might have more severe consequences in adults compared with young mice.
The down-regulation of tight junction gene expression in cultured keratinocytes from K5-R1/R2 mice together with the increased expression of these genes in wild-type keratinocytes in response to FGF7 treatment indicates that they are targets of FGFs. These results are consistent with the reduced expression of tight junction proteins in blood vessels and the defective blood–brain barrier function in FGF2/FGF5 double knockout mice (Reuss et al., 2003), with the FGF2-mediated preservation of the composition of tight junctions in organotypic cortical cultures of mice (Bendfeldt et al., 2007), and with the disassembly of adherens and tight junctions in endothelial cells upon inhibition of FGFR signaling (Murakami et al., 2008). These findings indicate that maintenance of junctional integrity is a general but as yet poorly characterized function of FGFs.
Keratinocyte hyperproliferation is mediated via the stroma
The enhanced water loss seen in K5-R1/R2 mice resulted in severe skin dryness and fragility, which is likely to be aggravated by the loss of sebaceous glands that release the moisturizing sebum. It was previously shown that low humidity stimulates keratinocyte proliferation and amplifies the hyperproliferative response to barrier disruption, causing dermal mast cell hypertrophy, their degranulation, and subsequent inflammation (Denda et al., 1998). In addition, it is likely to stress and damage keratinocytes, which results in activation and proliferation of γδ T cells (Jameson and Havran, 2007; Strid et al., 2009). The role of an impaired barrier and subsequent dryness in the skin phenotype is supported by the results obtained with acetone-treated wild-type mice and by the beneficial effect of moisturizing cream on the inflammatory phenotype of K5-R1/R2 mice. In addition, it may well be that irritants/allergens and bacteria can invade the dry and fragile skin, particularly at sites of minor injury, which results in immune cell activation and enhancement of the inflammatory response.
One of the responsible inflammatory mediators is most likely IL-1F8. We identified this cytokine as a keratinocyte mitogen, which suggests that γδ T cell–derived IL-1F8 contributes to the hyperproliferative phenotype through an intra-epidermal paracrine mechanism. Most importantly, it induced the expression of keratinocyte mitogens in fibroblasts. This is likely to be relevant for the in vivo situation because these mitogens were overexpressed in the dermis of K5-R1/R2 mice. Preliminary results also showed that intradermal injection of IL-1F8 induces expression of the same growth factors in vivo. Their up-regulation may then further promote keratinocyte hyperproliferation in a paracrine manner. The role of IL-1F8 in the induction of the hyperproliferative phenotype is consistent with the cutaneous inflammation, hyperkeratosis, and acanthosis seen in transgenic mice overexpressing IL-1F6 in keratinocytes (Blumberg et al., 2007). Because IL-1F6 and IL-1F8 signal through the same receptors (Towne et al., 2004), the results from Blumberg et al. (2007) and from our study identify the novel IL-1 family members as major players in inflammatory/hyperproliferative skin disease.
In summary, our study revealed novel roles of FGFR1 and FGFR2 in epidermal barrier function and cutaneous homeostasis through their regulation of tight junction components. The importance of the latter in human skin disease associated with hyperscaling and acanthosis is emerging, as reflected by the ichthyosis phenotype seen in patients with a claudin 1 mutation (Hadj-Rabia et al., 2004) and by the abnormal expression/distribution of tight junction proteins in psoriasis (Peltonen et al., 2007; Watson et al., 2007; Kirschner et al., 2009). Future studies will reveal if abnormal expression of FGFs and/or FGF receptors and their tight junction targets is associated with inflammatory skin diseases such as atopic dermatitis, which is also characterized by impaired barrier function (Proksch et al., 2006; Elias and Steinhoff, 2008) and which shows several similarities with the phenotype of K5-R1/R2 mice, particularly with regard to the inflammatory infiltrate and the epidermal thickening. In addition, the data presented in this manuscript suggest new therapeutic applications of FGFs in patients with impaired epidermal barrier function.
Materials and methods
Generation and maintenance of mice lacking FGFR1 and/or FGFR2 in keratinocytes
Fgfr1flox/flox mice were mated with Fgfr2flox/flox mice (provided by A. Ramirez and J. Jorcano, Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas, Madrid, Spain). After two generations of breeding, the Fgfr1flox/flox/Fgfr2flox/flox mice were mated with K5cre/wt transgenic mice. The male Fgfr1flox/wt/Fgfr2flox/wt K5Cre/wt mice obtained from this mating were then crossed with the Fgfr1flox/flox/Fgfr2flox/flox female mice. In addition, mice lacking FGFR1IIIb in all cells (provided by L.T. Williams, FivePrime Therapeutics, San Francisco, CA) were mated with Fgfr2flox/flox and K5cre/wt. All mice were in a C57BL/6 genetic background. Mice were housed and fed according to Swiss federal animal protection guidelines. All procedures were approved by the local veterinary authorities of Zurich, Switzerland.
Identification of genetically modified mice by PCR
The floxed Fgfr1 allele was detected by genotyping PCR using primers 5′-CGAATGGACAAGCCCAGTAAC-3′ and 5′-CTCCTGCTTCCTTCAGAGC-3′. PCR products of 200 bp or 300 bp were obtained for the wild-type or floxed allele, respectively.
Genotyping PCR for the Fgfr2 floxed allele was performed using primers FGFR2F5 (5′-ATAGGAGCAACAGGCGG-3′) and FGFR2F3 (5′-TGCAAGAGGCGACCAGTCAG-3′). PCR products of 140 bp or 200 bp were obtained for the wild-type or floxed alleles, respectively. The recombined allele was identified using primers FGFR2F5 and FGFR2rec (5′-CATAGCACAGGCCAGGTTG-3′). A 470-bp fragment was obtained.
The K5-Cre transgene was amplified using primers 5′-AACATGCTTCATCGTCGG-3′ and 5′-TTCGGATCATCAGCTACACC-3′. A 420-bp fragment was obtained with DNA from transgenic mice.
Analysis of TEWL
Mice were shaved, and, after 30 min, TEWL was determined using a Tewameter (Courage and Khazaka Electronic GmbH). The device was placed on the dorsal skin of mice, and 30 consecutive measurements were taken from four different places on the back as described by the manufacturer.
Separation of dermis and epidermis
Mice were sacrificed, then washed in 10 mM iodine solution, followed by 70% ethanol and PBS, and a piece of back skin was excised. It was washed in gentamycin solution (1:200; Sigma-Aldrich) and then floated in dispase solution (Invitrogen) in defined keratinocyte serum-free medium (Invitrogen), including defined keratinocyte growth supplements for 2 h at 37°C. The epidermal sheet was peeled off from the dermal layer using fine forceps. Isolated dermis and epidermis were immediately frozen in liquid nitrogen and used for the isolation of RNA or preparation of protein lysates.
FACS analysis of cells from isolated dermis or epidermis and preparative sorting of keratinocytes and γδ T cells
Dermis and epidermis from mouse back skin were separated after overnight incubation in 0.2% trypsin in DME at 4°C. The epidermis was cut into small pieces and incubated for 20 min at 37°C in 0.2% trypsin in DME. The dermis was treated with DME containing 2.5 mg/ml collagenase II (Worthington Biochemical Corporation), 2.5 mg/ml collagenase IV (Invitrogen), 0.5 mg/ml DNase I (Sigma-Aldrich), and 1 mg/ml hyaluronidase (Worthington Biochemical Corporation). A single cell suspension was prepared by passing the mix through a 70-µm strainer. DME/10% FCS was added to inactivate trypsin. Cells were washed with FACS buffer (PBS/0.2% BSA/5 mM EDTA), and an unspecific antibody binding was blocked with anti-CD32.
For preparative sorting of keratinocytes and γδ T cells, cells were stained for γδ T cell receptor, CD-45, and 7-amino-actinomycin D to determine viability. Analysis was performed using a cell sorter (Aria; BD). The fraction costained for the γδ T cell receptor and for CD45 contains predominantly γδ T cells, and the fraction that was negative for both markers consists predominantly of keratinocytes. RNA was isolated from the sorted cells using the RNeasy Micro kit (QIAGEN). cDNA was generated with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems).
Antibodies used for FACS
See Table I for the antibodies used.
Table I.
Antibodies used for FACS
| Antibodies | Source |
| Anti-CD32 | BD |
| Anti-CD45 | BD |
| Anti-γδ T cell receptor | BD |
| Anti-CD11b | eBioscience |
| Anti-Ly-6G | eBioscience |
| 1-A/1-E | BD |
| Anti-αβ cell receptor | eBioscience |
| Anti-F4/80 | Invitrogen |
| Anti-CD3 | BD |
Intradermal injection of IL-1F8
Wild-type mice were intradermally injected with 50 µl of a solution of 1 µg/ml IL-1F8 (R&D Systems) in 0.1% BSA in PBS or vehicle control. 24 h later, proliferating cells were analyzed using BrdU labeling as described below.
BrdU incorporation assay
Mice were injected intraperitoneally with BrdU (250 mg/kg in 0.9% NaCl; Sigma-Aldrich) and sacrificed 2 h after injection. Skin samples were fixed in 95% ethanol/1% acetic acid. Sections were incubated with a peroxidase-conjugated monoclonal antibody directed against BrdU (Roche) and stained with diaminobenzidine.
Analysis of tight junction protein expression in cultured keratinocytes
Primary keratinocytes from control and K5-R1/R2 mice were grown to confluency in defined keratinocyte growth medium with 10 ng/ml EGF and 10−10 M cholera toxin, and subsequently lysed. Alternatively, cells were starved overnight in medium without supplements and subsequently treated for 24 h with 10 ng/ml FGF7 (R&D Systems). Lysates were obtained before and after FGF7 treatment and analyzed by Western blotting for expression of tight junction components.
Measurement of TER
Three different immortalized keratinocyte lines from control and K5-R1/R2 mice were grown to confluency on permeable supports (Costar Snapwell; Sigma-Aldrich) with 0.4-µm polycarbonate filter membranes in keratinocyte growth medium and subsequently cultured in differentiation medium (1:1 DME/F-12 supplemented with 10% FCS, penicillin/streptomycin, 0.1 nM cholera toxin, 2 nM T3, 5 µg/ml transferrin, 0.4 µg/ml hydrocortisone, 25 µg/ml gentamycin, 5 µg/ml insulin, and 10 ng/ml EGF) for 7–14 d. TER measurements were performed daily using an epithelial tissue volt ohmmeter (World Precision Instruments) as described previously (Balda et al., 1996). For each cell line, 3–6 filters were analyzed daily.
Cell culture
Keratinocytes were isolated from pools of mice with different genotypes, as described previously (Braun et al., 2002), with the exception that cells were seeded at a density of 5 × 104 cells/cm2 on collagen IV (2.5 µg/cm2; Sigma-Aldrich)-coated dishes. The freshly isolated cells were incubated for 30 min at 37°C. Thereafter, the medium was replaced and cells were grown in defined keratinocyte serum-free medium (Invitrogen) supplemented with 10 ng/ml EGF and 10−10 M cholera toxin.
Analysis of FGF receptor-mediated signaling
Keratinocytes were suspended in defined keratinocyte medium at a cell density of 5 × 105 cells/ml and seeded in 6-well plates (400 µl/well). After overnight incubation, they were starved in defined medium without supplements for 24 h and then treated for 10 min with 10 ng/ml FGF7, FGF10, or 20 ng/ml EGF (all from R&D Systems). Cells were harvested on ice and analyzed by Western blotting for the levels of signaling proteins and their phosphorylated forms.
RNA isolation and RNase protection assay
RNA isolation and RNase protection assays were performed as described previously (Werner et al., 1994). Fgfr3 and Fgfr4 cDNAs were used as templates (Steiling et al., 2003).
Real-time RT-PCR
Real-time RT-PCR was performed according to the manual of the Light Cycler 480 (Roche). The reverse transcription product obtained from ∼0.5 ng RNA was used together with 10 µl of SYBRGreen reaction mix. The reaction was performed in 50 cycles (95°C for 10 min for initial denaturation; 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s for each cycle). Duplicate determinations were performed for each sample.
Primers used for RT-PCR
See Table II for the primers used.
Table II.
Primers used for RT-PCR
| Target gene | Forward sequence | Reverse sequence |
| FGFR1 | 5′-CAACCGTGTGACCAAAGTGG-3′ | 5′-TCCGACAGGTCCTTCTCCG-3′ |
| FGFR2 | 5′-ATCCCCCTGCGGAGACA-3′ | 5′-GAGGACAGACGCGTTGTTATCC-3′ |
| IL-1α | 5′-ATGATCTGGAAGAGACCATCC-3′ | 5′-GGCAACTCCTTCAGCAACA-3′ |
| IL-1β | 5′-CTGAAAGCTCTCCACCTC-3′ | 5′-TGCTGATGTACCAGTTGGGG-3′ |
| IL-1F8 | 5′-GCCTGTCATTCTTAGCTTGAT-3′ | 5′-TGTCTACTTCCTTAAGCTGC-3′ |
| TNF | 5′-CCCCAATGTGTCCGTCGTG-3′ | 5′-GCCTGCTTCACCACCTTCT-3′ |
| Claudin 8 | 5′-TCAGAATGCAGTGCAAGGTC-3′ | 5′-AGCCGGTGATGAAGAAGATG-3′ |
| Claudin 3 | 5′-GCGGCTCTGCTCACCTTAGT-3′ | 5′-GACGTAGTCCTTGCGGTCGTA-3′ |
| Claudin 1 | 5′-CTTCTCTGGGATGGATCG-3′ | 5′-GGGTTGCCTGCAAAGTACTGT-3′ |
| Occludin | 5′-TTGAAGTCCACCTCCTTACAGA-3′ | 5′-CCGGATAAAAAGAGTACGCTGG-3′ |
| SPRR2a | 5′-GAACCTGATTCTGAGACTCAA-3′ | 5′-GCACACTACAGGACGACAC-3′ |
| S100A8 | 5′-GCCGTCTGAACTGGAGAAG-3′ | 5′-GTGAGATGCCACACCCACTTT-3′ |
| S100A9 | 5′-CGCAGCATAACCACCATCAT-3′ | 5′-AAGATCAACTTTGCCATCAGC-3′ |
| HGF | 5′-ACTTGCAAGGCCTTCG-3′ | 5′-GCAAAAAGCTGTGTTCATGGG-3′ |
| TGF-α | 5′-CTGAGTGACTCACCCGTGGC-3′ | 5′-GCGGAGCTGACAGCAGTGGAT-3′ |
| ICAM-1 | 5′-TGTTTCCTGCCTCTGAAGC-3′ | 5′-CTTCGTTTGTGATCCTCCG-3′ |
| G-CSF | 5′-TGCACTATGGTCAGGACGAG-3′ | 5′-GGGGTGACACAGCTTGTAGG-3′ |
| Vimentin | 5′-CCTGTACGAGGAGGAGATGC-3′ | 5′-GTGCCAGAGAAGCATTGTCA-3′ |
| Keratin 14 | 5′-AACCACGAGGAGGAAATGG-3′ | 5′-CCGGAGCTCAGAAATCTCAC-3′ |
| TcR-Vγ3 | 5′-GGGTCGACTCCTGGATATCTCAGGATCAG-3′ | 5′-GGGTCGACTTGTTTCAGCAGAAGAAGGAAG-3′ |
| GAPDH | 5′-TCGTGGATCTGACGTGCCGCCTG-3′ | 5′-CACCACCCTGTTGCTGTAGCCGTAT-3′ |
| RPS29 | 5′-GGTCACCAGCAGCTCTACTG-3′ | 5′-GTCCAACTTAATGAAGCCTATGTCC-3′ |
HGF, hepatocyte growth factor; ICAM-1, intercellular adhesion molecule 1; TCR, T cell receptor.
Histological analysis
Skin samples were fixed overnight in 95% ethanol/1% acetic acid or in 4% PFA/PBS followed by paraffin embedding. Sections (7 µm) were stained with hematoxylin/eosin (H/E).
Quantification of hair follicles in mouse back skin
Hair follicles in the back skin were quantified by counting the number of dermal papillae in sections at 100× magnification. Seven serial images were counted for mice at P18, and 18 serial images were counted for mice at P36.
Electron microscopy
Mice were lethally anesthetized with 700 mg/kg pentobarbital and perfused with 4% PFA in PBS. Skin samples were kept overnight in fixation solution, then rinsed and stored in PBS. After washing in 0.1 M cacodylate buffer, pH 7.2, at 4°C, the specimens were either directly treated with 2% OsO4 for 2 h or first incubated in 1% tannin for 3 h and then treated with 2% OsO4 for 2 h. After washing, they were stained in 1% uranyl acetate, dehydrated through series of graded ethanols, and embedded in araldite resin. Ultrathin sections (30–60 nm) were processed with a diamond knife and placed on copper grids. Transmission electron microscopy was performed using a 902A electron microscope (Carl Zeiss, Inc.).
Immunofluorescence and immunohistochemistry
After deparaffinization, skin sections were blocked with PBS containing 3% BSA and 0.025% NP-40 for 1 h at room temperature, then incubated overnight at 4°C with the primary antibodies diluted in the same buffer. After three washes with PBST (1× PBS/0.1% Tween 20), slides were incubated at room temperature for 1 h with the Cy2- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.) and 1 µg/ml DAPI as a counterstain, washed with PBST again, and mounted with Mowiol (Sanofi-Aventis). Sections were photographed using a microscope (Imager.A1) equipped with a camera (Axiocam Mrm) and enhanced-contrast Plan-Neofluar objectives (10×/0.3 NA, 20×/0.5 NA; all from Carl Zeiss, Inc.). For data acquisition, we used the Axiovision 4.6 software (Carl Zeiss, Inc.).
For immunofluorescence analysis of cultured cells, cells were fixed for 10 min with ice-cold acetone/methanol (1/1) and subsequently treated as described above for sections. Stained cells were photographed using a confocal microscope (SP1-2; Leica) equipped with a 63 × 0.6–1.32 NA (Iris) Plan-Apochromat oil objective lens. For data acquisition, we used the LCS Software (Leica).
For immunohistochemistry, biotinylated secondary antibodies (Vectastain ABC kit; Vector Laboratories) and diaminobenzidine were used, followed by counterstaining with hematoxylin. Stained sections were photographed with a microscope (Axioskop 2) equipped with a camera (Axiocam HRc) and Plan-Neofluor (10×/0.3 NA, 20×/0.5 NA; all from Carl Zeiss, Inc.) objectives. For data acquisition, we used the Axiovision 4.2 software (Carl Zeiss Inc.).
Antibodies used for immunofluorescence
See Table III for the antibodies used.
Table III.
Antibodies used for immunofluorescence
| Primary antibody | Host | Source |
| Anti-CD3 IgG | Rabbit | DAKO |
| Anti-Ly6G IgG | Rat | BD |
| Anti-F4/80 IgG | Rat | BMA Biomedicals |
| Anti-keratin 6 IgG | Rabbit | Covance |
| Anti-keratin 10 IgG | Mouse | DAKO |
| Anti-keratin 14 IgG | Rabbit | Covance |
| Anti-filaggrin IgG | Rabbit | Covance |
| Anti-loricrin IgG | Rabbit | Covance |
| Anti-claudin3 IgG | Rabbit | Invitrogen |
| Anti-BrdU IgG-POD | Mouse | Roche |
| Anti-γδ-TCR IgG | Hamster | BD |
| Anti-PCNA IgG | Rabbit | Santa Cruz Biotechnology, Inc. |
| Anti-CD45 | Rat | BD |
| Secondary antibody | ||
| Anti–mouse-Cy3 IgG | Goat | Jackson ImmunoResearch Laboratories, Inc. |
| Anti–rabbit-Cy2 IgG | Goat | Jackson ImmunoResearch Laboratories, Inc. |
| Anti–rabbit-Cy3 IgG | Goat | Jackson ImmunoResearch Laboratories, Inc. |
| Anti–rat-Cy3 IgG | Donkey | Jackson ImmunoResearch Laboratories, Inc. |
TCR, T cell receptor.
Staining of mast cells with toluidine blue
After deparaffination and rehydration, skin sections were stained with toluidine blue solution (0.5% toluidine blue and 0.5 N HCl, pH 2.3) for 30 min. They were then washed with distilled water, dehydrated by a short incubation in 95% ethanol and two short incubations in 100% ethanol, and incubated twice for 3 min in xylene. Sections were then mounted with resinous mounting medium. Mast cells appear violet or purple.
Treatment of mouse skin with moisturizing cream
Moisturizing cream containing aluminum hydroxychloride and glycerol (Excipial Protect; Spirig Pharma AG) was applied to one side of the back skin of adult K5-R1/R2 mice, and the other side was left untreated.
Treatment of mouse skin with acetone
Adult female wild-type mice (C57Bl/6 background) were shaved, and the skin was rinsed with water-soaked tissues. One flank of the back was treated with 10 acetone wipes using delipidized, acetone-soaked swabs. The other flank was treated in the same manner with water and used as a control.
Western blot analysis
Cells were lysed in 2× Laemmli buffer. Frozen tissue was homogenized in a buffer containing phosphatase and protease inhibitors (Beyer et al., 2008).
Protein lysates were separated by SDS-PAGE and transferred to nitrocellulose filters. Antibody incubations were performed in 5% nonfat dry milk in TBS-T (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.05% Tween-20) or in 5% BSA in TBS-T, depending on the requirement of the different antibodies.
Antibodies used for Western blot analysis
See Table IV for the antibodies used.
Table IV.
Antibodies used for Western blot analysis
| Primary antibody | Host | Source |
| Anti-phospho-NF-κB IgG | Rabbit | Cell Signaling Technology |
| Anti-total-NF-κB IgG | Rabbit | Santa Cruz Biotechnology, Inc. |
| Anti-phospho-STAT3 IgG | Rabbit | Cell Signaling Technology |
| Anti-STAT3 IgG | Rabbit | Cell Signaling Technology |
| Anti-phospho-ERK IgG | Rabbit | Cell Signaling Technology |
| Anti-ERK IgG | Rabbit | Cell Signaling Technology |
| Anti-phospho-P38 IgG | Rabbit | Cell Signaling Technology |
| Anti-P38 IgG | Goat | Santa Cruz Biotechnology, Inc. |
| Anti-phospho-FRS2α IgG | Rabbit | Cell Signaling Technology |
| Anti-Claudin 1 IgG | Mouse | Invitrogen |
| Anti-Claudin 3 IgG | Rabbit | Invitrogen |
| Anti-Occludin IgG | Rabbit | Invitrogen |
| Anti-Lamin A IgG | Goat | Santa Cruz Biotechnology, Inc. |
| Anti-GAPDH IgG | Mouse | HyTest |
| Secondary antibody | ||
| Anti-goat-HRP IgG | Rabbit | EMD |
| Anti-mouse-HRP IgG | Goat | Promega |
| Anti-rabbit-HRP IgG | Goat | Promega |
| Anti-rat-HRP IgG | Goat | GE Healthcare |
Statistical analysis
Statistical analysis was performed using PRISM software (Graph Pad Software, Inc.). A Mann-Whitney U test was used for experiments examining differences between groups, except for the cream and acetone treatment experiments, where a paired t test was performed.
Online supplemental material
Fig. S1 shows that keratinocyte differentiation is only mildly affected in K5-R1/R2 mice. Fig. S2 shows the time course of the increase in mast cells and γδ T cells as well as analysis of CD45-positive cells at P36. Fig. S3 shows quantification of immune cells in the dermis and epidermis of adult control and K5-R1/R2 mice by FACS. Fig. S4 shows the time course of claudin-3, -8, and occludin expression during postnatal development. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910126/DC1.
Acknowledgments
We thank Christiane Born-Berclaz and Nicole Hallschmid (Eidgenössische Technische Hochschule Zurich) for excellent technical assistance, Dr. Maria Antsiferova (Eidgenössische Technische Hochschule Zurich) for help with the FACS analysis, and Drs. Masayuki Amagai and Akiharu Kubo (Keio University, Tokyo, Japan) for helpful suggestions with the tight junction characterization.
This work was supported by grants from the Eidgenössische Technische Hochschule Zurich (TH-08 06-3 and TH -41/04-2), the Swiss National Science Foundation (3100A0-109340 to S. Werner), and the National Institutes of Health (HD049808 to D.M. Ornitz). M. Meyer, A.-K. Müller and F. Böhm are/were members of the Zurich graduate program in Molecular Life Sciences.
Footnotes
Abbreviations used in this paper:
- ERK
- extracellular signal-regulated kinase
- FGFR
- FGF receptor
- FRS
- FGFR substrate
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- G-CSF
- granulocyte colony-stimulating factor
- H/E
- hematoxylin/eosin
- IL
- interleukin
- NF-κB
- nuclear factor κB
- PCNA
- proliferating cell nuclear antigen
- RPS29
- ribosomal protein S29
- TER
- transepithelial electrical resistance
- TEWL
- transepidermal water loss
References
- Balda M.S., Whitney J.A., Flores C., González S., Cereijido M., Matter K. 1996. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J. Cell Biol. 134:1031–1049 10.1083/jcb.134.4.1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barksby H.E., Lea S.R., Preshaw P.M., Taylor J.J. 2007. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin. Exp. Immunol. 149:217–225 10.1111/j.1365-2249.2007.03441.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken A., Mohammadi M. 2009. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8:235–253 10.1038/nrd2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer H.D., Florence C., Dammeier J., McGuire L., Werner S., Duan D.R. 1997. Mouse fibroblast growth factor 10: cDNA cloning, protein characterization, and regulation of mRNA expression. Oncogene. 15:2211–2218 10.1038/sj.onc.1201383 [DOI] [PubMed] [Google Scholar]
- Beer H.D., Gassmann M.G., Munz B., Steiling H., Engelhardt F., Bleuel K., Werner S. 2000. Expression and function of keratinocyte growth factor and activin in skin morphogenesis and cutaneous wound repair. J. Investig. Dermatol. Symp. Proc. 5:34–39 10.1046/j.1087-0024.2000.00009.x [DOI] [PubMed] [Google Scholar]
- Bendfeldt K., Radojevic V., Kapfhammer J., Nitsch C. 2007. Basic fibroblast growth factor modulates density of blood vessels and preserves tight junctions in organotypic cortical cultures of mice: a new in vitro model of the blood-brain barrier. J. Neurosci. 27:3260–3267 10.1523/JNEUROSCI.4033-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer T.A., Werner S., Dickson C., Grose R. 2003. Fibroblast growth factor 22 and its potential role during skin development and repair. Exp. Cell Res. 287:228–236 10.1016/S0014-4827(03)00139-3 [DOI] [PubMed] [Google Scholar]
- Beyer T.A., Xu W., Teupser D., auf dem Keller U., Bugnon P., Hildt E., Thiery J., Kan Y.W., Werner S. 2008. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 27:212–223 10.1038/sj.emboj.7601950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H., Dinh H., Trueblood E.S., Pretorius J., Kugler D., Weng N., Kanaly S.T., Towne J.E., Willis C.R., Kuechle M.K., et al. 2007. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J. Exp. Med. 204:2603–2614 10.1084/jem.20070157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakebusch C., Grose R., Quondamatteo F., Ramirez A., Jorcano J.L., Pirro A., Svensson M., Herken R., Sasaki T., Timpl R., et al. 2000. Skin and hair follicle integrity is crucially dependent on beta 1 integrin expression on keratinocytes. EMBO J. 19:3990–4003 10.1093/emboj/19.15.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner J.M., Kief S., Grund C., Rendl M., Houdek P., Kuhn C., Tschachler E., Franke W.W., Moll I. 2002. Organization and formation of the tight junction system in human epidermis and cultured keratinocytes. Eur. J. Cell Biol. 81:253–263 10.1078/0171-9335-00244 [DOI] [PubMed] [Google Scholar]
- Braun S., Hanselmann C., Gassmann M.G., auf dem Keller U., Born-Berclaz C., Chan K., Kan Y.W., Werner S. 2002. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol. Cell. Biol. 22:5492–5505 10.1128/MCB.22.15.5492-5505.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilenko D.M., Ring B.D., Yanagihara D., Benson W., Wiemann B., Starnes C.O., Pierce G.F. 1995. Keratinocyte growth factor is an important endogenous mediator of hair follicle growth, development, and differentiation. Normalization of the nu/nu follicular differentiation defect and amelioration of chemotherapy-induced alopecia. Am. J. Pathol. 147:145–154 [PMC free article] [PubMed] [Google Scholar]
- Denda M., Sato J., Tsuchiya T., Elias P.M., Feingold K.R. 1998. Low humidity stimulates epidermal DNA synthesis and amplifies the hyperproliferative response to barrier disruption: implication for seasonal exacerbations of inflammatory dermatoses. J. Invest. Dermatol. 111:873–878 10.1046/j.1523-1747.1998.00364.x [DOI] [PubMed] [Google Scholar]
- Elias P.M., Steinhoff M. 2008. “Outside-to-inside” (and now back to “outside”) pathogenic mechanisms in atopic dermatitis. J. Invest. Dermatol. 128:1067–1070 10.1038/jid.2008.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluhr J.W., Feingold K.R., Elias P.M. 2006. Transepidermal water loss reflects permeability barrier status: validation in human and rodent in vivo and ex vivo models. Exp. Dermatol. 15:483–492 10.1111/j.1600-0625.2006.00437.x [DOI] [PubMed] [Google Scholar]
- Furuse M., Hata M., Furuse K., Yoshida Y., Haratake A., Sugitani Y., Noda T., Kubo A., Tsukita S. 2002. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J. Cell Biol. 156:1099–1111 10.1083/jcb.200110122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C., Németh J., Angel P., Hess J. 2006. S100A8 and S100A9 in inflammation and cancer. Biochem. Pharmacol. 72:1622–1631 10.1016/j.bcp.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Greco V., Chen T., Rendl M., Schober M., Pasolli H.A., Stokes N., Dela Cruz-Racelis J., Fuchs E. 2009. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 4:155–169 10.1016/j.stem.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grose R., Fantl V., Werner S., Chioni A.M., Jarosz M., Rudling R., Cross B., Hart I.R., Dickson C. 2007. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 26:1268–1278 10.1038/sj.emboj.7601583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Degenstein L., Fuchs E. 1996. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 10:165–175 10.1101/gad.10.2.165 [DOI] [PubMed] [Google Scholar]
- Hadj-Rabia S., Baala L., Vabres P., Hamel-Teillac D., Jacquemin E., Fabre M., Lyonnet S., De Prost Y., Munnich A., Hadchouel M., Smahi A. 2004. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 127:1386–1390 10.1053/j.gastro.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Inai T., Kobayashi J., Shibata Y. 1999. Claudin-1 contributes to the epithelial barrier function in MDCK cells. Eur. J. Cell Biol. 78:849–855 [DOI] [PubMed] [Google Scholar]
- Jameson J., Havran W.L. 2007. Skin gammadelta T-cell functions in homeostasis and wound healing. Immunol. Rev. 215:114–122 10.1111/j.1600-065X.2006.00483.x [DOI] [PubMed] [Google Scholar]
- Kawano M., Komi-Kuramochi A., Asada M., Suzuki M., Oki J., Jiang J., Imamura T. 2005. Comprehensive analysis of FGF and FGFR expression in skin: FGF18 is highly expressed in hair follicles and capable of inducing anagen from telogen stage hair follicles. J. Invest. Dermatol. 124:877–885 10.1111/j.0022-202X.2005.23693.x [DOI] [PubMed] [Google Scholar]
- Kirschner N., Poetzl C., von den Driesch P., Wladykowski E., Moll I., Behne M.J., Brandner J.M. 2009. Alteration of tight junction proteins is an early event in psoriasis: putative involvement of proinflammatory cytokines. Am. J. Pathol. 175:1095–1106 10.2353/ajpath.2009.080973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komi-Kuramochi A., Kawano M., Oda Y., Asada M., Suzuki M., Oki J., Imamura T. 2005. Expression of fibroblast growth factors and their receptors during full-thickness skin wound healing in young and aged mice. J. Endocrinol. 186:273–289 10.1677/joe.1.06055 [DOI] [PubMed] [Google Scholar]
- Langbein L., Grund C., Kuhn C., Praetzel S., Kartenbeck J., Brandner J.M., Moll I., Franke W.W. 2002. Tight junctions and compositionally related junctional structures in mammalian stratified epithelia and cell cultures derived therefrom. Eur. J. Cell Biol. 81:419–435 10.1078/0171-9335-00270 [DOI] [PubMed] [Google Scholar]
- Logié A., Dunois-Lardé C., Rosty C., Levrel O., Blanche M., Ribeiro A., Gasc J.M., Jorcano J., Werner S., Sastre-Garau X., et al. 2005. Activating mutations of the tyrosine kinase receptor FGFR3 are associated with benign skin tumors in mice and humans. Hum. Mol. Genet. 14:1153–1160 10.1093/hmg/ddi127 [DOI] [PubMed] [Google Scholar]
- Magne D., Palmer G., Barton J.L., Mézin F., Talabot-Ayer D., Bas S., Duffy T., Noger M., Guerne P.A., Nicklin M.J., Gabay C. 2006. The new IL-1 family member IL-1F8 stimulates production of inflammatory mediators by synovial fibroblasts and articular chondrocytes. Arthritis Res. Ther. 8:R80 10.1186/ar1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Nguyen L.T., Zhuang Z.W., Zhang Z.W., Moodie K.L., Carmeliet P., Stan R.V., Simons M. 2008. The FGF system has a key role in regulating vascular integrity. J. Clin. Invest. 118:3355–3366 10.1172/JCI35298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatake Y., Hoshikawa M., Asaki T., Kassai Y., Itoh N. 2001. Identification of a novel fibroblast growth factor, FGF-22, preferentially expressed in the inner root sheath of the hair follicle. Biochim. Biophys. Acta. 1517:460–463 [DOI] [PubMed] [Google Scholar]
- Ornitz D.M., Itoh N. 2001. Fibroblast growth factors. Genome Biol. 2:S3005 10.1186/gb-2001-2-3-reviews3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen S., Riehokainen J., Pummi K., Peltonen J. 2007. Tight junction components occludin, ZO-1, and claudin-1, -4 and -5 in active and healing psoriasis. Br. J. Dermatol. 156:466–472 10.1111/j.1365-2133.2006.07642.x [DOI] [PubMed] [Google Scholar]
- Petiot A., Conti F.J., Grose R., Revest J.M., Hodivala-Dilke K.M., Dickson C. 2003. A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development. 130:5493–5501 10.1242/dev.00788 [DOI] [PubMed] [Google Scholar]
- Pirvola U., Ylikoski J., Trokovic R., Hébert J.M., McConnell S.K., Partanen J. 2002. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 35:671–680 10.1016/S0896-6273(02)00824-3 [DOI] [PubMed] [Google Scholar]
- Proksch E., Feingold K.R., Man M.Q., Elias P.M. 1991. Barrier function regulates epidermal DNA synthesis. J. Clin. Invest. 87:1668–1673 10.1172/JCI115183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proksch E., Fölster-Holst R., Jensen J.M. 2006. Skin barrier function, epidermal proliferation and differentiation in eczema. J. Dermatol. Sci. 43:159–169 10.1016/j.jdermsci.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Pummi K., Malminen M., Aho H., Karvonen S.L., Peltonen J., Peltonen S. 2001. Epidermal tight junctions: ZO-1 and occludin are expressed in mature, developing, and affected skin and in vitro differentiating keratinocytes. J. Invest. Dermatol. 117:1050–1058 10.1046/j.0022-202x.2001.01493.x [DOI] [PubMed] [Google Scholar]
- Ramirez A., Page A., Gandarillas A., Zanet J., Pibre S., Vidal M., Tusell L., Genesca A., Whitaker D.A., Melton D.W., Jorcano J.L. 2004. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 39:52–57 10.1002/gene.20025 [DOI] [PubMed] [Google Scholar]
- Reuss B., Dono R., Unsicker K. 2003. Functions of fibroblast growth factor (FGF)-2 and FGF-5 in astroglial differentiation and blood-brain barrier permeability: evidence from mouse mutants. J. Neurosci. 23:6404–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist T.A., Martin G.R. 1996. Fibroblast growth factor signalling in the hair growth cycle: expression of the fibroblast growth factor receptor and ligand genes in the murine hair follicle. Dev. Dyn. 205:379–386 [DOI] [PubMed] [Google Scholar]
- Sandilands A., Sutherland C., Irvine A.D., McLean W.H. 2009. Filaggrin in the frontline: role in skin barrier function and disease. J. Cell Sci. 122:1285–1294 10.1242/jcs.033969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J.A. 2006. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 116:1150–1158 10.1172/JCI28521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiling H., Werner S. 2003. Fibroblast growth factors: key players in epithelial morphogenesis, repair and cytoprotection. Curr. Opin. Biotechnol. 14:533–537 10.1016/j.copbio.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Steiling H., Wüstefeld T., Bugnon P., Brauchle M., Fässler R., Teupser D., Thiery J., Gordon J.I., Trautwein C., Werner S. 2003. Fibroblast growth factor receptor signalling is crucial for liver homeostasis and regeneration. Oncogene. 22:4380–4388 10.1038/sj.onc.1206499 [DOI] [PubMed] [Google Scholar]
- Strid J., Tigelaar R.E., Hayday A.C. 2009. Skin immune surveillance by T cells—a new order? Semin. Immunol. 21:110–120 10.1016/j.smim.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Szabowski A., Maas-Szabowski N., Andrecht S., Kolbus A., Schorpp-Kistner M., Fusenig N.E., Angel P. 2000. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell. 103:745–755 10.1016/S0092-8674(00)00178-1 [DOI] [PubMed] [Google Scholar]
- Towne J.E., Garka K.E., Renshaw B.R., Virca G.D., Sims J.E. 2004. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 279:13677–13688 10.1074/jbc.M400117200 [DOI] [PubMed] [Google Scholar]
- Tunggal J.A., Helfrich I., Schmitz A., Schwarz H., Günzel D., Fromm M., Kemler R., Krieg T., Niessen C.M. 2005. E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 24:1146–1156 10.1038/sj.emboj.7600605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H., Gunn M., Dell K., Tseng A., Jr, Williams L. 1992. A truncated form of fibroblast growth factor receptor 1 inhibits signal transduction by multiple types of fibroblast growth factor receptor. J. Biol. Chem. 267:1470–1476 [PubMed] [Google Scholar]
- Watson R.E., Poddar R., Walker J.M., McGuill I., Hoare L.M., Griffiths C.E., O’neill C.A. 2007. Altered claudin expression is a feature of chronic plaque psoriasis. J. Pathol. 212:450–458 10.1002/path.2200 [DOI] [PubMed] [Google Scholar]
- Werner S., Peters K.G., Longaker M.T., Fuller-Pace F., Banda M.J., Williams L.T. 1992. Large induction of keratinocyte growth factor expression in the dermis during wound healing. Proc. Natl. Acad. Sci. USA. 89:6896–6900 10.1073/pnas.89.15.6896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Weinberg W., Liao X., Peters K.G., Blessing M., Yuspa S.H., Weiner R.L., Williams L.T. 1993. Targeted expression of a dominant-negative FGF receptor mutant in the epidermis of transgenic mice reveals a role of FGF in keratinocyte organization and differentiation. EMBO J. 12:2635–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S., Smola H., Liao X., Longaker M.T., Krieg T., Hofschneider P.H., Williams L.T. 1994. The function of KGF in morphogenesis of epithelium and reepithelialization of wounds. Science. 266:819–822 10.1126/science.7973639 [DOI] [PubMed] [Google Scholar]
- Yu K., Xu J., Liu Z., Sosic D., Shao J., Olson E.N., Towler D.A., Ornitz D.M. 2003. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 130:3063–3074 10.1242/dev.00491 [DOI] [PubMed] [Google Scholar]
- Zhang H., Dessimoz J., Beyer T.A., Krampert M., Williams L.T., Werner S., Grose R. 2004. Fibroblast growth factor receptor 1-IIIb is dispensable for skin morphogenesis and wound healing. Eur. J. Cell Biol. 83:3–11 10.1078/0171-9335-00355 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ibrahimi O.A., Olsen S.K., Umemori H., Mohammadi M., Ornitz D.M. 2006. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 281:15694–15700 10.1074/jbc.M601252200 [DOI] [PMC free article] [PubMed] [Google Scholar]