Abstract
Background
The extracellular promastigote and the intracellular amastigote stages alternate in the digenetic life cycle of the trypanosomatid parasite Leishmania. Amastigotes develop inside parasitophorous vacuoles of mammalian phagocytes, where they tolerate extreme environmental conditions. Temperature increase and pH decrease are crucial factors in the multifactorial differentiation process of promastigotes to amastigotes. Although expression profiling approaches for axenic, cell culture- and lesion-derived amastigotes have already been reported, the specific influence of temperature increase and acidification of the environment on developmental regulation of genes has not been previously studied. For the first time, we have used custom L. infantum genomic DNA microarrays to compare the isolated and the combined effects of both factors on the transcriptome.
Results
Immunofluorescence analysis of promastigote-specific glycoprotein gp46 and expression modulation analysis of the amastigote-specific A2 gene have revealed that concomitant exposure to temperature increase and acidification leads to amastigote-like forms. The temperature-induced gene expression profile in the absence of pH variation resembles the profile obtained under combined exposure to both factors unlike that obtained for exposure to acidification alone. In fact, the subsequent fold change-based global iterative hierarchical clustering analysis supports these findings.
Conclusions
The specific influence of temperature and pH on the differential regulation of genes described in this study and the evidence provided by clustering analysis is consistent with the predominant role of temperature increase over extracellular pH decrease in the amastigote differentiation process, which provides new insights into Leishmania physiology.
Background
The life cycle of the trypanosomatid parasite Leishmania is digenetic because it is developed in two distinct hosts. Promastigote is the extracellular stage and differentiates inside the gut of female phlebotominae sand-fly vectors, which then transmit the parasite to the definitive mammalian host during blood meal intakes [1]. Once inside the dermis, some promastigotes interact with phagocytes and are internalised in parasitophorous vacuoles (phagolysosomes), where they differentiate into the intracellular amastigote stage and multiply [2,3]. Amastigotes are released and infect other phagocytes when the host cell collapses. Remarkable features of the new harsh environment are acidic pH (4.5-5.5) and the physiological temperature of the mammalian host (32-37°C).
Phagolysosomal conditions can be mimicked in vitro to grow axenic cultures of the amastigote stage. However, there is not agreement about the equivalence of these forms to amastigotes obtained from their natural environment. In fact, axenic amastigotes are considered as amastigote-like forms (AL) by several authors (e.g. [4,5]), as they show slightly different features from those of amastigotes obtained from host cells. In vitro research supported that concomitant exposure to elevated temperatures and acidic pH triggers differentiation of promastigotes to amastigotes [6,7]. Specifically, this could be achieved by combining pH 5.5 and 37°C in the presence of 5-7% CO2 [6] or at pH 4.5 and 37°C [8] in a host-free medium. Leishmania promastigotes also cope with temperature increase in the absence of pH variation and vice versa [9]. The isolated effects of each factor also induce developmentally regulated changes in the shape and gene expression of promastigotes, but neither of these environmental conditions alone leads to a complete differentiation of promastigotes to amastigotes. Moreover, there is no agreement about the effect of temperature increase. On the one hand, it has been reported that this factor stimulates the entry of promastigotes into stationary phase [10], whereas Shapira et al. [9] on the other hand, observed a different effect with both light and scanning electron microscopy: cell shape was round resembling amastigotes but the flagellum still clearly emerged from the cellular body. Regarding the effect of extracellular pH decrease in the absence of temperature variation, it has been stated that generation time increases and a specific protein of the amastigote stage is expressed under these conditions [11] and that acidification itself leads to the differentiation of promastigotes to metacyclic forms in 48 h; these cells then differentiate to amastigotes but only when the temperature is increased [12].
A descriptive differentiation sequence of promastigotes to amastigotes has been proposed: (1) differentiation signal, 0-4 h; (2) disappearance of cell motility, G1 arrest and aggregation, 5-9 h; (3) change of shape, 10-24 h; and (4) completion of subsequent differentiation processes, 25-120 h. The adaptations necessary for survival in the new harsh conditions inside the host cell are mainly due to gene expression modulation. The expression profiles of several genes during this complex differentiation process have been studied. For instance, the A2 gene is up-regulated in the first step, as well as an amastigote-specific proline transporter in the last step. In contrast, 3'-nucleotidase/nuclease (3'NT/Nase) is down-regulated and pentavalent antimonial resistance decreases, presumably due to sodium stibogluconate-resistance protein (SbGRP) expression down-regulation in the same step (reviewed in [6,7,11,13,14]). In addition, partial gene expression profiling of L. major, L. mexicana, L. infantum and L. donovani amastigotes (axenic and lesion-derived) with respect to promastigotes has been reported [15,19]. However, the effects on the transcriptome of particular factors that influence differentiation in vivo (mainly temperature increase and pH decrease) have not been studied to date. So in this study we have analysed, for the first time, the concomitant (TPS) and the isolated effects of temperature and pH shift (respectively, TS and PS) relative to control promastigote culture conditions (CC) on the transcriptome of L. infantum by custom genomic DNA microarrays.
TPS-treated promastigotes differentiate to AL with regard to the up-regulation of the amastigote-specific A2 gene and the absence of promastigote-specific glycoprotein 46 (gp46) expression as verified by indirect immunofluorescence assay (IFA). In addition, the up-regulation of several amastin genes and the down-regulation of 3'NT/Nase and SbGRP genes under TPS and TS is in agreement with previous data (reviewed in [13]). None of these genes have been found to be differentially regulated under PS. As a consequence, TPS-treated cells are AL and TS-treated ones are also progressing towards amastigote differentiation but PS-treated cells do not seem to undergo the same differentiation process. After performing IFA, transcriptome analysis was carried out and a large set of genes differentially regulated by the effect of both factors was found. A broader analysis of their influence on differentiation at the gene expression modulation level by multi-experimental Serial Analysis of Microarrays (SAM) and iterative hierarchical clustering analysis (HCL-ST) of genes with respect to their expression modulation has led us to conclude that temperature increase has a greater influence than pH decrease on the differentiation process of promastigotes to amastigotes.
Results and Discussion
Cell growth, gp46-IFA and microarray hybridisation analysis and validation
Growth curves of promastigotes cultured under CC (from the mid-logarithmic to the early stationary phase), TPS, TS and PS conditions are represented in Figure 1. Proliferation decrease is more noticeable under PS conditions than under TPS and TS. Therefore, pH decrease inhibits proliferation of parasites at both 37°C and 27°C, which is in agreement with previous findings [11]. Consequently, TPS-treated promastigotes show more pronounced proliferation detention than TS due to the effect of acidification (Figure 1). Taken together, these data are consistent with cell proliferation arrest during the differentiation process leading up to the amastigote stage, after which mature amastigotes are able to multiply.
Figure 1.
Average growth curves of control and temperature/pH-treated L. infantum promastigotes. Three replicates of the cultures were performed for each of the conditions assayed. RNA samples were extracted and processed for transcriptome analysis on day 4. Growth arrest is induced by pH decrease.
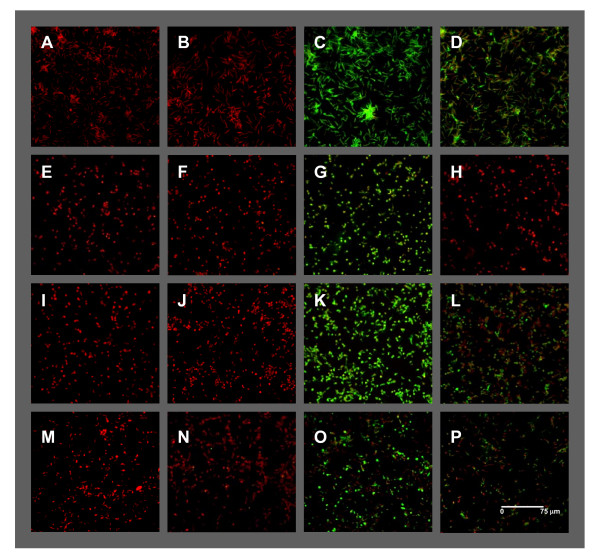
Surface glycoprotein gp46 is known to be promastigote-specific. In fact, it is also called promastigote surface antigen 2 (PSA2) [20]. This glycoprotein has not been detected in amastigotes, although transcripts have been detected at this stage [21]. We have used a monoclonal antibody against gp46 in IFA to assess its expression under CC, TPS, TS and PS conditions, and the absence of gp46 expression can only be observed in the case of TPS (Figure 2). These findings provide evidence for an AL stage after 4 days of TPS exposure. Consequently, TPS-treated cells undergo a more intensive differentiation process leading up to AL than TS and PS-treated cells.
Figure 2.
gp46 IFA. Samples of all the experimental conditions described in this article were collected on day 4 for IFA analysis. (A-D) CC; (E-H) TPS; (I-L) TS; and (M-P) PS. Incubations were performed with: PBS as negative control for the FITC-conjugated anti-mouse IgG secondary antibody (A, E, I, M); monoclonal anti-rabbit complement factor H primary antibody negative control (B, F, J, N); SIM110 monoclonal anti-SLA as positive control (C, G, K, O); and monoclonal anti-gp46 (D, H, L, P). As a summary, gp46 is expressed under CC, TS and PS but not in TPS-treated AL.
Total RNA was extracted and its integrity and absence of DNA contamination were checked by capillary electrophoresis in samples obtained on day 4 (Additional file 1). After mRNA amplification, cDNA was synthesised and labelled with Cy3 for CC and with Cy5 for each of the conditions assayed. DNA microarray hybridisations with these cDNA samples (TPS vs. CC, TS vs. CC and PS vs. CC) were carried out in triplicate. Subsequently, local background was substracted and raw data were normalized and t-test performed for three replicates. A total of 225 spots for TPS, 102 for TS and 117 for PS vs. CC were selected as they fulfilled the following selection criteria: (i) F ≥ 1.7 (Cy5/Cy3 ratio if Cy5 > Cy3) or ≤ -1.7 (-Cy3/Cy5 ratio if Cy3 > Cy5), (ii) total relative fluorescence intensity value > 5000 FU and (iii) p < 0.05 (Additional file 2). Clones corresponding to selected spots had their insert ends sequenced and were mapped against the L. infantum genome to identify overlapping genes. Normalized and contrasted microarray hybridisation results of those clones that contain known annotated genes are described in Tables 1 and 2 for TPS, 3 and 4 for TS and 5 for PS. Hypothetical and unknown genes found to be regulated differentially are described in (Additional file 3: Table S1, S2, S3, S4, S5 and S6), as well as clones that map against minicircle sequences. Gene expression data obtained by microarray hybridisation assays were validated by relative quantitative real time PCR (qRT-PCR) in 15% clones (12% genes excluding redundancy expected in a shotgun microarray strategy). Molecular function GO annotations are indicated in Tables 1, 2, 3, 4 and 5 in order to relate differentially regulated genes with direct acyclic graphs (DAGs) (Additional file 4) and molecular function multilevel sector charts (Figure 3). Once the individual effect of each factor on the transcriptome was analysed, a multi-experiment comparison (SAM) was performed to determine if there were statistically significant differences between PS, TS and TPS expression profiles for each of the differentially regulated genes found. Finally, an HCL-ST analysis including control spots allowed us to determine the relative distance between the experimental groups: TS is closer than PS to the TPS profile (Figure 4).
Table 1.
Up-regulated genes after 37°C/pH4.5 treatment (day 4) in L. infantum.
| Clone | F | Log2F ± SD | p | GenBank | e-value | Def. | Id. | Annotated Gene Function (GO terms in Figure S3., additional file 4) | qRT-PCR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GSS | Fw | Rv | +/- | F ± SD | |||||||
| Lin11D7 | 4.78 | 2.3 ± 0.2 | 0.002 | GS598854 | 6e-118 | 0 | b | LinJ31_V3.0460 | Amastin, putative (uTPS0) | + | 7.8 ± 0.2 |
| Lin19D1 | 1.88 | 0.9 ± 0.3 | 0.028 | GS598855 | 3e-18 | 0 | b | LinJ08_V3.0680 | Amastin-like protein (uTPS0) | N.D. | |
| LinJ08_V3.0690 | Amastin-like protein (uTPS0) | N.D. | |||||||||
| Lin22E12 | 2.79 | 1.5 ± 0.1 | 0.001 | GS598856 | 0 | 0 | b | LinJ31_V3.1850 | Amino acid permease (uTPS13) | N.D. | |
| Lin33G2 | 4.01 | 2.0 ± 0.8 | 0.044 | GS598857 | 6e-118 | 6e-118 | b | LinJ34_V3.4370 | Amastin-like surface protein, putative (uTPS0) | N.D. | |
| Lin34G1 | 1.81 | 0.9 ± 0.1 | 0.008 | GS598858 | 0 | 0 | a | LinJ16_V3.0790 | Chitinase (uTPS18, uTPS8) | N.D. | |
| Lin36B8 | 1.99 | 1.0 ± 0.2 | 0.015 | GS598859 | 0 | 0 | b | LinJ30_V3.3230 | 3-hydroxy-3-methyglutaryl-CoA reductase, putative (uTPS21, uTPS12) | N.D. | |
| Lin50G2 | 3.38 | 1.8 ± 0.1 | 0.000 | GS598860 | 0 | 4e-153 | b | LinJ34_V3.2660 | Amastin-like surface protein (uTPS0) | + | 1.8 ± 0.0 |
| Lin54G3 | 1.84 | 0.9 ± 0.3 | 0.027 | GS598861 | 0 | 0 | b | LinJ24_V3.1230 | Hypothetical protein, conserved | N.D. | |
| LinJ24_V3.1240 | Translation factor SUI1, putative (uTPS0) | + | 3.1 ± 0.1 | ||||||||
| Lin62D3 | 1.92 | 0.9 ± 0.4 | 0.040 | GS598862 | 0 | 0 | b | LinJ05_V3.0340 | Hypothetical protein, conserved | N.D. | |
| LinJ05_V3.0350 | Trypanothione reductase (uTPS10, uTPS14, uTPS20) | + | 3.8 ± 0.4 | ||||||||
| Lin62D10 | 1.76 | 0.8 ± 0.3 | 0.051 | GS598863 | 0 | 0 | b | LinJ17_V3.1150 | Esterase-like protein (uTPS5) | + | 18.5 ± 1.5 |
| Lin66A8 | 3.59 | 1.8 ± 0.4 | 0.013 | GS598864 | 0 | 0 | b | LinJ22_V3.0470 | Hypothetical protein, conserved | N.D. | |
| LinJ22_V3.0480 | Ubiquitin-conjugating enzyme-like protein (uTPS0) | + | 2.3 ± 0.2 | ||||||||
| Lin66F8 | 1.92 | 0.9 ± 0.2 | 0.017 | GS598865 | 0 | 3e-132 | a | LinJ33_V3.2470 | Succinyl-CoA:3-ketoacid-CoA transferase, mitochondrial precursor, putative (3-oxoacid-CoA transferase) | - | -1.3 ± 0.3 |
| LinJ33_V3.2480 | Hypothetical protein, conserved/RABreg (uTPS17) | N.D. | |||||||||
| Lin77H8 | 5.63 | 2.5 ± 0.5 | 0.016 | GS598866 | 0 | 0 | b | LinJ08_V3.0690 | Amastin-like protein (uTPS0) | N.D. | |
| Lin86H7 | 3.06 | 1.6 ± 0.2 | 0.004 | GS598867 | 0 | 2e-101 | b | LinJ08_V3.0700 | Amastin-like protein (uTPS0) | + | 9.5 ± 0.3 |
| Lin87H2 | 4.20 | 2.1 ± 0.3 | 0.008 | GS598868 | 3e-15 | 3e-33 | b | LinJ08_V3.0690 | Amastin-like protein (uTPS0) | N.D. | |
| Lin89D9 | 1.71 | 0.8 ± 0.3 | 0.043 | GS598869 | 0 | 0 | b | LinJ21_V3.0770 | ATP-binding cassette sub-family E, putative (uTPS9, uTPS11, uTPS24, uTPS28) | N.D. | |
| Lin90B6 | 1.71 | 0.8 ± 0.3 | 0.040 | GS598870 | 0 | 0 | b | LinJ30_V3.0640 | Ribosome biogenesis regulatory protein (RRS1), putative (uTPS0) | + | 16.4 ± 0.2 |
| LinJ30_V3.0650 | Histidyl-tRNA synthetase, putative | N.D. | |||||||||
| LinJ30_V3.0660 | Hypothetical protein, conserved | N.D. | |||||||||
| Lin90H2 | 1.76 | 0.8 ± 0.1 | 0.005 | GS598871 | 0 | 0 | b | LinJ30_V3.2200 | RNA-binding protein (uTPS3, uTPS6, uTPS15, uTPS16) | N.D. | |
| Lin91B12 | 5.24 | 2.4 ± 0.3 | 0.004 | GS598872 | 0 | 0 | b | LinJ34_V3.2660 | Amastin-like surface protein (uTPS0) | + | 1.8 ± 0.0 |
| Lin92H5 | 2.48 | 1.3 ± 0.5 | 0.041 | GS598873 | 0 | 0 | b | LinJ28_V3.2060 | Zinc transporter, putative (uTPS23) | + | 45.7 ± 0.5 |
| LinJ28_V3.2070 | Hypothetical protein, conserved | N.D. | |||||||||
| Lin93E3 | 1.83 | 0.9 ± 0.4 | 0.033 | 0 | 0 | b | LinJ10_V3.0410 | Pteridine transporter ft3, putative (uTPS0) | N.D. | ||
| Lin104C10 | 6.68 | 2.7 ± 0.1 | 0.001 | GS598874 | 0 | 0 | b | LinJ08_V3.1320 | Amastin-like protein (uTPS0) | N.D. | |
| Lin106A1 | 2.43 | 1.3 ± 0.0 | 0.000 | GS598875 | 0 | 0 | c | LinJ06_V3.1200 | Hypothetical protein, conserved | N.D. | |
| LinJ31_V3.0590 | Amino acid transporter aATP11, putative (uTPS13) | + | 2.6 ± 0.0 | ||||||||
| Lin113C3 | 1.87 | 0.9 ± 0.1 | 0.040 | GS598876 | 3e-74 | 0 | a | LinJ14_V3.1440 | Pteridine transporter (uTPS0) | + | 1.9 ± 0.0 |
| LinJ14_V3.1450 | Myo-inositol-1-phosphate synthase (uTPS0) | + | 2.1 ± 0.3 | ||||||||
| Lin118A11 | 2.90 | 1.5 ± 0.3 | 0.010 | GS598877 | 0 | 7e-28 | c | LinJ30_V3.0630 | Nitrate reductase, putative (uTPS0) | + | 3.7 ± 0.3 |
| LinJ36_V3.2480 | Glyceraldehyde-3-phosphate dehydrogenase, putative | N.D. | |||||||||
| Lin119F3 | 3.40 | 1.8 ± 0.2 | 0.005 | GS598878 | 0 | 0 | b | LinJ25_V3.2570 | Phosphoglycan beta-1,3-galactosyltransferase 6 (uTPS22) | N.D. | |
| Lin123D6 | 2.91 | 1.5 ± 0.1 | 0.002 | GS598879 | - | 0 | c | LinJ34_V3.2660 | Amastin-like surface protein (uTPS0) | + | 1.8 ± 0.0 |
| Lin137A10 | 1.98 | 1.0 ± 0.3 | 0.039 | GS598880 | 0 | 0 | b | LinJ24_V3.1230 | Hypothetical protein, conserved | N.D. | |
| LinJ24_V3.1240 | Translation factor SUI1, putative (uTPS0) | + | 3.1 ± 0.1 | ||||||||
| Lin142H9 | 1.74 | 0.8 ± 0.1 | 0.004 | GS598881 | 0 | 0 | b | LinJ31_V3.0460 | Amastin, putative (uTPS0) | + | 7.8 ± 0.2 |
| Lin146E3 | 2.35 | 1.2 ± 0.4 | 0.038 | GS598882 | 0 | 0 | b | LinJ31_V3.0590 | Amino acid transporter aATP11, putative (uTPS13) | + | 2.6 ± 0.0 |
| Lin156B2 | 1.82 | 0.9 ± 0.2 | 0.025 | GS598883 | 0 | 0 | b | LinJ33_V3.2960 | Hypothetical protein, conserved/Transcription regulator (uTPS 1, uTPS4, uTPS7) | N.D. | |
| Lin162A9 | 4.29 | 2.1 ± 0.2 | 0.004 | GS598884 | 0 | 0 | b | LinJ22_V3.0470 | Hypothetical protein, conserved | N.D. | |
| LinJ22_V3.0480 | Ubiquitin-conjugating enzyme-like protein (uTPS0) | + | 2.3 ± 0.2 | ||||||||
| Lin165E2 | 3.48 | 1.8 ± 0.2 | 0.004 | GS598885 | 0 | 0 | b | LinJ22_V3.0680 | 3'a2rel-related protein (uTPS0) | + | 4.0 ± 0.4 |
| Lin183A3 | 1.75 | 0.8 ± 0.1 | 0.010 | GS598886 | 0 | 0 | b | LinJ24_V3.2250 | Hypothetical protein, conserved/GPDE (uTPS26) | N.D. | |
| Lin185A12 | 4.53 | 2.2 ± 0.2 | 0.002 | GS598887 | 0 | 0 | b | LinJ34_V3.2660 | Amastin-like surface protein, putative (uTPS0) | + | 1.8 ± 0.0 |
| Lin188H2 | 3.20 | 1.7 ± 0.6 | 0.042 | GS598888 | 0 | 0 | c | LinJ08_V3.0680 | Amastin-like protein (uTPS0) | N.D. | |
| Lin194E2 | 3.22 | 1.7 ± 0.4 | 0.023 | GS598889 | 7e-56 | 4e-153 | b | LinJ08_V3.0710 | Amastin-like protein (uTPS0) | + | 9.5 ± 0.3 |
| Lin197A12 | 1.95 | 1.0 ± 0.2 | 0.016 | GS598890 | 0 | 0 | a | LinJ31_V3.2540 | Lipase, putative (uTPS19) | N.D. | |
| Lin201F8 | 2.00 | 1.0 ± 0.4 | 0.041 | GS598891 | 0 | 0 | b | LinJ31_V3.3330 | Phosphoglycan beta-1,3-galactosyltransferase 5 (uTPS22) | N.D. | |
| Lin206B6 | 5.40 | 2.4 ± 0.5 | 0.012 | GS598892 | 7e-133 | 0 | b | LinJ22_V3.0680 | 3'a2rel-related protein (uTPS0) | + | 4.0 ± 0.4 |
| Lin210C4 | 2.77 | 1.5 ± 0.1 | 0.003 | GS598893 | 0 | 0 | b | LinJ08_V3.0690 | Amastin-like protein (uTPS0) | N.D. | |
| Lin223F2 | 1.73 | 0.8 ± 0.3 | 0.044 | GS598894 | 0 | 0 | b | LinJ13_V3.0330 | Unknown/Tubulin associated GTPase (uTPS2, uTPS25, uTPS27) | N.D. | |
| Lin224G2 | 2.20 | 1.1 ± 0.2 | 0.023 | GS598895 | 0 | 0 | b | LinJ08_V3.0720 | Amastin-like protein (uTPS0)0 | N.D. | |
| Lin235G8 | 3.16 | 1.7 ± 0.2 | 0.003 | GS598896 | 0 | 0 | b | LinJ08_V3.1320 | Amastin-like protein (uTPS0) | N.D. | |
| Lin245E2 | 2.61 | 1.4 ± 0.4 | 0.040 | GS598897 | 0 | 0 | b | LinJ22_V3.0680 | 3'a2rel-related protein (uTPS0) | + | 4.0 ± 0.4 |
| Lin267D9 | 2.06 | 1.0 ± 0.2 | 0.010 | GS598898 | 9e-111 | 0 | b | LinJ16_V3.0590 | Carbamoyl-phosphate synthetase, putative (uTPS0) | + | 2.9 ± 0.4 |
| LinJ16_V3.0600 | Histone H3, putative | - | 1.3 ± 0.2 | ||||||||
| Lin274G6 | 5.77 | 2.5 ± 0.2 | 0.003 | GS598899 | 0 | 0 | b | LinJ08_V3.0680 | Amastin-like protein (uTPS0) | N.D. | |
| LinJ08_V3.0690 | Amastin-like protein (uTPS0) | N.D. | |||||||||
| Lin275A8 | 2.72 | 1.4 ± 0.2 | 0.006 | GS598900 | 0 | 4e-168 | b | LinJ08_V3.0720 | Amastin-like protein (uTPS0) | N.D. | |
| Lin276B6 | 1.76 | 0.8 ± 0.3 | 0.041 | GS598901 | 0 | 0 | b | LinJ31_V3.2540 | Lipase, putative (uTPS19) | N.D. | |
| Lin294A11 | 4.86 | 2.3 ± 0.1 | 0.001 | GS598902 | 0 | 0 | b | LinJ08_V3.1320 | Amastin-like protein (uTPS0) | N.D. | |
| Lin295D9 | 4.40 | 2.1 ± 0.1 | 0.000 | GS598903 | 0 | 0 | b | LinJ34_V3.1720 | Amastin-like surface protein, putative (uTPS0) | N.D. | |
| Lin310F2 | 2.34 | 1.2 ± 0.5 | 0.046 | GS598904 | 0 | 0 | b | LinJ23_V3.1220 | Hydrophilic surface protein (HASPB) (uTPS0) | N.D. | |
| cLinA2 | 6.45 | 2.7 ± 0.1 | 0.000 | S69693 | - | - | - | - | L. infantum A2 gene -- DNA microarray control spot | ||
| cLdoA2 | 2.51 | 1.3 ± 0.3 | 0.021 | - | - | - | - | - | L. donovani A2 gene -- DNA microarray control spot | ||
This table contains clones that map against up-regulated genes (not hypothetical or unknown) with the combined effect of temperature increase and pH decrease (TPS). The features described are: clone number; F; base-two logarithmic scale F and SD values; p; GenBank GSS accession numbers; e-values; Def. according to mapping outcomes a, b or c (see brief explanation in the text); Id.; annotated gene function (codes for Additional file 4: Figure S3,); qRT-PCR. When a given clone overlaps with more than one annotation, stage-specific regulation is only demonstrated if the qRT-PCR result is positive (+). Genes in bold are also up-regulated under TS.
Table 2.
Down-regulated genes after 37°C/pH4.5 treatment (day 4) in L. infantum.
| Clone | F | Log2F ± SD | P | GenBank GSS | e-value | Def. | Id. | Annotated Gene Function (GO terms in Figure S4., additional file 4) | qRT-PCR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fw | Rv | +/- | F ± SD | ||||||||
| Lin1G8 | -1.80 | -0.8 ± 0.1 | 0.009 | GS598905 | 5e-35 | 8e-31 | B | LinJ22_V3.1340 | Serine/threonine protein phosphatase, putative (dTPS6) | N.D. | |
| Lin4F4 | -2.27 | -1.2 ± 0.3 | 0.016 | GS598906 | 0 | 0 | a | LinJ31_V3.0430 | Cysteine peptidase, Clan CA, family C2, putative (dTPS0) | + | -3.3 ± 0.1 |
| Lin9B9 | -1.71 | -0.8 ± 0.2 | 0.015 | GS598907 | 5e-26 | 0 | a | LinJ36_V3.1010 | Dynein heavy chain, putative (dTPS0) | N.D. | |
| Lin15D6 | -2.05 | -1.0 ± 0.2 | 0.042 | GS598908 | 0 | 0 | a | LinJ31_V3.0610 | Amino acid transporter aATP11, putative (dTPS0) | N.D. | |
| Lin22B1* | -2.03 | -1.0 ± 0.2 | 0.016 | GS598909 | 0 | 0 | a | LinJ23_V3.1400 | Coronin, putative (dTPS0) | + | -4.9 ± 0.7 |
| Lin21H10 | -1.90 | -0.9 ± 0.1 | 0.001 | GS598910 | 0 | 0 | b | LinJ26_V3.1670 | Sphingolipid delta-4 desaturase, putative (dTPS22) | N.D. | |
| Lin24E10* | -1.80 | -0.8 ± 0.3 | 0.038 | GS598911 | 0 | 0 | b | LinJ22_V3.1310 | I/6 autoantigen-like protein (dTPS0) | + | -6.7 ± 0.9 |
| Lin27B2 | -1.80 | -0.8 ± 0.3 | 0.034 | GS598912 | 0 | - | c | LinJ35_V3.1230 | Short chain dehydrogenase, putative (dTPS1, dTPS7) | + | -3.2 ± 0.7 |
| Lin28C5 | -1.81 | -0.9 ± 0.1 | 0.005 | GS598913 | 0 | 2e-154 | b | LinJ26_V3.1670 | Sphingolipid delta-4 desaturase, putative (dTPS22) | N.D. | |
| Lin31H9* | -1.94 | -1.0 ± 0.1 | 0.006 | GS598914 | 0 | 0 | b | LinJ26_V3.1000 | Dynein heavy chain, putative (dTPS0) | + | -6.2 ± 0.8 |
| Lin36A9* | -2.15 | -1.1 ± 0.2 | 0.009 | GS598915 | 0 | 0 | b | LinJ26_V3.1000 | Dynein heavy chain, putative (dTPS0) | + | -6.2 ± 0.8 |
| Lin40G12* | -1.92 | -0.9 ± 0.2 | 0.013 | GS598916 | 2e-161 | 0 | b | LinJ23_V3.1560 | Lathosterol oxidase-like protein (dTPS28, dTPS30) | + | -14.3 ± 1.7 |
| Lin47D8 | -4.00 | -2.0 ± 0.5 | 0.023 | GS598917 | 0 | 0 | a | LinJ06_V3.1330 | Coproporphyrinogen III oxidase, putative (dTPS33) | + | -5.8 ± 0.1 |
| LinJ06_V3.1340 | Protoporphyrinogen oxidase-like protein (dTPS13, dTPS32) | + | -2.2 ± 0.4 | ||||||||
| Lin49B7 | -4.38 | -2.1 ± 0.0 | 0.000 | GS598918 | 0 | 4e-64 | a | LinJ34_V3.4160 | Phosphatidylinositol-3-kinase (tor2)-like protein (dTPS0) | N.D. | |
| Lin50H7 | -2.32 | -1.2 ± 0.1 | 0.005 | GS598919 | 7e-164 | 0 | b | LinJ28_V3.2380 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase-like protein (dTPS9, dTPS35) | + | -3.4 ± 0.3 |
| LinJ28_V3.2390 | Cyclin dependent kinase-binding protein, putative (dTPS0) | + | -127.4 ± 7.4 | ||||||||
| Lin60B1* | -3.84 | -1.9 ± 0.1 | 0.001 | GS598920 | 0 | 0 | c | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| Lin60E5 | -1.80 | -0.8 ± 0.3 | 0.035 | GS598921 | 0 | 0 | b | LinJ26_V3.0970 | Hypothetical protein, conserved/HPB (dTPS5, dTPS16) | N.D. | |
| Lin63F3 | -2.95 | -1.6 ± 0.4 | 0.018 | GS598922 | 0 | 0 | b | LinJ36_V3.6550 | Glucose transporter lmgt2, putative (dTPS47) | + | -8.1 ± 1.1 |
| LinJ36_V3.6560 | Glucose transporter, putative (dTPS47) | + | -6.3 ± 1.4 | ||||||||
| Lin66F10 | -2.18 | -1.1 ± 0.0 | 0.000 | GS598923 | 0 | 0 | b | LinJ36_V3.6550 | Glucose transporter lmgt2, putative (dTPS47) | + | -8.1 ± 1.1 |
| LinJ36_V3.6560 | Glucose transporter, putative (dTPS47) | + | -6.3 ± 1.4 | ||||||||
| Lin80B3 | -2.06 | -1.0 ± 0.3 | 0.024 | GS598924 | 0 | 0 | b | LinJ28_V3.3250 | Glucose-6-phosphate-N-acetyltransferase, putative (dTPS46) | N.D. | |
| Lin82C6 | -1.74 | -0.8 ± 0.3 | 0.038 | GS598925 | 0 | - | c | LinJ31_V3.0440 | Cysteine peptidase, Clan CA, family C2, putative (dTPS0) | + | -3.3 ± 0.1 |
| Lin84E8 | -2.26 | -1.2 ± 0.2 | 0.007 | GS598926 | 0 | 0 | a | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| LinJ31_V3.2380 | 3'-nucleotidase/nuclease precursor, putative (dTPS4, dTPS29) | + | -2.7 ± 0.6 | ||||||||
| Lin86H3 | -2.17 | -1.1 ± 0.1 | 0.004 | GS598927 | 0 | 8e-130 | b | LinJ31_V3.0950 | Sodium stibogluconate-resistance protein, putative (dTPS) | N.D. | |
| Lin89F9 | -2.93 | -1.6 ± 0.4 | 0.026 | GS598928 | 0 | 0 | b | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| LinJ31_V3.2380 | 3'nucleotidase/nuclease precursor, putative (dTPS4, dTPS29) | + | -2.7 ± 0.6 | ||||||||
| LinJ31_V3.2390 | Helicase-like protein | N.D. | |||||||||
| Lin92D7* | -2.27 | -1.2 ± 0.2 | 0.005 | GS598929 | 0 | 0 | b | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| LinJ31_V3.2380 | 3'-nucleotidase/nuclease precursor, putative (dTPS4, dTPS29) | + | -2.7 ± 0.6 | ||||||||
| Lin92G9 | -2.29 | -1.2 ± 0.1 | 0.004 | GS598930 | 0 | 0 | a | LinJ06_V3.1320 | Pteridine transporter, putative (dTPS0) | + | -2.1 ± 0.3 |
| LinJ06_V3.1330 | Coproporphyrinogen III oxidase, putative (dTPS33) | + | -5.8 ± 0.1 | ||||||||
| Lin97D1 | -11.57 | -3.5 ± 0.2 | 0.001 | GS598931 | 0 | 0 | a | LinJ06_V3.1320 | Pteridine transporter, putative (dTPS0) | + | -2.1 ± 0.3 |
| Lin97F6 | -1.80 | -0.8 ± 0.3 | 0.029 | GS598932 | 0 | 0 | b | LinJ26_V3.0460 | Hypothetical protein, conserved | N.D. | |
| Lin98C7* | -2.12 | -1.1 ± 0.5 | 0.044 | GS598933 | 0 | 0 | b | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| LinJ31_V3.2380 | 3'nucleotidase/nuclease precursor, putative (dTPS4, dTPS29) | + | -2.7 ± 0.6 | ||||||||
| Lin98F9 | -2.43 | -1.3 ± 0.3 | 0.016 | GS598934 | 2e-190 | 1e-101 | b | LinJ32_V3.3120 | Minichromosome maintenance (MMC) complex subunit, putative (dTPS8, dTPS38, dTPS48) | N.D. | |
| Lin98G10* | -2.13 | -1.1 ± 0.2 | 0.015 | GS598935 | 0 | 0 | b | LinJ30_V3.2780 | Superoxide dismutase, putative (dTPS0) | + | -3.7 ± 0.1 |
| Lin101B5 | -2.62 | -1.4 ± 0.3 | 0.011 | GS598936 | 0 | 0 | b | LinJ09_V3.0650 | Serine peptidase family S51, peptidase E, putative (dTPS19) | N.D. | |
| Lin105A3 | -2.22 | -1.1 ± 0.1 | 0.005 | GS598937 | 0 | 0 | b | LinJ36_V3.1320 | Fructose-1,6-bisphosphate aldolase (dTPS37) | N.D. | |
| Lin105B9 | -6.13 | -2.6 ± 0.2 | 0.001 | GS598938 | 0 | 0 | a | LinJ06_V3.1320 | Pteridine transporter, putative (dTPS0) | + | -2.1 ± 0.3 |
| Lin109F4 | -3.16 | -1.7 ± 0.5 | 0.024 | GS598939 | 0 | 0 | a | LinJ06_V3.1330 | Coproporphyrinogen III oxidase, putative (dTPS33) | + | -5.8 ± 0.1 |
| LinJ06_V3.1340 | Protoporphyrinogen oxidase-like protein (dTPS13, dTPS32) | + | -2.2 ± 0.4 | ||||||||
| Lin111C2 | -3.14 | -1.7 ± 0.1 | 0.001 | GS598940 | 0 | 0 | b | LinJ09_V3.0650 | Serine peptidase family S51, peptidase E, putative (dTPS19) | N.D. | |
| Lin111F3 | -2.09 | -1.1 ± 0.4 | 0.035 | GS598941 | 0 | 0 | a | LinJ31_V3.2210 | Prostaglandin F2α synthetase (dTPS31) | N.D. | |
| Lin125H5 | -1.95 | -1.0 ± 0.2 | 0.022 | GS598942 | 0 | 0 | b | LinJ36_V3.1590 | Serine/threonine protein kinase, putative (dTPS43) | N.D. | |
| Lin144F11 | -2.28 | -1.2 ± 0.1 | 0.004 | GS598943 | 0 | 0 | a | LinJ31_V3.2210 | Prostaglandin F2α synthetase (dTPS31) | N.D. | |
| Lin144H10 | -1.85 | -0.9 ± 0.1 | 0.007 | GS598944 | 0 | 0 | a | LinJ22_V3.1300 | Cyclophilin, putative (dTPS0) | + | -4.1 ± 0.6 |
| LinJ19_V3.1310 | I/6-autoantigen-like protein (dTPS0) | N.D. | |||||||||
| Lin144H11 | -2.32 | -1.2 ± 0.1 | 0.003 | GS598945 | 0 | 0 | b | LinJ36_V3.2700 | Membrane-bound acid phosphatase precursor (dTPS42) | N.D. | |
| Lin150E4 | -1.86 | -0.9 ± 0.4 | 0.048 | GS598946 | 0 | 0 | b | LinJ13_V3.1060 | Calmodulin, putative (dTPS16) | N.D. | |
| Lin153D1 | -1.80 | -0.8 ± 0.1 | 0.002 | GS598947 | 0 | 0 | b | LinJ27_V3.0530 | Amino acid permease, putative (dTPS26) | N.D. | |
| Lin155H12 | -2.52 | -1.3 ± 0.4 | 0.024 | GS598948 | 0 | 0 | a | LinJ36_V3.0250 | Peptidyl-prolyl cis-trans isomerase, putative (dTPS18, dTPS25) | N.D. | |
| Lin157D8 | -2.78 | -1.5 ± 0.2 | 0.005 | GS598949 | 0 | 0 | b | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| Lin158A10 | -2.40 | -1.3 ± 0.2 | 0.008 | GS598950 | 0 | 0 | b | LinJ23_V3.0870 | Hypothetical protein, conserved | N.D. | |
| Lin177E10 | -1.72 | -0.8 ± 0.3 | 0.042 | GS598951 | 7e-167 | 0 | b | LinJ16_V3.0600 | Histone H3, putative (dTPS8) | N.D. | |
| LinJ16_V3.0610 | Histone H3, putative (dTPS8) | N.D. | |||||||||
| Lin182F2 | -2.18 | -1.1 ± 0.3 | 0.022 | GS598952 | 0 | 0 | b | LinJ25_V3.0740 | Eukaryotic initiation factor 5a, putative (dTPS15) | N.D. | |
| LinJ25_V3.0750 | Eukaryotic initiation factor 5a, putative (dTPS15) | N.D. | |||||||||
| Lin187C10 | -4.02 | -2.0 ± 0.3 | 0.003 | GS598953 | 0 | 0 | b | LinJ06_V3.1320 | Pteridine transporter, putative (dTPS0) | + | -2.1 ± 0.3 |
| Lin193E6 | -2.00 | -1.0 ± 0.4 | 0.040 | GS598954 | 0 | 0 | b | LinJ23_V3.1230 | SHERP (dTPS0) | N.D. | |
| Lin194A4 | -1.70 | -0.8 ± 0.2 | 0.016 | GS598955 | 0 | 1e-11 | b | LinJ22_V3.1270 | Aquaporin, putative (dTPS3) | N.D. | |
| Lin197D2* | -3.32 | -1.7 ± 0.3 | 0.007 | GS598956 | 0 | 0 | b | LinJ07_V3.0150 | Acyl-CoA dehydrogenase, mitochondrial precursor, putative (dTPS21, dTPS27) | + | -2.4 ± 0.2 |
| LinJ07_V3.0170 | Maoc family protein | - | -1.1 ± 0.3 | ||||||||
| Lin206C10* | -1.99 | -1.0 ± 0.3 | 0.032 | GS598957 | 6e-69 | 1e-82 | b | LinJ07_V3.0940 | Cytochrome b5-like protein (dTPS0) | + | -1.7 ± 0.0 |
| Lin206H7* | -1.90 | -0.9 ± 0.2 | 0.015 | GS598958 | 0 | 0 | b | LinJ31_V3.1240 | Vacuolar-type proton translocating pyrophosphatase 1, putative (dTPS0) | + | -3.4 ± 0.7 |
| Lin208F2* | -2.10 | -1.1 ± 0.3 | 0.026 | GS598959 | 2e-86 | 3e-64 | b | LinJ31_V3.1240 | Vacuolar-type proton-translocating pyrophosphatase 1, putative | + | -3.4 ± 0.7 |
| Lin208H10 | -2.68 | -1.4 ± 0.4 | 0.010 | GS598960 | 0 | 0 | a | LinJ18_V3.1070 | Cysteine peptidase, Clan CA, family C2, putative (dTPS0) | + | -3.3 ± 0.1 |
| LinJ18_V3.1080 | Vacuolar protein sorting complex subunit, putative (dTPS 0) | - | -1.2 ± 0.3 | ||||||||
| Lin219A10* | -1.89 | -0.9 ± 0.1 | 0.004 | GS598961 | 0 | 0 | b | LinJ19_V3.0710 | Glycosomal malate dehydrogenase (dTPS1, dTPS30) | + | -2.3 ± 0.0 |
| Lin228D4* | -1.86 | -0.9 ± 0.2 | 0.014 | GS598962 | 0 | 0 | a | LinJ19_V3.0090 | Fibrillarin, putative (dTPS0) | + | -4.1 ± 0.8 |
| Lin229E6 | -9.63 | -3.3 ± 0.2 | 0.002 | GS598963 | 0 | 0 | a | LinJ06_V3.1320 | Pteridine transporter, putative (dTPS0) | + | -2.1 ± 0.3 |
| LinJ06_V3.1330 | Coproporphyrinogen III oxidase, putative (dTPS33) | + | -5.8 ± 0.1 | ||||||||
| Lin231G4 | -1.73 | -0.8 ± 0.2 | 0.013 | GS598964 | 0 | 0 | b | LinJ31_V3.1240 | Vacuolar-type proton translocating pyrophosphatase 1, putative (dTPS0) | + | |
| LinJ31_V3.1250 | Hypothetical protein, unknown function | N.D. | |||||||||
| Lin234C9 | -1.77 | -0.8 ± 0.3 | 0.038 | GS598965 | 0 | 0 | b | LinJ20_V3.1220 | Cysteine peptidase, Clan CA, family C2, putative (dTPS41) | + | -3.3 ± 0.1 |
| Lin242E2 | -2.85 | -1.5 ± 0.4 | 0.025 | GS598966 | 0 | 0 | b | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative (dTPS4, dTPS29) | + | -4.6 ± 0.4 |
| LinJ31_V3.2380 | 3'-nucleotidase/nuclease precursor, putative (dTPS4, dTPS29) | + | -2.7 ± 0.6 | ||||||||
| Lin252B11 | -2.12 | -1.1 ± 0.3 | 0.028 | GS598967 | 0 | 0 | b | LinJ17_V3.170/200 | Elongation factor 1αg(dTPS14, dTPS39, dTPS44p | N.D. | |
| Lin265E2 | -1.89 | -0.9 ± 0.3 | 0.042 | GS598968 | 1e-165 | 0 | b | LinJ29_V3.1880 | Paraflagellar rod protein 1D, putative (dTPS0) | N.D. | |
| Lin270H10* | -1.96 | -1.0 ± 0.2 | 0.010 | GS598969 | 0 | 0 | b | LinJ31_V3.1150 | Monoglyceride lipase, putative (dTPS0) | + | -1.9 ± 0.0 |
| Lin271C2* | -1.9 | -0.9 ± 0.3 | 0.043 | GS598970 | 0 | 0 | b | LinJ28_V3.0090 | Adenylate cyclase-like protein (dTPS9, dTPS12, dTPS23, dTPS24) | + | -2.3 ± 0.0 |
| Lin285H1 | -2.12 | -1.1 ± 0.2 | 0.012 | GS598971 | 0 | 0 | b | LinJ36_V3.6550 | Glucose transporter lmgt2, putative (dTPS47) | + | -8.1 ± 1.1 |
| LinJ36_V3.6560 | Glucose transporter, putative (dTPS47) | + | -6.3 ± 1.4 | ||||||||
| Lin294G4* | -2.00 | -1.0 ± 0.2 | 0.013 | GS598972 | 0 | 0 | b | LinJ31_V3.1640 | Dipthine synthase, putative | - | |
| LinJ31_V3.1660 | Putative 3-ketoacyl-CoA thiolase-like protein (dTPS0) | + | -3.6 ± 0.5 | ||||||||
This table contains clones that map against down-regulated genes (not hypothetical or unknown) with the combined effect of temperature increase and pH decrease (TPS). The features described are: clone number; F; base-two logarithmic scale F and SD values; p; GenBank GSS accession numbers; e-values; Def. according to mapping outcomes a, b or c (see brief explanation in the text); Id.; annotated gene function (codes for Additional file 4: Figure S4,); qRT-PCR. When a given clone overlaps with more than one annotation, stage-specific regulation is only demonstrated if the qRT-PCR result is positive (+). The asterisk indicates that the clone contains more gene sequences that have not been checked by qRT-PCR. Genes in bold are also down-regulated under TS.
Table 3.
Up-regulated genes after temperature increase up to 37°C (day 4) in L. infantum.
| Clone | F | Log2F ± SD | P | GenBank GSS | e-value | Def. | Id. | Annotated Gene Function | qRT-PCR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fw | Rv | +/- | F ± SD | ||||||||
| Lin11D7 | 2.37 | 1.2 ± 0.1 | 0.004 | GS598854 | - | 0 | c | LinJ31_V3.0460 | Amastin, putative | + | |
| Lin17C6 | 1.92 | 0.9 ± 0.1 | 0.006 | GS598973 | 0 | 0 | b | LinJ36_V3.0640 | Delta-8 fatty acid desaturase-like protein | N.D. | |
| Lin19D1 | 1.88 | 0.9 ± 0.3 | 0.028 | GS598855 | 3e-18 | 0 | b | LinJ08_V3.0680 | Amastin-like protein | N.D. | |
| LinJ08_V3.0690 | Amastin-like protein | N.D. | |||||||||
| Lin33G2 | 2.29 | 1.2 ± 0.8 | 0.046 | GS598857 | 6e-118 | 6e-118 | b | LinJ34_V3.4370 | Amastin-like surface protein, putative | N.D. | |
| Lin70F5 | 2.03 | 1.0 ± 0.4 | 0.045 | GS598974 | 0 | 0 | b | LinJ36_V3.7290 | Delta-8 fatty acid desaturase-like protein | N.D. | |
| Lin77H8 | 2.89 | 1.5 ± 0.4 | 0.022 | GS598975 | 3e-175 | 0 | b | LinJ08_V3.0690 | Amastin-like protein | N.D. | |
| Lin86H7 | 2.03 | 1.0 ± 0.2 | 0.005 | GS598867 | 0 | 2e-101 | b | LinJ08_V3.0700 | Amastin-like protein | + | 6.8 ± 0.9 |
| Lin87H2 | 1.89 | 0.9 ± 0.1 | 0.007 | GS598868 | 3e-15 | 3e-33 | b | LinJ08_V3.0690 | Amastin-like protein | N.D. | |
| Lin89D9 | 1.70 | 0.8 ± 0.3 | 0.040 | GS598869 | 0 | 0 | b | LinJ21_V3.0770 | ATP-binding cassette sub-family E, putative | N.D. | |
| Lin90B6 | 1.95 | 1.0 ± 0.3 | 0.032 | GS598976 | 0 | 0 | a | LinJ30_V3.0640 | Ribosome biogenesis regulatoy protein (RRS1), putative | + | 1.9 ± 0.2 |
| LinJ30_V3.0650 | Histidyl-tRNA synthetase, putative | N.D. | |||||||||
| LinJ30_V3.0660 | Hypothetical protein, conserved | N.D. | |||||||||
| Lin91B12 | 1.75 | 0.8 ± 0.1 | 0.003 | GS598872 | 0 | 0 | b | LinJ34_V3.2660 | Amastin-like surface protein | N.D. | |
| Lin100B2 | 1.84 | 0.9 ± 0.3 | 0.034 | GS598977 | 0 | 9e-27 | b | LinJ27_V3.2500 | Glycosomal phosphoenolpyruvate carboxykinase | N.D. | |
| Lin104B11 | 1.77 | 0.8 ± 0.2 | 0.022 | GS598978 | 0 | 0 | b | LinJ04_V3.0570 | Spermidine synthase 1, putative | N.D. | |
| Lin104C10 | 1.82 | 0.9 ± 0.2 | 0.015 | GS598979 | 0 | 0 | b | LinJ08_V3.1320 | Amastin-like protein | N.D. | |
| Lin106A1 | 2.43 | 1.3 ± 0.0 | 0.000 | GS598980 | 0 | 0 | c | LinJ06_V3.1200 | Hypothetical protein, conserved | N.D. | |
| LinJ31_V3.0590 | Amino acid transporter aATP11, putative | + | 2.4 ± 0.3 | ||||||||
| Lin109B3 | 1.89 | 0.9 ± 0.2 | 0.024 | GS598981 | 0 | 0 | b | LinJ21_V3.2130 | Centromere/microtubule binding protein (cbf5), putative | N.D. | |
| Lin113C3 | 2.99 | 1.6 ± 0.3 | 0.010 | GS598876 | 3e-74 | 0 | a | LinJ14_V3.1440 | Pteridine transporter | + | 2.5 ± 0.3 |
| LinJ14_V3.1450 | Myo-inositol-1-phosphate synthase | + | 4.2 ± 0.1 | ||||||||
| Lin137A10 | 1.98 | 1.0 ± 0.3 | 0.039 | GS598982 | 0 | 0 | b | LinJ24_V3.1230 | Hypothetical protein, conserved | N.D. | |
| LinJ24_V3.1240 | Translation factor SUI1, putative | + | 1.8 ± 0.1 | ||||||||
| Lin146E3 | 2.52 | 1.3 ± 0.3 | 0.043 | GS598882 | 0 | 0 | b | LinJ31_V3.0590 | Amino acid transporter aATP11, putative | + | 2.4 ± 0.3 |
| Lin162E6 | 1.92 | 0.9 ± 0.3 | 0.044 | GS598983 | 0 | 0 | a | LinJ14_V3.1430 | Hypothetical protein, conserved | N.D. | |
| LinJ14_V3.1440 | Pteridine transporter | + | 2.5 ± 0.3 | ||||||||
| LinJ14_V3.1450 | Myo-inositol-1-phosphate synthase | + | 4.2 ± 0.1 | ||||||||
| Lin168A2 | 1.87 | 0.9 ± 0.2 | 0.017 | GS598984 | 1e-78 | 0 | b | LinJ22_V3.0670 | Hypothetical protein | N.D. | |
| LinJ22_V3.0680 | 3'a2rel-related protein | + | 3.5 ± 0.6 | ||||||||
| Lin175D6 | 2.20 | 1.2 ± 0.4 | 0.023 | GS598985 | 0 | 0 | b | LinJ31_V3.0460 | Amastin, putative | + | 4.7 ± 1.2 |
| Lin185A10 | 2.04 | 1.0 ± 0.3 | 0.036 | GS598986 | 0 | 0 | a | LinJ28_V3.0620 | MAP kinase, putative | N.D. | |
| Lin185D7 | 1.75 | 0.0 ± 0.2 | 0.020 | GS598987 | 2e-161 | 0 | b | LinJ17_V3.0200 | Elongation factor 1-alpha | N.D. | |
| Lin188H2 | 3.20 | 1.7 ± 0.6 | 0.042 | GS598988 | 0 | 0 | c | LinJ08_V3.0680 | Amastin-like protein | N.D. | |
| Lin194E2 | 1.79 | 0.8 ± 0.2 | 0.025 | GS598989 | - | 0 | c | LinJ08_V3.0710 | Amastin-like protein | + | 6.8 ± 0.9 |
| Lin206B6 | 2.08 | 1.0 ± 0.3 | 0.035 | GS598990 | 7e-19 | 0 | b | LinJ22_V3.0680 | 3'a2rel-related protein | + | 3.5 ± 0.6 |
| Lin207A1 | 1.84 | 0.9 ± 0.2 | 0.015 | GS598991 | 0 | 0 | b | LinJ17_V3.0170 | Elongation factor 1-alpha | N.D. | |
| LinJ17_V3.0180 | Elongation factor 1-alpha | N.D. | |||||||||
| Lin210C4 | 1.71 | 1.8 ± 0.1 | 0.030 | GS598893 | 0 | 0 | b | LinJ08_V3.0690 | Amastin-like protein | N.D. | |
| Lin224G2 | 1.70 | 0.8 ± 0.2 | 0.014 | GS598895 | 0 | 0 | b | LinJ08_V3.0720 | Amastin-like protein | N.D. | |
| Lin235G8 | 2.20 | 1.1 ± 0.2 | 0.002 | GS598896 | 0 | 0 | b | LinJ08_V3.1320 | Amastin-like protein | N.D. | |
| Lin245E2 | 2.05 | 1.0 ± 0.3 | 0.032 | GS598897 | 0 | 0 | b | LinJ22_V3.0680 | 3'a2rel-related protein | + | 3.5 ± 0.6 |
| Lin274G6 | 1.84 | 0.9 ± 0.2 | 0.012 | GS598992 | 0 | 0 | b | LinJ08_V3.0680 | Amastin-like protein | N.D. | |
| LinJ08_V3.0690 | Amastin-like protein | N.D. | |||||||||
| Lin275A8 | 2.10 | 1.1 ± 0.1 | 0.003 | GS598900 | 0 | 4e-168 | b | LinJ08_V3.0720 | Amastin-like protein | N.D. | |
| Lin282B6 | 2.08 | 1.0 ± 0.4 | 0.042 | GS598993 | 0 | 0 | b | LinJ03_V3.0960 | Elongation initiation factor 2 alpha subunit, putative | N.D. | |
| Lin294A11 | 1.72 | 0.8 ± 0.1 | 0.001 | GS598902 | 0 | 0 | b | LinJ08_V3.1320 | Amastin-like protein | N.D. | |
| Lin295D9 | 2.99 | 1.6 ± 0.4 | 0.020 | GS598903 | 0 | 0 | b | LinJ34_V3.1720 | Amastin-like surface protein, putative | N.D. | |
This table describes clones that contain up-regulated genes under the sole influence of temperature increase (TS) that do not map with hypothetical or unknown genes. The features described are: clone number; fold change (F); base-two logarithmic scale F and standard deviation (SD) values; p-value (p); GenBank GSS accession numbers; e-values of forward (Fw) and reverse (Rv) end mappings against BLAST; clone definition (Def.) according to mapping outcomes a, b or c (see brief explanation in the text); GeneDB identifiers (Id.), the corresponding annotated gene functions; qRT-PCR results. When a given clone overlaps with more than one annotation, stage-specific regulation is only demonstrated if the qRT-PCR result is positive (+). Genes in bold are also up-regulated under TPS.
Table 4.
Down-regulated genes after temperature increase up to 37°C (day 4) in L. infantum.
| Clone | F | Log2F ± SD | P | GenBank GSS | e-value | Def. | Id. | Annotated Gene Function | qRT-PCR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fw | Rv | +/- | F ± SD | ||||||||
| Lin9E5 | -1.77 | -0.8 ± 0.3 | 0.033 | GS598994 | 4e-131 | 0 | b | LinJ35_V3.1150 | Oligosaccharyl transferase-like protein | N.D. | |
| Lin40G12 | -1.97 | -1.0 ± 0.2 | 0.008 | GS598916 | 2e-161 | 0 | b | LinJ23_V3.1550 | Hypothetical protein, unknown function | N.D. | |
| LinJ23_V3.1560 | Lathosterol oxidase-like protein | + | 5.0 ± 0.7 | ||||||||
| Lin49B7 | -4.38 | -2.1 ± 0.0 | 0.000 | GS598995 | 0 | 4e-64 | a | LinJ34_V3.4160 | Phosphatidylinositol-3-kinase (tor2)-like protein | N.D. | |
| Lin 60B1 | -2.41 | -1.3 ± 0.4 | 0.025 | GS598996 | 0 | 4e-162 | c | LinJ36_V3.7040 | Hypothetical protein, conserved | N.D. | |
| LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative | + | 7.2 ± 1.0 | ||||||||
| Lin63F3 | -2.23 | -1.2 ± 0.4 | 0.043 | GS598997 | 0 | 0 | a | LinJ36_V3.6550 | Glucose transporter lmgt2, putative | + | 6.1 ± 0.8 |
| LinJ36_V3.6560 | Glucose transporter, putative | + | 6.1 ± 0.8 | ||||||||
| Lin84E8 | -2.26 | -1.2 ± 0.2 | 0.007 | GS598998 | 0 | 0 | a | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative | + | 7.2 ± 1.0 |
| LinJ31_V3.2380 | 3'-nucleotidase/nuclease precursor, putative | + | 7.2 ± 1.0 | ||||||||
| Lin85H1 | -1.74 | -0.8 ± 0.2 | 0.025 | GS598999 | 1e-57 | 1e-20 | b | LinJ30_V3.3440 | CAS/CSE importin domain protein, putative | N.D. | |
| Lin93H3 | -2.26 | -1.2 ± 04 | 0.030 | GS599000 | 0 | 0 | a | LinJ31_V3.2370 | 3'-nucleotidase/nuclease, putative | + | 7.2 ± 1.0 |
| LinJ31_V3.2380 | 3'-nucleotidase/nuclease precursor, putative | + | 7.2 ± 1.0 | ||||||||
| Lin97D1 | -3.41 | -1.7 ± 0.3 | 0.001 | GS598931 | 0 | 0 | a | LinJ06_V3.1320 | Pteridine transporter, putative | + | 2.3 ± 0.3 |
| Lin98G10 | -2.13 | -1.1 ± 0.2 | 0.015 | GS599001 | 0 | 0 | b | LinJ30_V3.2770 | Hypothetical protein, conserved | N.D. | |
| LinJ30_V3.2780 | Superoxide dismutase, putative | + | 3.7 ± 0.0 | ||||||||
| Lin111C2 | -2.74 | -1.5 ± 0.1 | 0.002 | GS599002 | 0 | 0 | a | LinJ09_V3.0650 | Serine peptidase family S51, peptidase E, putative | N.D. | |
| Lin150E4 | -1.86 | -0.9 ± 0.3 | 0.036 | GS599003 | 0 | 8e-22 | b | LinJ13_V3.1060 | Calmodulin, putative | N.D. | |
| Lin155H12 | -2.34 | -1.2 ± 0.3 | 0.018 | GS599004 | 0 | 0 | a | LinJ36_V3.0250 | Peptidyl-prolyl cis-trans isomerase, putative | N.D. | |
| Lin157D8 | -2.27 | -1.2 ± 0.1 | 0.002 | GS599005 | 0 | - | c | LinJ31_V3.2380 | 3'-nucleotidase/nuclease precursor, putative | + | 7.2 ± 1.0 |
| Lin179B4 | -1.76 | -0.8 ± 0.1 | 0.004 | GS599006 | 0 | 0 | b | LinJ07_V3.0030 LinJ07_V3.0040 LinJ07_V3.0050 LinJ07_V3.0060 |
Hypothetical protein, conserved Hypothetical protein, conserved Hypothetical protein, conserved |
N.D. N.D. N.D. + |
|
| Alpha-adaptin-like protein | 5.3 ± 0.4 | ||||||||||
| Lin182F2 | -2.23 | -1.2 ± 0.1 | 0.008 | GS598952 | 0 | 0 | b | LinJ25_V3.0740 | Eukaryotic initiation factor 5a, putative | N.D. | |
| LinJ25_V3.0750 | Eukaryotic initiation factor 5a, putative | N.D. | |||||||||
| Lin187C10 | -4.72 | -2.2 ± 0.4 | 0.013 | GS598953 | 0 | 0 | b | LinJ06_V3.1320 | Pteridine transporter, putative | + | 2.3 ± 0.3 |
| Lin204A11 | -1.76 | -0.8 ± 0.3 | 0.038 | GS599007 | - | 1e-165 | c | LinJ09_V3.0650 | Serine peptidase, family S51, peptidase E, putative | N.D. | |
| Lin210B7 | -1.74 | -0.8 ± 0.2 | 0.016 | GS599008 | 0 | 0 | a | LinJ32_V3.3690 LinJ32_V3.3700 |
DEAD/DEAH box helicase, putative |
+ N.D. |
3.3 ± 0.8 |
| Hypothetical protein, conserved | |||||||||||
| Lin229E6 | -3.30 | -1.7 ± 0.3 | 0.005 | GS598963 | 0 | 0 | a |
LinJ06_V3.1320 LinJ06_V3.1330 |
Pteridine transporter, putative Coproporphyrinogen III oxidase, putative |
+ + |
2.3 ± 0.3 4.5 ± 0.6 |
| Lin242E2 | -2.37 | -1.2 ± 0.4 | 0.039 | GS599009 | 1e-137 | 0 | a |
LinJ31_V3.2370 LinJ31_V3.2380 |
3'-nucleotidase/nuclease, putative 3'-nucleotidase/nuclease precursor, putative |
+ + |
7.2 ± 1.0 7.2 ± 1.0 |
| Lin255E12 | -2.54 | -1.3 ± 0.3 | 0.011 | GS599010 | 0 | 0 | b | LinJ28_V3.0210 | Histone H2B variant | N.D. | |
This table contains clones that map against down-regulated genes (not hypothetical or unknown) with the single effect of temperature increase (TS). The features described are: clone number; F; base-two logarithmic scale F and SD values; p; GenBank GSS accession numbers; e-values; Def. according to mapping outcomes a, b or c (see brief explanation in the text); Id.; annotated gene function; qRT-PCR. When a given clone overlaps with more than one annotation, stage-specific regulation is only demonstrated if the qRT-PCR result is positive (+). Genes in bold are also down-regulated under TPS.
Table 5.
Differentially regulated genes after pH4.5 treatment (day 4) in L. infantum.
| Clone | F | Log2F ± SD | p |
GenBank GSS |
e-value | Def. | Id. | Annotated Gene Function | qRT-PCR | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fw | Rv | +/- | F ± SD | ||||||||
| Lin9E8 | 2.03 | 1.0 ± 0.1 | 0.003 | GS599011 | 0 | 0 | a | LinJ24_V3.0020 | Clathin coat assembly protein, putative | + | 7.6 ± 0.4 |
| LinJ24_V3.0030 | Hypothetical protein, conserved | N.D. | |||||||||
| LinJ24_V3.0040 | 60S ribosomal protein L17, putative | N.D. | |||||||||
| Lin10H12 | 2.26 | 1.2 ± 0.1 | 0.001 | GS599012 | 0 | 0 | a | LinJ31_V3.0860 | Triacylglycerol lipase-like protein | N.D. | |
| LinJ31_V3.0870 | Lipase precursor-like protein | N.D. | |||||||||
| Lin21H10 | 2.46 | 1.3 ± 0.1 | 0.001 | GS598910 | 0 | 0 | b | LinJ26_v3.1670 | Sphingolipid delta-4 desaturase, putative | N.D. | |
| Lin33G5 | 1.76 | 0.8 ± 0.0 | 0.000 | GS599013 | LinJ27_V3.1300 | 60S acidic ribosomal protein, putative | N.D. | ||||
| Lin37C10 | 2.74 | 1.5 ± 0.2 | 0.006 | GS599014 | 0 | 0 | b | LinJ33_V3.0280 | RNA binding protein rggm, putative | N.D. | |
| Lin58C1 | 2.32 | 1.2 ± 0.2 | 0.001 | GS599015 | 0 | 0 | b | LinJ22_V3.1360 | Hypothetical protein, unknown fuction | N.D. | |
| LinJ22_V3.1370 | 60S ribosomal protein L14 | N.D. | |||||||||
| LinJ22_V3.1380 | Dephospho-CoA kinase, putative | + | -2.9 ± 0.3 | ||||||||
| Lin63B7 | 1.83 | 0.9 ± 0.1 | 0.002 | GS599016 | 1e-100 | 1e-103 | b | LinJ15_V3.1200 | 60S acidic ribosomal protein P2 | N.D. | |
| Lin66A8 | 2.28 | 1.2 ± 0.1 | 0.006 | GS599017 | 0 | 0 | a | LinJ22_V3.0470 | Hypothetical protein, conserved | N.D. | |
| LinJ22_V3.0480 | Ubiquitin-conjugating enzyme-like protein | + | -3.1 ± 0.8 | ||||||||
| Lin80C3 | 3.15 | 1.7 ± 0.3 | 0.004 | GS599018 | 0 | 0 | b | LinJ28_V3.3250 | Glucose-6-phosphate N-acetyltransferase | N.D. | |
| Lin 95F10 | 2.26 | 1.2 ± 0.1 | 0.002 | GS599019 | 0 | 0 | a | LinJ28_V3.2360 | Ribosomal protein S29, putative | N.D. | |
| Lin100F8 | 2.12 | 1.1 ± 0.2 | 0.007 | GS599020 | 0 | 2e-161 | b | LinJ35_V3.3330 | 60S ribosomal protein L31, putative | N.D. | |
| LinJ35_V3.3340 | 60S ribosomal protein L31, putative | N.D. | |||||||||
| Lin107C12 | 2.90 | 1.5 ± 0.2 | 0.001 | GS599021 | 7e-130 | 5e-134 | a | LinJ11_V3.1180 | 40S ribosomal protein S15a, putative | N.D. | |
| Lin122C5 | 1.93 | 0.9 ± 0.1 | 0.005 | GS599022 | 0 | 0 | b | LinJ30_V3.3770 | CPSF-domain protein, putative | N.D. | |
| LinJ30_V3.3780 | 60S acidic ribosomal protein P2, putative | N.D. | |||||||||
| LinJ30_V3.3790 | 60S acidic ribosomal protein P2, putative | N.D. | |||||||||
| Lin135F6 | 2.63 | 1.4 ± 0.3 | 0.036 | GS599023 | 0 | 0 | b | LinJ29_V3.1920 | 40S ribosomal protein S15a, putative | N.D. | |
| LinJ29_V3.1930 | Hypothetical protein, conserved | N.D. | |||||||||
| Lin137A10 | 2.00 | 1.0 ± 0.2 | 0.019 | GS599024 | 0 | 0 | b | LinJ24_V3.1230 | Hypothetical protein, conserved | N.D. | |
| LinJ24_V3.1240 | Translation factor SUI1, putative | + | -5.0 ± 0.6 | ||||||||
| Lin144F11 | 1.72 | 0.8 ± 0.2 | 0.032 | GS599025 | 0 | 0 | a | LinJ31_V3.2210 | Prostaglandin F2α synthetase | N.D. | |
| Lin161C9 | 2.54 | 1.3 ± 0.1 | 0.003 | GS599026 | 0 | 1e-177 | b | LinJ26_V3.1670 | Sphingolipid delta-4 desaturase, putative | N.D. | |
| Lin162A9 | 1.97 | 1.0 ± 0.2 | 0.024 | GS599027 | 0 | 0 | b | LinJ22_V3.0470 | Hypothetical protein, conserved | N.D. | |
| LinJ22_V3.0480 | Ubiquitin-conjugating enzyme-like protein | + | -3.1 ± 0.8 | ||||||||
| Lin182F2 | 3.27 | 1.7 ± 0.2 | 0.009 | GS599028 | 0 | 0 | b | LinJ25_V3.0740 | Eukaryotic initiation factor 5a, putative | N.D. | |
| LinJ25_V3.0750 | Eukaryotic initiation factor 5a, putative | N.D. | |||||||||
| Lin200H12 | 2.54 | 1.3 ± 0.1 | 0.005 | GS599029 | 0 | 0 | a | LinJ14_V3.1340 | Hypothetical protein, unknown funcion | N.D. | |
| LinJ14_V3. 1350 | Ubiquitin/ribosomal protein S27a, putative | + | -4.8 ± 0.4 | ||||||||
| LinJ14_V3.1360 | Hypothetical protein, conserved | N.D. | |||||||||
| Lin218E3 | 1.82 | 0.9 ± 0.1 | 0.001 | GS599030 | 0 | 0 | b | LinJ31_V3.2210 | Prostaglandin F2α synthase | N.D. | |
| Lin247D7 | 2.41 | 1.3 ± 0.4 | 0.018 | GS599031 | 5e-109 | 0 | a | LinJ28_V3.0090 | Adenylate cyclase-like protein | + | -3.5 ± 0.6 |
| LinJ28_V3.0100 | Hypothetical protein, conserved | N.D. | |||||||||
| LinJ28_V3.0110 | Proteasome beta 3 subunit, putative | N.D. | |||||||||
| Lin254A4 | 1.93 | 0.9 ± 0.2 | 0.009 | GS599032 | 0 | 0 | b | LinJ04_V3.1250 | Actin | N.D. | |
| Lin254H7 | 1.73 | 0.8 ± 0.1 | 0.004 | GS599033 | 0 | 0 | b | LinJ04_V3.1250 | Actin | N.D. | |
| Lin261F8 | 2.84 | 1.5 ± 0.2 | 0.007 | GS599034 | 0 | 0 | b | LinJ21_V3.1310 | 40S ribosomal protein S23, putative | N.D. | |
| Lin266F6 | 1.79 | 0.8 ± 0.1 | 0.009 | GS599035 | 0 | 0 | b | LinJ27_V3.0300 | Acyl carrier protein, putative | N.D. | |
| Lin267B9 | 1.74 | 0.8 ± 0.2 | 0.010 | GS599036 | 0 | 0 | b | LinJ36_V3.0580 | Hypothetical protein, conserved | N.D. | |
| LinJ36_V3.0590 | Ubiquitin-like protein, putative | + | -2.3 ± 0.0 | ||||||||
| LinJ36_V3.0600 | Cdc2-related kinase | N.D. | |||||||||
| Lin269B5 | 2.75 | 1.5 ± 0.2 | 0.002 | GS599037 | 0 | 0 | b | LinJ29_V3.2970 | 40S ribosomal protein S19-like protein | N.D. | |
| Lin282B6 | 2.44 | 1.3 ± 0.2 | 0.009 | GS599038 | 0 | 0 | a | LinJ03_V3.0960 | Elongation initiation factor 2α subunit, putative | N.D. | |
| Lin290G8 | 1.80 | 0.8 ± 0.1 | 0.003 | GS599039 | 0 | 0 | a | LinJ17_V3.1520 | Otubain cysteine peptidase, Clan CA. family C65, putative | N.D. | |
| Lin43G10 | -5.36 | -2.4 ± 0.3 | 0.007 | GS599040 | 0 | 0 | c | LinJ28_V3.3060 | Heat-shock protein hsp70, putative | + | -2.1 ± 0.2 |
| Lin130C5 | -4.71 | -2.2 ± 0.3 | 0.041 | GS599041 | 0 | 0 | b | LinJ36_V3.3170 | Exosome complex exonuclease RRP41, putative | N.D. | |
| LinJ36_V3.3180 | Clathrin coat assembly protein-like protein | N.D. | |||||||||
| LinJ36_V3.3190 | Pre-mRNA branch-site protein p14 | + | -4.7 ± 2.3 | ||||||||
| LinJ36_V3.3200 | Hypothetical protein, conserved | N.D. | |||||||||
| Lin173E11 | -7.74 | -3.0 ± 0.4 | 0.002 | GS599042 | 6e-44 | 3e-147 | b | LinJ36_V3.2280 | ER-golgi transport protein erv25 precursor, putative | N.D. | |
| Lin177H3 | -5.08 | -2.3 ± 0.2 | 0.001 | GS599043 | 0 | 4e-60 | b | LinJ28_V3.3060 | Heat shock protein hsp70, putative | + | -2.1 ± 0.2 |
| Lin197E1 | -2.53 | -1.3 ± 0.1 | 0.007 | GS599044 | 0 | 0 | c | LinJ18_V3.0830 | Periodic tryptophan protein 2-like protein | - | 1.4 ± 0.4 |
| LinJ23_V3.1610 | Acetyltransferase-like protein | + | -2.1 ± 0.2 | ||||||||
| Lin228H5 | -7.90 | -3.0 ± 0.4 | 0.012 | GS599045 | 7e-196 | 0 | b | LinJ21_V3.0310 | Hexokinase, putative | N.D. | |
| Lin281H8 | -2.01 | -1.0 ± 0.1 | 0.001 | GS599046 | 8e-136 | 2e-102 | b | LinJ35_V3.1580 | Metacaspase, putative | N.D. | |
| Lihsp70 | -4.21 | -2.0 ± 0.2 | 0.004 | XM001470292 | - | - | - | - | L. infantum hsp70 - DNA microarray control spot | + | -2.1 ± 0.2 |
| Ldohsp70 | -4.57 | -2.2 ± 0.1 | 0.028 | - | - | - | - | - | L. donovani hsp70 - DNA microarray control spot | N.D. | |
| Lmahsp70 | -3.85 | -1.9 ± 0.2 | 0.017 | - | - | - | - | - | L. major hsp70 -DNA microarray control spot | N.D. | |
This table contains clones that map against up- and down-regulated genes (not hypothetical or unknown) under pH decrease (PS). The features described are: clone number; F; base-two logarithmic scale F and SD values; p; GenBank GSS accession numbers; e-values; Def. according to mapping outcomes a, b or c (see brief explanation in the text); Id.; annotated gene function; qRT-PCR. When a given clone overlaps with more than one annotation, stage-specific regulation is only demonstrated if the qRT-PCR result is positive (+). Genes in bold are also up-regulated under TPS.
Figure 3.
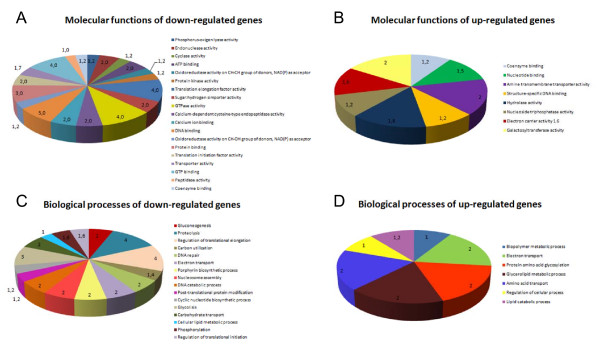
Multilevel sector charts of α-scores for GO molecular functions annotated on differentially regulated genes under TPS. (A) Molecular function GO terms annotated on down-regulated genes under TPS. (B) Molecular function GO terms annotated on up-regulated genes under TPS. (C) Biological process GO terms annotated on down-regulated genes under TPS. (D). Biological process GO terms annotated on up-regulated genes under TPS.
Figure 4.
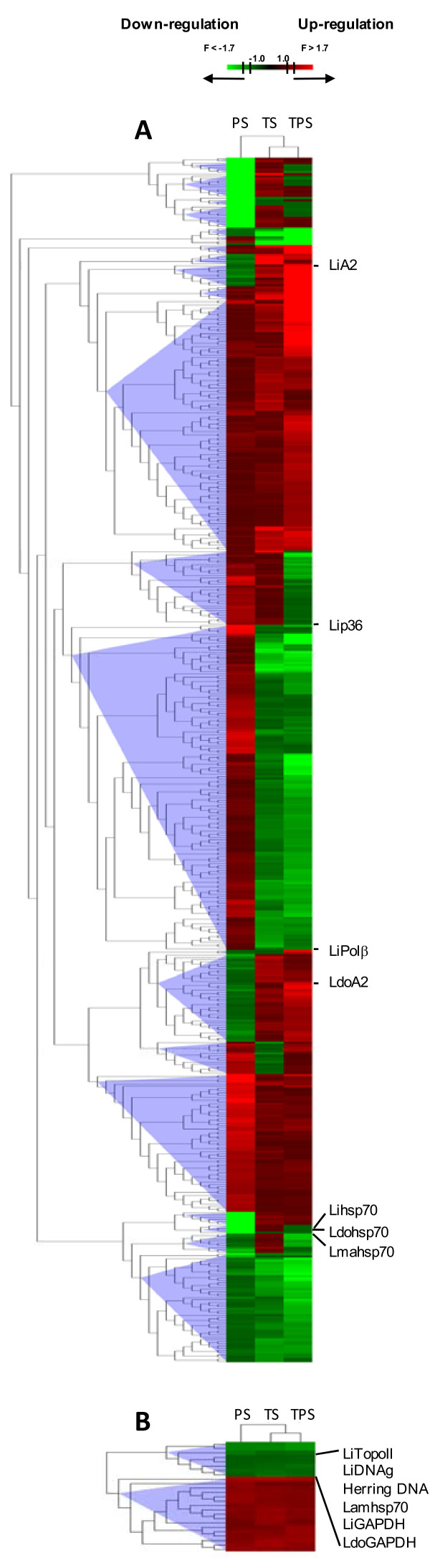
HCL-ST of genes differentially modulated under TPS, TS and/or PS. After performing SAM for all experimental groups, HCL-ST analysis was performed independently for (A) genes with (B) and without significant differences between groups according to SAM. Support Tree algorithm with a jackknifing resampling option and 100 iterations for the construction and clustering of gene expression matrix were applied in HCL-ST. Clones in (A) were grouped into 26 clusters and clones in (B) in two clusters depending on differential regulation. This analysis confirms that expression profile similarity is higher between TPS and TS than between TPS or TS and PS. Control spots LiA2, LdoA2, Lip36, Lipolb, Lihsp70, Ldohsp70 and Lmahsp70 show significant differences in gene expression between the experimental groups (A2 gene is up-regulated under TPS and hsp70 under PS) and LiTopoII, LiDNAg, Lamhsp70, LiGAPDH, LdoGAPDH and herring DNA do not. Clones with significant differences between the experimental conditions are identified in Additional file 6.
Concomitant temperature increase and acidification (TPS) leads promastigotes toward AL
It has been stated that acidification and the simultaneous effect of temperature increase induce the differentiation of promastigotes to amastigotes [6,7]. In spite of the amastigote-like round cell morphology induced under these conditions, we have observed that in a fraction of the population flagella are not hidden (Figure 2). Nevertheless, it is important to take into account that we have performed the assays in standard medium in which promastigotes are cultured (RPMI supplemented with HIFBS) instead of media used for axenic amastigote culture such as Schneider's medium in order to avoid the effect of this factor and focus this study on pH and temperature influence.
We have observed that TPS-treated cells differentiate into AL after 4 days of stimulation (Figure 2), when control promastigotes are reaching the stationary-phase (Figure 1). As mentioned before, TPS-treated cells proliferate to a lesser extent than TS-treated ones due to the effect of pH decrease. Expression profiling by DNA microarrays has revealed a set of up- and down-regulated genes (Tables 1, 2, Additional file 3: Table S1 and S2) that are fully discussed below in the TPS expression profile section and illustrated in Figures 3 and 5. Taken as a whole, TPS induces promastigote differentiation to AL, as indicated by gp46 IFA and agreement with previous reports on the differential expression regulation of the following genes [13]: A2 gene and a set of amastin genes (up-regulated); 3'NT/Nase cluster and SbGRP encoding gene (down-regulated).
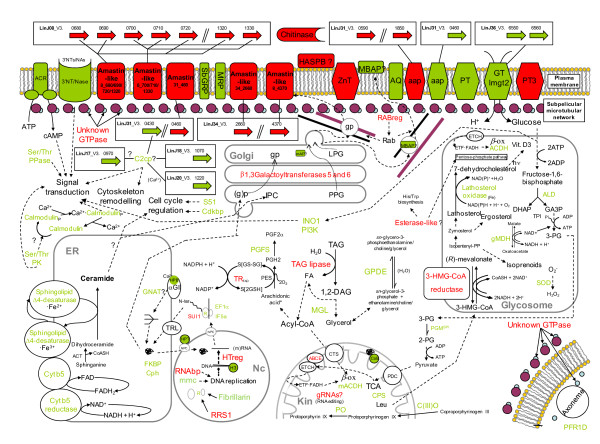
Figure 5.
Scheme representing differentially regulated genes under TPS and their subcellular localisation and/or functional relations. Up-regulated genes are represented in red colour (Cy5) and down-regulated in green (Cy3). Further explanations in the TPS expression profile subsection, which is included in the Results and Discussion section.
TS alone leads to a TPS-like expression profile
TS-stimulated differentiation processes have been studied only from a morphological point of view in L. infantum, but not at the differential gene expression level. For the first time, we have described in this research the influence of TS on the whole transcriptome of the parasite (Tables 3, 4, Additional file 3: Table S3 and S4). Analogies between TPS and TS expression profiles have been observed, namely in the differential regulation of the following genes (Tables 1, 2, 3 and 4, in bold): up-regulation of 3'a2rel-related protein, some amastin superfamily genes (see Figure 6 and Amastin Superfamily subsection below), ribosome biogenesis regulatory protein (RRS1), myo-inositol-1-phosphate synthase (INO1), amino acid transporter aATP11, three conserved hypothetical protein genes and eight clones that do not map with any annotated genic sequence; and down-regulation of 3'NT/Nase, pteridine transporter (PT) LinJ06_V3.1320, glucose transporters (GT), paraflagellar rod protein 1D (PFR1D), superoxide dismutase (SOD), phosphatidylinositol-3-kinase (tor2)-like (PI3K), peptidyl-prolyl cis-trans isomerase (FKBP) LinJ36_V3.0250, calmodulin, lathosterol oxidase, one hypothetical protein of unknown function, six conserved hypothetical proteins and seven clones that do not map with any annotated gene. These clones unmapped with genes strongly suggest that gene annotations on the L. infantum genomic sequence are incomplete, thus highlighting the advantages of using shotgun genomic DNA microarrays and the subsequent genomic library. As pointed out above, TPS-induced in vitro stimulation results in a differentiation process that resembles the differentiation of promastigotes to amastigotes inside the phagocytes of the mammalian host. Despite TS itself inducing analogous differentiation events and TS-treated cells being called AL [9], the A2 gene is not up-regulated (Table 3), all cells show a large flagellum and gp46 IFA is positive under TS (Figure 2). Nevertheless, SAM and the subsequent HCL-ST analysis of clones with regard to their fold-change values have revealed significant similarities between the transcriptome under TPS and TS (Figure 4). Genes of known function with the same expression pattern under TPS and TS are highlighted in bold in Tables 1, 2, 3 and 4 (those of unknown function in Additional file 3). The specific regulation of these genes by temperature increase is directly correlated to the differentiation to the amastigote stage. To sum up, even though TS-treated cells are not differentiated to the same extent as TPS, the similarities found between TPS and TS expression profiles when contrasted with the PS profile have led us to conclude that temperature has a greater influence than pH on the differentiation process leading up to the amastigote stage.
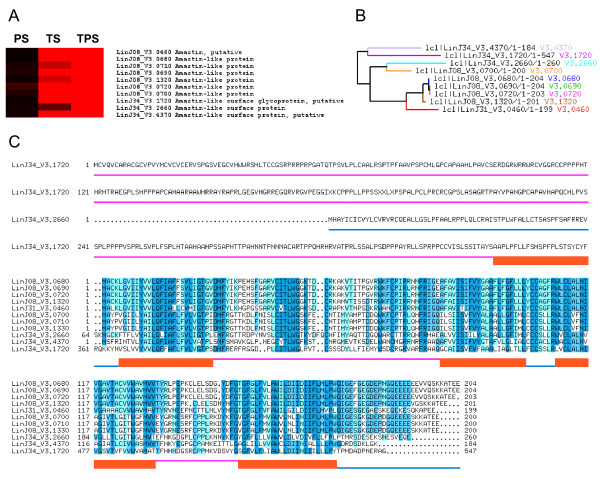
Figure 6.
Amino acid sequence and domain analysis of amastin genes found to be differentially regulated under TPS and TS. (A) MEV comparison of differential regulation under TPS, TS and PS. (B) Sequence similarity tree representing distances between amastin genes found to be up-regulated under TPS and TS. (C) Amino acid sequences were aligned with CLUSTALW2 software. The darker the position is highlighted the more conserved the residue is. The boundaries of inner, transmembrane and outer domain sequences predicted with TMHMM 2.0 software are represented below sequence alignments.
Acidification (PS) contributes little to the differentiation process
Some authors have considered that the induction of metacyclogenesis in promastigotes by acidic pH is a response common to a variety of Leishmania species [21,22]. Although there is no evidence concerning the metacyclic status of such promastigotes except for morphological considerations, proliferation seems to be inhibited by the single effect of acidification (pH 4.5-5.5) after 48 h according to [5] and our own observations. Figure 1 shows that promastigote growth is limited under these conditions, which is consistent with the generation time increase previously observed at pH 4.5 [11]. After an intermediate-term exposure to PS (day 4), two cell morphologies were observed: round and promastigote-like, both with emerging flagellum (Figure 2). Moreover, lack of A2 gene up-regulation (control gene spotted in each microarray) and an atypical gene expression profile have been found. There are some similarities in the expression profiles of TPS-obtained AL and PS-treated cells: up-regulation of triacylglycerol (TAG) lipase (TGL), translation factor SUI (TFSUI1)-also up-regulated under TS-, ubiquitin conjugating enzyme-like and five clones that do not map with any annotated gene; down-regulation of a conserved hypothetical protein and a gene still to be annotated; and the previous finding of an amastigote-specific protein induced by pH decrease [11]. In addition, 60S acidic ribosomal protein P2, 60S ribosomal protein L31 [23], ribosomal protein S29 and RNA binding protein rggm [24] are up-regulated in intracellular amastigotes according to Serial Analysis of Gene Expression (SAGE), which is due to PS (Table 5). In spite of this, the vast majority of differentially regulated genes under PS (Tables 5, Additional file 3: Table S5 and S6) have not been found to match up with those of the TPS and TS profiles. In fact, SAM output of differentially modulated genes between PS, TS and TPS was analysed by HCL-ST, which revealed that the most distant experimental group is PS (Figure 4). Moreover, there are opposite gene expression regulation events between TPS and PS: down-regulation under TPS and up-regulation under PS of glucose-6-phosphate N-acetyltransferase gene (GNAT), sphingolipid Δ4-desaturase, prostaglandin F synthetase (PGFS), eukaryotic translation initiation factor 5a (eIF5a) and two clones that do not map with any annotated sequence. Furthermore, there is also a lack of resemblance with the metacyclic promastigote profile [25], except for the up-regulation of 60S acidic ribosomal protein LinJ27_V3.1300 and some clones probably containing contig 957 guide RNA (gRNA) sequence (Additional file 3: Table S7 and S8). Considered together with the HCL-ST analysis of gene expression, these data suggest that intermediate-term exposure of promastigotes to PS leads to forms with features that do not match with any of the stages of the parasite's biological cycle (Figure 4) except for explained coincidences. Consequently, although pH has a role in differentiation, temperature is more relevant.
TPS-induced expression profile
Overview: Gene Ontology term annotations
All genes identified as potentially regulated under these conditions were re-annotated with BLAST2GO to describe globally the influence of TPS on the L. infantum transcriptome. Despite the useful overview provided by this analysis, which has revealed the functions of some hypothetical proteins, specific genes of trypanosomatid parasites like amastins or A2 cannot be correlated to any of the terms included in the database, as they do not show any known activity. The analysis of GO molecular function terms associated with a TPS-induced profile (Figure 3A and 3B) indicates an increase in galactosyltransferase (also observed by SAGE [24]), nucleoside triphosphatase activity and amine transmembrane transporter activities and a decrease in transcripts with associated GO molecular function term annotations such as cyclase, protein kinase and calcium-related cysteine peptidase (all related to signal transduction processes), translation initiation and elongation factor and oxidoreductase activities related to electron transport. These findings at the molecular function level can be clearly described at the biological process GO term level (Figure 3C and 3D): the down-regulation of several genes related to the regulation of translational initiation, elongation and post-translational modification indicates that protein biosynthesis and modification is more active in stationary-phase promastigotes rather than in AL. The same occurs with signal transduction, prostaglandin F and porphyrin biosynthesis. Genes related to biopolymer and lipid metabolic processes, glycosylation of proteins and regulation of cellular processes are up-regulated in TPS-induced AL. Nevertheless, there are some common biological process GO terms that are up- and down-regulated simultaneously, but this refers to different genes in each case: electron transport activity is referred mainly to cytochrome b5 reductase at CC (it is involved in electron transport to the sphingolipid-Δ4-desaturase reaction), while trypanothione reductase (TR) and the ABC transporter subfamily E (ribonuclease L-inhibitor) gene (ABCE) are both related to the same term; the amino acid transport term is also present at both stages, but nucleotide sequences of the corresponding aminoacid permeases are different, which suggests that a different transporter is used in each stage.
The resulting microarray data for the TPS-induced AL expression profile analysis is discussed in the next subsections according to the iterative HCL-ST (Figure 4) and BLAST2GO-based analyses (Figure 3 and Additional file 4). Moreover, it is illustrated schematically in Figure 5 with regard to the leishmanial surface, cytoskeleton, secretory pathway, metabolic and signalling processes. Direct acyclic graphs (DAGs) (Additional file 4) have been associated with genes shown in Tables 1, 2, 3, 4 and 5 by means of custom codes assigned in brackets after the name of each gene annotation.
Amastin superfamily
Several proteins from the uncharacterised surface amastin superfamily have been shown to be up-regulated basically in the amastigote stage of Trypanosoma cruzi, L. major and L. infantum [26,27]. The microarray-based transcriptome analysis contained in this study has revealed that eleven amastin genes are up-regulated under TPS, ten out of these under TS but none under PS. In fact, SAM highlights significant differences in the expression pattern of the eleven amastin genes and the subsequent amastin HCL-ST analysis supports the same expression pattern except for LinJ34_V3.2660 (Figure 6A). Furthermore, these amastin genes have been reported to be up-regulated in intracellular and axenic amastigotes by microarrays [28] and SAGE [24]. According to TMHMM predictions, these amastins contain 4 transmembrane, 3 inner and 2 outer domains, except for LinJ34_V3.1720, which contains a 300 amino acid long N-terminal (N-ter) region followed by an additional short transmembrane domain. Outer domains are variable among amastin superfamily members, although they are very similar in a given amastin group or class (Figure 6B and 6C). Amastins LinJ08_V3.0680/0690 and LinJ08_V3.0700/0710 were previously found to be up-regulated in metacyclic promastigotes [25], which supports that amastin genes are not amastigote markers. The expression rate of these genes increases as the life cycle progresses.
A2-A2rel cluster
A2 gene cluster was first identified in L. donovani, where A2 transcripts are abundant in amastigotes but hardly detectable in promastigotes [29]. These molecules were proposed as virulence factors that enhance survival of the amastigote inside the macrophage [30]. It has been suggested that a balance between A2 and A2rel proteins is required for the parasite's survival [31]. L. donovani and L. infantum A2 genes were spotted onto the microarrays as amastigote-specific control genes. We have observed an increase in the corresponding transcript levels under TPS in the hybridisation analysis (Table 3). In addition, our results indicate that TPS and TS elicit the up-regulation of 3'a2rel-related transcripts in L. infantum.
DNA repair and replication, gene expression and secretory pathway
A member of minichromosome maintenance complex protein (mmc) 2/3/5 family (PFAM annotation PF00493) is down-regulated and an RNA binding protein (RNAbp) up-regulated in TPS-induced AL. mmc and RNAbp are involved in DNA replication according to GO biological process annotation. The histone H3 gene is up-regulated under TPS, as well as in intracellular amastigotes according to SAGE [23]. It is also involved in nucleosome assembly and DNA repair according to GO annotation.
With regard to gene expression and protein processing, a hypothetical transcription regulator gene (HTreg) and RRS1 are up-regulated under TPS, while nucleolar fibrillarin is down-regulated. RRS1 is also up-regulated under TS. TFSUI1 is up-regulated under TPS, TS and PS (see above). The elongation factor 1α (EF1α) is down-regulated in both TPS-induced AL and in intracellular L. major promastigotes as previously reported [32] and IF5a is also down-regulated by TPS, suggesting a different translation regulation mechanism under TPS selective pressure. Peptidyl-prolyl cis-trans isomerases FKBP and cyclophilin (Cph) are also down-regulated in TPS-generated AL and FKBP under TS. FKBP and Cph are involved in protein folding inside the endoplasmic reticulum (ER) and we had already found the down-regulation of both genes in metacyclic peanut lectin non-agglutinating promastigotes [25]. As a consequence, Cph and FKBP gene expression decreases throughout the parasite's life cycle. In addition, a hypothetical protein related to calcium ion and protein binding GO molecular functions (αGII-HPB) localises to the dimeric α-glucosidase-II complex according to GO cellular component analysis and is down-regulated at the level of transcript under TPS. GNAT is also down-regulated and is involved in protein oligosaccharide biosynthesis inside the ER lumen, possibly in the glucosylation/deglucosylation cycle. We have found that a Rab GTPase regulator protein (RABreg, see further explanation in the Cytoskeleton remodelling subsection) is up-regulated under TPS, probably promoting vesicle transport from Golgi apparatus. In addition, β-1,3-galactosyltransferase-5/6 carries out galactosylation of proteophosphoglycan and lypophosphoglycan if required. These genes have been found to be up-regulated under TPS, as it was also reported for metacyclic peanut lectin non-agglutinating promastigotes [25] and intracellular amastigotes according to SAGE [24].
Energetic metabolism
TPS-obtained AL down-regulate transcript levels of two glycolitic genes: fructose-1,6-bisphosphate aldolase (ALD) and 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (PGMBPI). This agrees with the down-regulation of PGMBPI protein in L. donovani [33] and transcripts in L. infantum (unpublished data) mature intracellular amastigotes. The ALD gene was also found to be down-regulated at the post-transcriptional level in L. mexicana mature amastigotes [17] and at the protein level in immature L. donovani amastigotes. By contrast, ALD protein is up-regulated in L. donovani mature intracellular amastigotes [33], which differs from TPS-induced AL. Down-regulation of both genes is consistent with high energy requirements in the promastigote stage. ALD and PGMBPI are independent of catabolite regulation and are located in the glycosome and the cytosol respectively. Inhibition of glycolysis by ALD and PGMBPI down-regulation is consistent with the down-regulation of two GTs under TPS and TS. Both genes are located in tandem in chromosome 36 and custom CLUSTALW2 alignments (Additional file 5) illustrate that their sequences are identical except for N-terminal regions (N-ter) of coded peptides. qRT-PCR analysis is consistent with the up-regulation of both GT, as well as the up-regulation of GT lmgt2 in L. mexicana [17] and L. major [32] intracellular amastigotes. NAD+ supply for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reaction is assured by the up-regulation of the glycosomal malate dehydrogenase (gMDH) gene at CC with respect to TPS. The mitochondrial precursor of acyl-CoA dehydrogenase LinJ07_V3.0150 gene (mACDH) is also down-regulated under TPS, which suggests that β-oxidation (β-ox) of fatty acids (FA) is activated under such conditions, as well as glucose uptake and glycolysis.
ABCE is up-regulated under TPS and is involved in electron transport. In fact, the only ABCE family member studied to date is a multifunctional protein that includes a metal-binding domain (PF04068) adjacent to the 4Fe-4S binding domain (PF00037), as well as two ATP-binding sites/ATPase domains typical of ABC proteins (PF00005) but it lacks transporter domains. This kind of protein has been found in pluricellular eukaryotes but not in yeast and binds directly to RNase L to prevent it from binding 5'-phosphorylated 2',5'-linked oligo-adenylates [34]. The biological role of ABC and the meaning of its up-regulation at the level of transcript in TPS-obtained AL are still unknown in Leishmania spp. ABCE localises to the kinetoplast according to GO cellular component annotation.
Lipid metabolism
TGL is post-transcriptionally up-regulated under TPS and is involved in sn-2 and sn-3 hydrolysis of TAGs. CoA can be incorporated in the released FA and enter β-ox, where mACDH and 3-ketoacyl-CoA thiolase (thiolase I) are down-regulated. On the other hand, monoglyceride lipase (MGL) is down-regulated under TPS. Another gene with the same regulation pattern is a hypothetical protein with a glycerolphosphodiester phosphodiesterase (GPDE) function (GO molecular function analysis), which is related to glycerol derivative metabolism. An additional destination for FA is the sphingolipid biosynthesis pathway, in which sphingolipid-Δ4-desaturase oxidises dihydroceramide to ceramide in the presence of O2 and Fe as cofactor (Fe3+ reduced to Fe2+) [35]. The sphingolipid-Δ4-desaturase gene is down-regulated under TPS and is located in the ER membrane (GO cellular component analysis), as well as the cytochrome b5 reductase gene (cyt b5 reductase), which provides reduction power for desaturases through cytb5. After ceramide biosynthesis, a molecule of phosphoinositol can be added inside the Golgi apparatus resulting in inositol phosphoceramide (IPC) for anchoring inositol derivatives. Saturated acyl groups are also the precursors of polyinsaturated fatty acids like arachidonic acid, from which prostaglandins are derived. PGFS is down-regulated under TPS, while TR is up-regulated. The reaction prior to PGFS is catalysed by prostaglandin peroxide synthase (PES) and requires trypanothione in its reduced state. TR regenerates reduced molecules for PES reaction, as well as for many other redox processes. Thus, increases in PGFS and TR at different stages is not a contradictory fact, given the wide functional spectrum of the latter. PGFS is also up-regulated in procyclic promastigotes with respect to metacyclics [25] and has been associated with vector competence of procyclic promastigotes. Taken together, these data confirm that PGFS levels diminish throughout differentiation. Finally, 1,2-DAG can enter inositolphospholipid metabolism, where PI3K is down-regulated under TPS and TS. As PGFS, mACDH, thiolase I, sphingolipid Δ4-desaturase and PI3K are down-regulated, the destination of 1,2-DAG and FA excess generated by gene up-regulation of TAG lipase remains unclear.
The gene coding for 3-hydroxymethylglutaryl-CoA (HMG-CoA) reductase (HMGCR) is up-regulated under TPS. This is the rate-limiting step of sterol and isoprenoid biosynthesis. In view of this result, ergosterol biosynthesis may be increased in AL. HMGCR localises to the glycosome in Leishmania spp., where leucine (in trypanosomatids [36]) must be carried for priming steroid biosynthesis (reviewed in [37]). In spite of HMGCR increase in TPS-induced AL, the lathosterol oxidase gene has been found previously to be down-regulated in intracellular amastigotes ([28] and unpublished custom data) and the analysis reported in this study has revealed that the down-regulation of this gene is due to the specific influence of TPS and TS. Lathosterol oxidase yields 7-dehydrocholesterol. Leishmania parasites lack the enzyme cholesterol:NADP+ Δ7-oxidoreductase, that catalyses the conversion of 7-dehydrocholesterol into cholesterol (KEGG database for L. major [38]). Cholesterol functions are performed by ergosterol in these organisms. A question arises about the destination of 7-dehydrocholesterol in promastigotes (CC). Vitamin D3 (cholecalciferol) is synthesised by exciting 7-dehydrocholesterol with a photon (hν), that according to our gene expression results may occur inside the insect vector's gut, where promastigotes are undergoing a developmental process. Obviously, the biological meaning of this fact still remains unclear.
Porphyrin biosynthesis
The prosthetic heme group is required for many electron transport chain proteins (cytochromes), including cyt b5. Leishmania spp. does not have the ability to perform porphyrin biosynthesis de novo, because it lacks δ-aminolevulinate synthase, porphobilinogen synthase and deaminase and uroporphyrinogen decarboxylase. These parasites are able to acquire protophorphyrinogen IX or heme group directly from the mammalian host. Moreover, coproporphyrinogen III, protoporphyrinogen oxidases (C(III)O, PO) and a ferrochelatase-like protein are annotated in the genome of the parasite, which highlights its ability to perform heme group biosynthesis from the substrate coproporphyrinogen III. Interestingly, C(III)O and PO are located in tandem in chromosome 6 and are down-regulated under TPS according to our microarray hybridisation results. In fact, CC mimic the environment inside the gut of the phlebotominae sand-fly, where the parasites cannot acquire heme or protoporphyrin IX. In spite of this, we have not found ferrochelatase gene (located in chromosome 17) to be differentially regulated under the experimental conditions assayed. This observation is additional evidence backing the hypothesis of gene organization in DGCs depending on the post-transcriptional regulation in Leishmania spp (reviewed in [39]).
Redox homeostasis and oxidative stress
TR catalyses the reduction of reactive oxygen radical superoxide anion to hydrogen peroxide and is responsible for maintaining the glutathione orthologue trypanothione in its reduced form, essential for redox defence systems in trypanosomatids. For this reason, TR would be a useful chemotherapeutic target [40]. TRs are members of the NADPH-dependent flavoprotein oxidoreductase family and are structurally and mechanistically related to gluthathione reductase [41]. The disruption of the TR gene in Leishmania decreases the ability to survive oxidative stress inside macrophages [41]. The up-regulation of this gene detected under TPS is further evidence for TR demand in the amastigote stage.
Transport
Two different genes coding for 3'NT/Nase are located in tandem in chromosome 31 and are down-regulated under TPS and TS, as well as in intracellular amastigotes according to unpublished custom data and previous analyses (reviewed in [13]). CLUSTAL alignments show exact identity in the central region, and differences between N-ter and C-ter domains. 3'-NT/Nase is essential for Leishmania parasites because they are not able to synthesise purines de novo. It localizes to the plasma membrane, may play a role in purine acquisition and its substrates are 3'-ribonucleotides (3'AMP and 3'-IMP) and nucleic acids [42,44]. According to previous observations [45], these genes are up-regulated in promastigotes in the logarithmic phase of axenic culture and absence of expression is shown in the amastigote stage. Interestingly, we have found down-regulation of these genes under TPS and TS with respect to CC. Consequently, temperature down-regulates 3'-NT/Nase expression.
Apart from the GT genes already mentioned, aquaporin (AQ) and Zn transporter (ZnT) genes are down-regulated under TPS. The AQ gene is related to changes in cell volume and shape during the life cycle of the parasite and it also acts both as an osmotic sensor and in passive transport of solutes. It may be related to the osmotactic response shown by the promatigote stage inside the insect vector for migration to the foregut [46]. In addition, four different aminoacid permeases are differentially regulated: two are up-regulated (LinJ31_V3.0590 and LinJ31_V3.1580) and two down-regulated (LinJ27_V3.0530 and LinJ31_V3.0610). Furthermore, three different pteridine transporter genes are differentially regulated by the effect of temperature and pH: PT3 LinJ10_V3.0410 and PT LinJ14_V3.1440 are up-regulated under TPS, while PT LinJ06_V3.1320 is down-regulated, and this pattern is repeated under TS except for PT3. Trypanosomatids are auxotrophes for pteridines and therefore they depend on exogenous sources of these compounds. Finally, the vacuolar type proton-translocating pyrophosphatase gene (vH+-PPi) is down-regulated under TPS.
Signal transduction and cell cycle regulation
Protein kinase and cAMP signalling pathways have not been elucidated in Leishmania to date. A first approach is to find the TPS-generated down-regulation of a receptor-type adenylate cyclase-like (ACR), a serine/threonine protein phosphatase (Ser/Thr PPase) and three non-ligated cysteine peptidase/calpain C2 family (C2cp) genes. C2cp genes are also involved in cytoskeleton remodelling (see further explanation in the following section). The down-regulation under TPS of a serine/threonine protein kinase (Ser/Thr PK) may be related to the down-regulation of calmodulin, which binds calcium divalent cations after its activation by PK phosphorylation. Ser/Thr PK is also down-regulated under TS. The exact physiological functions of the calcium-calmodulin pathway have not been described in Leishmania either and the inositolphospholipid regulator pathway also remains uncharacterised. For this reason, there is no known biological meaning for the down-regulation of PI3K and the up-regulation of INO1 under TPS and TS found in the microarray hybridisation analysis. INO1 down-regulation has been observed in L. infantum axenic and intracellular amastigotes [28]. Apart from this, a cyclin dependent kinase binding protein (Cdkbp) and a serine peptidase E from the S51 family (S51) are also down-regulated and might be involved in S-phase or mitosis entry.
Cytoskeleton and flagellum
Several genes associated with the flagellar and paraflagellar rod structures are down-regulated under TPS: coronin, dynein heavy chain LinJ36_V3.2010 and LinJ26_V3.1000 and PFR1D, the latter also being up-regulated under TS. Dynein heavy chain LinJ26_V3.1000 down-regulation has also been described in AL [28] and intracellular amastigotes (unpublished data). Apart from that, we have found that an unknown tubulin-associated GTPase is up-regulated under TPS, as well as RABreg, an activator of prenylated RAB GTPases. An analogue of a RAB small GTPase is up-regulated in L. major amastigotes and may be related with pathogeny, as vesicle transport is essential for extracellular nutrient acquisition, release of virulence factors, microbicidal resistance and evasion of host immune responses [47]. In addition, calpains are involved in cytoskelleton remodelling and signal transduction in kinetoplastid parasites (reviewed in [48]). μ-calpain C2cp LinJ20_V3.1230 (see also the Signal transduction and cell cycle regulation subsection) is down-regulated under TPS and up-regulated in metacyclic promastigotes [25]. Moreover, L. mexicana [17] and L. donovani [33] mature intracellular amastigotes also down-regulate this gene. As a consequence, the greatest transcript levels of -calpain are reached in metacyclic promastigotes.
gRNAs
According to BLAST outcome mapping against GenBank database, 19 clones map against 4 different minicircle sequences (contig 200, 692, 878 and 957) (Additional file 3: Table S7) and according to the microarray hybridisation analysis, they must contain an up-regulated gene under TPS. Provided that each minicircle contains only a single gRNA gene for site-specific uridine insertion/deletion type RNA editing, 4 gRNA genes with unknown target are presumably up-regulated under TPS. The gRNA genes corresponding to contigs 878 (1 clone) and 957 (16 clones) have also been found to be up-regulated under PS (Additional file 3: Table S8) and were previously found as up-regulated in metacyclic promastigotes [25].
Other genes
Leishmania promastigotes use chitinase to break the chitinous peritrophic membrane inside the gut of the sand-fly vector [49]. Chitinase gene is up-regulated in TPS-induced AL, which is consistent with chitinase overexpression reported in amastigotes, as well as the associated enhanced lesion development observed in mice [50], suggesting an additional or different function for this gene. The microarray hybridisation analysis has also revealed that SbGRP is down-regulated in TPS-induced AL forms, as well as in L. mexicana [17], L. major [32] and L. infantum (unpublished data) intracellular amastigotes. In fact, a decrease in pentavalent antimonial resistance is a feature of AL, together with round morphology typical of amastigotes, the up-regulation of A2 cluster and the down-regulation of 3'NT/Nase, as reviewed previously [13].
The membrane bound acid phosphatase gene (MBAP) is down-regulated under TPS as revealed by microarray analysis. There is evidence to confirm that it is essential for cell survival, because it plays a critical role in nutrition. It is located in small vesicles between the Golgi apparatus and the flagellar pocket (secretory pathway) [51]. MBAP levels have been described as being higher in procyclic promastigotes rather than in metacyclics. It was reported that its activity is higher in virulent clones and consequently, it was supposed that it was involved in virulence in spite of the higher levels of MBAP protein found in logarithmic phase promastigotes according to [52], but further experiments have demonstrated the opposite. L. mexicana MBAP knockout parasites show that it is neither involved in the infection process nor required for amastigote survival in the infected host cell [51]. This supports our results concerning MBAP gene down-regulation in TPS-induced AL forms.
Hybridisation analysis has revealed the down-regulation of a hypothetical protein (CoB) with copper ion binding/transport and chaperone GO molecular functions under TPS. CoB is located in the mitochondrial lumen according to GO cellular component term analysis. Two genes from the HASP/SHERP cluster are differentially regulated by TPS: a small hydrophilic ER-associated protein (SHERP) is down-regulated by TPS; and hydrophilic surface protein (HASPB) is up-regulated under TPS, as well as in intracellular L. donovani promastigotes at the post-translational level [33]. In contrast, down-regulation was reported in L. major [16]. Despite these genes being previously presumed to be metacyclic promastigote-specific in L. major [16], we have reported a different pattern for HASPB in L. infantum. Finally, this analysis also revealed the up-regulation of an esterase-like protein, carbamoyl-phosphate synthetase (CPS), a short chain dehydrogenase and ubiquitin-conjugating enzyme-like proteins in TPS-induced AL, and the latter also under TS.
Conclusions
Absence of gp46 expression observed by means of IFA and up-regulation of the amastigote-specific A2 gene has been found in TPS-treated cells. As a consequence, we know that TPS leads to differentiation into AL. The up-regulation of several amastin genes and the down-regulation of 3'NT/Nase and SbGRP genes under TPS and TS point to a developmental process towards amastigote differentiation by the combined effect of temperature increase and acidification and the single effect of temperature. By contrast, none of these genes have been found to be differentially regulated under PS, which suggests that pH decrease itself does not prompt amastigote differentiation in the parasite. A wider analysis of TPS-, TS- and PS-induced expression profiles throughout HCL-ST clustering analysis of gene expression supports temperature shift alone or combined with acidification as triggering differentiation towards the amastigote stage whereas acidification itself does not. In fact, we have described examples of known annotated genes taken directly from the microarray output, namely the up-regulation of RRS1, INO1 and aATP11 and the down-regulation of 3'NT/Nase, PT, GT, SOD, PI3K, FKBP, calmodulin and lathosterol oxidase. These observations have led us to conclude that temperature increase is more relevant than pH decrease in the differentiation process to the amastigote stage with regard to transcriptome variation in L. infantum. In addition, we have provided the first description of transcriptome variation induced by the specific influence of temperature increase and acidification.
Methods
Parasite cultures and RNA isolation
Cultures of L. infantum isolate M/CAN/ES/98/10445 (zymodeme MON-1) from early passages after axenization were grown at a starting density of 4 × 106 mid-logarithmic phase promastigotes/ml in RPMI 1640 medium supplemented with L-glutamine (Cambrex, Karlskoga, Sweden), 10% heat inactivated foetal bovine serum (HIFBS) (Cambrex) and 100 μg/ml streptomycin - 100 IU/ml penicillin (Cambrex) at 27°C/pH7.2 (CC), 37°C/pH4.5 (TPS), 37°C/pH7.2 (TS) or 27°C/pH4.5 (PS). Cell density was counted daily and promastigotes were harvested at 2000 g for 10 min on day 4. RNA isolations were performed from 2 × 108 cells/ml of TRIzol® reagent (Invitrogen, La Jolla, CA) following the manufacturer's instructions. Three biological replicates of the cultures were carried out for each of the conditions described.
gp46 IFA
Cells were fixed with acetone:methanol (1:1) at -20°C for 10 min at a density of 2 × 106/5 μl drop. Then, they were incubated with purified anti-gp46 monoclonal IgG antibody at 37°C for 30 min in a hydration chamber, washed three times with PBS by mild agitation for 10 min, incubated with fluorescein isotiocyanate (FITC)-conjugated goat anti-mouse IgG antibody (Serotec, Raleigh, NC) and 0.1% Evans' Blue (Fisher, Pittsburgh, PA). Washes were repeated and preparations mounted with 90% glycerol. Negative control of the primary antibody was anti-rabbit complement factor H monoclonal IgG and the first incubation was carried out with PBS for the negative control of the secondary antibody. SIM 110 monoclonal IgG antibody against soluble leishmanial antigens (SLA) was used as a positive control. Anti-gp46, anti-rabbit factor H and SIM110 antibodies were kindly provided by Mercedes Domínguez (Centro Nacional de Microbiología, Virología e Inmunología Sanitarias, Instituto de Salud Carlos III, Majadahonda, Spain).
L. infantum DNA microarray construction and hybridisation
L. infantum DNA microarray construction and hybridisation assays were carried out as described previously [25]. To summarize, microarrays were generated by long template PCR amplification from a complete shotgun DNA-pUC18 genomic library with m13-pUC18 primers and spotting onto epoxy-coated slides. RNA quality was assessed by capillary electrophoresis, mRNA was amplified, cDNA was synthesised and indirectly labelled and L. infantum DNA microarrays blocked, hybridised and washed as detailed in [25]. Hybridisation assays were performed as follows: 37°C/pH4.5 vs. 27°C/pH7.2 (TPS vs. CC), 37°C/pH7.2 vs. 27°C/pH7.2 (TS vs. CC) and 27°C/pH4.5 vs. 27°C/pH7.2 (PS vs. CC). Hybridised microarrays were scanned and fluorescence intensity was analysed for Cy3 (532 nm) and Cy5 (635 nm) with local feature background subtraction (GenePix 4100A scanner and software, Axon Instruments, Foster City, CA). LOWESS per pin algorithm was used to normalise raw data (AlmaZen software, BioAlma, Tres Cantos, Spain). After that, comparative analysis of the replicates by paired t-test and selection of spots with meaningful values of stage-specific regulation were performed as described [25].
DNA sequencing and analysis
Clones corresponding to selected spots were sequenced and mapped following a strategy that has already been described in detail [25]. Briefly, insert ends were dideoxi-sequenced with m13-pUC18 primers and aligned against the L. infantum genome project sequence in General Feature Format (GFF) deposited in a GBrowse database. Forward and reverse reads were mapped to define the boundaries of the clones in the genome of L. infantum. Depending upon the insert length, the success of sequencing reactions of both ends and genome sequence complexity, three possibilities arose: when one pair of convergently oriented alignments separated by up to 11 Kbp were found, the clone mapping outcome was defined as type a; when more than one pair of alignments fulfilled those conditions, the best pair of alignments was used to define the boundaries of the clone, resulting in a type b outcome; and when those requirements were not fulfilled (incongruent pair of alignments or unpaired alignments), the outcome was defined as type c. Some of the clones were annotated by a custom Glimmer 3.0 analysis because they did not map against genes previously annotated on the L. infantum genome project sequence. Stage-specifically regulated genes were re-annotated and analysed with BLAST2GO to establish molecular function and biological process GO term distribution among them based on α-scores [53].
Multi-experiment SAM and the subsequent iterative hierarchical clustering-support tree analysis (HCL-ST) were carried out with TIGR's MultiExperiment Viewer 4.3 (MEV) by introducing normalised microarray hybridisation data matrixes (including medians and standard deviations of intensity and F values) of clones with significant differential regulation in each individual experiment. SAM p-value cutoff was 0.05, the same as for the previous independent t-tests for each experiment. HCL-ST was performed independently for significant and non-significant genes. ST algorithm with a jackknifing resampling option and 100 iterations for the construction and clustering of the gene expression matrix were applied in HCL-ST analysis. CLUSTALW2 was used for sequence alignments of amastin, GT and 3'NT/Nase genes differentially regulated by the effect of pH and/or temperature and CBS's TMHMM 2.0 for the prediction of transmembrane helices in these proteins.
qRT-PCR
qRT-PCR reactions were performed to determine whether a gene overlapping with a type c sequence end is developmentally regulated or to ascertain which gene is developmentally regulated in the clones overlapping more than one gene. We described previously the procedure applied [25]. The reference gene was 18S rRNA. When there were two copies of a gene in tandem in a given clone but one of the copies lacked a segment of the 5' end or differed in a specific sequence (GT and 3'NT/Nase genes), two pairs of primers were designed for qRT-PCR. A complete list of primers used for qRT-PCR is provided in Additional file 5.
Abbreviations
aap: amino acid permease; aATP11: amino acid transporter 11; ABCE: ABC transporter subfamily E (ribonuclease L-inhibitor) gene; ACR: receptor-type adenylate cyclase; ACT: acyl-CoA transferase; AL: amastigote-like; ALD: fructose-1,6-bisphosphate aldolase; AQ: aquaporin; CC: culture control conditions; C2cp: cysteine protease family C2; cdkbp: cyclin-dependent kinase binding protein; C(III)O: coproporphyrinogen oxidase; CoA: coenzyme A; CoB: cofactor binding protein; Cph: cyclophilin; CPS: carbamoyl phosphate synthetase; Cy: cyanin; cyt b5: cytochrome b5; DAG: direct acyclic graph; 1,2-DAG, 1,2-diacylglycerol; DGC: directional gene cluster; DHAP: dihydroxyacetone phosphate; EF1α, elongation factor 1 α; eIF5a: eukaryotic translation initiation factor 5a; ER: endoplasmic reticulum; ETCH: electron transport chain; F: Fold change; FA: fatty acid; FITC: fluorescein isotiocyanate; FKBP: FK506-binding protein; FU: Fluorescence Units; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; αGII-HPB: hypothetical calcium ion binding protein from α-glycosidase II complex; gMDH: glycosomal malate dehydrogenase; GNAT: glucose-6-phosphate N-acetyltransferase; GO: Gene Ontology; gp46: 46 KDa surface glycoprotein; GPDE: glycerolphosphodiester phosphodiesterase; gRNA: guide RNA; GT: glucose transporter; H3: histone H3; HASPB: hydrophilic surface protein B; HCL-ST: Hierarchical clustering-Support Tree; HIFBS: heat inactivated foetal bovine serum; HMG-CoA: 3-hydroxymethylglutaryl-CoA; HMGCR: HMG-CoA reductase; HPT: hypothetical protein transport protein; HTreg: hypothetical transcription regulator; IFA: immunofluorescence analysis; IPC: inositol phosphoceramide; INO1: myo-inositol-1-phosphate synthase; LOWESS: Locally Weighted Scatter Plot Smoothing algorithm; LPG: lypophosphoglycan; mACDH: mitochondrial acyl-CoA dehydrogenase; MBAP: membrane bound acid phosphatase; MEV: Multi Experiment Viewer; MGL: monoglyceride lipase; mmc: minichromosome maintenance complex protein; MRP: multidrug resistance protein; NPC: nuclear pore complex; 3'NT/Nase: 3'-nucleotidase/nuclease; β-ox: β-oxidation of FA; PES: prostaglandin peroxide synthase complex; PFR1D: paraflagellar rod protein 1D; PG: phosphoglycerate; PGFS: prostaglandin F synthase; PGMBPI: bisphosphoglycerate-independent phosphoglycerate mutase; PI3K: phosphatidylinositol triphosphate kinase; PO: protoporphyrinogen oxidase; PPG: proteophosphoglycan; PS: pH shift; PSA2: promastigote-specific surface antigen 2; PT: pteridine transporter; qRT-PCR: relative quantitative real time PCR; R: set of ribosomal proteins; RABreg: Rab GTPase regulator; RNAbp: RNA-binding protein; RRS1: ribosome assembly protein; S51: serine peptidase A family S51; SAGE: Serial Analysis of Gene Expression; SAM: Serial Analysis of Microarrays; Ser/Thr PK: serine/threonine protein kinase; SbGRP: sodium stibogluconate resistance protein; Ser/Thr PPase: Ser/Thr protein phosphatase; SHERP: small hydrophilic ER-associated protein; SLA: soluble leishmanial antigen; SOD: superoxide dismutase; TAG: triacylglycerol; TCA: tricarboxylic acid cycle; TFSUI1: translation factor SUI1; TGL: TAG lipase; TPS: temperature-pH shift; TS: temperature shift; TR: trypanothione reductase; vH+-PPi: vacuolar-type proton-translocating pyrophosphatase; ZnT: Zn transporter.
Authors' contributions
PJA, AA, ASG, MM, VP and VL contributed to the design of shotgun genomic DNA microarrays. PJA and ASG constructed the genomic library and PJA, AA, ASG and MM contributed to the microarray construction. PJA, AA, MM and EG contributed to the optimisation of the hybridisation procedure. The experimental design of biological sample preparation and expression profile analysis of temperature and acidification was carried out by PJA, AT and VL. PJA and AT contributed to gp46 IFA. Microarray hybridisation data acquisition and analysis was performed by PJA and AA with the assistance of MM, EG and VP. Sequence analysis, clone assembly and mapping against L. infantum genome, gene annotations with Glimmer software, minicircle sequence analyses, GO annotations and BLAST2GO analysis were performed by MJG. qRT-PCR analyses were performed by PJA with the assistance of AT. PJA, AA and VL contributed to the interpretation of data. The manuscript was drafted by PJA, AA and VL. All authors revised the manuscript thoroughly and made important contributions to the intellectual content of this manuscript. VL approved the version to be published.
Supplementary Material
Electropherograms of total RNA samples. Figure S1. 18S, 23Sα and 23Sβ spikes, absence of DNA contamination and RNA degradation.
Scatter plots of normalised and contrasted microarray hybridisation data. Figure S2. Spots that fulfill criteria to be considered as differentially regulated are highlighted.
Differentially regulated hypothetical and unknown genes and unresolved clones. Tables S1, S2, S3, S4, S5, S6, S7 and S8. Tables S7 and S8 describe clones containing minicircle sequences.
DAGs (BLAST2GO output). Figures. S3 and S4. GO codes for functions directly annotated on differentially regulated genes found in this study. Each GO code is associated to a custom code to find annotations on genes in Tables 1, 2, 3, 4 and 5.
3'NT/Nase and GT alignments and oligonucleotides for qRT-PCR. Figure S6 (GT and 3'NT/Nase alignments) and Additional file 3: Table S9 (oligonucleotides for qRT-PCR).
Fold change clusters of differentially regulated genes including clone names. Figure S5. Supplementary information for Figure 4 identifying the profiles with the clone numbers.
Contributor Information
Pedro J Alcolea, Email: pjalcolea@cib.csic.es.
Ana Alonso, Email: amalonso@cib.csic.es.
Manuel J Gómez, Email: gomezrmj@inta.es.
Alicia Sánchez-Gorostiaga, Email: asanchez@cnb.csic.es.
Mercedes Moreno-Paz, Email: morenopm@inta.es.
Eduardo González-Pastor, Email: gonzalezpje@inta.es.
Alfredo Toraño, Email: atorano@isciii.es.
Víctor Parro, Email: parrogv@inta.es.
Vicente Larraga, Email: vlarraga@cib.csic.es.
Acknowledgements
We thank Juan Pérez Mercader, Merecedes Domínguez, Rafael Giraldo and Francisco Ferrezuelo for their unconditional support and Marina Postigo, Marta Godoy and Eduardo Gil for their excellent technical assistance. This work has been supported by the Ministerio de Educación y Ciencia (MEC) project AGL2003-06152-C02-01 and the internal budget of the Centro de Astrobiología. PJA thanks Consejo Superior de Investigaciones Científicas (CSIC) for I3P-BPD2003-1 grant and AA thanks CSIC for the JAE-Doc027 (2008) contract.
References
- Sacks DL, Perkins PV. Identification of an infective stage of Leishmania promastigotes. Science. 1984;223(4643):1417–1419. doi: 10.1126/science.6701528. [DOI] [PubMed] [Google Scholar]
- Chang KP, Dwyer DM. Multiplication of a human parasite (Leishmania donovani) in phagolysosomes of hamster macrophages in vitro. Science. 1976;193(4254):678–680. doi: 10.1126/science.948742. [DOI] [PubMed] [Google Scholar]
- Alexander J, Russell DG. The interaction of Leishmania species with macrophages. Advances in parasitology. 1992;31:175–254. doi: 10.1016/S0065-308X(08)60022-6. [DOI] [PubMed] [Google Scholar]
- Pral EM, da Moitinho ML, Balanco JM, Teixeira VR, Milder RV, Alfieri SC. Growth phase and medium ph modulate the expression of proteinase activities and the development of megasomes in axenically cultivated Leishmania (Leishmania) amazonensis amastigote-like organisms. J Parasitol. 2003;89(1):35–43. doi: 10.1645/0022-3395(2003)089[0035:GPAMPM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Somanna A, Mundodi V, Gedamu L. In vitro cultivation and characterization of Leishmania chagasi amastigote-like forms. Acta Trop. 2002;83(1):37–42. doi: 10.1016/S0001-706X(02)00054-2. [DOI] [PubMed] [Google Scholar]
- Debrabant A, Joshi MB, Pimenta PF, Dwyer DM. Generation of Leishmania donovani axenic amastigotes: their growth and biological characteristics. International journal for parasitology. 2004;34(2):205–217. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Saar Y, Ransford A, Waldman E, Mazareb S, Amin-Spector S, Plumblee J, Turco SJ, Zilberstein D. Characterization of developmentally-regulated activities in axenic amastigotes of Leishmania donovani. Molecular and biochemical parasitology. 1998;95(1):9–20. doi: 10.1016/S0166-6851(98)00062-0. [DOI] [PubMed] [Google Scholar]
- Antoine JC, Prina E, Lang T, Courret N. The biogenesis and properties of the parasitophorous vacuoles that harbour Leishmania in murine macrophages. Trends Microbiol. 1998;6(10):392–401. doi: 10.1016/S0966-842X(98)01324-9. [DOI] [PubMed] [Google Scholar]
- Shapira M, McEwen JG, Jaffe CL. Temperature effects on molecular processes which lead to stage differentiation in Leishmania. Embo J. 1988;7(9):2895–2901. doi: 10.1002/j.1460-2075.1988.tb03147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina P, Kaur S. Chronic heat-shock treatment driven differentiation induces apoptosis in Leishmania donovani. Mol Cell Biochem. 2006;289(1-2):83–90. doi: 10.1007/s11010-006-9151-5. [DOI] [PubMed] [Google Scholar]
- Zilberstein D, Blumenfeld N, Liveanu V, Gepstein A, Jaffe CL. Growth at acidic pH induces an amastigote stage-specific protein in Leishmania promastigotes. Molecular and biochemical parasitology. 1991;45(1):175–178. doi: 10.1016/0166-6851(91)90040-D. [DOI] [PubMed] [Google Scholar]
- Abou el Naga IF, el Moneim Sharaf IA. Metacyclogenesis of Leishmania major in an acidic medium. J Egypt Soc Parasitol. 2000;30(2):431–440. [PubMed] [Google Scholar]
- Zilberstein D. In: Leishmania after the genome. Myler PJ, Fassel N, editor. Norfolk: Caister Academic Press; 2008. Physiological and Biochemical Aspects of Leishmania Development. Chapter 6. [Google Scholar]
- Zilberstein D, Shapira M. The role of pH and temperature in the development of Leishmania parasites. Annu Rev Microbiol. 1994;48:449–470. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- Almeida R, Gilmartin BJ, McCann SH, Norrish A, Ivens AC, Lawson D, Levick MP, Smith DF, Dyall SD, Vetrie D. et al. Expression profiling of the Leishmania life cycle: cDNA arrays identify developmentally regulated genes present but not annotated in the genome. Mol Biochem Parasitol. 2004;136(1):87–100. doi: 10.1016/j.molbiopara.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Akopyants NS, Matlib RS, Bukanova EN, Smeds MR, Brownstein BH, Stormo GD, Beverley SM. Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Mol Biochem Parasitol. 2004;136(1):71–86. doi: 10.1016/j.molbiopara.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Holzer TR, McMaster WR, Forney JD. Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146(2):198–218. doi: 10.1016/j.molbiopara.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Saxena A, Lahav T, Holland N, Aggarwal G, Anupama A, Huang Y, Volpin H, Myler PJ, Zilberstein D. Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol Biochem Parasitol. 2007;152(1):53–65. doi: 10.1016/j.molbiopara.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicoll F, Drummelsmith J, Muller M, Madore E, Boilard N, Ouellette M, Papadopoulou B. A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics. 2006;6(12):3567–3581. doi: 10.1002/pmic.200500853. [DOI] [PubMed] [Google Scholar]
- Kahl LP, McMahon-Pratt D. Structural and antigenic characterization of a species- and promastigote-specific Leishmania mexicana amazonensis membrane protein. J Immunol. 1987;138(5):1587–1595. [PubMed] [Google Scholar]
- Handman E, Osborn AH, Symons F, van Driel R, Cappai R. The Leishmania promastigote surface antigen 2 complex is differentially expressed during the parasite life cycle. Mol Biochem Parasitol. 1995;74(2):189–200. doi: 10.1016/0166-6851(95)02500-6. [DOI] [PubMed] [Google Scholar]
- Zakai HA, Chance ML, Bates PA. In vitro stimulation of metacyclogenesis in Leishmania braziliensis, L. donovani, L. major and L. mexicana. Parasitology. 1998;116(Pt 4):305–309. doi: 10.1017/S0031182097002382. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhao Y, Ni B, Yao C, Zhou Y, Xu W, Wang Z, Qiao Z. Comparison of the expression profiles of promastigotes and axenic amastigotes in Leishmania donovani using serial analysis of gene expression. Parasitol Res. 2008;103(4):821–828. doi: 10.1007/s00436-008-1048-7. [DOI] [PubMed] [Google Scholar]
- Guerfali FZ, Laouini D, Guizani-Tabbane L, Ottones F, Ben-Aissa K, Benkahla A, Manchon L, Piquemal D, Smandi S, Mghirbi O. et al. Simultaneous gene expression profiling in human macrophages infected with Leishmania major parasites using SAGE. BMC Genomics. 2008;9:238. doi: 10.1186/1471-2164-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcolea PJ, Alonso A, Sanchez-Gorostiaga A, Moreno-Paz M, Gomez MJ, Ramos I, Parro V, Larraga V. Genome-wide analysis reveals increased levels of transcripts related with infectivity in peanut lectin non-agglutinated promastigotes of Leishmania infantum. Genomics. 2009;93(6):551–564. doi: 10.1016/j.ygeno.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Teixeira SM, Russell DG, Kirchhoff LV, Donelson JE. A differentially expressed gene family encoding "amastin," a surface protein of Trypanosoma cruzi amastigotes. The Journal of biological chemistry. 1994;269(32):20509–20516. [PubMed] [Google Scholar]
- Rochette A, McNicoll F, Girard J, Breton M, Leblanc E, Bergeron MG, Papadopoulou B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol Biochem Parasitol. 2005;140(2):205–220. doi: 10.1016/j.molbiopara.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Rochette A, Raymond F, Corbeil J, Ouellette M, Papadopoulou B. Whole-genome comparative RNA expression profiling of axenic and intracellular amastigote forms of Leishmania infantum. Mol Biochem Parasitol. 2009;165(1):32–47. doi: 10.1016/j.molbiopara.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Charest H, Matlashewski G. Developmental gene expression in Leishmania donovani: differential cloning and analysis of an amastigote-stage-specific gene. Mol Cell Biol. 1994;14(5):2975–2984. doi: 10.1128/mcb.14.5.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Matlashewski G. Loss of virulence in Leishmania donovani deficient in an amastigote-specific protein, A2. Proc Natl Acad Sci USA. 1997;94(16):8807–8811. doi: 10.1073/pnas.94.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Matlashewski G. Characterization of the A2-A2rel gene cluster in Leishmania donovani: involvement of A2 in visceralization during infection. Mol Microbiol. 2001;39(4):935–948. doi: 10.1046/j.1365-2958.2001.02286.x. [DOI] [PubMed] [Google Scholar]
- Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152(1):35–46. doi: 10.1016/j.molbiopara.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Rosenzweig D, Smith D, Opperdoes F, Stern S, Olafson RW, Zilberstein D. Retooling Leishmania metabolism: from sand fly gut to human macrophage. Faseb J. 2008;22(2):590–602. doi: 10.1096/fj.07-9254com. [DOI] [PubMed] [Google Scholar]
- Bisbal C, Salehzada T, Silhol M, Martinand C, Le Roy F, Lebleu B. The 2-5A/RNase L pathway and inhibition by RNase L inhibitor (RLI) Methods Mol Biol. 2001;160:183–198. doi: 10.1385/1-59259-233-3:183. [DOI] [PubMed] [Google Scholar]
- Ternes P, Franke S, Zahringer U, Sperling P, Heinz E. Identification and characterization of a sphingolipid delta 4-desaturase family. J Biol Chem. 2002;277(28):25512–25518. doi: 10.1074/jbc.M202947200. [DOI] [PubMed] [Google Scholar]
- Ginger ML, Chance ML, Goad LJ. Elucidation of carbon sources used for the biosynthesis of fatty acids and sterols in the trypanosomatid Leishmania mexicana. Biochem J. 1999;342(Pt 2):397–405. doi: 10.1042/0264-6021:3420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConville MJ, De Souza DP, Saunders EC, Pyke J, Naderer T, Ellis MA, Sernee FM, Ralton JE, Likic VA. In: Leishmania after the genome. Myler PJ, Fassel N, editor. Norfolk: Caister Academic Press; 2008. Analysis of the Leishmania Metabolome. [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Molec Biochem Parasitol. 2007;156(2):93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Steenkamp DJ. Thiol metabolism of the trypanosomatids as potential drug targets. IUBMB Life. 2002;53(4-5):243–248. doi: 10.1080/15216540212649. [DOI] [PubMed] [Google Scholar]
- Dumas C, Ouellette M, Tovar J, Cunningham ML, Fairlamb AH, Tamar S, Olivier M, Papadopoulou B. Disruption of the trypanothione reductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. The EMBO journal. 1997;16(10):2590–2598. doi: 10.1093/emboj/16.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacci JB Jr, Campbell TA, Gottlieb M. Leishmania donovani: regulated changes in the level of expression of the surface 3'-nucleotidase/nuclease. Exp Parasitol. 1990;71(2):158–168. doi: 10.1016/0014-4894(90)90018-8. [DOI] [PubMed] [Google Scholar]
- Gbenle GO. Characterization of a filtrable 3'-nucleotidase of Leishmania donovani. Biochim Biophys Acta. 1993;1203(1):162–167. doi: 10.1016/0167-4838(93)90051-r. [DOI] [PubMed] [Google Scholar]
- Debrabant A, Bastien P, Dwyer DM. A unique surface membrane anchored purine-salvage enzyme is conserved among a group of primitive eukaryotic human pathogens. Mol Cell Biochem. 2001;220(1-2):109–116. doi: 10.1023/A:1010809420104. [DOI] [PubMed] [Google Scholar]
- Sopwith WF, Debrabant A, Yamage M, Dwyer DM, Bates PA. Developmentally regulated expression of a cell surface class I nuclease in Leishmania mexicana. Int J Parasitol. 2002;32(4):449–459. doi: 10.1016/S0020-7519(01)00372-1. [DOI] [PubMed] [Google Scholar]
- Figarella K, Uzcategui NL, Zhou Y, LeFurgey A, Ouelette M, Bhattacharjee H, Mukhopadhyay R. Biochemical characterization of Leishmania major aquaglyceroporin LmAQP1: possible role in volume regulation and osmotaxis. Molecular Microbiology. 2007;65(4):1006–1017. doi: 10.1111/j.1365-2958.2007.05845.x. [DOI] [PubMed] [Google Scholar]
- Chenik M, Chaabouni N, Ben Achour-Chenik Y, Ouakad M, Lakhal-Naouar I, Louzir H, Dellagi K. Identification of a new developmentally regulated Leishmania major large RAB GTPase. Biochem Biophys Res Commun. 2006;341(2):541–548. doi: 10.1016/j.bbrc.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ersfeld K, Barraclough H, Gull K. Evolutionary relationships and protein domain architecture in an expanded calpain superfamily in kinetoplastid parasites. Journal of molecular evolution. 2005;61(6):742–757. doi: 10.1007/s00239-004-0272-8. [DOI] [PubMed] [Google Scholar]
- Schlein Y, Jacobson RL, Shlomai J. Chitinase secreted by Leishmania functions in the sandfly vector. Proc Biol Sci. 1991;245(1313):121–126. doi: 10.1098/rspb.1991.0097. [DOI] [PubMed] [Google Scholar]
- Joshi MB, Rogers ME, Shakarian AM, Yamage M, Al-Harthi SA, Bates PA, Dwyer DM. Molecular characterization, expression, and in vivo analysis of LmexCht1: the chitinase of the human pathogen, Leishmania mexicana. J Biol Chem. 2005;280(5):3847–3861. doi: 10.1074/jbc.M412299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzel I, Weise F, Wiese M. Deletion of the gene for the membrane-bound acid phosphatase of Leishmania mexicana. Mol Biochem Parasitol. 2000;111(1):77–86. doi: 10.1016/S0166-6851(00)00306-6. [DOI] [PubMed] [Google Scholar]
- Katakura K, Kobayashi A. Acid phosphatase activity of virulent and avirulent clones of Leishmania donovani promastigotes. Infect Immun. 1988;56(11):2856–2860. doi: 10.1128/iai.56.11.2856-2860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (Oxford, England) 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electropherograms of total RNA samples. Figure S1. 18S, 23Sα and 23Sβ spikes, absence of DNA contamination and RNA degradation.
Scatter plots of normalised and contrasted microarray hybridisation data. Figure S2. Spots that fulfill criteria to be considered as differentially regulated are highlighted.
Differentially regulated hypothetical and unknown genes and unresolved clones. Tables S1, S2, S3, S4, S5, S6, S7 and S8. Tables S7 and S8 describe clones containing minicircle sequences.
DAGs (BLAST2GO output). Figures. S3 and S4. GO codes for functions directly annotated on differentially regulated genes found in this study. Each GO code is associated to a custom code to find annotations on genes in Tables 1, 2, 3, 4 and 5.
3'NT/Nase and GT alignments and oligonucleotides for qRT-PCR. Figure S6 (GT and 3'NT/Nase alignments) and Additional file 3: Table S9 (oligonucleotides for qRT-PCR).
Fold change clusters of differentially regulated genes including clone names. Figure S5. Supplementary information for Figure 4 identifying the profiles with the clone numbers.