Abstract
Synthetic protocells provide a new means to probe, mimic and deconstruct cell behavior; they are a powerful tool to quantify cell behavior and a useful platform to explore nanomedicine. Protocells are not simple particles; they mimic cell design and typically consist of a stabilized lipid bilayer with membrane proteins. With a finite number of well characterized components, protocells can be designed to maximize useful outputs. Energy conversion in cells is an intriguing output; many natural cells convert transmembrane ion gradients into electricity by membrane-protein regulated ion transport. Here, a synthetic cell system comprising two droplets separated by a lipid bilayer is described that functions as a biological battery. The factors that affect its electrogenic performance are explained and predicted by coupling equations of the electrodes, transport proteins and membrane behavior. We show that the output of such biological batteries can reach an energy density of 6.9 × 106 J·m−3 which is ≈ 5 % of the volumetric energy density of a lead-acid battery. The configuration with maximum power density has an energy conversion efficiency of 10 %.
Keywords: Synthetic cell, protocell, biological energy conversion, ion transport, biomimetic
1. Introduction
It is now possible to create protocells - synthetic cells with a minimum set of components to reproduce cell function – which provide a novel method to explore and reproduce biological systems such as biological energy conversion processes. There is great interest in understanding and exploiting the energy conversion processes of natural cells, including light harvesting, ion transport and electron transfer.[1–5] Some of these processes may lead to purely biological approaches to energy conversion, but they often inspire biomimetic solid-state analogues. In either case, protocells are at the forefront of bionanotechnology, as they create a platform to exploit individual molecular machines, e. g., the ideal synthetic biological channel is of little use if not integrated into an appropriate synthetic biological system.
Protocells are not just simple particles; they mimic natural cell design and typically consist of a lipid bilayer (a membrane formed from two layers of lipid molecules held together by hydrophobic/hydrophilic interactions) with membrane proteins and often include means to stabilize the lipid bilayer, such as a mesoporous silica core.[6] Recent work has shown that protocells can interact with native cells in ways that particles do not.[7] With a finite number of well characterized components, protocells can be designed to maximize useful outputs; it is left only to the imagination as to what components may be included and what output examined.
Energy conversion is an intriguing output; there are many solid state devices for energy conversion, including piezoelectric,[8, 9] thermoelectric[10, 11] and photovoltaic devices.[12, 13] Piezoelectric devices convert mechanical strain into electric potential by strain-induced charge separation in the crystal lattice of piezoelectric materials, such as quartz crystals[8, 9] or ZnO nanowires.[14] However, the energy density of these devices is very low (104 J·m−3)[15, 16] so they are mainly used in small electronic systems with low power needs (~mW or less); [17, 18] the performance is highly dependent on the mechanical and electrical impedance of the system.[19, 20] Thermoelectric devices can convert thermal gradients into electricity, but require materials with good electrical conductivity and poor thermal conductivity, such as bismuth telluride (Bi2Te3).[10, 11, 21–23] However, thermoelectric device have typical energy conversion efficiencies of 6 % (assuming ΔT = 100 °C from room temperature and the dimensionless figure of merit ZT = 1.5) to 10 % (assuming ΔT = 500 °C from room temperature and ZT = 0.6). Photovoltaic devices directly convert light energy into electricity;[24, 25] multi-junction inorganic photovoltaic devices have been reported to reach an energy conversion efficiency of ≈ 42 %[12, 13] while thin-film photovoltaic devices from NREL have reached 19 % to 20 % efficiency.[26, 27] High efficiency photovoltaic devices rely on multiple energy capture strategies to utilize photons across broad energy levels that often mimic biological light capturing strategies[28].
In comparison to solid state materials, individual cell components have high energy conversion efficiencies, the Na+/K+-ATPase can actively pump Na+ and K+ ions against their gradients by using the free energy from Adenosine Triphosphate (ATP) hydrolysis with an efficiency of 70–85%;[29, 30] F1ATPase is reported to convert energy from ATP hydrolysis to the rotation of the F1 motor with an efficiency approaching 100%[31–34] (this number does not take into account energy losses in the rest of the ATP cycle for those concerned with preserving the laws of thermodynamics).
Protocells offer an intriguing way to exploit biological energy conversion. In the simplest case, a battery can be made out of two droplets with high and low salt concentration; this system would not have the necessary components to recharge itself, but would function like a disposable battery – energy would be released once the ion flow was initiated, and would continue until steady-state concentrations were reached. A rechargeable/renewable biological battery could also be made using the same approach, but would require additional molecular components to recharge the ion concentration gradient, which could be accomplished using the light driven ATP cycle (FoF1-ATP synthase incorporated with bacteriorhodopsin[1–3, 35] or the redox chain of carotene–porphyrin–naphthoquinone[36]) or the conventional, chemically driven, ATP cycle and ATP-fueled ion pumps. [1–3, 29]
Many natural cells can generate electricity in the form of action potentials;[37] a single squid giant axon can generate an action potential of ~100mV for ~1ms,[38] while an electrocyte from the electric eel can produce ~150mV for ~3ms.[39, 40] The transmembrane biological ion concentration gradient across the cell membrane is converted into electricity by temporally activated membrane-protein-regulated ion transport.[37] Action potential formation depends on the concerted system-level interactions of numerous ion channels (pore forming membrane proteins that regulate ion flow in the direction of the electrochemical gradient) and at least one type of ion pump[41] (membrane proteins that pump ions against the direction of the electrochemical gradient), but these systems remain complex. A synthetic cell that produces direct current (DC) electricity[42] requires just a single type of channel. One approach to making such an electrogenic cell is shown in Figure 1 and is based on suspending aqueous droplets in an immiscible medium[43, 44] such as oil/lipid mixture; the ion channels, incorporated in the lipid bilayer formed at the interface of droplets, which regulate the ion flux and form the electric potential difference across the bilayer.
Figure 1.
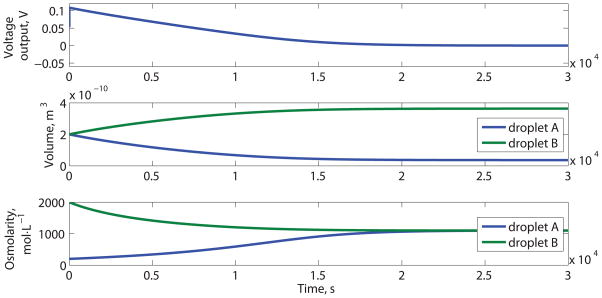
Schematic and performance of a DC biobattery. (A) Device based on cation selective α-HL, adapted from Ref [42], not drawn to scale. (B) Voltage output, volumes and ion concentrations of droplets over time
We report below on the basis and limits for the behavior of droplet protocells and show how they can be harnessed to produce useful levels of DC power. Our model reveals that the electrogenic mechanism in this device is the combination of selective ion transport through ion channels and electrode reactions in the different solutions. New behaviors must be anticipated when biological systems are used as a replacement for solid state devices. [45–48] For example, it is quite important to choose appropriate metrics to evaluate such systems; we characterized the droplet system performance using energy density, which is dependent on output voltage, output current, time, and device volume. Using only one of these parameters, such as voltage, to characterize the system will lead to erroneous conclusions.
2. The Electrogenic Mechanism
The DC ion transport biobattery is based on droplets of different salt concentrations connected by a bilayer containing ion selective channels as shown, not to scale, in Figure 1A (adapted from [42]). Ions will flow from high to low concentration through the ion channels (or synthetic channel analogues). Stable lipid bilayers form at the interface between droplets, based on monolayers formed at the interface of water droplets in an organic solvent[43, 49] or oil/lipid mixtures.[42, 50] The chemical energy stored in the ion concentration gradient between droplets is converted into electricity using membrane transport[42] and electrodes to convert the ion flux into an electron flux.
S. Aureus α-hemolysin (α-HL) is one platform to engineer synthetic channel analogues for this application, as this exotoxin protein is normally weakly anion selective but can be engineered to be cation selective by point mutation (substitution of individual amino acids) of the α-HL monomer.[51, 52] Treating the amino acid residues with sulfhydryl reagents can further enhance the ion selectivity of the α-HL mutant; [51, 53] when G133C α-HL mutant (see Figure S1 in supporting information) is treated with 2-sulfonatoethyl methanethiosulfonate (MTSES), the –SH group in the cysteine side chain is converted into a strongly negative charged moiety –SS-CH2CH2SO3−; this modification improves the cation selectivity to 170 (gK/gCl) when the bathing solutions are 0.1 mol·L−1 : 0.3 mol·L−1 KCl. [51]
Electrodes are necessary to interface the droplets with an external circuit; the electrodes close the circuit by converting ion flow in the electrolytes into electron flow in the external circuit, or vice versa; the reactions at the silver/silver chloride (Ag/AgCl) electrodes depend on the local Cl− concentration. As a result, the total voltage output of the DC biobattery is the sum of the potential from the ion transport through the membrane and the electrode potentials. Since these types of devices are intended to power or interface with external circuits, delivering recognizable currents is an important feature; using, or assuming, very large external impedance forces the device into a regime with high output voltage but negligible current and negligible energyoutput.
The total voltage output (Et) consists of two components: first, the electric potential (Er), due to ion movement through the membrane and channels, based here on α-HL mutants; second, the electrode potential (Ec), due to the different activities of the two electrodes in solutions of different ion concentration.
| (1) |
The Ag/AgCl electrode potential, Ec, is a function of chloride concentration:
| (2) |
where aCl−, Anode and aCl−, Cathode are the chemical activities of Cl− around the two electrodes; γB is the activity coefficient of the ions in droplet B (trans side); mB is the molality (mol·kg−1) of droplet B; γA and mA are for droplet A (cis side); R is the gas constant; F is the Faraday constant and T is temperature.
The potential (Er) associated with current (I) through the α-HL channels in the bilayer is determined by a simplified version of Hodgkin-Huxley model: [38]
| (3) |
where C is the membrane capacitance; gK and gCl are the K+ and Cl− conductance’s of the α-HL mutants; VK and VCl are the reversal potential of K+ and Cl− ions respectively.
The activity coefficient (γ) changes with the concentration of the solution. Generally, this relationship is described by the Debye-Huckel’s limiting law[54] (Equation S2 of supporting information). However, the Debye-Huckel equation is only good for a dilute solution. For a more concentrated solution (> 0.1 mol·kg−1), the appropriate form is: [55]
| (4) |
where z+ and z− are ionic valences; A is a temperature-related constant;[55] Is is the ionic strength of the solution; BSi, β, C and D are ion-species-specific empirical parameters, found by fitting experimental data.[55] The experimental relationship of the mean activity coefficient of KCl and the ionic strength is shown in Figure S2; this relationship demonstrates dramatically dissimilar behaviors in the dilute and concentrated solutions; the higher order coefficients, e.g. CI2 + DI3, have to be introduced to fully capture the behavior. The basis for this equation and the experimental data are in the supporting information.
To predict the device performance over a range of operating parameters, we need a general equation relating ion selectivity of the synthetic channels to solution ion concentration. We know the solution ion concentrations surrounding the α-HL channel influence the ion selectivity by affecting the local electrostatic environment. Experimental data shows that the ion selectivity of α-HL channels decreases with increasing solution ion concentrations.[51, 56] When the ion concentration in the solution is high, the α-HL channels become practically unselective between cations and anions[56] as the local changes due to the mutation will be mostly screened out by the strong electrostatic contributions from ions in solution. Based on the available experimental data[51, 57] and the observations enumerated above, a phenomenological equation to relate ion selectivity to solution ion concentration is defined as:
| (5) |
where dS (mol·L−1) is the solution concentration difference across the lipid bilayer; a1 and b1 are ion selectivity parameters determined by regression methods from the experimental data; the parameter a1 increases as the channel becomes more selective; the parameter b1 increases as the channel becomes more responsive to a change in the solution ion concentration. For MTSES-treated α-HL mutant G133C, the values found for parameters a1 and b1 are 273 and 2.40, based on data reported in [51].
The total voltage output (Et) of the DC biobattery consists of the electric potential (Er, Equation 3) from the membrane-protein-regulated ion transport and the electrode potential (Ec, Equation 2). These potentials are coupled electrically by Kirchhoff’s laws through the external resistance (R). With very large external resistance, the potential reaches a maximum and the current output reaches a minimum.
The energy output density of the DC biobattery is the energy output to the external resistance during the useful lifetime (see Supporting Information for the numerical definition) of this device, divided by the volume of the biobattery. Over time, the voltage output (Et) of the DC biobattery decreases from the initial value, due to the decrease in the ion concentration difference between the droplets (Figure 1B), which is the result of both ion transport through the membrane proteins and water exchange through the lipid bilayer. The decrease in the difference between the ion concentrations causes both the potential from selective ion transport (Er) and the electrode potential (Ec) to decrease. Eventually, the two droplets reach equilibrium at the same ion concentration, which is the end of life for this DC biobattery (but it could be reused by restoring the salt concentration to the initial conditions).
The energy conversion efficiency is the ratio of the energy output to the free energy stored in the ion concentration gradient. The unitary free energy stored in the concentration gradient, dGo(J·mol−1), is given by:
| (6) |
where SA and SB (mol·L−1) are the ion concentrations of the two droplets; R and T are the gas constant and the temperature. The total free energy (dG, J·m−3) available in the gradient is:
| (7) |
where Nion (mol) is the number of ions available and V (m3) is the volume. The ion concentration gradient can be replenished by the active transport of ions through ion pumps[29, 58]; Equation 7 also defines the minimum energy cost for restoring (recharging) the ion concentration gradient. The electrode reactions (Figure 1A) are reversible; therefore this energy is not included in the energy efficiency calculations.
3. Parameters Affecting Electrogenic Behavior
The configuration shown in Figure 1A can produce 1.37 × 102 J·m−3 of electrical energy (Figure 1B); this design is based on many of the parameters reported in,[42] but uses parameters for the cation selective α-HL mutant (MTSES treated G133C), since anion selective mutants produce a potential that conflicts with the potential produced by the electrodes.
The results shown in Figure 1 are based on calculations with highly cation-selective α-HL mutants (MTSES treated G133C) inserted between droplets with a large external resistance (1GΩ); the maximum voltage output is 108.2 mV. This voltage output (Et) is close to the summation of the electrode potential (Ec=53.3mV, calculated from Equation 2) and the reversal potential of the potassium ions (59.5mV). The reversal potential (Erev, K) of the potassium ions is determined from the ion concentration of the two droplets using the Nernst Equation.[59]
| (8) |
[K]A and [K]B are the K+ concentrations in the droplet A and B, respectively. The Nernst Equation is the simplified version of Goldman-Hodgkin-Katz voltage equation,[60] when the inserted channels are of high cation selectivity (see the supporting information for the detailed explanation). With a hypothetical mutant with even higher cation selectivity than MTSES treated G133C, the voltage output of DC biobattery would approach, but not exceed, 112.8 mV (53.3 mV + 59.5 mV), using the same initial ion concentrations as Figure 1.
The droplet with the higher ion concentration will expand in volume over time, while the droplet with lower concentration will shrink over time (Figure 1B, the middle panel). This phenomenon was predicted in the model and observed experimentally (Supporting Infromation, Figure S3) for two droplets connected with a bilayer, containing solutions with different salt concentrations; transport in this system is due wholly to water transport across the bilayer. The droplet bilayer permeability to water was found to be 19 μm·s−1, which is similar to other reports for membrane water permeability (≈ 22 μm·s−1).[61–64] The experimental details are included in the supporting information; these experiments were used to validate the principles and parameters of the model.
With a detailed model, we can predict the ideal configuration for these devices and calculate the theoretical maximum energy density and energy conversion efficiencies. As is typical for galvanic batteries, the highest energy density (3.46×103 J·m−3) was found when the external resistance was matched to the internal resistance; at that point, the output voltage is half of the open circuit voltage. Simply matching the external resistance to that of the device shown in Fig 1A results in a 25-fold improvement in the energy output. Further numerical optimization of the configuration, which can not be done based on closed form expressions, produced a design with significant additional improvements to the energy density.
The electrogenic performance of the DC biobattery depends on eight parameters: the ion selectivity and density of the α-HL mutant channels, the volumes of the two droplets, the lipid bilayer area, the initial ion concentrations of the two droplets, and the external resistance.
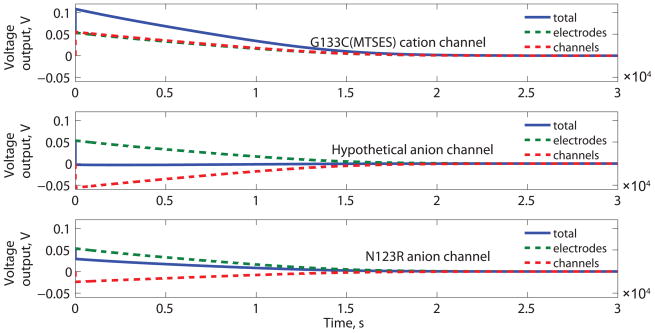
The energy output density depends on the α-HL mutant channel ion selectivity (Figure 2B). The ion selectivity of the membrane protein influences both the direction and the amplitude of electric potential due to ion transport through channels (Er) for anion and cation selective α-HL mutants. The direction of the electrode potential (Ec) is fixed by the droplet concentrations. Cation specific α-HL mutants result in Er having the same direction as the Ec (based on concentration ratios shown in Figure 1A), while anion specific α-HL mutants result in Er having the opposite direction as the Ec.
Figure 2.
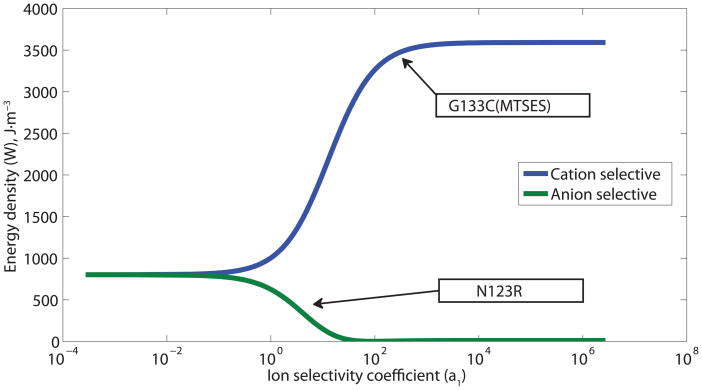
Performance of a DC biobattery with cation and anion selective α-HL mutants. (A) The cation selective channel (MTSES G133C) uses parameters based on published MTSES treated G133C α-HL mutants. For comparison, calculations using two anion-selective channels are shown: a hypothetical anion selective channel that is as strongly anion specific as the MTSES G133C is cation specific and parameters based on published N123R channels[42]. For each case, the total potential (Et) is shown along with the contributions due to the channels (Er) and the electrodes (Ec). (B) The output energy density (W) as a function of ion selectivity (a1) of the membrane proteins for both cation and anion specific channels. The selectivity reported for specific mutants, MTSES treated G133C and N123R, is highlighted with arrows.
For a cation selective channel (e.g. G133C (MTSES)[51] Figure 2), a more selective α-HL mutant channel will produce more energy, because the electric potential Er from the unbalanced ion transport depends on the ion selectivity of the membrane protein. However, the increase in Er is limited by the reversal potential of K+ ions between the two droplets which depends on the concentration difference. There is a limit to the improvement in energy density related to channel selectivity; calculations based on a hypothetical mutant with very high ion selectivity (a1 = 1.0 × 106, b1 = 2.40, as defined in Equation 5), showed that this device could produce just 3.8 % higher energy density than one based on MTSES-G133C.
On the contrary, if the α-HL mutants are anion selective (e.g. N123R[42]), the contributions of Ec and Er subtract from one another (Figure 2A); increasing the selectivity of the anion selective α-HL mutant results in the production of less energy. As a result, the DC biobattery with cation selective channels will always have higher energy output density than the one with anion selective channels (Figure 2B); the device with anion selective channels has even lower output than a simple concentration cell,[48] made from two connected salt water droplets of different concentrations with silver/silver chloride electrodes, because of the reversed contributions from anion selective channels and the electrodes (see Supporting Information for detailed analysis).
The performance scales linearly with volume. Increasing the droplet size does increase the energy output, but not the volumetric energy density. A larger droplet has longer lifetime, as there are more ions available to be exchanged. The volumetric energy density is the same when two droplets of 0.02 nL or 2000 nL were compared; both produced 3.46×103 J·m−3.
The optimal configuration for the three parameters that control membrane and external resistance: nα-HL, A1 and Rext, are related by:
| (9) |
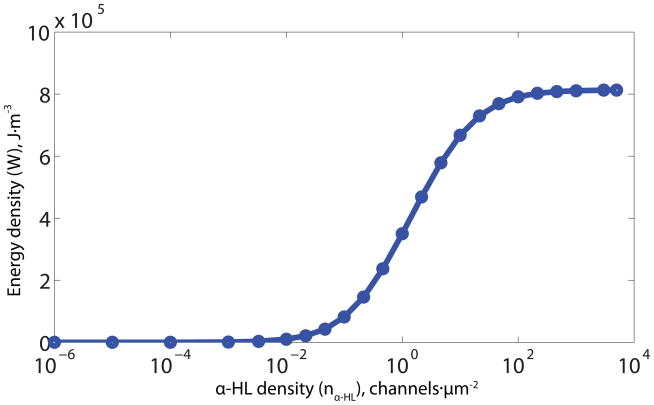
where g0 (S·channel−1) is the unitary conductance of the α-HL channel; A1 (m2) is the bilayer area; nα-HL (channels·m−2) is the density of α-HL channels; Rext (Ω) is the external resistance; C1 is a dimensionless channel-density-dependent constant determined using the Nelder-Mead algorithm[65] at various channel densities that defines a ridge shown in Figure 3A of equal energy output density. When the channel density is low (< 0.2 channels·μm−2), C1 is close to 1. When the channel density is higher than 0.2 channels·μm−2, C1 increases gradually over 1; for instance, when the G133C (MTSES) α-HL mutant density is 100 channels·μm−2, C1= 6.95.
Figure 3.
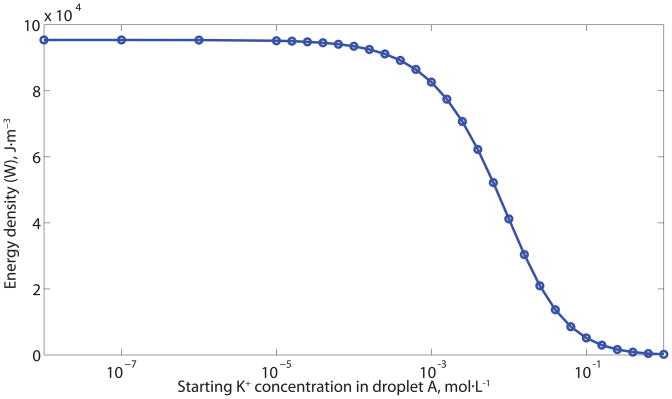
The output energy density (W) as a function of device parameters. (A) Output energy density as a function of α-HL channel density (nα-HL), external resistance (Rext) and the area of the bilayer (A1). (B) The output voltage and energy density as a function of external impedance for a devices based on anion and cation specific channels and with low number of channels (≈ 100) and low channel density (≈ 0.003 channels· m−2). This channel density is comparable to that in the reported experimental setup [42], but is much lower than the values we report in Table 1. (C) Local maximum of energy density as a function α-HL channel density (nα-HL). (D) Output energy density as a function of starting K+ concentration in the lower-concentration droplet (droplet A). At every given K+ concentration, the maximum energy output density was found by Nelder-Mead optimization[65]. The other parameters are the same as Figure 1.
The importance of impedance matching is well known for traditional galvanic cells, which have maximum power output when the internal resistance equals the external resistance. Figure 3B illustrates the changes in voltage output and energy density for anion and cation bearing designs with a low number of total channels (≈ 100 channels) and low channel density (≈ 0.003 channels·um−2); this channel density is comparable to that in the reported experimental setup; [42] the output voltage reaches a maximum with large external impendence, while the energy density (energy output), is a maximum when the external resistance matches the internal impedance. Based on the relationship shown in Equation 9, it is possible to tune droplet biobatteries for the load by independently adjusting the area of the lipid bilayer and the droplet volume, which implies modifying the droplet shape, to achieve maximum energy output density for a given external resistance.
The energy density reaches a plateau (see Figure 3C) at ≈102 channels·μm−2. Although increases in channel density generate increases in transient ion flux, the higher flux also shortens the device lifetime. With low channel densities, the lifetime is reduced by water exchange through the lipid bilayer; the optimal configuration balances these phenomena. The upper limit for the density of α-HL channels in the membrane was set as 5 × 103 channels·μm−2, estimated by assuming 50 % close packing[51, 66] of proteins in the membrane, but this constraint turned out to be unimportant as it was not reached. Channel densities on the order of 50% of close packing have been observed with purified S. Aureus α-hemolysin reconstituted into vesicles by electron microscopy with negative staining[67] and on α-HL observed in supported lipid bilayers using Atomic Force Microscopy (AFM).[68]
The ideal droplet size ratio and initial concentrations are related (Figure 3D). The voltage output increases as the concentration ratio of the two droplets ([K+]B/[K+]A) increases while the lifetime decreases as the concentration difference between the two droplets (|[K+]B−[K+]A|) increases. However, the voltage output cannot grow infinitely, since the lipid bilayer will break down reversibly or irreversibly at high transmembrane potential,[69–71] since the electric field influences the line tension and the surface tension of the lipid bilayer, which in turn affects the possibility of pore formation in the bilayer and the bilayer stability.[71–73] The electric breakdown potential for the lipid bilayer is in the range of 200–700mV,[70–72, 74] depending on the composition of the bilayer, solution environment, and thermal fluctuation.[71, 72, 75] For these reasons, the membrane potential was constrained in the optimization to be less than 400mV. The volume ratio also affects the lifetime; optimal values, to maximize energy density, were found by nonlinear constrained numerical optimization,[65] the results are shown in Table 1. The volume of electrode material needed can be calculated once the total ion flux is known; the charge associated with electron flow must balance the charge associated with ion flow. Table 1 also shows that the volume of electrode material needed is small but not negligible – it represents less than 4% of the droplet volume.
Table 1.
Optimal design parameters to produce maximum energy density or maximum energy conversion efficiency
| Design goal | Maximum energy density | Maximum energy conversion efficiency |
|---|---|---|
| KCl concentration in droplet A (mol·L−1) | < 10−4 | 1.00 |
| KCl concentration in droplet B (mol·L−1) | 3.89 | 1.05 |
| Droplet A volume (nL)a | 254 | 200 |
| Volume ratio (droplet B/droplet A) | 0.575 | 1.00 |
| External resistance (Ω)b | 267 | 267 |
| α-HL channel density (channels·μm−2) | >102 | >102 |
| α-HL charge selectivity (parameter a1 in Eq. 5) | >103 | >103 |
| Bilayer area (m2) | 3.41× 10−8 | 4.07 × 10−8 |
| Silver chloride required (mol)c | 3.7 × 10−7 | 5.6 × 10−9 |
| Volume ratio of electrode in droplet A | 3.7 × 10−2 | 7.3 × 10−4 |
| Energy density (J·m−3) | 6.9 × 106 | 1.7 × 103 |
| Energy conversion efficiency | 10 % | 48 % |
Energy is proportional to volume
Fixed according to Equation 9
The electrode reactions are reversible
The lifetime scales linearly with the total volume. With an external resistance of 267 Ω, as given in Table 1, a device with 400 nL droplets has a lifetime of 550 s. A device with droplets that are 3 times larger in diameter has a volume of 10.8 μL and a lifetime of 1.54 × 104 s, with the same energy output density. The energy output of a device maximized for high energy density (6.92 × 106 J·m−3), with two pairs of droplets with diameters of 3.9 cm and 3.3 cm, could power a typical mp3 player drawing 20 mW for ≈ 10 h.
4. Optimization of Electrogenic Behavior
The design was numerically optimized by varying device parameters using a non-linear optimization algorithm to search for the optimal configuration of the biobattery to maximize the objective functions. Objective functions are the mathematical relationships that quantify the “goodness” of the design, such as energy density or energy conversion efficiency. The optimization algorithm would vary the parameters and calculate the device performance over its lifetime; at that point the objective function would be evaluated and the optimization routine would compare the current value of the objective function to previous values and decide to either stop the optimization (if a maximum was reached) or vary additional parameters in an effort to maximize the objective function.
The upper limit for energy output was found by numerically optimizing the device to maximize energy output density. A device maximized for high energy density results in a design that can produce 6.92 × 106 J·m−3 energy (Table 1), with an energy conversion efficiency of 10 %; the energy density is ≈5 % that of a lead-acid battery,[76, 77] putting it into the realm of practical applications.
Higher energy conversion efficiency (Table 1) can be obtained at the expense of reduced energy density. The maximum energy conversion efficiency was found to be 48 % when droplet A and B have similar concentrations (A is 1.00 mol·L−1; B is 1.05 mol·L−1); the device would have high α-HL channel density as shown in Table 1. Unfortunately, this configuration has impractically-low energy density (1.74 × 103 J·m−3), but it does show that higher efficiencies are possible.
Water permeation can significantly shorten the lifetime of small droplet devices when the salt concentrations are not equal (such as for batteries). A small droplet battery optimally configured for the maximum energy output density (initial droplet sizes: 254 nL and 146 nL, Table 1), even with no ion channels inserted, will loose half its energy by “self discharge” in 1.3 h due to water transport through the lipid bilayer (see the Supporting Information for the calculations). However, the storage time could be prolonged by simply moving the droplets away so that there was no osmolarity-driven water exchange between droplets. Larger droplets, such as those given in the mp3-player example, are not significantly impacted by water exchange since the volume is substantially greater in relation to the bilayer area and the “self discharge” time would be on the order of 18 years.
5. Conclusions and Outlook
In summary, protocells offer a new method to study cell systems by mimicking natural biological processes. Simplified protocell systems can be modeled and the output can be predicted by coupling the necessary equations and numerical evaluation. Optimized designs for DC biobattery protocells may be able to produce energy densities sufficient for practical applications.
Protocells intended to replace solid state devices must be designed with the practical limitations in mind: First, they must be designed to operate with application-appropriate current output. Second, the energy contributions of the electrodes must be accurately accounted for, and the electrodes designed with adequate lifetime for the application. Third, the lipid bilayer and inserted proteins must be stable for extended periods. The droplet bilayer configuration provides one way to stabilize a bilayer, the lipid bilayer can be stabilized by forming bilayers on top of a porous support, such as mesoporous silica[78] or polymerized hydrogel.[79] Finally, one should not neglect the water transport through the lipid bilayer, as water transport will equalize the ion concentration gradient over time and remove the driving force for the battery, even with no functional ion channels present.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health through the NIH Roadmap for Medical Research, as part of the National Center for Design of Biomimetic Nanoconductors, award number PHS 2 PN2 EY016570B. Supporting Information is available online from Wiley InterScience or from the author. The full description of the materials and software used in this paper requires the identification of certain materials and software suppliers. The inclusion of such information should in no way be construed as indicating that such materials or software are endorsed by NIST or are recommended by NIST or that they are necessarily the best materials or software for the purposes described.
Contributor Information
Jian Xu, School of Engineering and Applied Science, Yale University, New Haven, CT 06511 (USA).
Prof. Fred J. Sigworth, Department of Cellular and Molecular Physiology, Yale University, New Haven, CT 06520 (USA)
Dr. David A. LaVan, Email: david.lavan@nist.gov, Ceramics Division, Materials Science and Engineering Laboratory, National Institute of Standards and Technology, Gaithersburg, MD 20899 (USA)
References
- 1.Choi HJ, Montemagno CD. Nano Lett. 2005;5:2538–2542. doi: 10.1021/nl051896e. [DOI] [PubMed] [Google Scholar]
- 2.Bald D, Amano T, Muneyuki E, Pitard B, Rigaud JL, Kruip J, Hisabori T, Yoshida M, Shibata M. J Biol Chem. 1998;273:865–870. doi: 10.1074/jbc.273.2.865. [DOI] [PubMed] [Google Scholar]
- 3.Pitard B, Richard P, Dunach M, Rigaud JL. European journal of biochemistry/FEBS. 1996;235:779. doi: 10.1111/j.1432-1033.1996.t01-1-00779.x. [DOI] [PubMed] [Google Scholar]
- 4.Miyasaka T, Watanabe T, Fujishima A, Honda K. J Am Chem Soc. 1978;100:6657–6665. [Google Scholar]
- 5.Remy A, Gerwert K. Nat Struct Biol. 2003;10:637–644. doi: 10.1038/nsb954. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Fan H, Doke N, Loy DA, Assink RA, LaVan DA, Brinker CJ. J Am Chem Soc. 2000;122:5258–5261. [Google Scholar]
- 7.Liu J, Stace-Naughton A, Jiang X, Brinker CJ. J Am Chem Soc. 2009;131:1354–1355. doi: 10.1021/ja808018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunde RL, Jarvi EJ, Rosentreter JJ. Talanta. 1998;46:1223–1236. doi: 10.1016/s0039-9140(97)00392-5. [DOI] [PubMed] [Google Scholar]
- 9.Fung YS, Wong YY. Anal Chem. 2001;73:5302–5309. doi: 10.1021/ac010655y. [DOI] [PubMed] [Google Scholar]
- 10.Riffat SB, Ma X. Applied Thermal Engineering. 2003;23:913–935. [Google Scholar]
- 11.Bell LE. Science. 2008;321:1457. doi: 10.1126/science.1158899. [DOI] [PubMed] [Google Scholar]
- 12.King RR, Law DC, Edmondson KM, Fetzer CM, Kinsey GS, Yoon H, Sherif RA, Karam NH. Appl Phys Lett. 2007;90:183516. [Google Scholar]
- 13.Barnett A, Kirkpatrick D, Honsberg C, Moore D, Wanlass M, Emery K, Schwartz R, Carlson D, Bowden S, Aiken D. Progress in Photovoltaics: Research and Applications. 2008;17:75–83. [Google Scholar]
- 14.Yang R, Qin Y, Dai L, Wang ZL. Nature Nanotechnology. 2008. [DOI] [PubMed] [Google Scholar]
- 15.Roundy S, Leland ES, Baker J, Carleton E, Reilly E, Lai E, Otis B, Rabaey JM, Wright PK, Sundararajan V. IEEE Pervasive computing. 2005;4:28–36. [Google Scholar]
- 16.Roundy S, Wright PK, Rabaey JM. Energy scavenging for wireless sensor networks: with special focus on vibrations. Springer; New York: 2004. [Google Scholar]
- 17.Paradiso JA, Starner T. IEEE Pervasive Computing. 2005;4:18–27. [Google Scholar]
- 18.Iino A, Suzuki K, Kasuga M, Suzuki M, Yamanaka T. Ultrasonics. 2000;38:54–59. doi: 10.1016/s0041-624x(99)00192-4. [DOI] [PubMed] [Google Scholar]
- 19.Sodano HA, Inman DJ, Park G. Journal of Intelligent Material Systems and Structures. 2005;16:799. [Google Scholar]
- 20.Cho JH, Richards RF, Bahr DF, Richards CD, Anderson MJ. Appl Phys Lett. 2006;89:104107. [Google Scholar]
- 21.Sapp SA, Lakshmi BB, Martin CR. Adv Mater. 1999;11 [Google Scholar]
- 22.Yamashita O, Tomiyoshi S, Makita K. J Appl Phys. 2003;93:368. [Google Scholar]
- 23.Cole T. Science. 1983;221:915–920. doi: 10.1126/science.221.4614.915. [DOI] [PubMed] [Google Scholar]
- 24.Crandall R, Luft W. Progress in Photovoltaics: Research and Applications. 1995;3:315–331. [Google Scholar]
- 25.Little RG, Nowlan MJ. Progress in Photovoltaics: Research and Applications. 1997;5:309–315. [Google Scholar]
- 26.Ramanathan K, Contreras MA, Perkins CL, Asher S, Hasoon FS, Keane J, Young D, Romero M, Metzger W, Noufi R. Progress in Photovoltaics: research and applications. 2003;11:225–230. [Google Scholar]
- 27.Repins I, Contreras MA, Egaas B, DeHart C, Scharf J, Perkins CL, To B, Noufi R. Progress in Photovoltaics: research and applications. 2008;16:235–239. [Google Scholar]
- 28.LaVan DA, Cha JN. Proc Natl Acad Sci U S A. 2006;103:5251–5255. doi: 10.1073/pnas.0506694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skou JC. Biosci Rep. 1998;18:155–169. doi: 10.1023/a:1020196612909. [DOI] [PubMed] [Google Scholar]
- 30.Cornelius F. In: ATPases (Biomembranes. A Multi-Volume Treatise) Lee AG, editor. Vol. 5. JAI Press Inc; London, England: 1996. p. 133. [Google Scholar]
- 31.Lanyi JK, Pohorille A. Trends Biotechnol. 2001;19:140–144. doi: 10.1016/s0167-7799(01)01576-1. [DOI] [PubMed] [Google Scholar]
- 32.Noji H, Yasuda R, Yoshida M, Kinosita K. 1997 [Google Scholar]
- 33.Elston T, Wang H, Oster G. Nature. 1998;391:511. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- 34.Junge W, Sielaff H, Engelbrecht S. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 35.Pitard B, Richard P, Dunach M, Girault G, Rigaud JL. European journal of biochemistry/FEBS. 1996;235:769. doi: 10.1111/j.1432-1033.1996.00769.x. [DOI] [PubMed] [Google Scholar]
- 36.Yrach G, Rigaud J, Durantini E, Moore A, Gust D, Moore T. Nature. 1998;392:479–482. doi: 10.1038/33116. [DOI] [PubMed] [Google Scholar]
- 37.Katz B. Nerve, Muscle, and Synapse. McGraw-Hill Companies; New York: 1966. [Google Scholar]
- 38.Hodgkin A, Huxley A. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keynes R, Ferreira H. J Physiol. 1953;119:315–351. doi: 10.1113/jphysiol.1953.sp004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altamirano M. J Cell Physiol. 1955;46:249–277. doi: 10.1002/jcp.1030460205. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, Lavan DA. Nature Nanotechnology. 2008;3:666–670. doi: 10.1038/nnano.2008.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holden MA, Needham D, Bayley H. J Am Chem Soc. 2007;129:8650–8655. doi: 10.1021/ja072292a. [DOI] [PubMed] [Google Scholar]
- 43.Funakoshi K, Suzuki H, Takeuchi S. Anal Chem. 2006;78:8169–8174. doi: 10.1021/ac0613479. [DOI] [PubMed] [Google Scholar]
- 44.Bayley H, Cronin B, Heron A, Holden MA, Hwang WL, Syeda R, Thompson J, Wallace M. Mol Biosyst. 2008;4:1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owens B. Batteries for implantable biomedical devices. Plenum Press; New York, USA: 1986. [Google Scholar]
- 46.Piccolino M. Trends Neurosci. 2000;23:147–151. doi: 10.1016/s0166-2236(99)01544-1. [DOI] [PubMed] [Google Scholar]
- 47.Kohler U, Antonius C, Bauerlein P. J Power Sources. 2004;127:45–52. [Google Scholar]
- 48.Sanger MJ, Greenbowe TJ. Journal of Research in Science Teaching. 1997;34:377–398. [Google Scholar]
- 49.Malmstadt N, Nash MA, Purnell RF, Schmidt JJ. Nano Lett. 2006;6:1961–1965. doi: 10.1021/nl0611034. [DOI] [PubMed] [Google Scholar]
- 50.Hwang WL, Holden MA, White S, Bayley H. J Am Chem Soc. 2007;129:11854–11864. doi: 10.1021/ja074071a. [DOI] [PubMed] [Google Scholar]
- 51.Merzlyak PG, Capistrano MFP, Valeva A, Kasianowicz JJ, Krasilnikov OV. Biophys J. 2005;89:3059–3070. doi: 10.1529/biophysj.105.066472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu LQ, Cheley S, Bayley H. J Gen Physiol. 2001;118:481–493. doi: 10.1085/jgp.118.5.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krasilnikov OV, Merzlyak PG, Yuldasheva LN, Rodrigues CG, Bhakdi S, Valeva A. Mol Microbiol. 2000;37:1372–1378. doi: 10.1046/j.1365-2958.2000.02080.x. [DOI] [PubMed] [Google Scholar]
- 54.Debye P, Huckel E. Lowering of freezing point and related phenomena. Physik. 1923:185–206. [Google Scholar]
- 55.Hamer WJ, Wu YC. Chem Ref Data. 1972;1:1047–1075. [Google Scholar]
- 56.Krasilnikov OV, Merzlyak PG, Yuldasheva LN, Capistrano MF. Eur Biophys J. 2005;34:997–1006. doi: 10.1007/s00249-005-0485-9. [DOI] [PubMed] [Google Scholar]
- 57.Krasilnikov OV, Merzliak PG, Yuldasheva LN, Nogueira RA, Rodrigues CG. BBA-Biomembranes. 1995;1233:105–110. doi: 10.1016/0005-2736(94)00231-d. [DOI] [PubMed] [Google Scholar]
- 58.Pintschovius J, Fendler K, Bamberg E. Biophys J. 1999;76:827–836. doi: 10.1016/S0006-3495(99)77246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nernst W. Z Phys Chem. 1888;3:613–637. [Google Scholar]
- 60.Goldman DE. J Gen Physiol. 1943;27:37–60. doi: 10.1085/jgp.27.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orbach E, Finkelstein A. J Gen Physiol. 1980;75:427–436. doi: 10.1085/jgp.75.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berg J, Tymoczko J, Stryer L. Biochemistry. W. H. Freeman and Company; New York, USA: 2001. [Google Scholar]
- 63.Novotny J, Jakobsson E. Am J Physiol. 1996;270:C1751–C1763. doi: 10.1152/ajpcell.1996.270.6.C1751. [DOI] [PubMed] [Google Scholar]
- 64.Novotny J, Jakobsson E. Am J Physiol. 1996;270:C1764–C1772. doi: 10.1152/ajpcell.1996.270.6.C1764. [DOI] [PubMed] [Google Scholar]
- 65.Nelder JA, Mead R. Computer Journal. 1965;7:308–313. [Google Scholar]
- 66.Hille B. Ion channels of excitable membranes. Sinauer Associates, Inc; Sunderland, MA, USA: 2001. [Google Scholar]
- 67.Fussle R. J Cell Biol. 1981;91:83–94. doi: 10.1083/jcb.91.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Czajkowsky DM, Sheng S, Shao Z. J Mol Biol. 1998;276:325–330. doi: 10.1006/jmbi.1997.1535. [DOI] [PubMed] [Google Scholar]
- 69.Chang DC, Reese TS. Biophys J. 1990;58:1–12. doi: 10.1016/S0006-3495(90)82348-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Benz R, Beckers F, Zimmermann U. J Membr Biol. 1979;48:181–204. doi: 10.1007/BF01872858. [DOI] [PubMed] [Google Scholar]
- 71.Wilhelm C, Winterhalter M, Zimmermann U, Benz R. Biophys J. 1993;64:121. doi: 10.1016/S0006-3495(93)81346-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sung W, Park PJ. Biophys J. 1997;73:1797–1804. doi: 10.1016/S0006-3495(97)78210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winterhalter M, Helfrich W. Physical Review A. 1987;36:5874–5876. doi: 10.1103/physreva.36.5874. [DOI] [PubMed] [Google Scholar]
- 74.Tien HT, Ottova A. IEEE Transactions on Dielectrics and Electrical Insulation. 2003;10:717–727. [Google Scholar]
- 75.Needham D, Hochmuth RM. Biophys J. 1989;55:1001–1009. doi: 10.1016/S0006-3495(89)82898-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hariprakash B, Mane AU, Martha SK, Gaffoor SA, Shivashankar SA, Shukla AK. Electrochemical and Solid-State Letters. 2004;7:A66. [Google Scholar]
- 77.Juergens T, Nelson RF. J Power Sources. 1995;53:201–205. [Google Scholar]
- 78.Brinker J, Lu Y, Sellinger A, Fan H. Adv Mater. 1999;11:579–585. [Google Scholar]
- 79.Malmstadt N, Jeon TJ, Schmidt JJ. Adv Mater. 2008;20:84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.