Abstract
The cottontail rabbit papillomavirus (CRPV) animal model is used in several laboratories worldwide to investigate immunogenicity, carcinogenicity and life cycle aspects of papillomaviruses. It is the only animal model in which the full life cycle of the virus from initiation of infection to malignant progression can be studied. A major strength of the model is that the viral DNA is infectious. This feature allows for the study of mutant genomes without the need to create infectious mutant virus. Results from laboratory to laboratory have not always been consistent. Different laboratories use different methods for creating infections from DNA and it was postulated that the different challenge methods could play a role in the differential outcomes. Because different laboratories use different strains of CRPV, it was also desirable to test if the difference in CRPV genomes contributed to the differential outcomes. In this study, three of the CRPV strains used most widely (Washington B, Orth CRPV and Hershey CRPV) were cloned into PUC19; the E8ATG ko mutants for each strain were also generated. We employed the infection technique reported previously in which scarification is done first and is followed with delivery of DNA by pipette three days later. The papilloma outgrowth generated by these three wild type constructs and their E8ATG ko mutants was compared. No significant difference was found among the three strains or their E8ATG ko mutants. E8ATGko mutants induced significantly smaller but persistent papillomas when compared to their respective wild type CRPVs. The gene gun was also used to create infections with both Hershey CRPV DNA and the corresponding E8 ATG ko and was found to lead to less vigorous growth as well as some regressions. Further studies suggested that gene gun delivery might have induced an immune response which then resulted in compromised growth of papillomas. It was concluded that the E8 gene is not required for We suggest that standardized infection methods should be used in laboratories so that inconsistencies in conclusions will be minimized.
1. INTRODUCTION
Papillomaviruses are double stranded DNA tumor viruses with genomes of about 8Kb. More than 100 human viruses have been identified to date and of these, a number are associated with cancers. The most notable are human papillomaviruses (HPV) 16 and 18 which infect mucosal sites and are implicated in most of the cases of cervical cancer worldwide. The cutaneous viruses are common in patients with the congenital disorder Epidermodysplasia verruciformis and often lead to skin malignancies in those individuals. Other papillomaviruses have been implicated in skin cancers in immunocompetent individuals (zur Hausen, 2000). There is an ongoing need to learn more about these viruses. Papillomaviruses require differentiating cells to complete their life cycle and thus cannot be propagated in tissue culture. A subset of the viruses can be grown in raft cultures and this tool has provided a way to explore many aspects of the viral life cycle. However, a robust animal model is required to investigate contributions of the immune system to control of infection; such a model is also essential for studying malignant progression.
The cottontail rabbit papillomavirus (CRPV) animal model is a powerful tool to study the immunogenicity and oncogenicity of papillomaviruses. It is the only animal model available in which the entire life cycle of the virus from initiation of infection to malignant progression can be investigated. The CRPV model system has been in use in this laboratory for many years and is also employed by several other investigators around the world. (Brandsma, et al., 1991; Brandsma, 2005; Jeckel, et al, 2002; Zeltner, et al., 1994; Breitburd, et al., 1997; Breitburd F, 2007; Bodily, et al., 1999; Duan, et al., 2000). The strength of the model has been encumbered, however, by the failure to use consistent procedures to initiate infection. Whereas most laboratories utilize the gene gun™ to create infections from DNA, this laboratory uses direct application of DNA by pipette to prescarified sites (Cladel, et al., 2008a). The technique yields consistent infections at an efficiency approaching 100%. In contrast, gene gun technology for inducing infection has been less successful in this laboratory.
The need for a consistent delivery technique is critical to the outcome of experiments. This is demonstrated clearly, for example, by the conflicting results obtained by this laboratory and that of Nonnenmacher, et al, (2006) with respect to the requirement for an intact E8 gene for CRPV infection. These investigators reported that the E8 gene is essential for infection, whereas this laboratory reported earlier that the gene is dispensable for infection (Hu, et al., 2002) and that its absence does not prevent production of high titer infectious virus (Christensen, 2005).
The present study was designed to address several questions: 1) Could the strain of virus be a factor in inter-laboratory discrepancies? 2) Are gene gun and pipette delivery infections comparable? 3) Do the E8 ko mutants of different viral strains behave similarly? 4) Does gene gun delivery of CRPV DNA potentially provoke an immune response that can compromise the infection?
Results of this study suggested that the strain of CRPV made little difference in outcome. The three viral strains were found to grow at about the same rate and their E8 mutants were equally infectious. These results clearly confirmed earlier reports from this laboratory that the E8 gene is not required for infection (Hu, et al, 2002). These findings support the conclusion that differences in observations between laboratories using the CRPV model could be traced to differences in infection technique.
2. MATERIALS AND METHODS
2.1 Plasmid DNA
Hershey CRPV (H. CRPV), Washington B. CRPV (W. CRPV) and Orth CRPV (O. CRPV) genomes were cloned into PUC19 at the Sal1 restriction site, transfected into E.coli and purified, first by the Qiagen maxiprep system and then by CsCl density centrifugation as described previously(Cladel, et al, 2008a). The H. CRPV genome was cloned and sequenced in this laboratory from a pool of virus obtained from Kansas cottontail rabbits (Kreider, et al, 1995). Washington B and Orth CRPVs were archival samples obtained many years ago from Felix Wettstein and Gerard Orth respectively. The original Orth genome was cloned at EcoR1 in pBR322. It was recloned into PUC 19 at Sal1 in this laboratory. The Wettstein genome was originally cloned in plasmid pLA2 at Sal1. It was recloned into PUC 19 in this laboratory. All three genomes, H. CRPV, W. CRPV and O. CRPV, were in antisense orientation with respect to the vector.
Sequence similarity appears to be high from the limited sequencing we have done. There are differences, however. One region of divergence is in the area of the putative E5 gene (Brandsma, et al., 1992; data cited in this paper). The differences result in a premature stop for H. CRPV as well as frameshifts and thus amino acid alterations in the other two genomes. We also found differences in the URRs and in the E6 genes. (Table 1). We have not sequenced the entire O. CRPV and W. CRPV clones and thus there may be other variations.
Table 1.
Nucleotide differences found in the Orth and Washington B clones based on partial sequencing of the Orth and Washington B genomes. Sequence positions are referenced to the H. CRPV sequence. Frameshifts and amino acid changes are noted.
| n.t. position | Strain | Location | AA change? |
|---|---|---|---|
| 394 A>C | Orth | E6 and E8 | Thr to Pro in E6 No change in E8 |
| 569 C>A | Wash. B | E6 | Ser to Tyr |
| 7432 T>C | Orth | URR | n.a. |
| 7475C>T | Orth | URR | n.a. |
| 7486 T>C | Orth | URR | n.a. |
| 7609 A | Missing in Orth | URR | n.a. |
| 16 G>A | Wash. B | URR | n.a. |
| 33 G>A | Wash. B | URR | n.a. |
| Between 58, 59 G | Insertion ; Wash. B | URR | n.a. |
| 67 G>T | Wash. B | URR | n.a. |
| 69 A>T | Wash. B | URR | n.a. |
| 75 G>A | Wash. B | URR | n.a. |
| 79 G>C | Wash. B | URR | n.a. |
| 94 G>A | Wash. B | URR | n.a. |
| 4336-8 del TTT | Orth | E5 | del Phe |
| 4336-7 del TT | Wash. B | E5 | frameshift |
| Between 4362, 4363 add ACA |
Orth | E5 | add His |
| 4372-3 del AC | Orth | E5 | frameshift |
| 4421 T>C | Orth | E5 | within the frameshift |
E8 ATG ko mutants were prepared using site directed mutagenesis. Complementary primers 192 (CAT AAA GGG TGG CCG TAC GGG ACC TGC AGA GAC) and 193(GTC TCT GCA GGT CCC GTA CGG CCA CCC TTT ATG) were used to mutagenize the E8 ATG to ACG. This mutation does not change the coding sequence in the overlapping E6 gene. There were no sequence differences in this region of the three genomes and this made it possible to use the same primer set for each of the three genomes. The Avr2 (741) - Cel2 (7068) portion of each genome was sequenced to verify the ATG to ACG mutation and to assure that no other mutations had been introduced into this fragment of the genome. The Avr2-Cel2 fragment from each wild type genome was then excised and replaced by the respective fragment containing the E8 ATG mutation. Thus the three E8 mutant genomes were also in reverse orientation with respect to the PUC 19 vector.
2.2 DNA Infections
New Zealand white rabbits were purchased from Covance (Denver, PA) and maintained in the animal facility at the Pennsylvania State University College of Medicine. The studies were approved by the Institutional Animal Care and Use Committee of the Pennsylvania State University. All rabbits were anesthetized using an intramuscular combination of Xyla-ject (Xylazine) at 5 mg/kg body weight and Ketaset (Ketamine HCl) at 40 mg/kg body weight prior to viral DNA inoculations.
Infection by pipette application of plasmid DNA to prescarified sites was done as previously reported (Cladel, et al, 2008a). This work had shown that infectivity was improved by at least two orders of magnitude when the DNA was delivered to sites that had been scarified three days prior to infection. 5µg of plasmid DNA in 50µl TE was applied per site unless other wise noted.
For the gene gun infections, bullets were prepared according to the protocol of the Gene Gun manufacturer (Bio-Rad, Hercules, CA) using 1.6mm gold particles and DNA at a ratio of 0.5mg gold to 1 microgram DNA. Gold particles were purchased from Seashell Technologies (LaJolla, CA) and tubing from Saint-Gobain Performance Plastics (Mickleton, NJ). Ten µg plasmid DNA was delivered in ten shots per site at a pressure of 375psi. Scarification three days prior to infection or no scarification was done depending upon the experiment.
2.3 Comparison of infectivities of three viral strains and their E8 ATG mutants
On day -3, three groups of four rabbits each were scarified on five sites each on the left and right sides as per our normal procedure (Cladel, et al, 2008a). On day zero, each group of animals was infected on the left sides with one of the wild type genomes and on the right sides with the respective E8 mutant. Animals were monitored daily and papilloma measurements were taken weekly by the same technician to maintain consistency in week to week size evaluations. Geometric mean diameters (GMD’s) were calculated. Data were entered into SigmaPlot and mean and SEM for each time point for each group were computed. Plots were generated using SigmaPlot.
2.4 Comparison of infectivities of Gene Gun and pipette delivery of wild type H. CRPV and its E8 mutant on scarified and non-scarified sites
Ten animals were treated as shown in Table 2A. Ten µg DNA was delivered to each site. Animals were monitored daily and papilloma measurements were taken weekly by the same technician to maintain consistency in week to week size evaluations. Geometric mean diameters (GMD’s) were calculated. Data were entered into SigmaPlot and mean and SEM for each time point for each group were computed. Plots were generated using SigmaPlot. Table 2B shows the numbers of positive sites at week 11 after infection (week 8 of papilloma growth).
Table 2.
| Table 2A Comparison of gene gun and pipette infections of H. CRPV and H. CRPV E8ko in rabbits. Each of ten rabbits was infected as follows: L(left side)1, L3: E8ko via gene gun; L2, L4, H. CRPV via gene gun; R(right side)1, R3, E8ko via pipette; R2, R4, H. CRPV via pipette. Sites L1, R1 and L2, R2 received no scarification prior to infection. Sites L3, R3 an L4, R4 were scarified three days prior to infection. | ||||
|---|---|---|---|---|
| Infections by gene gun |
DNA | Infections by pipette delivery |
DNA | Scarification at day -3? |
| L1 | E8M | R1 | E8M | No |
| L2 | wt.H. | R2 | wt. H. | No |
| CRPV | CRPV | |||
| L3 | E8 M | R3 | E8M | Yes |
| L4 | wt. H. | R4 | wt. H. | Yes |
| CRPV | CRPV | |||
| Table 2B Positive papilloma sites at week 11 following infection on rabbits infected with H. CRPV and the E8ko as shown in table 1A. Note that all animals experienced the gene gun although only half of the infections were initiated by gene gun. | |||||
|---|---|---|---|---|---|
| Gene gun Delivery Site Scarification? |
Genome | Paps/ total infection sites Week 11 |
Pipette Delivery Site Scarification? |
Genome | Paps/ total infection sites Week 11 |
| L1 no | E8M | 1/10 | R1 no | E8M | 2/10 |
| L2 no | W.T. | 1/10 | R2 no | W.T. | 4/10 |
| L3 yes | E8M | 1/10 | R3 yes | E8M | 2/10 |
| L4 yes | W.T. | 5/10 | R4 yes | W.T. | 8/10 |
2.5 Experimental plan to test contributions of host immunity on subsequent CRPV infection
Four groups of four animals each (three animals for the no immunization group) were established. (Table 3). Each animal was scarified at five sites on the left side and five on the right on day -3 as per normal protocol (Cladel, et al, 2008a). On day zero, 10µg of E8M H. CRPV plasmid DNA was delivered by gene gun to the sites on the left side and the same amount of wild type DNA to the sites on the right. Group A received no additional treatment, Group B received 20 shots of vector DNA, Group C, 20 shots of H. CRPV plasmid DNA and Group D 20 shots of E8 ATG ko Mutant DNA. All additional shots were done on the skin on the back of the neck. The area was shaved but not scarified prior to the shots. Animals were monitored daily and papilloma measurements were taken weekly by the same technician to maintain consistency in week to week size evaluations. Geometric mean diameters (GMD’s) were calculated. Data were entered into SigmaPlot and mean and SEM for each time point for each group were computed. Plots were generated using SigmaPlot.
Table 3.
Influence of inoculations on unscarified neck sites on papilloma development on scarified sites. Each of four animals was infected by Gene Gun on the left sides with E8koCRPV and on the right sides with H. CRPV. Infection sites were scarified three days prior to infection. Group A received no gene gun shots to unscarified neck sites. Group B received vector shots at these sites, Group C received wild type H. CRPV shots and Group D received E8ko shots.
| GROUP | Papilloma outgrowth after DNA challenge by Gene-gun |
|
|---|---|---|
| E8 ATG koCRPV |
WT CRPV | |
| A (N=3) no shots |
4/12 (33.3%) |
12/12 (100%) |
| B (N=4) 20 shots of vector |
9/16 (56.25%) |
14/16 (87.5%) |
| C (N=4) 20 shots of wt CRPV |
4/16 (25%) |
8/16 (50%) |
| D (N=4) 20 shots of E8 ATG ko |
3/16 (18.75%) |
16/16 (100%) |
2.6 Statistics
Papilloma size was determined by calculating the cubic root of the product of length × width × height of individual papillomas in millimeters to obtain a geometric mean diameter (GMD). Data were entered into SigmaPlot and means and standard errors (SEM) for each test group were calculated.
3. RESULTS
3.1 Growth rates of the three viral strains and their respective E8 ATG mutants
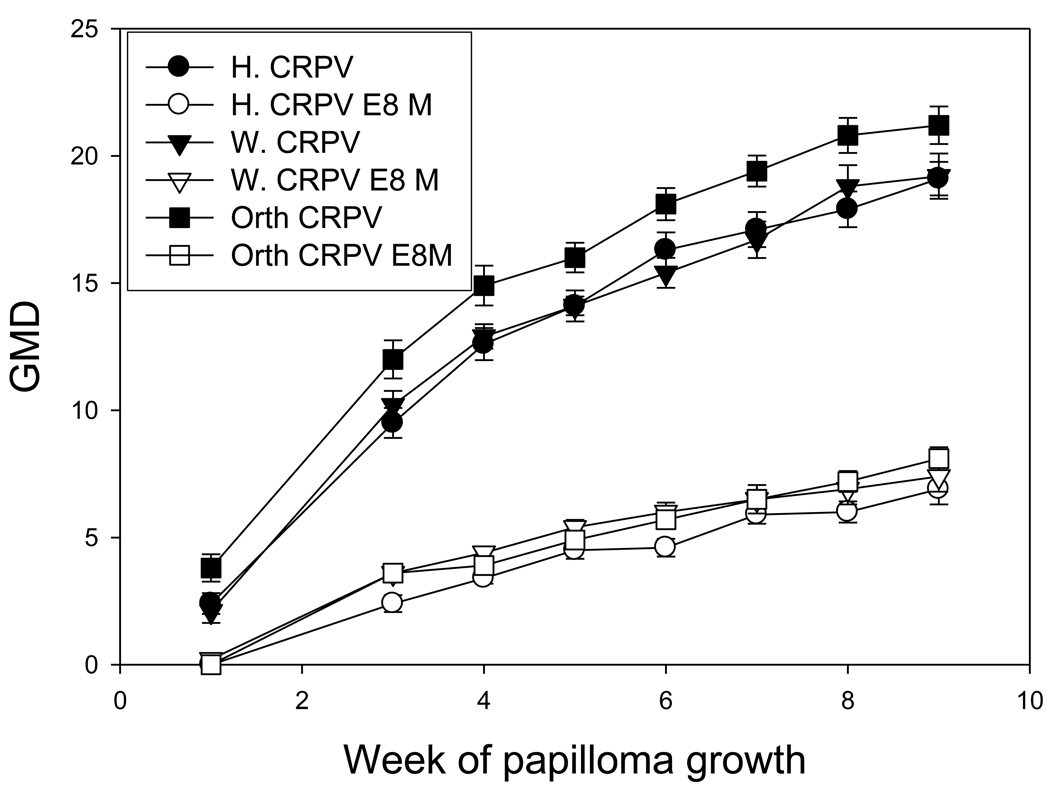
Hershey, Washington B and Orth strains of CRPV and their respective E8 ATG ko mutants were challenged on three groups of four rabbits each as described in the Methods section. Each construct had 5 infection sites on each animal (20 total challenge sites for each construct). From three weeks following DNA challenge, the tumor outgrowth was monitored weekly until the termination of the experiment. 100% of the challenge sites infected with wild type CRPV DNA or E8 ko DNA grew tumors. Papillomas induced by the three wild type CRPV strains were comparable and so were their respective E8 mutants (P>0.05, unpaired student t test, Figure 1). E8 mutants induced significantly smaller tumors when compared with their respective wild type DNA (P<0.05, unpaired student t test, Figure 1). However, 100% of the E8 mutants induced papillomas which remained persistent and grew slowly but steadily.
Figure 1.
Comparison of the three vial strains and their E8 ATG ko mutants. Growth was comparable for the three viral strains. The respective E8 mutants grew at similar rates but slower than wild type virus.
3.2 Comparison between gene gun infections and infections by direct application of viral DNA
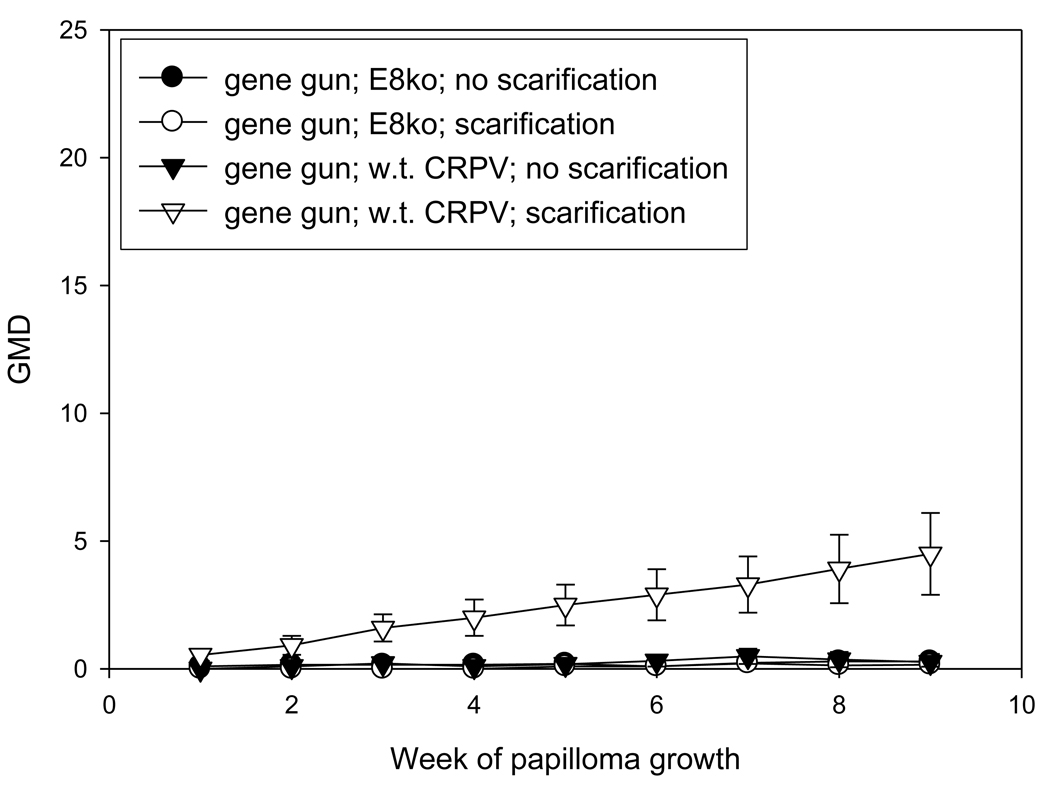
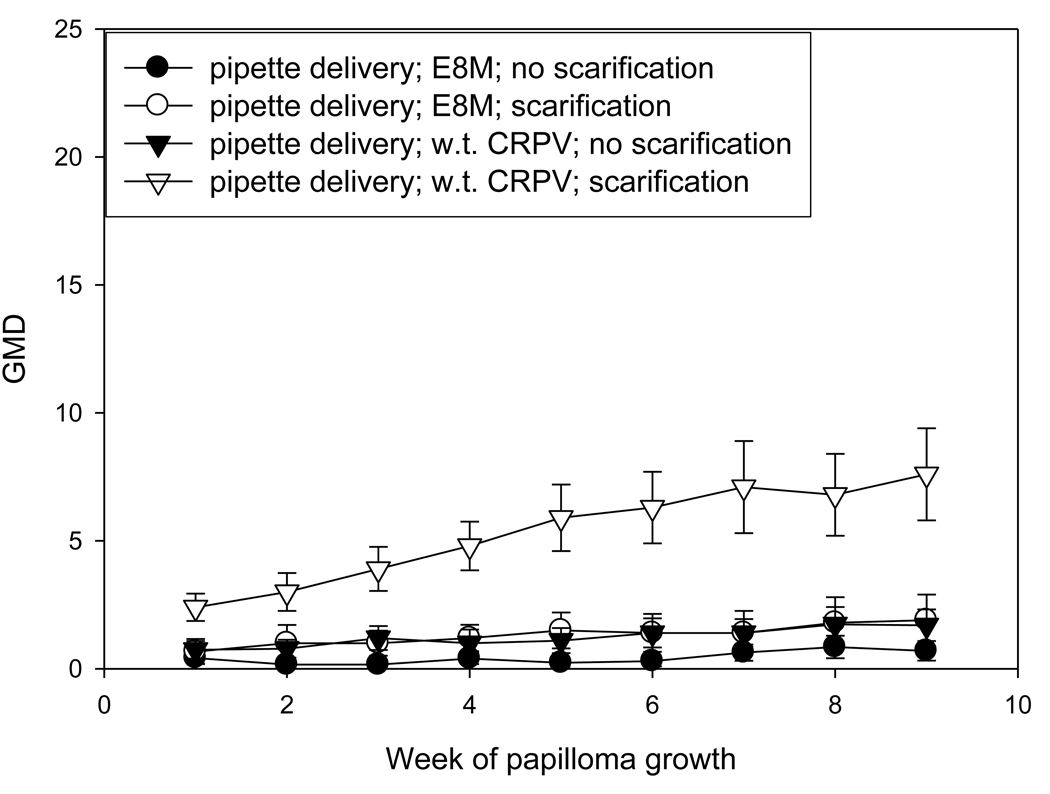
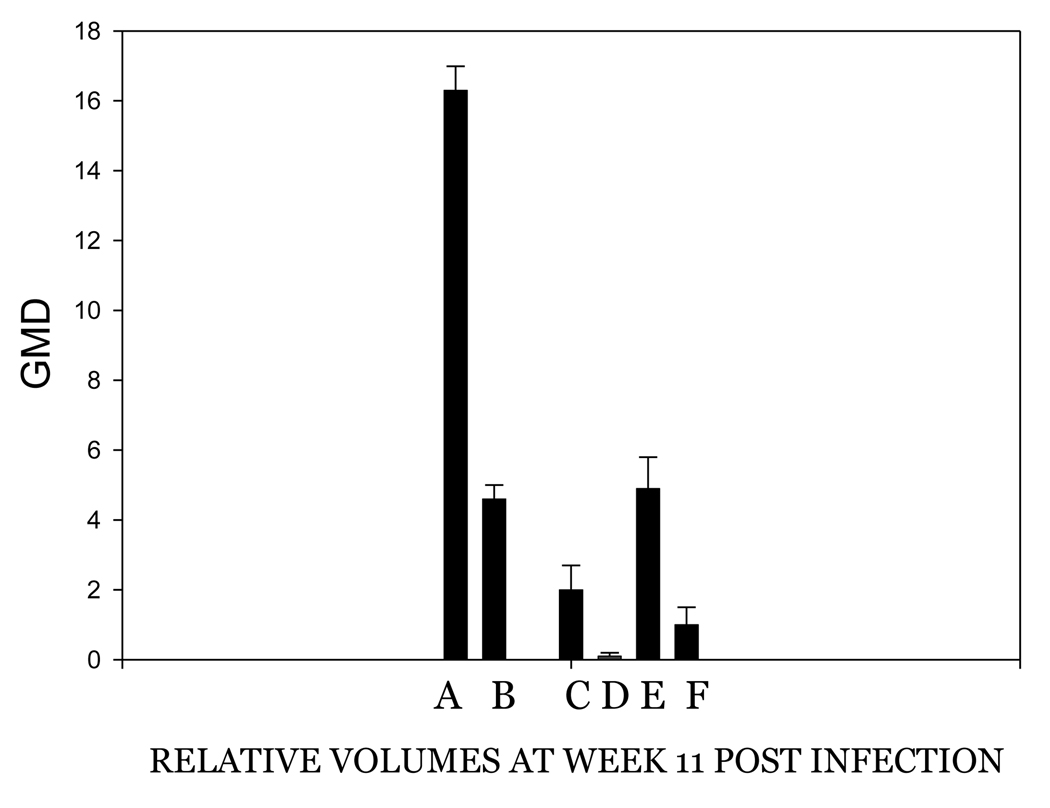
The investigators who reported the requirement for an intact E8 ATG for successful infection used the gene gun for the delivery of DNA (Nonnenmacher, et al 2006). It was therefore desirable to test whether the infection method was a critical factor in the failure of E8 mutant infections. Because prescarification has been shown to play a very important role in the success and consistency of DNA infection via pipette, an experiment was designed to compare the efficiency of infections by gene gun and pipette application of H. CRPV plasmid DNA and H. CRPV E8 ko plasmid DNA on both scarified and unscarified sites. In this experiment, each animal received each treatment in order to allow for direct comparisons between treatments. Scarification three days prior to infection was again demonstrated to yield more reliable infections than no prior scarification. Gene gun infections produced fewer papillomas than did direct application infections. (P< 0.05, student t test). Multiple regressions were also noted, a process that is rarely observed with H. CRPV delivered by pipette. Finally, gene gun delivery of DNA influenced the growth of papillomas initiated via direct delivery of DNA (Figures 2A and 2B). Papilloma volumes were variable and growth was slower than that observed in animals in which the DNA was delivered by pipette only. Table 2B compares the numbers of papillomas found at week 8 of papilloma growth with each treatment in this experiment. Figure 3 shows H. CRPV and E8 ATG ko papilloma sizes at week 11 when there was no gene gun influence and compares these with papilloma sizes in animals experiencing the gene gun. All sites were infected by pipette delivery. The data in this figure for animals without gene gun influence were extracted from the experiment comparing the three viral strains and is comparable to measurements found in other experiments at week 11 (data not shown). The influence of the gene gun is clearly visible in the smaller papillomas generated in animals treated with the gene gun. It is hypothesized that the gene gun delivery of viral DNA may provoke an immune response in the animals, thereby encouraging regressions and/or slow and variable growth of papillomas.
Figure 2.
Figure 2A
Influence of the gene gun on the growth of both wild type H. CRPV and E8ko papillomas initiated by gene gun. Sites were either scarified at day -3 or were not scarified prior to infection. The scale is the same as that used in Figure 1 in order to facilitate comparison.
Figure 2B
Influence of the gene gun on the growth of both wild type CRPV and E8ko papillomas initiated by pipette. Sites were either scarified at day -3 or were not scarified prior to infection. The scale is the same as that used in Figure 1 in order to facilitate comparison. Note that all animals also had gene gun infections (figure 2A) and thus were influenced by the gene gun even though the infections represented in this graph were initiated by pipette.
Figure 3.
Papilloma volumes at week 8 of papilloma growth. A. Wild type H. CRPV, pipette delivery, no gene gun influence. B. E8ko H. CRPV, pipette delivery, no gene gun influence. C. Wild type, gene gun infection. D. E8ko gene gun infection. E. Wild type, pipette delivery in animals also experiencing the gene gun. F. E8ko, pipette delivery in animals also experiencing the gene gun.
3.3 Induction of host immunity via additional shots of E8ATG ko CRPV and wild type CRPV
It was suggested from the work above that gene gun delivery might prime a host immune response that reduced the efficiency of papilloma initiation and growth. We hypothesized that additional shots with either w.t. H. CRPV or E8 ko H. CRPV would enhance this effect. To this end, sites were scarified three days prior to infection and then infected via gene gun with E8 ATG ko H. CRPV DNA on one side of the animals and with H. CRPV DNA on the other side. Animals were then administered additional shots via gene gun on unscarified sites on the neck as noted in the methods section. We chose to initiate infections by gene gun rather than by pipette delivery as we knew the gene gun infections were less robust and we felt that we might have a better opportunity to detect an additional immune response initiated by the shots to unscarified neck sites. A parallel experiment using pipette delivery of DNA to scarified sites and gene gun “immunization” at the unscarified neck sites would have been desirable if the animals had been available.
There were three animals in group A, the control animals that received no neck shots. Two of the rabbits were partially protected (animals 1961 and 1965). The third (1979) grew papillomas at a rate approximating that for pipette delivery of DNA. For this control group of three animals, it was concluded that the DNA used to initiate infections was itself sufficient to mount an immunological response in two of the three animals.
Group B animals received neck shots of vector DNA. Two of these animals (1978 and 1960) had wild type papillomas approaching the size of those initiated via pipette delivery of DNA. The E8 ko papillomas of animal 1960 approached the sizes of those resulting from pipette delivery. Animals 1982 and 1964 had smaller papillomas and it was concluded that those animals mounted an immunological response upon infection.
Group C animals were treated with wild type H. CRPV DNA at the neck sites. Two of the four animals in this group (1959 and 1981) were free from tumors following infection with both wild type and E8 ko H. CRPV DNA. A third animal had very small wild type papillomas and no E8 ko papillomas (1977). The fourth animal, 1963, grew papillomas with no evidence of immunological compromise. Group D animals were treated at neck sites with H. CRPV E8 ko DNA. All papillomas appeared to be compromised by the gene gun infections. It was not possible to conclude that the extra shots enhanced this response, however. The results of this experiment corroborate earlier findings that gene gun infections, alone, may result in an immunological response that serves to compromise papilloma growth. Additional shots, as reported in this study, appeared to amplify the response. This was especially evident in Group 3, the group treated with additional shots of wild type H. CRPV DNA, in which two animals were completely protected from infection and a third one was partially protected. Gene-gun challenge has been reported to result in more regressions following CRPV DNA infection (Salmon, et al, 2000). A possible increased T cell-mediated immune response induced by gene gun infection was suggested as the explanation. Findings from this study further confirmed that gene gun delivery compromised tumor outgrowth in rabbits and that extra shots of H. CRPV DNA at neck sites significantly suppressed papilloma growth (Table 3, P<0.05 vs. vector control group, Fisher’s exact test).
It was of interest to note whether neck sites for groups C and D developed papillomas since these animals were treated by the technique used by other laboratories to initiate infections. Of the four animals treated with E8 ko H. CRPV DNA, there were very small papillomas on the neck sites of animals 1976 and 1980. These papillomas regressed and then reappeared. Of those receiving extra shots of wild type genome, animal 1977, the one with small w.t. papillomas, had a few very tiny papillomas on the neck. A second animal, 1963, developed a large neck papilloma mass. This animal is the one that did not experience any apparent immunological response to the gene gun infection. These data demonstrate the influence of animal to animal variation in helping to determine outcome of infection. The animals in all of the experiments reported here were outbred. The data also demonstrate that the failure to scarify sites prior to delivery of CRPV DNA by gene gun often results in no obvious infection.
4. DISCUSSION
One of the major strengths of the CRPV model lies in the ability to generate infections from viral DNA. This allows mutations to be introduced into the genome and the effects to be tested in vivo without the need to first generate virus. This infection strategy has been an important research tool in this laboratory (Cladel, et al., 2008a; Hu, et. al., 2002; Hu, et. al, 2002; Hu et al., 2006 Hu, et. al., 2009,) and has been the basis of research in other laboratories as well (Brandsma, et al, 1991; Brandsma, et al, 1992; Jeckel, et al, 2002; Bodily, et al, 1999; Duan, et al, 2000; Nonnenmacher, et al, 2006; Salmon, et al, 2000). The method of DNA delivery differs, however, from laboratory to laboratory and may be responsible for differences in outcome of experiments and subsequent interpretation of data. In addition, different laboratories use different viral strains. The purpose of this study was to examine the influence of both viral strain and delivery method on the initiation and continuation of DNA infections.
Papillomaviruses take advantage of abrasions in skin tissue to gain access to epithelial stem cells in the hair follicle where infection is initiated ((Jeckel, et al, 2002; Schmitt, et al, 1996). The events associated with wound healing are felt to play a role in the establishment of infection. The finding that scarification three days prior to infection greatly improves infections of both virus and viral DNA supports this theory (Cladel, et al, 2008a). In this technique, DNA is applied by pipette on day 0 to sites prescarified at day -3. DNA is scraped into the site gently with the tip of a 25 gauge needle. Efficiency of infection with this technique approaches 100%. Viral titer is improved at least two orders of magnitude. Other laboratories have used the gene gun, tattoo gun or Bioject to initiate infection with viral DNA (Brandsma, et a., 1991; Brandsma, et al, 1992; Jeckel, et al, 2002; Salmon, et al, 2000; Nonnenmacher, et al, 2006). The gene gun has been used extensively for the delivery of DNA vaccines (Han, et al., 2000; Han, et al, 2000; Han, et a., 1999; Hu, et al, 2002; Hu, et al, 2006; Hu, et al, 2008; Cladel, et al, 2008b) and is the main method for vaccination of rabbits in this laboratory. This vaccine system is effective in eliciting T cell-mediated immune responses (Alvarez, et al., 2005; Christensen, 2005; Sasaki, et al., 2002). However, on several occasions when CRPV DNA has been delivered by gene gun to initiate infections in this laboratory, success has been limited (data not reported). It was hypothesized that gene gun delivery may stimulate an immune response that subsequently interferes with the development and/or maintenance of papillomas. In this laboratory, a DNA challenge method which leads to consistent growth and high yield of papillomas has been established and standardized. Different infection methods may help to explain the disparities between the data of Nonnenmacher, et al (2006) and those of laboratory with respect to the requirement of the E8 gene for function.
Nonnenmacher, et al (2006) reported that the E8 gene of CRPV is “essential for wart formation”. However, studies in this laboratory showed that the E8 mutant produces high titer virus and proved that the E8 gene is not essential (Christensen, 2005). Nonnenmacher et al (2006) argued that conditions for infection used in the Hershey laboratory provided wounding and that the wounding substituted for the need for the E8 protein. They postulated that the E8 gene is necessary to trigger cell proliferation and that the wounding technique substituted for the function of E8. Papillomas initiated by the standardized technique with CRPV E8 ATG ko virus or viral DNA grow steadily, albeit slowly, and do not experience regressions. If the E8 gene were necessary to provide the proliferative environment necessary for the establishment and maintenance of infection, the E8 mutant genome infections would fail to survive once the site had healed from its initial scarification. Indeed, the conditions provided by the standardized infection technique mirror more closely those of the natural infection than do those of the gene gun. Wounding is the route by which a papillomavirus gains access to the basal layers of the epithelium and to the hair follicles; wound healing provides the milieu which the virus needs to begin its life cycle. The standardized technique establishes both the route and the healing milieu, and under these conditions, the E8 gene is not required for successful infection nor for maintenance of infection. The data in this study would suggest that the reason that Nonnenmacher et al (2006) concluded that the E8 gene was essential for infection was because the gene gun technique may have elicited an immune response that abrogated or masked infection in their animals. The effect was more evident for gene gun infection with the E8 mutant than with the wild type genome. Previous work in their laboratory (Salmon et al, 2000) supports this postulate. In that work, regressions were higher in gene gun infections than in those initiated by direct DNA application and the authors hypothesized that this could be due to direct transfection of Langerhans cells and resultant stimulation of T-cell responses. In view of this observation, it is curious that subsequent investigators in the same laboratory chose to use the gene gun for infections rather than to use direct application of DNA.
Another variable between laboratories using the CRPV model is the strain of virus. A genome from a mixture of Kansas cottontail papillomas has been isolated and cloned in this laboratory and is known as Hershey (H.) CRPV. This is the strain that has been in use in this laboratory for many years. Two other strains in use in other laboratories are the Washington B (W) and the Orth (O) strains. These latter strains share considerable homology with H. CRPV but do have a number of differences in the putative E5 region, in the E6 gene and in the URR. In this study, it was shown that strain of virus could not account for differences found from laboratory to laboratory. Infection profiles for three strains of CRPV and their E8 mutants were monitored and little difference was found between strains.
These basic experiments point out the importance of technique in the outcome of experiments. The results also argue strongly for standardization of methodologies in laboratories using the same model systems so that results can be more readily compared from laboratory to laboratory. This laboratory has been striving to establish techniques that can be easily adopted by other laboratories working with the CRPV model. Previous work (Cladel, et. al, 2008a) describes a method of infection that has been found to work consistently and uniformly.
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute grant R01 CA47622 from the National Institutes of Health and the Jake Gittlen Memorial Golf Tournament.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez D, Harder G, Fattouh R, Sun JF, Goncharova S, Stampfli MR, Coyle AJ, Bramson JL, Jordana M. Cutaneous antigen priming via gene gun leads to skin-selective Th2 immune-inflammatory responses. Journal of Immunology. 2005;174:1664–1674. doi: 10.4049/jimmunol.174.3.1664. [DOI] [PubMed] [Google Scholar]
- Bodily JM, Hoopes DJ, Roeder BL, Gilbert SG, Pettit GR, Herald CL, Rollins DN, Robison RA. The inhibitory effects of bryostatin 1 administration on the growth of rabbit papillomas. Cancer Lett. 1999;136:67–74. doi: 10.1016/s0304-3835(98)00310-3. [DOI] [PubMed] [Google Scholar]
- Brandsma JL. The cottontail rabbit papillomavirus model of high-risk HPV-induced disease. Methods Mol.Med. 2005;119:217–235. doi: 10.1385/1-59259-982-6:217. [DOI] [PubMed] [Google Scholar]
- Brandsma JL, Yang ZH, Barthold SW, Johnson EA. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proc.Natl.Acad.Sci.U.S A. 1991;88:4816–4820. doi: 10.1073/pnas.88.11.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsma JL, Yang ZH, DiMaio D, Barthold SW, Johnson E, Xiao W. The putative E5 open reading frame of cottontail rabbit papillomavirus is dispensable for papilloma formation in domestic rabbits. J Virol. 1992;66:6204–6207. doi: 10.1128/jvi.66.10.6204-6207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitburd F, Nonnenmacher M, Salmon J, Orth G. S.Campo. Papillomavirus research: from natural history to vaccines and beyond. Norwich, UK: Caister Academic Press; 2007. Rabbit viral papillomas and carcinomas: model diseases for human papillomavirus infections and carcinogenesis; pp. 339–420. Ref Type: Electronic Citation. [Google Scholar]
- Breitburd F, Salmon J, Orth G. The rabbit viral skin papillomas and carcinomas: a model for the immunogenetics of HPV-associated carcinogenesis. Clin.Dermatol. 1997;15:237–247. doi: 10.1016/s0738-081x(97)00009-6. [DOI] [PubMed] [Google Scholar]
- Christensen ND. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir.Chem.Chemother. 2005;16:355–362. doi: 10.1177/095632020501600602. [DOI] [PubMed] [Google Scholar]
- Cladel NM, Hu J, Balogh K, Mejia A, Christensen ND. Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection. J Virol.Methods. 2008a;148:34–39. doi: 10.1016/j.jviromet.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel NM, Hu J, Balogh KK, Christensen ND. CRPV genomes with synonymous codon optimizations in the CRPV E7 gene show phenotypic differences in growth and altered immunity upon E7 vaccination. PLoS ONE. 2008b;3:e2947. doi: 10.1371/journal.pone.0002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Paris W, De Marte J, Roopchand D, Fleet TL, Cordingley MG. Topical effects of cidofovir on cutaneous rabbit warts: treatment regimen and inoculum dependence. Antiviral Res. 2000;46:135–144. doi: 10.1016/s0166-3542(00)00080-2. [DOI] [PubMed] [Google Scholar]
- Han R, Cladel NM, Reed CA, Peng XW, Budgeon LR, Pickel M, Christensen ND. DNA vaccination prevents and/or delays carcinoma development of papillomavirus-induced skin papillomas on rabbits. Journal of Virology. 2000;74:9712–9716. doi: 10.1128/jvi.74.20.9712-9716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Reed CA, Cladel NM, Christensen ND. Immunization of rabbits with cottontail rabbit papillomavirus E1 and E2 genes: protective immunity induced by gene gun-mediated intracutaneous delivery but not by intramuscular injection. Vaccine. 2000;18:2937–2944. doi: 10.1016/s0264-410x(00)00110-9. [DOI] [PubMed] [Google Scholar]
- Han RC, Cladel NM, Reed CA, Peng XW, Christensen ND. Protection of rabbits from viral challenge by gene gun-based intracutaneous vaccination with a combination of cottontail rabbit papillomavirus E1, E2, E6, and E7 genes. Journal of Virology. 1999;73:7039–7043. doi: 10.1128/jvi.73.8.7039-7043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cladel N, Peng X, Balogh K, Christensen ND. Protective immunity with an E1 multivalent epitope DNA vaccine against cottontail rabbit papillomavirus (CRPV) infection in an HLA-A2.1 transgenic rabbit model. Vaccine. 2008;26:809–816. doi: 10.1016/j.vaccine.2007.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Balogh K, Budgeon L, Christensen ND. Impact of genetic changes to the CRPV genome and their application to the study of pathogenesis in vivo. Virology. 2007:384–390. doi: 10.1016/j.virol.2006.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Budgeon LR, Reed CA, Pickel MD, Christensen ND. Protective cell-mediated immunity by DNA vaccination against papillomavirus L1 capsid protein in the cottontail rabbit papillomavirus model. Viral Immunology. 2006b;19:492–507. doi: 10.1089/vim.2006.19.492. [DOI] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Pickel MD, Christensen ND. Amino acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus E6 influence spontaneous regression of cutaneous papillomas. J.Virol. 2002;76:11801–11808. doi: 10.1128/JVI.76.23.11801-11808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Han R, Cladel NM, Pickel MD, Christensen ND. Intracutaneous DNA vaccination with the E8 gene of cottontail rabbit papillomavirus induces protective immunity against virus challenge in rabbits. J.Virol. 2002;76:6453–6459. doi: 10.1128/JVI.76.13.6453-6459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeckel S, Huber E, Stubenrauch F, Iftner T. A transactivator function of cottontail rabbit papillomavirus e2 is essential for tumor induction in rabbits. J.Virol. 2002;76:11209–11215. doi: 10.1128/JVI.76.22.11209-11215.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreider JW, Cladel NM, Patrick SD, Welsh PA, DiAngelo SL, Bower JM, Christensen ND. High efficiency induction of papillomas in vivo using recombinant cottontail rabbit papillomavirus DNA. J.Virol.Methods. 1995;55:233–244. doi: 10.1016/0166-0934(95)00062-y. [DOI] [PubMed] [Google Scholar]
- Nonnenmacher M, Salmon J, Jacob Y, Orth G, Breitburd F. Cottontail rabbit papillomavirus E8 protein is essential for wart formation and provides new insights into viral pathogenesis. J.Virol. 2006;80:4890–4900. doi: 10.1128/JVI.80.10.4890-4900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Nonnenmacher M, Caze S, Flamant P, Croissant O, Orth G, Breitburd F. Variation in the nucleotide sequence of cottontail rabbit papillomavirus a and b subtypes affects wart regression and malignant transformation and level of viral replication in domestic rabbits. J. Virol. 2000;74:10766–10777. doi: 10.1128/jvi.74.22.10766-10777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Xin KQ, Okudela K, Okuda K, Ishii N. Immunomodulation by apoptosis-inducing caspases for an influenza DNA vaccine delivered by gene gun. Gene Therapy. 2002;9:828–831. doi: 10.1038/sj.gt.3301696. [DOI] [PubMed] [Google Scholar]
- Schmitt A, Rochat A, Zeltner R, Borenstein L, Barrandon Y, Wettstein FO, Iftner T. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J Virol. 1996;70:1912–1922. doi: 10.1128/jvi.70.3.1912-1922.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeltner R, Borenstein LA, Wettstein FO, Iftner T. Changes in Rna Expression Pattern During the Malignant Progression of Cottontail Rabbit Papillomavirus-Induced Tumors in Rabbits. Journal of Virology. 1994;68:3620–3630. doi: 10.1128/jvi.68.6.3620-3630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J.Natl.Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]