KNL targets PP1 to kinetochores, where it antagonizes Aurora B activity.
Abstract
Regulated interactions between kinetochores and spindle microtubules are essential to maintain genomic stability during chromosome segregation. The Aurora B kinase phosphorylates kinetochore substrates to destabilize kinetochore–microtubule interactions and eliminate incorrect attachments. These substrates must be dephosphorylated to stabilize correct attachments, but how opposing kinase and phosphatase activities are coordinated at the kinetochore is unknown. Here, we demonstrate that a conserved motif in the kinetochore protein KNL1 directly interacts with and targets protein phosphatase 1 (PP1) to the outer kinetochore. PP1 recruitment by KNL1 is required to dephosphorylate Aurora B substrates at kinetochores and stabilize microtubule attachments. PP1 levels at kinetochores are regulated and inversely proportional to local Aurora B activity. Indeed, we demonstrate that phosphorylation of KNL1 by Aurora B disrupts the KNL1–PP1 interaction. In total, our results support a positive feedback mechanism by which Aurora B activity at kinetochores not only targets substrates directly, but also prevents localization of the opposing phosphatase.
Introduction
To ensure accurate chromosome segregation during cell division, chromosomes must attach correctly to spindle microtubules, with kinetochores from the paired sister chromatids attached to microtubules from opposite spindle poles (bi-orientation). Attachments are regulated through changes in the phosphorylation state of kinetochore proteins that interact directly with spindle microtubules, including the Ndc80 complex (Cheeseman et al., 2006; DeLuca et al., 2006; Ciferri et al., 2008), the Dam1 complex (Cheeseman et al., 2002; Gestaut et al., 2008), and the kinesin-13 family member MCAK (mitotic centromere-associated kinesin; Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004). The key regulatory kinase responsible for targeting these substrates is Aurora B, which has an established role in eliminating incorrect attachments (Tanaka, 2002; Lampson et al., 2004; Ruchaud et al., 2007). Although previous work has focused on the crucial function of Aurora B, proper chromosome segregation requires a dynamic interplay between phosphorylation and dephosphorylation. Phosphorylation of Aurora B substrates at kinetochores destabilizes incorrect attachments, resetting the kinetochore to provide a new opportunity to bi-orient. However, this process requires that Aurora B substrates are subsequently dephosphorylated to stabilize correct attachments. In support of this idea, mutations that mimic constitutive substrate phosphorylation in vivo are as damaging as those that prevent phosphorylation (Cheeseman et al., 2002; DeLuca et al., 2006; Guimaraes et al., 2008). Coordination of kinase and phosphatase activities at kinetochores is therefore required to establish correct attachment of all chromosomes. Defining the function of the phosphatase opposing Aurora B at kinetochores is critical to understand the mechanisms that ensure accurate chromosome segregation.
Evidence from fungi and other organisms suggests that protein phosphatase 1 (PP1; Glc7) opposes Aurora B activity (Francisco et al., 1994; Hsu et al., 2000; Cheeseman et al., 2002; Pinsky et al., 2006; Emanuele et al., 2008; Vanoosthuyse and Hardwick, 2009), and in mammalian cells, at least two isoforms of PP1 (PP1α and PP1γ) localize to kinetochores (Trinkle-Mulcahy et al., 2003, 2006). However, PP1 plays multiple diverse roles in the cell, and it has been a challenge to define its specific function in chromosome segregation. A nonessential PP1 regulatory subunit, Fin1, was recently identified at the kinetochore in budding yeast (Akiyoshi et al., 2009), but this protein is not conserved. In metazoans, it is unknown how PP1 is targeted to kinetochores, what the functional significance of this targeting is, and whether phosphatase activity at kinetochores is regulated. In this study, we demonstrate that PP1 localizes to kinetochores through a direct interaction with a conserved motif in the kinetochore protein KNL1. Recruitment of PP1 is required to oppose Aurora B activity at kinetochores by dephosphorylating Aurora B substrates and stabilizing microtubule attachments. Furthermore, the interaction between PP1 and KNL1 is regulated through phosphorylation of KNL1 by Aurora B, which provides a mechanism to coordinate kinase and phosphatase activities at kinetochores.
Results
KNL1 directly associates with PP1
Because PP1 catalytic subunits regulate multiple cellular processes, previous analyses of PP1 function in regulating chromosome segregation have been complicated by the pleotrophic defects associated with PP1 inhibition. The functional specificity of PP1 derives largely from associations with a wide array of regulatory proteins (Cohen, 2002), so we first asked which regulatory protein recruits PP1 to kinetochores. To identify PP1-associated proteins, we generated a clonal human cell line stably expressing GFPLAP-PP1γ, which localizes to kinetochores (Trinkle-Mulcahy et al., 2003), and used these cells to isolate PP1γ using a one-step purification procedure. As expected, this purification identified several established PP1-interacting proteins (Fig. S1 A). Because kinetochore-targeted PP1γ represents a small percentage of the total cellular PP1γ (Trinkle-Mulcahy et al., 2003), most copurifying proteins are unlikely to function in kinetochore targeting. Indeed, we tested several known regulatory subunits identified in these purifications including PPP1R7, PPP1R2, PPP1R11, and PPP1R12A, but each of these failed to localize to kinetochores when expressed as a GFPLAP fusion (unpublished data). We therefore examined the list of interacting proteins to identify known kinetochore components that might function to target PP1 to kinetochores. The only established kinetochore component identified in our purifications was hKNL1 (also known as Blinkin, AF15q14, D40, and CASC5), a member of the conserved KMN (KNL1/Mis12 complex/Ndc80 complex) network of kinetochore proteins (Fig. S1 A; Desai et al., 2003; Nekrasov et al., 2003; Kerres et al., 2004; Kiyomitsu et al., 2007; Przewloka et al., 2007; Cheeseman et al., 2008). Our previous work has demonstrated that stringent multistep tandem affinity purifications of the KMN network isolate a stable 10-subunit network (Cheeseman et al., 2004). To test whether PP1 interacts with the KMN network, we isolated the GFPLAP-tagged Nuf2 subunit using a one-step purification procedure to allow the identification of more weakly associated proteins. Purification of Nuf2 isolated the PP1 catalytic subunit in addition to other expected kinetochore proteins (Fig. S1 A), which suggests that PP1 does interact with the KMN network. Similarly, we found that FLAG-ggKNL1 immunoprecipitates purified from chicken DT40 cells also isolated PP1 (Fig. S1 B).
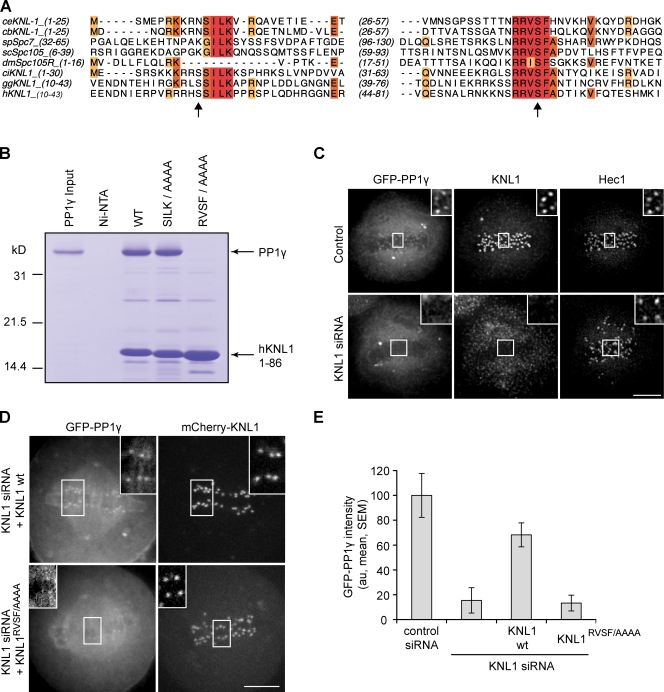
To determine which KMN network subunit interacts directly with PP1, we searched for docking motifs that are commonly found in PP1-interacting proteins. The RVxF motif is the most common of these motifs, but other sequences such as [S/G]ILK have also been described (Egloff et al., 1997; Hendrickx et al., 2009). Strikingly, both the SILK and RVSF motifs are present and highly conserved at the N terminus of KNL1 throughout eukaryotes, despite a weak overall sequence conservation (Fig. 1 A; Cheeseman et al., 2004). This finding strongly suggests that KNL1 interacts directly with PP1. To test this possibility, we performed binding assays with recombinant hPP1γ and an 86-residue N-terminal fragment of hKNL1 that contains the SILK and RVSF motifs. hKNL11–86 binds directly to PP1γ, and mutation of these motifs to alanines showed that binding depends on the RVSF, but not the SILK motif (Fig. 1 B). hPP1γ also binds Caenorhabditis elegans KNL-11–68 in an RVSF-dependent manner (unpublished data), which indicates that the interaction is conserved. These data demonstrate that KNL1 directly interacts with PP1 through the conserved RVSF motif.
Figure 1.
KNL1 recruits PP1γ to kinetochores. (A) Sequence alignment of C. elegans (ce), Caenorhabditis briggsae (cb), Schizosaccharomyces pombe (sp), Saccharomyces cerevisiae (sc), D. melanogaster (dm), Ciona intestinalis (ci), Gallus galls (gg), and human (h) KNL1 homologues showing conservation of N-terminal [S/G]ILK and RVSF motifs. Arrows indicate Aurora B phosphorylation sites (Welburn et al., 2010). Colors indicate conserved amino acids. (B) Binding of KNL1 to PP1γ depends on the RVSF motif. A Coomassie-stained gel shows binding of PP1γ to Ni-NTA agarose resin alone, or resin bound to either His-tagged hKNL11–86 wild type or mutants for the conserved PP1-binding motifs. (C) HeLa cells stably expressing GFPLAP-PP1γ were fixed and stained for KNL1 and Hec1. PP1γ fails to localize to kinetochores after depletion of KNL1 by siRNA. (D) HeLa cells were transfected with KNL1 siRNA and either siRNA-resistant wild-type (wt) KNL1 or the KNL1RVSF/AAAA mutant, together with GFP-PP1γ. The KNL1RVSF/AAAA mutant fails to restore kinetochore localization of PP1. (E) For cells treated as in D, the intensity of GFP-PP1γ at kinetochores was calculated relative to control cells for each of the conditions indicated. Each bar represents a mean (± SEM) over multiple cells (n ≥ 5), with ≥40 kinetochores analyzed per cell. Images in C and D are maximum intensity projections of confocal stacks; insets show enlarged views (indicated by the boxed regions) of optical sections showing individual kinetochores. Intensity scaling is consistent between all insets, but insets are scaled differently from the original images to show individual kinetochores more clearly. au, arbitrary units. Bars, 5 µm.
KNL1 recruits PP1 to the outer kinetochore
The biochemical interaction between PP1 and KNL1 suggests that KNL1 may function as a kinetochore-targeting subunit for PP1. To test whether KNL1 is required for PP1 localization to kinetochores in human cells, we depleted KNL1 by RNAi. We were unable to detect GFP-PP1γ or GFP-PP1α at kinetochores in KNL1-depleted cells (Fig. 1 C and Fig. S2, A and B). Because KNL1 has an established role in kinetochore assembly (Desai et al., 2003; Cheeseman et al., 2008), we considered the possibility that the failure to recruit PP1 might be caused by a general perturbation of the kinetochore. The RVSF/AAAA mutant prevents the interaction between KNL1 and PP1 (Fig. 1 B) while leaving KNL1 otherwise intact. We therefore examined PP1 localization in cells depleted of endogenous KNL1 by siRNA and expressing either siRNA-resistant wild-type KNL1 or the KNL1RVSF/AAAA mutant. PP1 localizes to kinetochores in cells expressing wild-type KNL1 but not in cells expressing the KNL1RVSF/AAAA mutant (Fig. 1, D and E), which indicates that the RVSF motif in KNL1 is required for recruitment of PP1 to kinetochores. PP1 was not completely restored to normal levels at kinetochores in these experiments, likely because we were not able to restore KNL1 to endogenous levels after depletion by siRNA (Fig. S3).
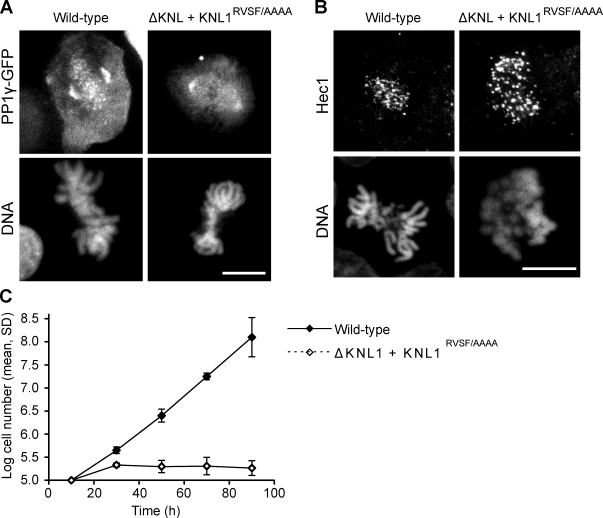
To test whether this role for KNL1 in targeting PP1 to kinetochores is conserved, we generated a chicken DT40 cell line in which wild-type ggKNL1 expression was replaced by a mutant with the RVSF motif mutated to alanines. PP1 fails to localize to kinetochores in KNL1RVSF/AAAA mutants (Fig. 2 A), which confirms that the RVSF motif is required for PP1 recruitment. KNL1 is required for normal kinetochore assembly, and depletion of ggKNL1 leads to reduced kinetochore localization of Hec1 in DT40 cells. In contrast, in HeLa cells, Hec1 localization is normal in the absence of KNL1 (Cheeseman et al., 2008). KNL1RVSF/AAAA mutant DT40 cells showed normal localization of Ndc80/HEC1, which indicates that disrupting this motif does not generally affect outer kinetochore assembly (Fig. 2 B). However, we found that the mutant cells are inviable (Fig. 2 C). Together, these data demonstrate that the interaction between KNL1 and PP1 is required for PP1 localization to kinetochores and for KNL1 function in vivo.
Figure 2.
The KNL1–PP1 interaction is required for cell viability. A tetracycline-repressible ggKNL1 chicken DT40 cell line (Cheeseman et al., 2008) was rescued by constitutive expression of a ggKNL1 mutant with the RVSF motif mutated to alanines. (A) PP1 fails to localize to kinetochores in the KNL1RVSF/AAAA mutant cells. Images show PP1γ-GFP localization in wild type or KNL1RVSF/AAAA mutants. (B) KNL1RVSF/AAAA mutants do not generally affect outer kinetochore assembly. Immunofluorescence images show Hec1 localization in wild type or KNL1RVSF/AAAA mutants. (C) KNL1RVSF/AAAA mutant cells are inviable. To determine viability, cells were counted after expression of the wild-type protein was inactivated at t = 0 by tet addition. Cell numbers are averages over multiple experiments (n = 2 for wild type, n = 3 for KNL1RVSF/AAAA). Bars, 10 µm.
Kinetochore-localized PP1 opposes Aurora B activity
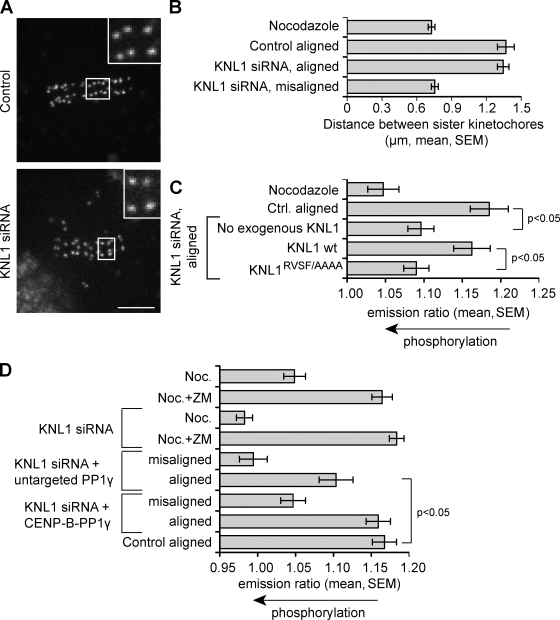
The identification of the KNL1–PP1 interaction provides a means to probe PP1 function specifically at kinetochores without disrupting its other cellular activities. To test whether PP1 opposes Aurora B activity, we measured phosphorylation of an Aurora B substrate at kinetochores using a biosensor that reports quantitative changes in phosphorylation by Aurora B in living cells through changes in fluorescence resonance energy transfer (FRET; Fuller et al., 2008). We previously demonstrated that phosphorylation of a kinetochore-targeted sensor depends on the spatial separation of kinetochores from Aurora B, which is located at the inner centromere (Liu et al., 2009). Phosphorylation increases when centromere tension is low and decreases when centromere tension is high and kinetochores are pulled away from the inner centromere. To test whether phosphorylation levels also depend on PP1 recruitment, we examined sensor phosphorylation in KNL1-depleted cells. We focused our analysis on kinetochores aligned at the metaphase plate, which are under normal tension in these cells based on interkinetochore distance measurements (Fig. 3, A and B). Phosphorylation is increased on aligned kinetochores in KNL1-depleted cells relative to aligned kinetochores in metaphase control cells (Fig. 3 C). In cells depleted of endogenous KNL1, expression of siRNA-resistant wild-type KNL1 reduces phosphorylation on aligned kinetochores to levels similar to control cells, but expression of the KNLRVSF/AAAA mutant does not (Fig. 3 C). These data demonstrate that recruitment of PP1 by KNL1 is required to dephosphorylate Aurora B substrates at the outer kinetochore.
Figure 3.
PP1 opposes Aurora B activity at kinetochores. HeLa cells expressing a kinetochore-targeted Aurora B phosphorylation sensor were treated as indicated and imaged live. (A) Images of YFP emission in control and KNL1-depleted cells are maximum intensity projections of confocal stacks; insets show enlarged views (indicated by boxed regions) of optical sections showing individual kinetochores. Bar, 5 µm. (B) Interkinetochore distances were measured for the indicated conditions. Aligned and misaligned kinetochores were analyzed separately. Centromeres aligned at the metaphase plate are under full tension in KNL1-depleted cells. Each bar represents a mean (± SEM) of >70 kinetochores from ≥12 cells. (C) The YFP/TFP emission ratio was analyzed to measure phosphorylation changes at kinetochores; an increased YFP/TFP emission ratio indicates dephosphorylation. Aligned kinetochores were analyzed in KNL1-depleted cells, either without exogenous KNL1 or expressing siRNA-resistant wild-type KNL1 or KNL1RVSF/AAAA. Depletion of KNL1 increases phosphorylation on aligned kinetochores, and dephosphorylation is restored by wild-type KNL1 but not by KNL1RVSF/AAAA. (D) To target exogenous PP1 to kinetochores in KNL1-depleted cells, cells were transfected with CENP-B–PP1γ-mCherry and analyzed as in C. ZM indicates the Aurora B inhibitor ZM447439. Expression of CENP-B–PP1γ-mCherry restores dephosphorylation at aligned kinetochores to levels similar to control cells, but untargeted PP1γ-mCherry does not. Each bar in C and D represents a mean (± SEM) over ≥8 cells, with ≥30 kinetochores analyzed per cell for aligned kinetochores, or a total of at least 60 kinetochores for misaligned kinetochores.
To further test whether the increase in phosphorylation observed after KNL1 depletion is caused by the loss of PP1, we introduced exogenous PP1γ fused to centromere protein B (CENP-B; CENP-B–PP1γ), which targets the phosphatase to centromeres independently of KNL1. In KNL1-depleted cells, expression of CENP-B–PP1γ reduces phosphorylation on aligned kinetochores to levels similar to control cells, whereas expression of untargeted PP1γ does not (Fig. 3 D). These results indicate that phosphorylation levels of Aurora B substrates at kinetochores depend on PP1 recruitment to kinetochores by KNL1.
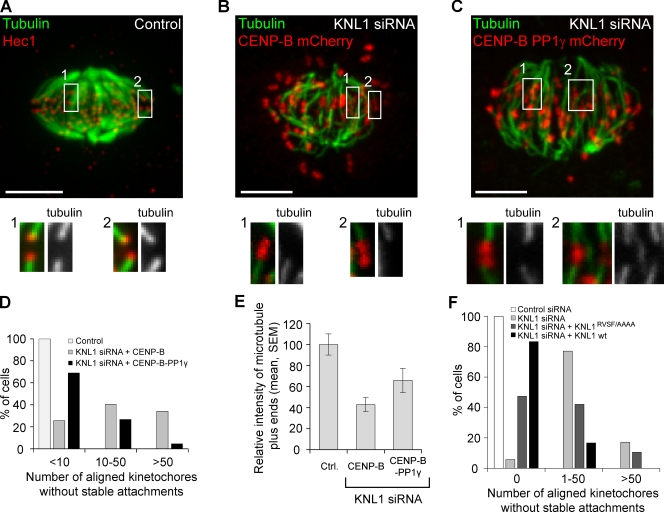
Because kinetochore–microtubule interactions depend on phosphorylation of Aurora B substrates at kinetochores, our findings predict destabilization of kinetochore microtubules in KNL1-depleted cells due to loss of phosphatase activity. To monitor the stability of kinetochore–microtubule interactions, we analyzed cold-stable microtubules in KNL1-depleted cells and found that some kinetochores align at the center of the spindle with stable microtubule fibers, whereas other aligned kinetochores lack stable fibers. In addition, the cold-stable fibers in KNL1-depleted cells stained less brightly for tubulin compared with control cells (Fig. 4, A–E), which indicates that these fibers contain fewer microtubules (King and Nicklas, 2000). The finding that it is possible to generate tension across the centromere, as indicated by interkinetochore distance (Fig. 3 A, B), without fully stabilizing kinetochore microtubules is consistent with our previous results using inner centromere protein (INCENP) fusion proteins to increase Aurora B activity at the kinetochore (Liu et al., 2009). Because KNL1 binds microtubules directly (Cheeseman et al., 2006) and is important for outer kinetochore assembly (Cheeseman et al., 2008), PP1 recruitment is likely only one of multiple functions of KNL1 at kinetochores. To determine whether the defects in kinetochore microtubules are caused by loss of PP1, we also analyzed KNL1-depleted cells expressing CENP-B–PP1γ. In KNL1-depleted cells, expression of CENP-B–PP1γ but not untargeted PP1γ restored cold-stable fibers to most of the aligned kinetochores (Fig. 4, C and D; and Fig. S4, A and B). However, CENP-B–PP1γ did not restore tubulin staining to normal levels (Fig. 4, C and E). In cells depleted of endogenous KNL1, the kinetochore microtubule defect was partially rescued by expression of the KNL1RVSF/AAAA mutant, but some kinetochores still lacked cold-stable fibers, compared with expression of wild-type KNL1 (Figs. 4 F and S4 C). These results indicate that recruitment of PP1 to kinetochores contributes to stabilizing bi-oriented attachments, but other functions of KNL1 are also necessary for kinetochores to bind the normal complement of microtubules.
Figure 4.
PP1 at kinetochores stabilizes microtubule attachments. HeLa cells were treated as indicated, fixed, and analyzed for cold-stable microtubules. (A–E) Cells were transfected with KNL1 siRNA and either CENP-B–PP1γ-mCherry to target exogenous PP1 to kinetochores, or CENP-B–mCherry as a control. An untransfected control cell (A) was analyzed the same way and stained for Hec1 to label kinetochores. Images (A–C) are maximum intensity projections of confocal stacks; insets show enlarged views (indicated by the numbered boxed regions) of optical sections showing individual kinetochores. Bars, 5 µm. (D) Cells were classified by the number of aligned kinetochores lacking cold-stable microtubule fibers (n ≥ 45 cells in each group). (E) The intensity of microtubule plus ends at attached kinetochores (n > 60 from multiple cells) was measured in each condition. (F) Cells were transfected with KNL1 siRNA and either siRNA-resistant wild-type KNL1 or the KNL1RVSF/AAAA mutant (n ≥ 25 cells in each group), and analyzed as in D. Note that the binning in D and F is different because the effect of mutating the RVSF motif is more subtle than the effect of depleting KNL1.
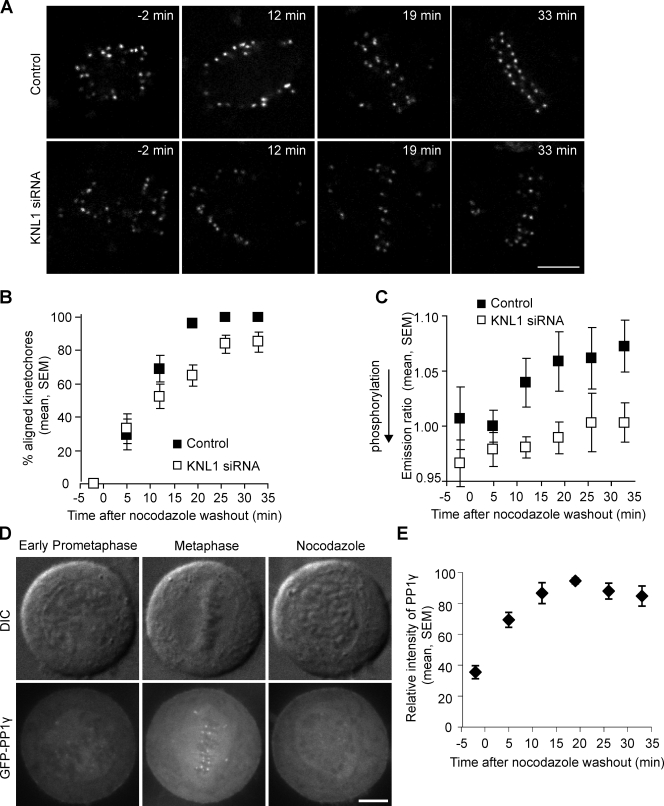
Centromere tension leads to stabilization of attachments and dephosphorylation of Aurora B substrates. To determine the contribution of kinetochore-targeted PP1 to dephosphorylation dynamics, we treated cells with a low dose of nocodazole (30 ng/ml), which releases tension at the centromere without completely depolymerizing microtubules. After removal of nocodazole, we imaged the cells during recovery as tension was established (Fig. S5). In control cells, sensor dephosphorylation occurs within 20 min of nocodazole washout as chromosomes align and tension is established. In contrast, in KNL1-depleted cells, chromosomes align more slowly, full alignment is never achieved, and dephosphorylation is slow and incomplete even when most centromeres are under tension (Fig. 5, A–C). These results demonstrate that kinetochore PP1 is required to reverse Aurora B phosphorylation at kinetochores as centromere tension is established.
Figure 5.
PP1γ is recruited to kinetochores and dephosphorylates an Aurora B substrate as centromere tension is established. (A–C) HeLa cells transfected with a kinetochore-targeted Aurora B phosphorylation sensor, with or without KNL1 siRNA, were imaged live during recovery from nocodazole (30 ng/ml). Cells were followed for 33 min, which was sufficient time for control cells to reach metaphase. Cells rarely entered anaphase during this time, and any anaphase cells were excluded from the analysis. Representative images (A) show YFP emission. At each time point, the percentage of kinetochores aligned at the metaphase plate was determined (B) and the YFP/TFP emission ratio was calculated (C). Each data point represents seven cells, >15 kinetochores per cell. (D) Images of HeLa cells stably expressing GFPLAP-PP1γ in early prometaphase, metaphase, or treated with nocodazole. (E) The relative intensity of GFP-PP1γ at kinetochores was calculated during recovery from nocodazole (30 ng/ml). n = 6 cells, multiple kinetochores per cell. Bars, 5 µm.
PP1 recruitment to kinetochores is regulated through phosphorylation of KNL1 by Aurora B
PP1γ association with kinetochores is dynamic (Trinkle-Mulcahy et al., 2003), which suggests that its kinetochore targeting might be regulated. We found that GFP-PP1α and GFP-PP1γ levels at kinetochores are low in early prometaphase or in nocodazole-treated cells, but increase dramatically at metaphase (Figs. 5 D, 6 E, and S2 C). In addition, PP1γ is recruited to kinetochores during recovery from nocodazole with kinetics similar to those of chromosome alignment (Fig. 5, B and E). These results demonstrate that kinetochore targeting of PP1 is regulated, but is reciprocal to many other kinetochore proteins such as components of the spindle checkpoint that bind to unattached kinetochores and dissociate at metaphase (Musacchio and Salmon, 2007).
Figure 6.
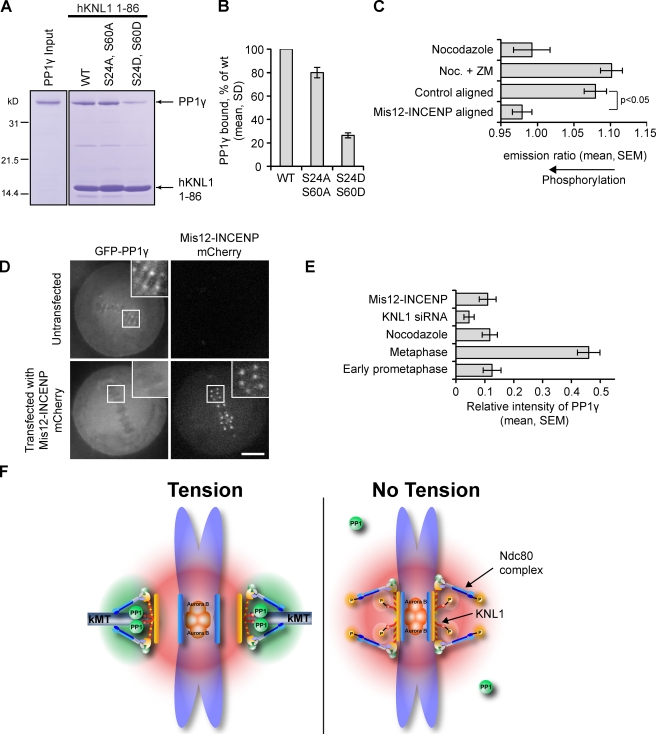
Phosphorylation of KNL1 by Aurora B disrupts the interaction with PP1γ. (A) A Coomassie-stained gel shows binding of PP1γ to Ni-NTA agarose resin bound to either His-tagged hKNL11–86 wild type, or phosphomimetic or phosphoinhibitory mutants in the SILK and RVSF motifs. (B) Binding was quantified from Coomassie-stained gels in three independent experiments. (C) Hela cells expressing a kinetochore-targeted Aurora B phosphorylation sensor were either treated with nocodazole to release interkinetochore tension, or transfected with Mis12-INCENP-mCherry to redistribute Aurora B to the kinetochore. ZM indicates the Aurora B inhibitor ZM447439. Cells were imaged live, and the YFP/TFP emission ratio was calculated to measure phosphorylation changes at kinetochores. The decrease in YFP/TFP emission ratio indicates increased phosphorylation at aligned kinetochores in cells expressing Mis12-INCENP-mCherry. Each bar represents a mean of ≥10 cells, with ≥30 kinetochores analyzed per cell. (D) HeLa cells stably expressing GFPLAP-PP1γ were transfected with (bottom) or without (top) Mis12-INCENP-mCherry to increase Aurora B activity at kinetochores. Insets show kinetochores at higher magnification (boxed regions). Bar, 5 µm. (E) Cells were transfected with Mis12-INCENP or KNL1 siRNA, or untransfected cells were analyzed at metaphase or at prometaphase, or treated with nocodazole. The relative intensity of GFP-PP1γ at kinetochores was calculated (n ≥ 9 cells, ≥40 kinetochores per cell) in each case. (F) Model comparing centromeres in low- and high-tension states. In the low-tension state, Aurora B phosphorylates KNL1 and Hec1, which leads to reduced binding of both PP1 and microtubules to kinetochores. In the high-tension state, Aurora B is spatially separated from kinetochore substrates, so KNL1 and Hec1 are dephosphorylated and the binding of PP1 and microtubules is increased.
KNL1 localization to kinetochores is constant from G2 until late anaphase (Cheeseman et al., 2008), whereas PP1 is recruited to kinetochores in metaphase (Figs. 5 D and S2 C), which suggests that the interaction between KNL1 and PP1 may be regulated. Indeed, conserved sites in the SILK and RVSF motifs in KNL1 are phosphorylated by Aurora B both in vitro and in vivo (Welburn et al., 2010). Phosphorylation of Ser/Thr residues in or near an RVxF docking motif can disrupt PP1 binding (Cohen, 2002), which suggests that phosphorylation of KNL1 might regulate PP1 targeting to kinetochores. Indeed, Ser-Asp phospho-mimetic mutations in the SILK and RVSF motifs of KNL1 reduced in vitro binding to PP1γ by >70%, whereas Ser-Ala mutations had only a negligible effect on binding (Fig. 6, A and B). This result provides a mechanism for Aurora B to regulate kinetochore targeting of its opposing phosphatase by phosphorylating the N terminus of KNL1. To test this model, we increased Aurora B activity at the kinetochore by manipulating the kinase localization. Aurora B targets to the inner centromere through interactions with INCENP (Adams et al., 2000), and expression of a Mis12-INCENP fusion protein redistributes Aurora B to kinetochores (Liu et al., 2009). In cells expressing Mis12-INCENP, a kinetochore-targeted phosphorylation sensor is highly phosphorylated, even on aligned kinetochores that are under tension, which shows that local Aurora B activity at kinetochores is increased (Fig. 6 C). Even when all chromosomes are aligned at metaphase in these cells, PP1γ localization is reduced to levels similar to early prometaphase or nocodazole-treated cells (Fig. 6, D and E). This result indicates that increased Aurora B activity at kinetochores prevents PP1 targeting. In total, our findings reveal a dynamic balance of kinase and phosphatase activities at kinetochores through a regulated interaction between KNL1 and PP1.
Discussion
Previous work has strongly implicated Aurora B kinase as a master regulator for controlling chromosome segregation by inactivating key microtubule-binding activities at the kinetochore (Carmena et al., 2009; Santaguida and Musacchio, 2009). A critical component of this regulation is to ensure that the phosphorylation of downstream targets is tightly controlled. We recently demonstrated that Aurora B can respond to centromere tension through spatial separation of the kinase from its substrates, which reduces phosphorylation levels (Liu et al., 2009). Thus, an important component of the regulatory cycle can be accounted for by the spatial distribution of the kinase and its substrates. However, the complementary component of this cycle, in which Aurora B substrates are dephosphorylated, has been largely ignored. PP1 has been established genetically as opposing Aurora B kinase, but unlike the tension-sensitive models for Aurora B function, PP1 has been proposed to function constitutively. Despite increasing interest in mitotic phosphatases (Trinkle-Mulcahy and Lamond, 2006; Bollen et al., 2009; De Wulf et al., 2009), the precise mechanism by which PP1 opposes Aurora B kinase at kinetochores has remained unknown.
Aurora B localization is restricted to centromeres during chromosome alignment and to the spindle midzone during anaphase, but PP1 is important for a broad range of cellular processes. Thus, it has been difficult to dissect the role of PP1 in chromosome segregation even using conditional mutants in fungi. Our findings demonstrate that KNL1 serves as a targeting subunit that recruits PP1 to the outer kinetochore through a conserved RVSF motif to oppose Aurora B activity. This discovery provides a way to specifically perturb PP1 at kinetochores without affecting its other cellular functions. Through experiments with KNL1 mutants that specifically disrupt interactions with PP1 in both HeLa cells and chicken DT40 cells, our results demonstrate that this interaction is essential for cell viability and is critical to control the balance between Aurora B kinase and opposing phosphatase activities at kinetochores. The RVSF motif is required for PP1 binding to KNL1 in vitro, for recruitment of PP1 to kinetochores in vivo, and for dephosphorylation of Aurora B substrates at kinetochores.
Regulation of kinetochore–microtubule interactions requires phosphorylation of downstream targets of Aurora B, but mutations that mimic constitutive phosphorylation also result in severe consequences. Our results provide a mechanism to reverse the phosphorylation of Aurora B substrates by targeting its opposing phosphatase to the kinetochore. Because of the position of KNL1 within the kinetochore, this targeting establishes two distinct regions of kinase and phosphatase localization (Fig. 6 F), with Aurora B at the inner centromere and PP1 at the outer kinetochore, where the critical Aurora B targets that bind to microtubules are localized.
Importantly, our results also demonstrate that PP1 does not constitutively interact with KNL1. Instead, this interaction is regulated through phosphorylation of KNL1 by Aurora B. Regulated PP1 recruitment suggests a positive feedback mechanism in which changes in local kinase activity at the kinetochore lead to reciprocal changes in phosphatase activity (Fig. 6 F). When centromere tension is low, Aurora B phosphorylates kinetochore substrates, including Hec1 and KNL1, which both destabilizes microtubule attachments and reduces PP1 recruitment to kinetochores. In contrast, centromere tension at bi-oriented chromosomes separates Aurora B from kinetochores, leading to substrate dephosphorylation. We propose that the initial dephosphorylation is caused by low levels of PP1, and kinetochore phosphatase activity is amplified through KNL1 dephosphorylation and PP1 recruitment. Dephosphorylation of Aurora B substrates also stabilizes microtubule attachments, which maintains centromere tension. Complementary mechanisms of kinase concentration at the inner centromere and phosphorylation-dependent recruitment of phosphatase to the outer kinetochore thereby create two spatially distinct domains of kinase and phosphatase activity to determine the phosphorylation state of kinetochore proteins. In total, our results have revealed a balance of regulatory activities that controls the fidelity of chromosome segregation.
Materials and methods
Cell culture and transfection
HeLa cells were cultured in growth medium: DME with 10% fetal bovine serum and penicillin-streptomycin, at 37°C in a humidified atmosphere with 5% CO2. Cells were transfected with plasmid DNA using Fugene (Roche) or with siRNA oligo using Oligofectamine (Invitrogen), according to the manufacturers’ instructions. Cells were used for analysis 2 d after transfection with plasmid DNA or 1 d after transfection with KNL1 siRNA. For transfection of both KNL1 siRNA and plasmid DNA, the siRNA transfection was performed 1 d after the plasmid transfection, and cells were used on the following day. The siRNA sequence targeting KNL1 (5′-GGAAUCCAAUGCUUUGAG-3′) was synthesized by Thermo Fisher Scientific. A clonal cell line stably expressing low levels of GFPLAP-PP1γ was generated using retroviral infection followed by FACS.
Plasmids
CENP-B–mCherry was created by inserting residues 1–167 of CENP-B into pEGFP-N1 (Takara Bio Inc.) and then inserting mCherry to replace GFP. Untargeted mCherry-PP1γ was created by replacing GFP with mCherry in vector pEGFP-C1 and inserting PP1γ at the C terminus of mCherry. CENP-B–mCherry-PP1γ was created by placing residues 1–167 of human CENP-B at the N terminus of mCherry-PP1γ in vector pCDNA3.1. Mis12-INCENP-mCherry has been described previously (Liu et al., 2009). The KNL1 mutant plasmids were created using the QuikChange Site-directed Mutagenesis kit (Agilent Technologies). The siRNA-resistant full-length mCherry-KNL1, under the control of the cytomegalovirus promoter, was mutated without changing the amino acid sequence, and the RVSF motif was mutated to AAAA.
The kinetochore-targeted FRET sensor used in this study was created by modifying the Mis12-targeted sensor described previously (Liu et al., 2009). Because Mis12 targeting depends on KNL1, we fused the sensor to the C terminus of Hec1 because kinetochore localization of Hec1 does not depend exclusively on KNL1 in human cells (Cheeseman and Desai, 2008).
Nocodazole washout assay
Cells were incubated in a low concentration of nocodazole (30 ng/ml) in growth medium for 2–3 h to release tension between sister kinetochores. To remove nocodazole, cells were washed three times with fresh L15 medium. The time of the first wash was recorded as time zero. One image was acquired before washing out nocodazole, and after the washout, one image was acquired every 7 min, starting at t = 5 min, to minimize photobleaching.
Immunofluorescence
For analysis of cold-stable microtubules, cells were incubated for 10 min on ice in L-15 medium (Invitrogen) with 20 mM Hepes, pH 7.3, then fixed for 10 min at room temperature with 4% formaldehyde in 100 mM Pipes, pH 6.8, 10 mM EGTA, 1 mM MgCl2 and 0.2% Triton X-100. Cells expressing GFP-PP1 were fixed in PBS with 4% formaldehyde for 10 min. For triple staining of GFP-PP1γ, KNL1, and Hec1, images were acquired as z stacks with 0.2 µm spacing using a 100×, 1.35 NA objective on an image restoration microscope (DeltaVision; Applied Precision and Olympus) and processed by iterative constrained deconvolution (SoftWoRx; Applied Precision). Maximal intensity projections of the entire z stack are shown, and optical sections show individual kinetochores more clearly (Fig. 1 C, insets). Other images were acquired with a spinning disk confocal: a microscope (DM4000; Leica) with a 100× 1.4 NA objective, an XY piezo z stage (Applied Scientific Instrumentation), a spinning disk microscope (Yokogawa), an electron multiplier charge-coupled device camera (ImageEM; Hamamatsu Photonics), and an LMM5 laser merge module (Spectral Applied Research) controlled by IP Laboratory (BD) or Metamorph (MDS Analytical Technologies) software. The following antibodies were used: 1:1,000 rat anti-tubulin monoclonal (AbD Serotec), 1:1,000 mouse anti-Hec1 monoclonal (9G3; Abcam), 1:1,000 rabbit anti-KNL1 polyclonal (Cheeseman et al., 2008), and 1:500 Alexa Fluor 594, Alexa Fluor 488, and Alexa Fluor 647 secondary antibodies (Invitrogen).
To measure the intensities of microtubule plus ends at kinetochores, a line (width = five pixels) was drawn across the plus end of an individual kinetochore–microtubule fiber, close to the kinetochore. The maximal intensity across the line was determined using the Plot profile function of ImageJ (National Institutes of Health). After subtracting background, the intensities were averaged over multiple microtubule fibers.
Live imaging and data analysis
For live imaging of the kinetochore-targeted FRET sensors (Liu et al., 2009), Hela cells were plated on 22 × 22-mm No. 1.5 glass coverslips (Thermo Fisher Scientific) coated with Poly-d-lysine (Sigma-Aldrich). Coverslips were mounted in custom-designed Rose chambers, using L-15 medium without phenol red (Invitrogen). Temperature was maintained at 35–37°C using an environmental chamber (PeCon GmbH). Images were acquired with the spinning disk confocal microscope described previously. Teal fluorescent protein (TFP) was excited at 440 nm, and TFP and YFP emissions were acquired simultaneously with a beam splitter (Dual-View; Optical Insights, LLC). Custom software written in Matlab (Mathworks) was used for image analysis. Individual kinetochores were defined automatically from confocal image stacks (5 planes, 0.5 µm spacing), and the YFP/TFP emission ratio was calculated at each kinetochore identified by our algorithm (Fuller et al., 2008). For measurements of dephosphorylation dynamics after nocodazole washout, a single image was taken at each time point to minimize photobleaching. All results are means over multiple kinetochores from multiple cells.
To measure the intensity of GFP-PP1γ in the nocodazole washout assay, we selected individual kinetochores manually and measured the intensity of each kinetochore using ImageJ software. After subtracting background, intensities were averaged over all kinetochores at a single time point in a single cell. The average intensity at each time point was normalized for the expression level by dividing by the peak intensity for that cell. The normalized values were then averaged over multiple cells. To compare the intensity of GFP-PP1γ under different conditions (prometaphase, metaphase, nocodazole, KNL1 siRNA, and Mis12-INCENP expression), we manually selected an intensity threshold to cover all kinetochores using ImageJ software. After background subtraction, the average kinetochore intensity was normalized by dividing by the GFP intensity in the cytoplasm to remove variation caused by different expression levels of GFP-PP1γ in different cells. Multiple cells were then averaged for each condition.
Immunoprecipitation and mass spectrometry
GFPLAP-tagged hPP1γ and hNuf2 were isolated from HeLa cells using one-step immunoprecipitations (Cheeseman and Desai, 2005). DT40 cells, in which expression of KNL1 was replaced with that of KNL1-FLAG, were used for immunoprecipitation with anti-FLAG antibodies (Okada et al., 2006). Purified proteins were identified by mass spectrometry by using an ion trap mass spectrometer (LTQ XL; Thermo Fisher Scientific) with MudPIT and SEQUEST software (Washburn et al., 2001; Cantin et al., 2007).
Expression and purification of recombinant proteins and in vitro binding assays
hKNL1 mutants were generated using the QuikChange Multi Site-Directed Mutagenesis kit. 6xHis-hKNL1 fragments (amino acids 1–86 in pRSETa) and GST-hPP1γ (in pGEX-6P-1) were transformed into BL21 cells and induced for 5 h at 20°C using 0.1 mM IPTG. hPP1γ-expressing cultures were grown in the presence of 1 mM MnCl2. 6xHis-hKNL1 and GST-hPP1γ were purified using Ni-NTA agarose and glutathione agarose resin, respectively. 6xHis-hKNL1 was eluted using 250 mM imidazole, and hPP1γ was released from the GST-tag using PreScission protease. Proteins were purified into Magic buffer (50 mM Hepes, pH 7.5, 300 mM NaCl, 30 mM imidazole, 2 mM MnCl2, 10 mM β-mercaptoethanol, and 0.1% Tween). For in vitro binding assays, 6xHis-hKNL1 fragments were bound to Ni-NTA agarose beads, washed 2× with Magic buffer, and incubated with hPP1γ at room temperature for 1 h. Ni-agarose beads were washed 3× with Magic buffer and resuspended in SDS-PAGE sample buffer.
DT40 experiments
DT40 cells were cultured at 38°C in Dulbecco’s modified medium supplemented with 10% fetal calf serum, 1% chicken serum, and penicillin/ streptomycin. The RVSF site in the chicken KNL1 cDNA was mutated to AAAA using the QuikChange Multi Site-Directed Mutagenesis kit and stably introduced into KNL1 conditional knockout cells, in which expression of wild-type KNL1 can be shut off by the addition of tetracycline (Cheeseman et al., 2008). Tetracycline was added to the culture at time t = 0, and the number of cells not stained with trypan blue was counted at the indicated time points. Expression of mutant KNL1 was confirmed in the presence of tetracycline by Western blotting. Chicken PP1 cDNA was cloned into a pEGFP vector (Takara Bio Inc.) and was stably introduced into KNL1 conditional knockout cells expressing the KNL1 mutant protein.
For immunofluorescence, cells were collected onto slides with a cytocentrifuge (Cytospin 3; Shandon) and fixed in 3% paraformaldehyde in PBS for 15 min at room temperature or 100% methanol for 15 min at −20°C, then permeabilized in 0.5% NP-40 in PBS for 10 min at room temperature, rinsed three times in 0.5% BSA, and incubated for 1 h at 37°C with primary antibody. Binding of anti-ggNdc80 antibody (Hori et al., 2003) was then detected with FITC-conjugated goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) diluted in PBS/0.5% BSA. Chromosomes and nuclei were counterstained with DAPI in Vectashield Antifade (Vector Laboratories). Immunofluorescence images were collected with a cooled EM charge-coupled device camera (Roper Industries) mounted on an inverted microscope (IX71; Olympus) with a 100× 1.4 NA objective. Subsequent analysis and processing of images were performed using Metamorph software (MDS Analytical Technologies).
Online supplemental material
Fig. S1 shows mass spectrometric analysis. Fig. S2 shows kinetochore localization of PP1α. Fig. S3 shows expression of siRNA-resistant KNL1. Fig. S4 shows analysis of cold-stable microtubules in KNL1-depleted cells. Fig. S5 shows changes in centromere tension during recovery from nocodazole. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001006/DC1.
Acknowledgments
We thank B.E. Black, G. Vader, and J. Welburn for critical reading of the manuscript and helpful comments; and K.V. Le and M. Takahashi for assistance with preparing reagents.
This work was supported by grants from the National Institutes of Health (GM083988 to M.A. Lampson and GM088313 to I.M. Cheeseman), the Searle Scholars Program (to M.A. Lampson and I.M. Cheeseman), the Smith Family Foundation (to I.M. Cheeseman), the Massachusetts Life Sciences Center (to I.M. Cheeseman), and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Science and Technology (MEXT) of Japan to T. Fukagawa. I.M. Cheeseman is a Thomas D. and Virginia W. Cabot Career Development Professor of Biology.
Footnotes
Abbreviations used in this paper:
- CENP-B
- centromere protein B
- FRET
- fluorescence resonance energy transfer
- INCENP
- inner centromere protein
- TFP
- teal fluorescent protein
References
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. 2000. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10:1075–1078 10.1016/S0960-9822(00)00673-4 [DOI] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C.R., Ranish J.A., Biggins S. 2009. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23:2887–2899 10.1101/gad.1865909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J.R. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Bollen M., Gerlich D.W., Lesage B. 2009. Mitotic phosphatases: from entry guards to exit guides. Trends Cell Biol. 19:531–541 10.1016/j.tcb.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Cantin G.T., Shock T.R., Park S.K., Madhani H.D., Yates J.R., III 2007. Optimizing TiO2-based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Anal. Chem. 79:4666–4673 10.1021/ac0618730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., Ruchaud S., Earnshaw W.C. 2009. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 21:796–805 10.1016/j.ceb.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R., III, Oegema K., Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., Dos Reis G., Maiolica A., Polka J., De Luca J.G., De Wulf P., et al. 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 133:427–439 10.1016/j.cell.2008.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P.T. 2002. Protein phosphatase 1—targeted in many directions. J. Cell Sci. 115:241–256 [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- De Wulf P., Montani F., Visintin R. 2009. Protein phosphatases take the mitotic stage. Curr. Opin. Cell Biol. 21:806–815 10.1016/j.ceb.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M.P., Johnson D.F., Moorhead G., Cohen P.T., Cohen P., Barford D. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16:1876–1887 10.1093/emboj/16.8.1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S., Stukenberg P.T. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L., Wang W., Chan C.S. 1994. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14:4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D.R., Graczyk B., Cooper J., Widlund P.O., Zelter A., Wordeman L., Asbury C.L., Davis T.N. 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10:407–414 10.1038/ncb1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes G.J., Dong Y., McEwen B.F., Deluca J.G. 2008. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr. Biol. 18:1778–1784 10.1016/j.cub.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16:365–371 10.1016/j.chembiol.2009.02.012 [DOI] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362 10.1242/jcs.00645 [DOI] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Kerres A., Vietmeier-Decker C., Ortiz J., Karig I., Beuter C., Hegemann J., Lechner J., Fleig U. 2004. The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol. Biol. Cell. 15:5255–5267 10.1091/mbc.E04-06-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.M., Nicklas R.B. 2000. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 113:3815–3823 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Renduchitala K., Khodjakov A., Kapoor T.M. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237 10.1038/ncb1102 [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., Rosasco S.E., Barrett-Wilt G.A., Shabanowitz J., Hunt D.F., Walczak C.E., Stukenberg P.T. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nekrasov V.S., Smith M.A., Peak-Chew S., Kilmartin J.V. 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:4931–4946 10.1091/mbc.E03-06-0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T.J. 2004. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 15:2895–2906 10.1091/mbc.E04-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kotwaliwale C.V., Tatsutani S.Y., Breed C.A., Biggins S. 2006. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26:2648–2660 10.1128/MCB.26.7.2648-2660.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka M.R., Zhang W., Costa P., Archambault V., D’Avino P.P., Lilley K.S., Laue E.D., McAinsh A.D., Glover D.M. 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One. 2:e478 10.1371/journal.pone.0000478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U. 2002. Bi-orienting chromosomes on the mitotic spindle. Curr. Opin. Cell Biol. 14:365–371 10.1016/S0955-0674(02)00328-9 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Lamond A.I. 2006. Mitotic phosphatases: no longer silent partners. Curr. Opin. Cell Biol. 18:623–631 10.1016/j.ceb.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andrews P.D., Wickramasinghe S., Sleeman J., Prescott A., Lam Y.W., Lyon C., Swedlow J.R., Lamond A.I. 2003. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol. Biol. Cell. 14:107–117 10.1091/mbc.E02-07-0376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andersen J., Lam Y.W., Moorhead G., Mann M., Lamond A.I. 2006. Repo-Man recruits PP1γ to chromatin and is essential for cell viability. J. Cell Biol. 172:679–692 10.1083/jcb.200508154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K.G. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn M.P., Wolters D., Yates J.R., III 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- Welburn J.P.I., Vleugel M., Liu D., Yates J.R., Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. In press [DOI] [PMC free article] [PubMed] [Google Scholar]