Enhanced polymerization of actin in latrunculin A–treated aip1Δ cells shows that filament assembly does not occur from monomeric actin alone in vivo.
Abstract
Although actin filaments can form by oligomer annealing in vitro, they are assumed to assemble exclusively from actin monomers in vivo. In this study, we show that a pool of actin resistant to the monomer-sequestering drug latrunculin A (lat A) contributes to filament assembly in vivo. Furthermore, we show that the cofilin accessory protein Aip1 is important for establishment of normal actin monomer concentration in cells and efficiently converts cofilin-generated actin filament disassembly products into monomers and short oligomers in vitro. Additionally, in aip1Δ mutant cells, lat A–insensitive actin assembly is significantly enhanced. We conclude that actin oligomer annealing is a physiologically relevant actin filament assembly pathway in vivo and identify Aip1 as a crucial factor for shifting the distribution of short actin oligomers toward monomers during disassembly.
Introduction
The budding yeast Saccharomyces cerevisiae has proven to be an excellent organism for studies of actin dynamics mechanisms and regulation. Endocytic internalization in this organism requires the stepwise association of adapters, coat proteins, motor proteins, and actin nucleation–promoting factors and culminates in Arp2/3-nucleated assembly of actin filaments at the endocytic site (Kaksonen et al., 2006). Actin assembly drives plasma membrane invagination and initiates a feedback loop for rapid and coordinated disassembly of endocytic components and actin filaments. Thus, the budding yeast endocytic pathway provides a biological context within which the full range of assembly and disassembly activities can be studied using a combination of approaches including genetics and real-time imaging.
A study in yeast previously showed that defects in actin filament disassembly led to defects in the rate of actin filament assembly at endocytic sites (Okreglak and Drubin, 2007). Therefore, studies of actin filament disassembly provide insight into actin filament assembly mechanisms.
The major mediator of actin filament disassembly at endocytic sites is the small actin-binding protein cofilin, whose localization to disassembling actin filaments is dependent on coronin (Gandhi et al., 2009). Defects in cofilin function lead to defects in endocytic internalization and late steps of trafficking to the vacuole (Okreglak and Drubin, 2007). In vitro, cofilin stimulates actin filament severing but does not accelerate release of actin monomers from filaments (Andrianantoandro and Pollard, 2006). Biochemical studies indicate that cofilin functions with Aip1 to stimulate rapid actin filament disassembly (Rodal et al., 1999). However, the importance of this stimulatory activity for endocytosis and the general cellular actin economy remains to be established.
Although actin assembly and disassembly at endocytic sites are resolvable temporally, these events cannot be resolved spatially by light microscopy. However, elimination of Sla2p, an essential link between F-actin and clathrin, results in elongated actin tails anchored to arrested endocytic machinery at the plasma membrane (Kaksonen et al., 2003). Actin within these tails continuously assembles via Arp2/3-nucleated polymerization stimulated by nucleation promotion factors associated with the rims of endocytic pits. Rapid F-actin turnover occurs distal to the membrane and is controlled by actin-bound nucleotide hydrolysis and Pi release, which are delayed relative to actin polymerization (Okreglak and Drubin, 2007). Assembly and disassembly within actin tails are resolvable spatially and readily quantified in live cells.
Models for actin assembly in vivo invoke a strict monomer requirement. However, this idea has been challenged by in vitro studies (Kawamura and Maruyama, 1970; Murphy et al., 1988), which indicate that actin filament annealing is energetically favorable (Andrianantoandro et al., 2001). The experiments described in this study identify a new link between disassembly and assembly reactions, providing evidence that Aip1 converts actin oligomers produced by cofilin-mediated actin filament disassembly into monomers and that actin oligomers contribute to filament assembly in vivo.
Results and discussion
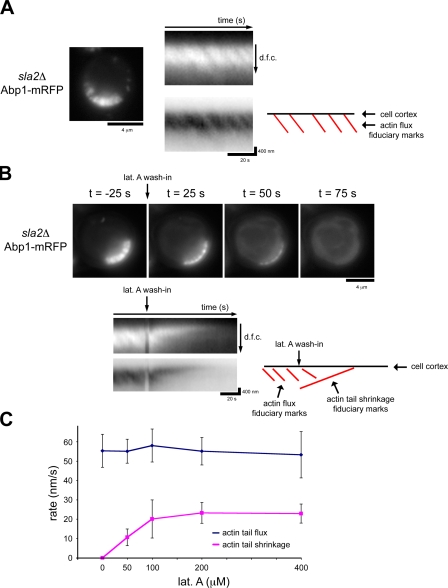
Within actin tails present in sla2Δ cells, regions of Arp2/3-dependent actin assembly and cofilin-mediated filament disassembly are spatially separated, allowing observation by live cell fluorescence microscopy of actin flux through the tails. To quantify actin flux rates, we imaged the actin-binding protein Abp1–monomeric RFP (mRFP) and used kymographs to follow fiduciary marks present in the tails as a result of small discontinuities in actin assembly (Fig. 1 A and Video 1). Abp1-mRFP completely colocalizes with actin in sla2Δ cells (Okreglak and Drubin, 2007). Flux rates determined using this approach were consistent with those obtained previously using fluorescence recovery after photobleaching of GFP-actin (Kaksonen et al., 2003; Okreglak and Drubin, 2007). The actin tails are ∼0.83 µm (± 0.14 µm) in length, and flux rates are ∼55.2 nm/s (± 8.5 nm/s). Following fiduciary marks using Abp1-mRFP bypasses the need to use GFP-actin, which is not fully functional.
Figure 1.
Actin assembly continues in sla2Δ cells treated with lat A. (A) A single frame from time-lapse imaging of Abp1-mRFP in sla2Δ cells. Kymographs show irregular actin assembly indicated by the oblique dark lines that traverse the actin tails. The details of these fiduciary marks are highlighted by a kymograph in which black and white have been inverted and by a schematic in which the red lines highlight each actin flux fiduciary mark. The kymographs are oriented so that the cell exterior is on the top and the cell interior is oriented downward. dfc, distance from cortex. (B) Single frames from time-lapse imaging of Abp1-mRFP in sla2Δ cells treated with 400 µM lat A. A kymograph shows the rapid, directional disassembly toward the cell cortex. The schematic highlights actin flux fiduciary marks as well as the actin tail shrinkage fiduciary marks. (C) Quantification of actin flux rates and tail shrinkage rates in sla2Δ cells treated with a range of concentrations of lat A. 16 cells were quantified before and after 50 µM lat A treatment, 13 cells for 100 µM, 14 cells for 200 µM, and 14 cells for 400 µM lat A. Three measurements for before and after lat A treatment were averaged per cell. Error bars represent SD.
To analyze the role of actin disassembly in actin flux through tails, we blocked assembly using latrunculin A (lat A) and quantified disassembly rates. Lat A binds actin monomers (Coué et al., 1987) and actin monomers bound to profilin (Yarmola et al., 2000) and potently inhibits assembly. We imaged sla2Δ cells upon treatment with high levels of lat A and measured actin flux rates before lat A treatment and actin disassembly rates after lat A treatment. Actin tails in sla2Δ cells treated with 400 µM lat A rapidly disassembled (Fig. 1 B and Video 2). Kymograph representations illustrate that disassembly occurs directionally from the cell interior to the cell cortex, which is consistent with cofilin localization distal to actin assembly sites (Okreglak and Drubin, 2007). Untreated actin tails maintain a constant length and therefore are at steady state with balanced actin assembly and disassembly rates. We reasoned that with efficient monomer sequestration by lat A, actin disassembly rates should equal the flux rate. However, surprisingly, we found that mean actin disassembly rates in sla2Δ cells treated with 400 µM lat A were significantly slower (22.9 ± 4.9 nm/s; n = 14) than actin flux rates before lat A treatment (53.2 nm/s ± 12.0 nm/s; n = 14). We tested a range of lat A concentrations and found that tail disassembly rates never matched actin subunit flux rates (Fig. 1 C). These unexpected data suggest that the apparent actin disassembly rate in the presence of lat A might reflect the combination of actin disassembly and an ongoing basal level of actin polymerization. Indeed, close examination of actin tail kymographs revealed that actin assembly continues in the presence of high concentrations of lat A (Fig. 1 B).
Actin assembly in the presence of lat A could be the consequence of insufficient cytoplasmic lat A concentration as a result of plasma membrane efflux pumps. We identified the multidrug resistance pump Pdr5p as the major efflux pump for lat A by testing sensitivity in mutants using a plate assay (unpublished data) and deleted it in an sla2Δ strain so actin disassembly rates could be measured in the presence of the highest achievable intracellular lat A concentration. Quantification of actin tail disassembly rates upon lat A treatment (unpublished data) revealed no effect on apparent actin tail disassembly rate even though the cells were more sensitive to lat A. The observation that actin tail assembly continues even upon exposure of cells to the highest achievable intracellular lat A levels and while net filament disassembly is occurring led us to suspect that a pool of lat A–resistant actin contributes to filament assembly.
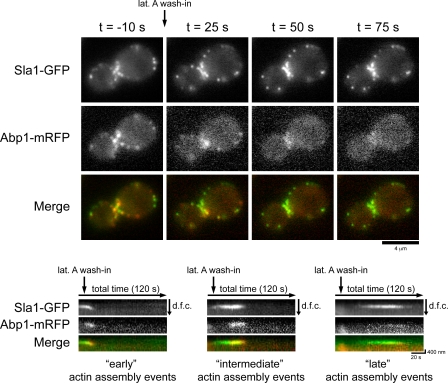
To determine whether a lat A–resistant actin pool contributes to assembly at endocytic sites in wild-type cells, we simultaneously imaged Abp1-mRFP and the endocytic coat marker Sla1-GFP in cells treated with 200 µM lat A. Previous data showed that prolonged lat A treatment blocks endocytic internalization and stabilizes early-arriving endocytic components at the plasma membrane (Kaksonen et al., 2003). In addition, abolishing actin assembly at endocytic sites prevents endocytic coat disassembly because actin filaments recruit endocytic coat disassembly factors (Toret et al., 2008). Actin rapidly disassembled in response to lat A (Fig. 2 and Video 3). However, kymographs of individual endocytic sites marked by Sla1-GFP and Abp1-mRFP (Fig. 2, bottom) showed that actin assembly was clearly detectable at endocytic sites at early (0–20 s), intermediate (20–45 s), and late (up to 90 s) time points after lat A treatment. Actin assembly early after lat A treatment is able to drive internalization and disassembly of the endocytic marker Sla1-GFP (Fig. 2, left). However, actin assembly initiated at intermediate or late time points after lat A addition was not capable of driving internalization but did facilitate Sla1-GFP disassembly (Fig. 2, middle and right). These data indicate that actin is able to assemble at endocytic sites in high lat A concentrations as with actin tail assembly in sla2Δ cells.
Figure 2.
Lat A–insensitive actin assembly at endocytic sites in wild-type cells. Single frames from simultaneous two-color imaging of Abp1-mRFP and Sla1-GFP in cells treated with 200 µM lat A are shown at the indicated times relative to lat A treatment. Kymographs show the spatiotemporal dynamics of actin assembly events at endocytic sites at early, intermediate, or late time points after lat A addition. The kymographs are oriented so that the cell exterior is on the top and the cell interior is oriented downward. dfc, distance from cortex.
We considered two hypotheses that could explain the continuation of actin assembly in the presence of lat A. The first is that lat A binds slowly to actin monomers. The second is that lat A–insensitive actin assembly at endocytic sites and in actin tails is the result of annealing by actin oligomers, which are not sequesterable by lat A. Actin oligomer generation is in part the result of F-actin disassembly pathways. In vitro data showed that Aip1 catalyzes actin filament disassembly substoichiometrically in a strictly cofilin-dependent manner (Rodal et al., 1999). Aip1 may function to enhance cofilin-mediated actin filament disassembly (Kueh et al., 2008), or it might prevent reannealing of cofilin-severed actin filaments by capping the products of cofilin-mediated severing reactions (Balcer et al., 2003), or both. Either or both of these activities might affect oligomer assembly in vivo but are not expected to alter the binding kinetics of lat A to actin monomer.
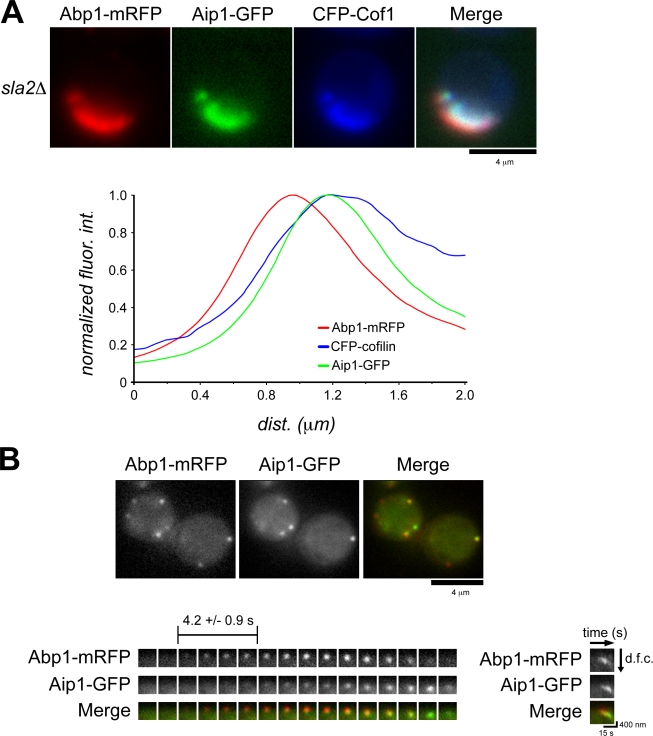
We tested whether Aip1 contributes to lat A–insensitive actin assembly in sla2Δ cells. We first verified that Aip1 and cofilin localize to regions of actin tail disassembly in sla2Δ cells. Normalized fluorescence intensity of Aip1-GFP and CFP-cofilin relative to Abp1-mRFP showed similar localization within actin tails in sla2Δ cells (Fig. 3 A). Simultaneous two-color imaging of Abp1-mRFP and Aip1-GFP in wild-type cells (Fig. 3 B and Video 4) indicated that like cofilin, Aip1 is recruited temporally after actin assembly initiation at endocytic sites. This delay (4.2 ± 0.9 s) (Fig. 3 B) is very similar to that previously reported for cofilin (3.6 ± 0.6 s; Okreglak and Drubin, 2007). Analysis of Aip1 spatiotemporal dynamics with kymographs indicates that Aip1, like cofilin, associates with actin patches during the internalization/disassembly phase of endocytic patch lifetime (Fig. 3 B). Despite its presence in actin tails and patches, deletion of the gene encoding Aip1 resulted in no discernable actin dynamics defects in actin tails of sla2Δ cells (Fig. S1 A) or at endocytic sites (Fig. S1 B; Rodal et al., 1999). Therefore, Aip1 may be important for a feature of actin filament turnover that is not manifested by changes in bulk actin dynamics.
Figure 3.
Aip1 localizes to sites of active actin filament disassembly. (A) Two-color imaging showing CFP-cofilin and Aip1-GFP localization in sla2Δ cells expressing Abp1-mRFP. Quantification of normalized fluorescence intensities (fluor int) along the length of the actin tails moving from the outside of the cell toward the cell interior is presented in the graph. (B) Single frames from simultaneous two-color imaging of Abp1-mRFP and Aip1-GFP. The maturation of a single actin patch over time is shown by the montage, where each frame represents a 1-s exposure. The mean temporal delay in Aip1-GFP at the patch is indicated (n = 51). A kymograph illustrates the spatiotemporal relationship between Aip1-GFP and Abp1-mRFP. The kymograph is oriented so that the cell exterior is on the top and the cell interior is oriented downward. dfc, distance from cortex.
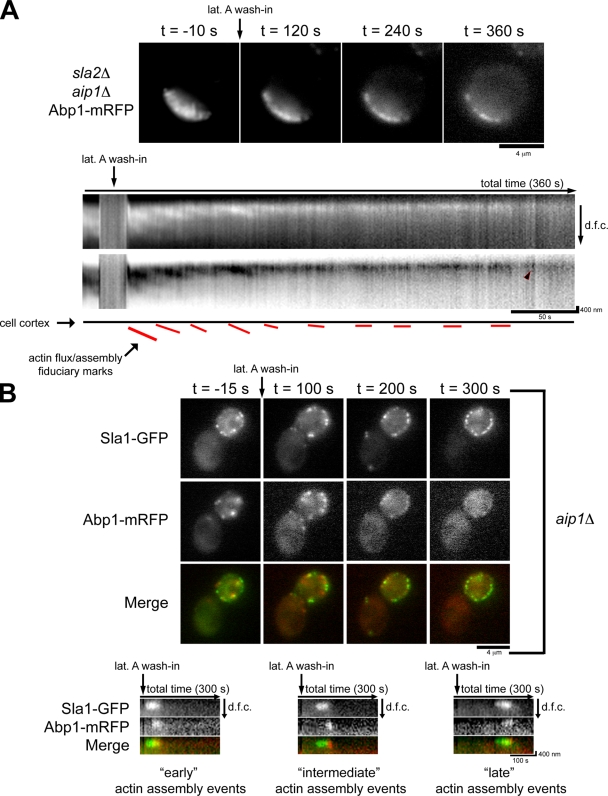
We next tested the impact of Aip1 on the lat A–insensitive actin assembly pathway in actin tails. Although actin tails in sla2Δ cells disassemble within 90 s of lat A treatment (Fig. 1 B and Video 2), actin tails in sla2Δ aip1Δ cells take >360 s to disassemble (Fig. 4 A and Video 5). Strikingly, kymographs indicate that flux is ongoing even 100 s after lat A treatment and that assembly bursts at the cortex occur even after complete tail disassembly (Fig. 4 A, arrowhead).
Figure 4.
aip1Δ cells show an increase in lat A–insensitive actin assembly. (A) Single frames from time-lapse imaging of Abp1-mRFP in sla2Δ aip1Δ cells treated with 400 µM lat A. Kymographs show the delayed disassembly of the actin tails, and an arrowhead shows a burst of actin assembly ∼300 s after treatment with lat A. The schematic highlights actin flux fiduciary marks in the presence of lat A. (B) Single frames from simultaneous two-color imaging of Abp1-mRFP and Sla1-GFP in aip1Δ cells treated with 200 µM lat A are shown at the indicated times relative to lat A treatment. Kymographs show the spatiotemporal dynamics of actin assembly events at endocytic sites at early, intermediate, or late time points after lat A addition. The kymographs are oriented so that the cell exterior is on the top and the cell interior is oriented downward. dfc, distance from cortex.
We next tested the impact of Aip1 on lat A–resistant actin assembly at endocytic sites. Sla1-GFP and Abp1-mRFP were simultaneously imaged in aip1Δ mutant cells treated with 200 µM lat A (Fig. 4 B and Video 6). As previously reported (Okada et al., 2006), compared with wild-type cells, actin patches in aip1Δ cells persist for a longer time after lat A treatment (Fig. 2 and Video 3). However, examination of the actin patch spatiotemporal dynamics associated with endocytic sites by kymographs (Fig. 4 B) indicates that rather than slowing actin filament disassembly, the time during which actin patches can assemble in the presence of lat A is prolonged. Indeed, actin assembly events associated with endocytic sites can take place up to 240 s after lat A addition (Fig. 4 B, right). Collectively, these results suggest that Aip1 is important for the presence of actin species in vivo that can be readily sequestered by lat A and that this effect is manifested in actin dynamics associated with sites of endocytosis in wild-type cells and actin tails in sla2Δ cells. Additionally, the lack of an appreciable effect on actin dynamics associated with endocytic sites or on actin flux rates when cells lack Aip1 indicates that a defect in generation of a lat A–sensitive actin pool does not affect bulk actin dynamics.
To rule out a role for increased actin levels in lat A–insensitive actin assembly seen in aip1Δ and sla2Δ aip1Δ cells, we quantified total cellular actin concentrations and found no difference between wild-type and aip1Δ cells (Fig. S2 A) or between sla2Δ and sla2Δ aip1Δ cells (unpublished data). We also tested whether aip1Δ cells have altered plasma membrane permeability (Fig. S2 B) or xenobiotic drug efflux rates (Fig. S2 C) by measuring the accumulation and efflux of rhodamine-6G. These data indicate that Aip1 has no role in xenobiotic diffusion or efflux across the plasma membrane and suggest that the increase in lat A–insensitive actin assembly in aip1Δ cells is specifically the result of a reduction in the relative amount of lat A–sequesterable actin present in the cell.
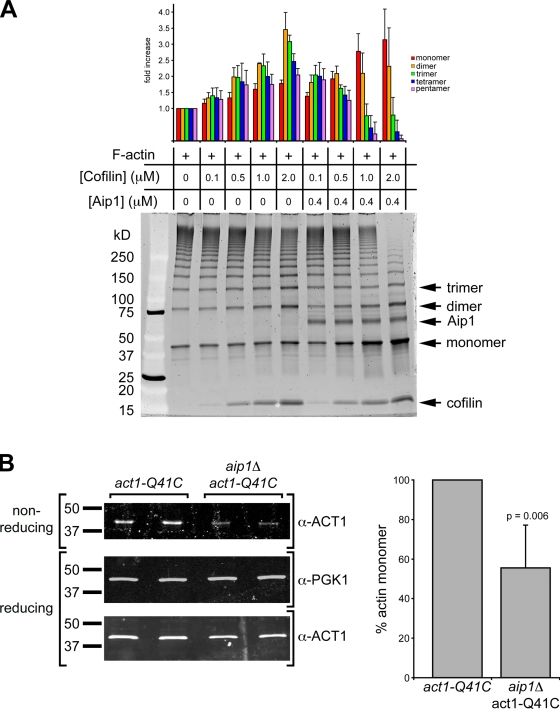
We hypothesized that Aip1 might be important for converting oligomeric depolymerization products to actin monomers, which are sequesterable by lat A. To test this hypothesis, we used a previously developed in vitro assay (Kim et al., 2000) to probe the polymeric species present in F-actin solutions. This assay has greater sensitivity than traditional actin sedimentation assays, which are unable to accurately distinguish actin monomer from other low molecular mass actin oligomer species. We purified yeast actin with a cysteine substitution at position Q41, which can be cross-linked to C341 in an adjacent actin subunit within an actin protofilament (Kim et al., 2000). A previous study showed that this modification does not affect polymerization dynamics or interactions with actin disassembly factors (Kim et al., 2000). Cross-linking with the thiol-specific cross-linker MTS-2-MTS (1,2-ethanediyl bismethanethiosulfonate) followed by SDS-PAGE analysis under nonreducing conditions resulted in a defined ladder of actin species that represents different oligomeric actin forms present in F-actin solutions under apparent steady-state conditions (Fig. 5 A, lane 1). To estimate the cross-linking efficiency, we used the SDS-PAGE gels to quantify the concentration of actin monomer present after cross-linking. This value (0.67 µM) is similar to the apparent critical concentration (0.4 µM) measured previously for yeast actin (Chen et al., 1993), which suggests that cross-linking was efficient, particularly given the contribution of small oligomeric species of actin to the apparent monomer pool after denaturation and analysis by SDS-PAGE. We used this assay to probe the contribution of cofilin and Aip1 to generation of low molecular mass actin species readily sequestered by lat A. Cofilin and Aip1 act synergistically to stimulate actin disassembly (Rodal et al., 1999). It has been proposed that this reaction could be the result of active and rapid disassembly stimulation (Kueh et al., 2008) or actin filament capping (Balcer et al., 2003). However, neither of these proposed mechanisms addresses the nature of the species present after the disassembly reactions. We hypothesized that Aip1, in a cofilin-dependent manner, specifically stimulates an increase in low molecular mass actin species, which are readily sequestered by lat A. Cofilin activity is strongly dependent on concentration (Andrianantoandro and Pollard, 2006). Therefore, we compared the status of actin species present after disassembly in reactions containing increasing concentrations of cofilin both with and without Aip1 (Fig. 5 A). The experimental conditions were chosen to approximate a physiologically relevant ratio of actin and Aip1 while increasing cofilin concentrations (Goode, B., and A. Goodman, personal communication). Aip1 strongly stimulated actin monomer and small oligomer accumulation at steady state. Intriguingly, although the highest concentration of cofilin tested in our in vitro assay strongly stimulated the production of actin monomer, an appreciable amount of low molecular mass actin oligomers are still present in the reaction, which may account for the pool of actin that participates in the lat A–insensitive actin assembly pathway observed in sla2Δ (Fig. 1 B and Video 2) and wild-type cells (Fig. 2 and Video 3). The relative fold increase of each actin species is quantified in Fig. 5 A.
Figure 5.
Aip1 stimulates the production of low molecular mass actin species. (A) Intermediates from in vitro F-actin disassembly reactions were captured by specific cross-linking between actin subunits along the protofilament axis. Low molecular mass intermediates are marked as monomer, dimer, and trimer. The amount of each species was quantified relative to the reaction with no cofilin or Aip1, and the results are presented in the graph. Quantification is the mean of three independent in vitro reactions run on 4–12% gradient gels stained with Sypro ruby. (B) Equal amounts of cross-linked cell lysates from act1-Q41C and aip1Δ act1-Q41C cells were loaded in duplicate on nonreducing and reducing SDS-PAGE gels, and actin levels were assessed using antiyeast actin antibodies to show monomer and total actin levels, respectively. Blots were probed for Pgk1p as loading controls. Immunoblot signals are from IRDye-labeled secondary antibodies acquired with an infrared imaging system (Odyssey). Actin monomer concentration was quantified after normalization to the Pgk1 signal. Actin levels are normalized to actin monomer levels in extracts from act1-Q41C cells, and the results of four independent cross-linking reactions are presented in the graph. Error bars represent SD.
We tested whether the increase in actin monomer concentration catalyzed by Aip1 was of physiological relevance by comparing the actin monomer concentration by chemical actin cross-linking in living act1-Q41C and aip1Δ act1-Q41C cells. Consistent with differences in in vitro actin disassembly reactions presented in Fig. 5 A, we found that aip1Δ cells have ∼45% less actin monomer (Fig. 5 B). We verified the decrease in actin monomer concentration in aip1Δ cells by assessing the actin monomer concentration in extracts prepared from wild-type and aip1Δ cells using DNase I inhibition assays (Blikstad et al., 1978) and found that cell extracts from aip1Δ cells have ∼15% less DNase I inhibitory activity than extracts from wild-type cells (Fig. S3). This value is likely an underestimation of the actual change in actin monomer concentration as a result of DNase I inhibition by oligomeric actin (Morrison and Dawson, 2007). Collectively, these results suggest that Aip1 is important for generating actin monomers, the species that is sequesterable by lat A, and that a defect in generating actin monomer has no readily detected effect on bulk actin dynamics in vivo. These data and the increase in lat A–insensitive actin assembly in sla2Δ aip1Δ and aip1Δ cells imply that actin oligomers participate in actin filament assembly in vivo. In addition, we provided evidence that actin oligomer assembly occurs even when the full complement of actin disassembly factors is present in the cell.
Our data indicate that a previously unappreciated actin assembly pathway exists in living cells and participates in assembly of Arp2/3-generated actin filament arrays. This observation raises key questions about the role of the assembly competent actin oligomer pool in vivo. In photobleaching experiments of actin tails, no recovery is seen distal to the plasma membrane, indicating that filament assembly is restricted to membrane-proximal regions (Kaksonen et al., 2003). In addition, Aip1 is reported to cap filament barbed ends generated by cofilin-dependent filament severing in vivo (Tsuji et al., 2009), which may bias oligomeric actin assembly events toward regions of actin filaments lacking Aip1 and cofilin. However, the detailed interplay between actin monomer and oligomer in Arp2/3-promoted nucleation and elongation of uncapped filament barbed ends near membranes remains to be elucidated.
Materials and methods
Yeast strains, media, and plasmids
Yeast strains used in this study are listed in Table S1. Unless otherwise noted, all strains were grown at 25°C in synthetic dextrose media supplemented with appropriate amino acids. The CFP-cofilin construct was made as previously described (Okreglak and Drubin, 2007), but pDH3 (Yeast Resource Center, University of Washington, Seattle, WA) was used as a PCR template to generate CFP.
Quantitative analysis of actin dynamics in vivo
All images were acquired on an epifluorescence microscope (IX71; Olympus) equipped with a 100× NA 1.4 objective and a camera (Orca II; Hamamatsu Photonics). Cells were immobilized on concanavalin A–coated glass coverslips and imaged using appropriate filter sets and neutral density filters. Actin flux rates were quantified by following fiduciary marks generated by discontinuous actin assembly at the cell cortex using kymographs generated with ImageJ (National Institutes of Health). Lat A–induced actin disassembly was performed as described previously (Okreglak and Drubin, 2007). To measure actin tail shrinkage rates in the presence of lat A, time-lapse data were corrected for photobleaching, a fluorescence threshold was applied to kymographs, and the position of the cytoplasmic end of the actin tail over time was determined.
Protein purification and expression
Yeast Q41C actin, cofilin, and Aip1 were purified as described previously (Lappalainen and Drubin, 1997; Rodal et al., 1999; Kim et al., 2000). All proteins were gel filtered on columns (Sephadex 200; GE Healthcare). G-actin was maintained under constant dialysis in argon-flushed dialysis chambers and used within 1 wk of purification.
Actin cross-linking and disassembly reactions
Q41C yeast actin was passed over homemade columns (Sephadex G-50) in 1.5-ml eppendorf tubes. Actin polymerization was induced using 20× initiation mix (500 mM KCl, 40 mM MgCl2, and 10 mM ATP) added to a 6 µM G-actin and allowed to reach steady state for 45 min. Cofilin and Aip1 were added to concentrations indicated in Fig. 5 A, and reactions were allowed to proceed for 15 min. All reactions were adjusted to the same final volume to minimize dilution effects. MTS-2-MTS was added to 1.5 mM, and cross-linking was quenched after 1 h with 3 mM N-ethylmaleimide. Samples were loaded onto 4–12% gradient gels (Bio-Rad Laboratories) and run under nonreducing conditions. Gels were stained with Sypro ruby (Invitrogen) and imaged on a variable mode imager (Typhoon 9400; GE Healthcare). PAGE quantification was performed using ImageQuant software (GE Healthcare).
Actin cross-linking in living cells
act1-Q41C and act1-Q41C aip1Δ cells were grown to OD600 = 0.5 in YPD, and 8 ml of cells was centrifuged at 1,500 g. The cells were washed two times with PBS supplemented with 1 M sorbitol (S-PBS) and resuspended to 8 OD600/ml in S-PBS. MTS-2-MTS was added to 1 mM, and cross-linking was allowed to proceed at room temperature for 2 min. The cross-linking reaction was quenched for 1 min by adding N-ethylmaleimide to 10 mM, after which ice-cold TCA was added to 15% final concentration. The cells were lysed by bead beating with 425–600-µm acid-washed glass beads (Sigma-Aldrich) for 15 min at 4°C. The precipitated cell lysate was spun at 14,000 g for 15 min and washed once with −20°C acetone. Pellets were air dried, resuspended in 80 µl TURB (62.5 mM Tris–HCl, pH 6.8, 3 M urea, and 1% SDS), and boiled for 3 min at 98°C. The samples were spun at 1,500 g for 5 min at 4°C, and 60 µl supernatant was collected. 30 µl of the supernatant was reduced with 5% β-mercaptoethanol and reboiling. Equal amounts of reduced and nonreduced samples were loaded on 10% SDS-PAGE gels, transferred to PVDF membrane, and immunodetection of relevant proteins was performed according to the manufacturer’s protocol for detection using an infrared imaging system (Odyssey; LI-COR Biosciences). Quantification of immunoblot data was performed using ImageJ.
DNase I inhibition assays
Wild-type and aip1Δ cells were grown to OD600 = 0.5 in YPD, centrifuged at 1,500 g, and washed once with FSB (10 mM Tris, pH 7.5, 100 mM KCl, 1 mM KCl, 100 µM CaCl2, and 1.2 M sorbitol). The cell pellet was resuspended in 450 µl FSB to which 50 µl 10 mg/ml zymolyase 100T (Seikagaku Corporation) was added. Cells were spheroplasted at 30°C for 30 min, spun at 1,500 g for 5 min, and washed once with FSB containing 30 µM phalloidin. Cells were resuspended in 100 µl FSB + 30 µM phalloidin, and ∼0.5-µm glass beads were added. The slurry was vortexed for 30 s, incubated for 30 s on ice, and this cycle was repeated three more times. The lysate was centrifuged at 1,500 g for 5 min. The resulting supernatant was further clarified by spinning once at 5,000 g for 5 min and again at 5,000 g for 10 min. Protein concentration was quantified in triplicate using protein assay (Bio-Rad Laboratories), and extracts were normalized to 10 mg/ml using FSB + 30 µM phalloidin. Extracts were used immediately and were not allowed to remain on ice for >1 h. 200 µl of 100 nM DNase I (Boehringer Ingelheim) was added to 800 µl of 25 µg/ml herring sperm DNA. A260 values were measured for 1 min immediately after mixing using a spectrophotometer (Thermo Fisher Scientific). 1 µl cell extract was used for the inhibition assay.
Plasma membrane permeability assays
Rhodamine-6G uptake was measured as described previously (van den Hazel et al., 1999) with the following modifications. 20 OD600 of wild-type and aip1Δ cells were harvested and washed three times with 50 mM Hepes, pH 7.2. The cell pellet was resuspended in 4 ml of deenergizing buffer A (50 mM Hepes, pH 7.2, 1 µM antimycin A [Sigma-Aldrich], and 5 mM 2-deoxy-glucose [Sigma-Aldrich]) and incubated at room temperature for 2 h with gentle agitation. 5 µM rhodamine-6G was added to the cells, and every 10 min, 100 µl of this cell mixture was harvested and washed three times with 1 ml of buffer A. The cells were centrifuged and resuspended in 1 ml Hepes, pH 7.2, and rhodamine-6G accumulation was measured using a spectrofluorimeter (FluoroMax-3; Horiba Jobin Yvon) with an excitation λ = 529 nm (slit 4) and an emission λ = 553 nm (slit 8). An aliquot before drug treatment was used to determine background fluorescence.
Rhodamine-6G efflux was measured as described previously (Kolaczkowski et al., 1996) with the following modifications. 5 OD600 of wild-type and aip1Δ cells was harvested and washed with H2O four times. The cell pellet was resuspended in 400 µl of deenergizing buffer B (50 mM Hepes, pH 7.2, and 5 mM 2-deoxy-glucose) supplemented with 5 µM rhodamine-6G and incubated at room temperature for 2 h with gentle agitation. 50 ml of this cell mixture was harvested and washed three times with 50 mM Hepes, pH 7.2, and resuspended in 2 ml 50 mM Hepes, pH 7.2. 1 ml of this cell mixture was placed into a quartz cuvette, and efflux was initiated with 1 ml of 1 M glucose. Efflux was measured by an increase in fluorescence every 5 s after addition of glucose using the aforementioned equipment and settings.
Online supplemental material
Fig. S1 shows that Aip1 does not contribute significantly to actin dynamics in vivo. Fig. S2 shows that aipΔ cells have no defects in actin levels, drug uptake, or drug efflux. Fig. S3 shows DNase I inhibition indicating that aip1Δ cells have less actin monomer than wild-type cells. Video 1 shows actin tail dynamics marked by Abp1-mRFP in sla2Δ cells. Video 2 shows actin tail dynamics in sla2Δ cells expressing Abp1-mRFP and treated with 400 µM lat A. Video 3 shows actin assembly (Abp1-mRFP) at endocytic sites marked by Sla1-GFP in wild-type cells treated with 200 µM lat A. Video 4 shows Aip1-GFP and Abp1-mRFP dynamics simultaneously imaged in wild-type cells. Video 5 shows actin tail dynamics in aip1Δ sla2Δ cells expressing Abp1-mRFP and treated with 400 µM lat A. Video 6 shows actin assembly (Abp1-mRFP) at endocytic sites marked by Sla1-GFP in aip1Δ cells treated with 200 µM lat A. Table S1 shows yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909176/DC1.
Acknowledgments
We are very grateful to Emil Reisler for providing the Q41C actin mutant, Isabelle Le Blanc, Helen Stimpson, and Alphée Michelot for their critical reading of the manuscript, and the rest of the Drubin/Barnes laboratory for helpful discussion.
This work was supported by the National Institutes of Health (RO1 grant GM42759 to D.G. Drubin).
Footnotes
Abbreviations used in this paper:
- lat A
- latrunculin A
- mRFP
- monomeric RFP
References
- Andrianantoandro E., Pollard T.D. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Blanchoin L., Sept D., McCammon J.A., Pollard T.D. 2001. Kinetic mechanism of end-to-end annealing of actin filaments. J. Mol. Biol. 312:721–730 10.1006/jmbi.2001.5005 [DOI] [PubMed] [Google Scholar]
- Balcer H.I., Goodman A.L., Rodal A.A., Smith E., Kugler J., Heuser J.E., Goode B.L. 2003. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13:2159–2169 10.1016/j.cub.2003.11.051 [DOI] [PubMed] [Google Scholar]
- Blikstad I., Markey F., Carlsson L., Persson T., Lindberg U. 1978. Selective assay of monomeric and filamentous actin in cell extracts, using inhibition of deoxyribonuclease I. Cell. 15:935–943 10.1016/0092-8674(78)90277-5 [DOI] [PubMed] [Google Scholar]
- Chen X., Cook R.K., Rubenstein P.A. 1993. Yeast actin with a mutation in the “hydrophobic plug” between subdomains 3 and 4 (L266D) displays a cold-sensitive polymerization defect. J. Cell Biol. 123:1185–1195 10.1083/jcb.123.5.1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coué M., Brenner S.L., Spector I., Korn E.D. 1987. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213:316–318 10.1016/0014-5793(87)81513-2 [DOI] [PubMed] [Google Scholar]
- Gandhi M., Achard V., Blanchoin L., Goode B.L. 2009. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol. Cell. 34:364–374 10.1016/j.molcel.2009.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., Drubin D.G. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 115:475–487 10.1016/S0092-8674(03)00883-3 [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C.P., Drubin D.G. 2006. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7:404–414 10.1038/nrm1940 [DOI] [PubMed] [Google Scholar]
- Kawamura M., Maruyama K. 1970. Electron microscopic particle length of F-actin polymerized in vitro. J. Biochem. 67:437–457 [DOI] [PubMed] [Google Scholar]
- Kim E., Wriggers W., Phillips M., Kokabi K., Rubenstein P.A., Reisler E. 2000. Cross-linking constraints on F-actin structure. J. Mol. Biol. 299:421–429 10.1006/jmbi.2000.3727 [DOI] [PubMed] [Google Scholar]
- Kolaczkowski M., van der Rest M., Cybularz-Kolaczkowska A., Soumillion J.P., Konings W.N., Goffeau A. 1996. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J. Biol. Chem. 271:31543–31548 10.1074/jbc.271.49.31543 [DOI] [PubMed] [Google Scholar]
- Kueh H.Y., Charras G.T., Mitchison T.J., Brieher W.M. 2008. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J. Cell Biol. 182:341–353 10.1083/jcb.200801027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P., Drubin D.G. 1997. Cofilin promotes rapid actin filament turnover in vivo. Nature. 388:78–82 10.1038/40418 [DOI] [PubMed] [Google Scholar]
- Morrison S.S., Dawson J.F. 2007. A high-throughput assay shows that DNase-I binds actin monomers and polymers with similar affinity. Anal. Biochem. 364:159–164 10.1016/j.ab.2007.02.027 [DOI] [PubMed] [Google Scholar]
- Murphy D.B., Gray R.O., Grasser W.A., Pollard T.D. 1988. Direct demonstration of actin filament annealing in vitro. J. Cell Biol. 106:1947–1954 10.1083/jcb.106.6.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ravi H., Smith E.M., Goode B.L. 2006. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell. 17:2855–2868 10.1091/mbc.E06-02-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178:1251–1264 10.1083/jcb.200703092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal A.A., Tetreault J.W., Lappalainen P., Drubin D.G., Amberg D.C. 1999. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145:1251–1264 10.1083/jcb.145.6.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toret C.P., Lee L., Sekiya-Kawasaki M., Drubin D.G. 2008. Multiple pathways regulate endocytic coat disassembly in Saccharomyces cerevisiae for optimal downstream trafficking. Traffic. 9:848–859 10.1111/j.1600-0854.2008.00726.x [DOI] [PubMed] [Google Scholar]
- Tsuji T., Miyoshi T., Higashida C., Narumiya S., Watanabe N. 2009. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PLoS One. 4:e4921 10.1371/journal.pone.0004921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hazel H.B., Pichler H., do Valle Matta M.A., Leitner E., Goffeau A., Daum G. 1999. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 274:1934–1941 10.1074/jbc.274.4.1934 [DOI] [PubMed] [Google Scholar]
- Yarmola E.G., Somasundaram T., Boring T.A., Spector I., Bubb M.R. 2000. Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J. Biol. Chem. 275:28120–28127 [DOI] [PubMed] [Google Scholar]