Abstract
NF-κB is activated by many stimuli and NF-κB binding sites have been identified in a wide variety of genes. Yet, NF-κB-dependent gene expression must be stimulus- and cell-type-specific. In others words, the cellular response to different NF-κB activating stimuli, such as TNFα, IL-1, and LPS, must be different; and the response of different cell types, such as lymphocytes, fibroblasts, or epithelial cells, to the same NF-κB-inducing stimulus must also be different. Finally, kinetics of gene expression must be accounted for, so that all NF-κB-dependent genes are not activated simultaneously even if cell type and stimulus are constant. Here, we explore the mechanistic framework in which such regulatory aspects of NF-κB-dependent gene expression have been analyzed because they are likely to form the basis for physiological responses.
NF-κB transcription factors are activated by many different stimuli. Dimer choice, activation kinetics, and other mechanisms ensure that only a subset of potential target genes respond in each case.
NF-κB was first reported in 1986 as a DNA-binding protein that recognizes, in a DNA sequence-specific manner, an important motif within the intronic enhancer of the immunoglobulin (Ig) κ light chain gene (Sen and Baltimore 1986). NF-κB DNA-binding activity was observed following stimulation of a pre-B cell line and represented the first example of inducible DNA binding as a primary response to cell stimulation. Subsequent studies revealed that NF-κB activity is induced in most cell types in response to a wide variety of stimuli, with major roles in cell activation, survival, and differentiation (Hayden and Ghosh 2004; Hoffmann et al. 2006; Ghosh and Hayden 2008; Vallabhapurapu and Karin 2009). In the classical model, NF-κB is found in the cytoplasm of unstimulated or resting cells in association with an inhibitory IκB protein. In response to cell stimulation, IκB is phosphorylated, ubiquitinated, and degraded, freeing NF-κB to translocate to the nucleus, bind its recognition sites in promoters and enhancers, and activate gene transcription.
Although this classical model of NF-κB has been well-documented and widely discussed, it has long been known that NF-κB function and regulation involves considerable complexity and diversity. This complexity and diversity, which are thought to be important for facilitating the differential and highly selective regulation of NF-κB target genes, are apparent at a number of levels, beginning with the existence of multiple NF-κB family members (Ghosh et al. 1998). Most vertebrates that have been studied contain five genes encoding the NF-κB family members RelA (p65), c-Rel, RelB, p50, and p52. These family members can bind DNA in a variety of heterodimeric species and all but RelB can bind as homodimers. The=existence of multiple family members and numerous dimeric species provides strategies for the selective regulation of target genes through differential expression of specific family members and dimeric species, differential DNA recognition, and differential transactivation mechanisms.
A second level of complexity arises from the existence of carboxy-terminal ankyrin repeat domains within the p105 and p100 precursors of p50 and p52, respectively. The ankyrin repeats are often removed immediately by proteolytic processing, but they are retained in a subset of stable dimers in the cytoplasm, providing an opportunity to regulate NF-κB activity via the regulation of proteolytic processing (Ghosh et al. 1998; Hoffmann et al. 2006; Ghosh and Hayden 2008; Vallabhapurapu and Karin 2009). The existence of multiple IκB proteins in vertebrates provides a third level of complexity. A primary function of classical IκBs is to sequester NF-κB dimers in the cytoplasm, but other IκBs appear to regulate NF-κB activity in the nucleus by functioning as transcriptional coregulators (Ghosh and Hayden 2008).
Although the existence of multiple NF-κB and IκB family members with variable properties provides intrinsic mechanisms for facilitating the selective regulation of NF-κB target genes, selectivity is further achieved through extrinsic mechanisms. In particular, variations in the kinetics of NF-κB activation can strongly influence target gene selection in response to a stimulus. In addition, the differential organization of chromatin at NF-κB target genes can regulate susceptibility to activation by NF-κB complexes (Natoli et al. 2006; Natoli 2009). The developmental history of a cell can further dictate which target genes are susceptible to activation, because of differences in chromatin structure established during development and differences in the expression of other transcription factors and coregulatory proteins that influence the expression of NF-κB target genes. Together, these intrinsic and extrinsic mechanisms allow NF-κB to regulate distinct but often overlapping sets of genes in different cell types and in response to different stimuli.
In this article, we highlight progress toward understanding the mechanisms by which the selective regulation of NF-κB target genes can be achieved. Rather than attempting to summarize the enormous body of literature relevant to this topic, we instead focus on a few key topics and on specific challenges that must be overcome to fully understand the determinants of selectivity. A primary focus is on the mechanisms by which closely related NF-κB family members and differences in the duration of activation can contribute to selectivity.
TARGET GENE SELECTIVITY OF DISTINCT NF-κB DIMERS
One fundamental contributor to the selectivity of an NF-κB response is the regulation of distinct sets of target genes by different NF-κB heterodimeric and homodimeric species (Fig. 1). The variable phenotypes of mice lacking different NF-κB family members provides strong evidence that different dimers regulate distinct sets of genes (Gerondakis et al. 1999, 2006). Some of the phenotypic differences observed in mice lacking specific NF-κB family members are caused by differences in the expression patterns of the family members. For example, RelA appears to be ubiquitously expressed, whereas c-Rel is largely restricted to hematopoietic tissues (Ghosh et al. 1998; Gerondakis et al. 1999, 2006; Hoffmann et al. 2006). Although expression differences are important to consider, phenotypic differences are apparent in cell types that express all family members, such as hematopoietic cells (Gerondakis et al. 1999, 2006). In these cells, many NF-κB target genes appear to be expressed normally in the absence of individual family members, suggestive of redundancy or compensation, but some target genes show strong dependence on a particular family member for expression.
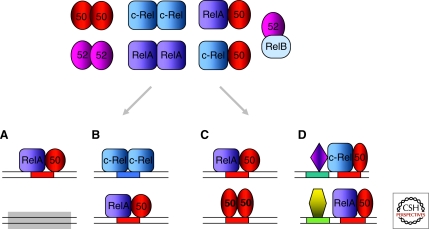
Figure 1.
Mechanisms that contribute to the specificity of gene activation by NF-κB proteins. Homo- and heterodimerization between Rel-homology region (RHR) containing proteins generates a diverse range of κB sequence element binding proteins (top). Typically, several of these may be present in the cell nucleus after an activating stimulus, although the composition varies according to cell type, stimulus, and duration of signaling. The spectrum of genes activated is determined by several factors discussed in the text. (A) Tissue- or signal-specific marking may allow NF-κB binding to some genes (top) but not others (bottom). Gray box represents chromatin constraints that may preclude NF-κB binding. (B) The sequence of the κB element in promoters, represented by blue and red boxes, within DNA may bind specific RHR family proteins. (C) The same κB element (red box) may bind more than one RHR dimer under different circumstances, leading to different transcriptional outcomes. (D) The proximity of κB elements (red box) to other transcription factor binding sites (blue and green boxes) may differentially regulate gene expression by combinatorial mechanisms.
The differential regulation of specific target genes by closely related NF-κB family members has been studied in greatest depth with RelA and c-Rel. These two family members are of particular interest because, in addition to the amino-terminal Rel homology region (RHR), which defines the NF-κB/Rel family and consists of the DNA-binding domain and dimerization domain, RelA and c-Rel contain potent transactivation domains at their carboxyl terminus (Ghosh et al. 1998; Ghosh and Hayden 2008; Vallabhapurapu and Karin 2009). (RelB also contains a transactivation domain, but RelB-containing heterodimers appear to be activated primarily by the nonclassical pathway [Vallabhapurapu and Karin 2009].) The RHRs of RelA and c-Rel are more closely related than are the RHRs of any other pair of NF-κB family members. Furthermore, structural studies have revealed that the residues that contact DNA are identical in these two family members (Chen and Ghosh 1999). Many NF-κB target genes in nonhematopoietic cell types are thought to require RelA for activation because c-Rel is absent. However, in hematopoietic cells that express RelA and c-Rel homodimers, as well as various heterodimeric species, both redundant and selective functions appear to exist.
Both c-Rel- and RelA-deficient mice show immune defects, demonstrating that the functions of the two proteins are not entirely redundant (Gerondakis et al. 1999, 2006). However, considerable redundancy appears to exist, as only a small number of NF-κB target genes have been identified that show strong dependence on either c-Rel or RelA. Microarray experiments performed in T cells revealed only a few genes that show strong dependence on c-Rel, although weak dependence was broadly observed (Bunting et al. 2007). In murine macrophages, Il12b and Il23a represent two of fewer than two dozen inducible genes identified in microarray experiments that show substantially reduced expression in the absence of c-Rel (Sanjabi et al. 2000, 2005; Carmody et al. 2007; K.J.W. and S.T.S., unpubl. data). RelA-dependent target genes have rarely been described in hematopoietic cells. However, one study revealed RelA-dependence in dendritic cells of several inflammatory cytokine genes, including Il6 (Wang et al. 2007).
A major unanswered question is whether genes that require either RelA or c-Rel for expression are regulated by homodimers of these family members or heterodimers. For example, c-Rel-dependent genes may require either c-Rel:c-Rel homodimers or any of a variety of heterodimeric species, such as c-Rel: p50 or c-Rel:p52. Because the functions of p50 and p52 appear to be partially or largely redundant, it is difficult to rely on analyses of p50- or p52-deficient mice for determining whether heterodimeric species are critical for the activation of c-Rel-dependent genes. Il12b, which is c-Rel-dependent, appears to be expressed normally in p50−/−p52−/− mice, consistent with the hypothesis that a c-Rel:c-Rel homodimer is responsible for selective regulation (Franzoso et al. 1997). However, because mechanisms can be envisioned that compensate for the simultaneous loss of these two family members, additional mechanistic insights are needed to determine whether c-Rel homodimers are indeed responsible for the selective regulation of Il12b. Unfortunately, chromatin immunoprecipitation (ChIP) experiments may not be informative, as both functional and nonfunctional dimers may associate with a given motif. For example, RelA, c-Rel, and p50 all appear to associate with the Il12b promoter, despite the c-Rel-dependence of transcription and the evidence that p50 and p52 may play no functional role (Sanjabi et al. 2005). These findings illustrate the challenge of understanding the selective regulation of NF-κB target genes by distinct dimeric species.
One strategy for distinguishing between the functions of homodimers and heterodimers may be to analyze substitution mutations that selectively disrupt the formation of homodimers or heterodimers. Mutations in v-Rel have been described that prevent the formation of v-Rel:v-Rel homodimers but not v-Rel:p50 heterodimers (Liss and Bose 2002). Analogous mutations in c-Rel or RelA may prove to be highly beneficial for distinguishing the targets of heterodimers from those of homodimers.
MECHANISMS OF TARGET GENE SELECTIVITY BY DIFFERENT NF-κB FAMILY MEMBERS
Although several examples of target genes that require specific NF-κB family members have been reported, surprisingly little is known about the range of mechanisms by which selective regulation is achieved. As mentioned previously, the RHRs of RelA and c-Rel are closely related but their transactivation domains show little similarity. At first glance, this observation suggests that the transactivation domains are likely to be responsible for each protein’s unique functions. However, the transactivation domains of both c-Rel and RelA are poorly conserved through vertebrate evolution, suggesting that, like transactivation domains in other transcription factors, their mechanisms of activation may not be highly specific. This lack of conservation is difficult to evaluate because structural features that are not reflected in sequence conservation may be conserved through evolution and may support highly specific protein–protein interactions. A number of post-translational modifications have been reported within the transactivation domains of both RelA and c-Rel, but the relevance of these modifications for the selective functions of the proteins have been examined in physiological assays in only a few instances.
One post-translational modification of RelA that has been examined extensively is the phosphorylation of serine 276 (S276) (Zhong et al. 1998, 2002; Dong et al. 2008). However, this residue is located within the RHR rather than the transactivation domain. Susceptibility to S276 phosphorylation is influenced by the RelA transactivation domain. S276 phosphorylation leads to the recruitment of the p300 coactivator, which is necessary for the activation of a subset of NF-κB target genes (Zhong et al. 1998, 2002). Interestingly, although the S276 residue is conserved in c-Rel, p300 overexpression does not enhance the transactivation capacity of c-Rel, in contrast to the strong enhancement observed with RelA (Wang et al. 2007). Indeed, an analysis of RelA-dependent genes expressed in dendritic cells suggested that the RelA requirement was because of RelA’s unique ability to associate with p300 (Wang et al. 2007). Thus, genes that require the function of the p300 coactivator for their expression may show a selective requirement for RelA-containing dimers.
Although differential interactions with coregulatory proteins may contribute to target gene selectivity of RelA and c-Rel, differences in binding properties may be of equal importance. Although the RHRs of RelA and c-Rel are highly conserved, and although the residues that contact DNA are identical, critical differences in binding properties appear to exist. Surprisingly, however, knowledge of these differences and of their functional relevance is limited. One major reason for our limited knowledge is that detailed studies of the range of potential recognition sites for each dimeric species have not been performed. A second reason is that the studies that have been completed suggest that, although recognition sequence differences can be identified, the differences are subtle and quantitative. A third and final reason for our limited knowledge is that it is very difficult to determine which of the many DNA motifs capable of binding each dimer with a range of affinities can support the regulation of endogenous target genes.
Remarkably, one of the only studies to characterize consensus DNA recognition motifs for NF-κB dimers was published almost two decades ago (Kunsch et al. 1992). In this study, an in vitro binding site selection analysis was performed to identify preferred recognition sequences for p50:p50, RelA:RelA, and c-Rel:c-Rel dimers. Fewer than two dozen high-affinity recognition sequences were described for each complex, revealing considerable similarity, but with clear differences. In particular, a few of the sequences that stably associated with c-Rel:c-Rel dimers were unable to bind to either RelA:RelA or p50:p50 dimers in electrophoretic mobility shift assays (EMSAs).
Although these results raised the possibility that c-Rel:c-Rel dimers may selectively bind some DNA motifs and selectively regulate target genes containing these motifs, these initial findings have not been significantly extended since their publication in 1992. In particular, comparable studies of multiple heterodimeric species have not been performed and detailed studies examining the precise binding affinities of homodimers and heterodimers for a range of sequences (in side-by-side experiments) have not been performed. Knowledge of binding affinities would not by itself reveal the degree to which selective binding can contribute to the selective regulation of target genes by various NF-κB dimers, but this knowledge would represent a critical step toward this goal. It is important to add that, although biochemical experiments can be used to identify intrinsic preferences, NF-κB, like most transcription factors, has the potential to bind DNA cooperatively with other factors. Cooperative binding can lead to functional activity at low affinity sites, because the specificity and affinity of the NF-κB-DNA interaction can be enhanced by the binding partner.
As an alternative strategy for understanding the selective regulation of NF-κB target genes, one of our laboratories analyzed c-Rel/RelA chimeric proteins to identify the residues of c-Rel that are responsible for its unique ability to activate the endogenous Il12b gene in LPS-stimulated murine macrophages (Sanjabi et al. 2005). By using retroviral vectors to express a series of chimeric proteins in c-Rel−/− macrophages, an 86-residue region of the c-Rel RHR was found to be sufficient for rescuing Il12b activation when inserted in place of the corresponding residues of RelA. DNA-binding analysis of these chimeric proteins revealed that this 86-residue region, which contains only 46 residues that differ between RelA and c-Rel, is responsible for the ability of c-Rel homodimers to bind with high affinity to a broader range of nonconsensus NF-κB recognition sequences than RelA homodimers, consistent with the earlier studies of Kunsch et al. (1992). Thus, these results provide strong evidence that differential DNA binding plays a major role in the selective activation of Il12b by c-Rel. Importantly, no other regions of c-Rel were important for Il12b activation in the chimeric protein analysis, suggesting that the unique DNA-binding properties of c-Rel may be solely responsible for the selective activation of at least one c-Rel target gene.
Another mechanism of selective regulation by different NF-κB family members emerged from the initial observation that NF-κB recognition sequences in specific promoters are strictly conserved through evolution, despite evidence that nucleotide substitutions could easily be tolerated while maintaining high-affinity binding (Leung et al. 2004). Furthermore, the NF-kB recognition sites were often found in pairs within promoters. Dependence versus independence on the p50 or p52 subunits of NF-κB, as shown by an analysis of p50−/−p52−/− MEFs, revealed that subunit dependence was dictated by the specific recognition sequences and by cooperation between the pairs of sites. Most significantly, alterations in the highly conserved nucleotides within these DNA motifs altered subunit dependence, but apparently without altering binding affinity. Instead, the different recognition sequences and pairs of recognition sequences varied in their responsiveness to specific coactivator proteins. Although much remains to be learned about the mechanisms by which different NF-κB dimers, recognition sequences, and coactivator proteins can contribute to selectivity of the NF-κB response, this study illustrates the degree of complexity that is likely to be involved.
DURATION OF NF-κB ACTIVATION
The preceding sections focused on the mechanisms by which the differential expression patterns of NF-κB family members, as well as their variable binding and transactivation properties, may contribute to the selective regulation of NF-κB target genes. Another fundamental regulatory feature of NF-κB activation via the canonical pathway is its transience. The duration of NF-κB activation may contribute to the selectivity of the transcriptional response by dictating which other factors can cooperate with NF-κB to activate gene transcription, by regulating the relative levels of different NF-κB proteins, and by changes in association with transcriptional coactivators and corepressors (Fig. 2). NF-κB duration may also regulate physiological responses by maintaining cell viability or cytokine and chemokine gene expression. Thus, the kinetics of NF-κB activation and inactivation is a central regulator of its function. Several pathways contribute to transient NF-κB activation, as discussed below.
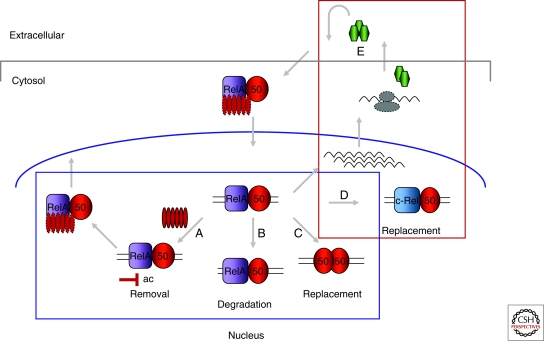
Figure 2.
Mechanisms that regulate duration of NF-κB activation. NF-κB activation via the classical pathway is initiated by degradation of IκB protein (cytosol center), resulting in NF-κB (schematized by a p50/RelA heterodimer here, but could involve many of the homo-/heterodimeric pairs shown in Fig. 1) translocation to the nucleus and gene activation after DNA binding. Mechanisms summarized within the blue box reduce the duration of NF-κB-mediated gene expression. (A) Newly synthesized IκB proteins can remove DNA-bound NF-κB and export the complex out to the cytosol; (B) post-translational modifications of RelA, such as methylation and phosphorylation, can target it for proteasome-mediated degradation within the nucleus. The fate of the heterodimeric partner in the complex is not known; (C) transcription-activating heterodimers (such as p50/RelA or p50/c-Rel) can be replaced by p50 homodimers that have been implicated in repressing transcription (note however, that there is also evidence that p50 homodimers may activate transcription in association with nonclassical IκB-like proteins such as Bcl3 and IκBξ). Mechanisms summarized in the red box can extend the duration of NF-κB-mediated gene expression. (D) De novo c-Rel gene transcription and translation can lead to long-term induction of c-Rel-containing heterodimers, and (E) NF-κB target genes such as TNFα (green hexagon) activated during the initial stimulus may feed back in an autocrine fashion to maintain nuclear NF-κB.
IκB-dependent Export
One major pathway is via nuclear export of IκBα and IκBε-associated nuclear proteins. IκBα and IκBε are NF-κB target genes that are rapidly activated at the transcriptional level in response to NF-κB induction in all cell types tested. This leads to de novo IκBα and IκBε protein synthesis to replenish IκBs degraded in response to the NF-κB-inducing stimulus. Newly synthesized IκB proteins translocate to the nucleus, associate with nuclear NF-κB proteins, and export the transcription factors to the cytoplasm via nuclear export sequences present at their amino-termini (Tam et al. 2000; Huang and Miyamoto 2001; Johnson et al. 1999; Kearns et al. 2006). Early observations that IκBα can disrupt DNA-bound NF-κB complexes in vitro suggested that this mechanism may apply to promoter-bound NF-κB in the nucleus (Zabel and Baeuerle 1990). Although disruption of promoter complexes would be sufficient to stop NF-κB-dependent transcription, it is believed that nuclear export serves the essential function of recreating a pool of cytoplasmic NF-κB/IκB complexes to respond to ongoing or subsequent stimuli.
Three aspects of IκB-dependent transcription termination are noteworthy with respect to the selectivity issue. First, IκBα gene transcription precedes IκBε transcription (Kearns et al. 2006); therefore, IκBα protein is resynthesized first and is likely to be the major player in NF-κB down-regulation. However, the kinetics of IκBα and IκBε synthesis may vary depending on the cell type and stimulus, and should therefore be evaluated in specific circumstances. Second, IκBβ does not contain a nuclear export sequence (NES) and has been proposed to be the only IκB that truly sequesters NF-κB in the cytosol (Tam and Sen 2001). However, newly synthesized IκBβ can translocate to the nucleus and associate with promoter-bound NF-κB proteins without disrupting DNA-protein interactions (Suyang et al. 1996). This mechanism has been proposed to “protect” DNA-bound NF-κB from IκBα-mediated disruption, thereby prolonging NF-κB-dependent gene expression. Third, acetylation of RelA by CBP/p300 complexes has been shown to pre-vent IκBα association, and thereby proposed to extend the duration of functional RelA complexes on some promoters (Chen et al. 2001; Chen et al. 2002; Kiernan et al. 2003). However, the proportion of RelA that gets acetylated is quite low; thus, the contribution of this pathway to the duration of NF-κB activation needs to be evaluated in specific situations. It is also likely that this mechanism applies to only a subset of promoters, such as those in which independently recruited p300/CBP is available to acetylate Rel proteins after cell stimulation. In our view, acetylation may provide protection to a subset of NF-κB-dependent promoters from rapid shut down by de novo synthesized IκBs.
Finally, NF-κB proteins themselves can contribute to their differential dwell time in the nucleus. RelA contains an NES in its carboxy-terminal domain, whereas c-Rel does not (Tam et al. 2001; Harhaj and Sun 1999). Therefore, different NF-κB homo- and heterodimers vary in their propensity to be nuclear. Specifically, RelA homodimers carry two NESs even in the absence of associated IκBα that can lead to their export from the nucleus. In contrast, c-Rel homodimers have no NESs and therefore require help to be exported from the nucleus. Although the RelA NES can mediate its export from the nucleus, IκBα and ε help reduce nuclear re-entry of RelA containing complexes by partially obscuring the nuclear localization signal. However, IκBα association does not completely prevent nuclear entry of NF-κB complexes; consequently, IκBα-associated complexes shuttle between the nucleus and the cytosol (Tam et al. 2000). The preponderance of IκBα-associated complexes in the cytosol in biochemical or immunohistochemical analyses therefore reflects a “snapshot” of a dynamic state in which the rate of export greatly exceeds the rate of import of these complexes. In this view, the NF-κB complexes vary in their nuclear propensity in approximate proportion to the number and strength of NLSs and NESs.
Nuclear Degradation of RelA
Transient NF-κB-dependent gene expression even in the absence of IκBα first suggested that other mechanisms may contribute to the duration of nuclear NF-κB (Saccani et al. 2004). Natoli and colleagues showed that a proportion of RelA in cells is degraded by the proteasome while bound to gene promoters. Since then, the combined effort of several laboratories has shown that ubiquitination is mediated by a SOCS1-containing E3 ligase complex in conjunction with COMMD1, a ubiquitously expressed negative regulator of NF-κB (Maine et al. 2007; Mao et al. 2009). Another E3 ligase, PDLIM2, has also been shown to ubiquitinate and target RelA to PML bodies where it is degraded by the proteasome (Tanaka et al. 2007). The relative contribution of each pathway for targeting RelA degradation may vary according to cell type and stimulus.
Recently, methylation of lysines 314 and 315 in RelA by the methyl transferase Set 7/9 has been implicated in marking promoter-bound RelA for degradation. Yang et al. (2009) found that knockdown of Set 7/9 in the U206 cell line resulted in higher levels of IL-6 and IL-8 gene expression in response to TNFα; in contrast, expression of a different NF-κB target gene, A20, was unaffected. Using chromatin immunoprecipitation, they located Set 7/9 at the IL-8 promoter in unactivated cells, but not at the A20 promoter regardless of the state of cell activation. This led to the hypothesis that prebound Set 7/9 at certain promoters is one mechanism by which promoters are marked for down-regulation by RelA degradation. At present, it is not clear why certain promoters are cleared in this manner, or whether this mechanism varies depending on the cell type or stimulus.
Degradation of nuclear RelA has also been proposed to be the basis for extended NF-κB-dependent gene expression in macrophages that lack IKK1 (Lawrence et al. 2005). In this case, Karin and colleagues showed that phosphorylation of nuclear RelA was reduced, as was proteasome-mediated nuclear degradation (Vallabhapurapu and Karin 2009). The generality of these observations to other cell types, or to other NF-κB-inducing stimuli in macrophages, remains to be determined. Taken together, these observations indicate that multiple mechanisms ensure that the duration of the NF-κB response is strictly regulated.
Secondary Stimulation Controls NF-κB Duration
Bacterial LPS is a well known inducer of canonical NF-κB via activation of Toll-like receptor 4 (TLR4). Typically, LPS induces long-lasting nu-clear NF-κB compared with induction by TNFα or IL-1. The basis for this in mouse embryo fibroblasts (MEFs) appears to be that LPS treatment induces production of endogenous TNFα, which acts in an autocrine fashion to maintain long-term nuclear NF-κB activity. Werner et al. (2005) found that long-term IKK activation and nuclear NF-κB expression was significantly reduced in TNFα-deficient MEFs. Concomitantly, transcription of a subset of late-activated NF-κB-dependent genes, such as IL-12β and IL-1α and β, was also attenuated in these cells.
De Novo c-Rel Synthesis as a Means of Prolonging the NF-κB Response
Despite nuclear degradation of RelA after an activating stimulus, the level of RelA protein for the most part does not vary significantly in activated cells. Presumably, this is because the bulk of induced protein is not targeted for nuclear degradation. The small decrease in RelA is likely made up by constitutive protein synthesis from pre-existing RelA mRNA. Thus, post-translational mechanisms account for most of RelA dynamics in response to cellular activation. However, this is not the case for c-Rel. In T lymphocytes activated via the T-cell receptor (or its pharmacologic analog phorbol ester plus calcium ionophore), c-Rel gene transcription is induced, leading to de novo synthesis of c-Rel protein (Venkataraman et al. 1996). This newly synthesized c-Rel accumulates in the nucleus with delayed kinetics compared with post-translationally induced NF-κB via the canonical pathway. This is likely to be the basis for early observations in Jurkat T cells, that “late” NF-κB DNA binding activity consisted of c-Rel containing complexes. The immunosuppressive drugs cyclosporin A (CsA) and FK506 inhibit inducible c-Rel gene transcription, which has been proposed to be mediated by the transcription factor NFAT (Grumont et al. 2004). Because this kind of c-Rel expression is controlled at the level of transcription, it does not occur with all forms of NF-κB-inducting stimuli. Notably, TNFα, IL-1, or LPS treatment, which do not induce long-term calcium mobilization, do not induce de novo c-Rel synthesis. Therefore, the nature of the signal determines when cells adopt this mode of “long-term” NF-κB (specifically, c-Rel) activation.
These observations have been extended to B lymphocytes activated via the B-cell antigen receptor (BCR). Surface immunoglobulin cross-linking induces two waves of nuclear NF-κB activity (B. Damdinsuren and R. Sen, unpubl. data). The first phase consists of both RelA and c-Rel and is mediated via the canonical post-translational pathway. This phase is transient, lasting between 4 and 6 hours, and coincides with reduced cellular IκB levels. It is followed by a slower second phase, composed mainly of c-Rel protein that is mediated by de novo c-Rel transcription and translation. As found in T cells, this long-term c-Rel induction is suppressed by CsA. Gene expression analyses in BCR-activated B lymphocytes indicate that phase I NF-κB induces auto-regulatory genes such as NFKBIZ and NFKBIE, and genes important for G1 progression such as c-myc. Phase II c-Rel is essential for cell survival and entry into S phase of the cell cycle. These observations show that early and late NF-κB induces distinct sets of genes and emphasize the importance of the duration of NF-κB activation as a key determinant of cellular responses.
PATTERNS OF NF-κB-DEPENDENT GENE EXPRESSION
In the preceding sections, we summarized mechanisms by which NF-κB target genes can be selectively regulated by different NF-κB family members and dimeric species, and by variations in the duration of classical NF-κB activation. Here, we briefly consider the impact of lineage-specific and stimulus-specific marking of target genes on the selectivity of an NF-κB response, and we further discuss the kinetics of NF-κB-dependent gene expression. A recurring theme is the central role played by the specific architecture of the control regions for each NF-κB target gene in defining the combination of inducible transcription factors required for gene activation, and in orchestrating changes in chromatin structure that modulate susceptibility to activation.
Lineage-specific Marking
The interleukin 2 (Il2) gene serves as a good example to explore the mechanisms that distinguish which NF-κB-dependent genes will be induced in specific cell types in response to NF-κB-inducing signals. Il2 is a T lymphocyte-specific gene that is activated in response to antigen receptor (CD3) plus costimulatory (CD28) signals. Its promoter has been extensively studied and contains functionally important binding sites for inducible transcription factors NF-κB, NF-AT and AP-1. The most important NF-κB binding site comprises the so-called CD28 response element (CD28RE) that is essential for high level gene induction (Himes et al. 1996; Shapiro et al. 1997; Rao et al. 2003). Two properties determine importance of the CD28RE. First, CD28 cross-linking, via its ability to activate the PI3 kinase pathway in T cells, leads to efficient IKK2 activation (Park et al. 2009). Consequently, IκB proteins are phosphorylated and degraded, releasing IκB-bound NF-κB protein to translocate to the nucleus. Second, the CD28RE preferentially binds c-Rel containing NF-κB proteins; therefore, CD28 costimulation is largely ineffective in c-Rel-deficient T cells. Because most of the cytoplasmic c-Rel in naïve CD4 T cells is bound to IκBβ (Banerjee et al. 2005), efficient CD28-dependent degradation of IκBβ is essential for c-Rel translocation to the nucleus to activate IL-2 gene transcription.
Although TCR signals are sufficient to induce NFAT and AP-1, the requirement for CD28 indicates that c-Rel is essential for promoter activation, particularly in naïve CD4+ T cells. However, these three transcription factors are also induced in BCR-activated B cells. Yet, Il2 gene transcription is T-cell specific. These observations emphasize that tissue-specific NF-κB target genes must be preselected during differentiation to respond to NF-κB inducing stimuli. This preselection is likely to consist of the changes in chromatin structure during T-cell differentiation that confer susceptibility to activation on TCR engagement. These developmental changes in chromatin structure and the mechanisms by which they are regulated remain poorly understood for the Il2 gene and other NF-κB target genes. However, in simplistic terms, because different loci become susceptible to NF-κB activation in different cell lineages, the determinants of susceptibility must consist of specific DNA motifs within each locus that bind developmentally regulated transcription factors capable of catalyzing changes in chromatin structure and susceptibility to activation.
It is important to add that the combinatorial activation of the Il2 gene by multiple inducible transcription factors illustrates that stimuli that induce only a subset of these proteins will not be sufficient to activate the Il2 gene in T cells. For example, TNFα stimulation of T cells activates NF-κB, but not NFAT; consequently, TNFα does not activate Il2 gene transcription.
Stimulus-specific Marking
In addition to lineage-specific marking (such as Il2 in T vs. B cells), genes can also be marked for stimulus-specific gene expression. One of the best examples of this kind of marking comes from the work of Natoli and colleagues (Saccani et al. 2002). They showed that certain NF-κB-dependent promoters, such as Il6, Il12b, and MCP-1, were marked by p38-MAP kinase-dependent phosphorylation of histone H3 in human dendritic cells treated with LPS. Pharmacologic inhibition of p38 MAP kinase reduced H3S10 phosphorylation, and delayed transcription activation of these genes, despite normal NF-κB induction. Not all NF-κB-dependent inducible genes in these cells were marked in this way, suggesting multiple regulatory mechanisms in play. Importantly, TNFα treatment induced NF-κB but not p38 in these cells, resulting in reduced expression of p38-marked NF-κB-dependent genes without affecting other NF-κB-dependent inducible transcription. How p38-dependent H3S10 phosphorylation helps to selectively confer NF-κB responsiveness at certain promoters remains to be clarified; nor is it clear how certain genes come to be marked in this fashion.
Stimulus-specific activation can also be regulated by the remodeling of promoter-encompassing nucleosomes catalyzed by stimulus-specific transcription factors. Many rapidly induced NF-κB target genes can be activated without a requirement for nucleosome remodeling by ATP-dependent nucleosome remodeling complexes of the SWI/SNF family (Ramirez-Carrozzi et al. 2006, 2009). These genes usually contain CpG-island promoters, which may be directly responsible for remodeling-independent activation through their intrinsic assembly into unstable nucleosomes (Ramirez-Carrozzi et al. 2009). Importantly, remodeling-independent activation is generally associated with genes known to be activated by a broad and diverse range of stimuli. In contrast, remodeling-dependent activation is generally observed at genes that are activated more selectively. These genes contain non-CpG-island promoters that assemble into stable nucleosomes, leading to a requirement for nucleosome remodeling by SWI/SNF complexes. In response to LPS stimulation of macrophages via TLR4, for example, genes that depend on both NF-κB and IRF3 for activation almost always require nucleosome remodeling. IRF3 is essential for nucleosome remodeling, thereby restricting activation of these genes to stimuli capable of inducing IRF3 in addition to NF-κB.
In this stimulus specificity example, as in the lineage-specific regulation of Il2 transcription, promoter architecture plays a central role in selective regulation. In this case, the presence or absence of a CpG-island largely dictates whether activation will be remodeling-independent or remodeling-dependent. Then, for remodeling-dependent genes, the presence of DNA recognition motifs for specific transcription factors capable of promoting the recruitment of nucleosome remodeling complexes dictates which NF-κB-inducing stimuli will be capable of inducing gene transcription.
Kinetics of NF-κB-dependent Gene Expression
Appropriate expression of NF-κB-dependent genes can be broadly categorized in terms of three parameters: (a) the rate of gene induction, (b) the duration of functional promoter-bound complexes, and (c) the rate of mRNA degradation. A rapidly induced gene will generate mRNA quickly but accumulation of mRNA will depend on its stability and the rate of continued RNA synthesis. Conversely, a gene may be induced slowly, but its expression level may be maintained by stability of the promoter complex and the mRNA that is generated. We shall briefly consider what is known about each of these parameters.
Rate of Gene Induction
Promoter architecture is the key determinant of the rate of transcription induction by induced NF-κB. For example, the promoter of the well-accepted NF-κB target gene IκBα contains 6 NF-κB binding sites located relatively close to the transcription start site (Rupec et al. 1999). To date, no other inducible factors have been implicated in IκBα transcription. Simplistically, we can therefore consider the IκBα promoter to be activated purely based on the presence or absence of NF-κB in the nucleus. This is consistent with the biology of IκBα, which is induced in response to all NF-κB-inducing stimuli, and in all cell types examined, to initiate feedback inhibition of NF-κB activity. In contrast, most other NF-κB-inducible genes are subject to more complex regulation, such as lineage- and/or stimulus-specificity discussed previously.
Therefore, most NF-κB-inducible promoters are more complex. This complexity may be reflected in the number of NF-κB binding sites present, their disposition relative to each other as well as the transcription initiation site, and most importantly in the requirement for other constitutive, or inducible, transcription factors to coordinate with inducible NF-κB to activate gene expression. These variables unfortunately have not been systematically investigated in enough genes to draw general conclusions. Nevertheless, it is easy to see how a promoter that requires a “late” inducible factor plus NF-κB could be activated later than a promoter that requires only NF-κB, or NF-κB plus a constitutively present transcription factor. Finally, some NF-κB-inducible genes may be regulated by distal NF-κB-dependent transcriptional enhancers, such as the immunoglobulin κ light chain gene.
Duration of Gene Transcription
The duration of gene transcription will also be determined by the duration of a functional promoter complex. As described previously, transient presence of NF-κB components in the nucleus is one way in which the complex can be disrupted. Mechanistically, this can occur by IκBα or IκBε proteins migrating into the nucleus and actively disrupting promoter complexes by removing DNA-bound NF-κB. Alternately, IκBα proteins may only remove predissociated NF-κB. It is very likely that the nature of the promoter complex determines the pathway for NF-κB eviction and thereby the effectiveness of transcription termination. For example, NF-κB interactions with its nearest neighbors, which will vary from promoter to promoter, may determine the stability of the promoter complex to IκB-mediated disruption. Thereby, newly synthesized IκBs will disrupt some promoters earlier than others. Another way to destabilize promoters could be based on the nuclear half-lives of other inducible transcription factors, or their post-translational modifications.
NF-κB-dependent transcription may also be terminated by replacing transcriptionally active NF-κB proteins by transcriptionally inactive ones. In particular, p50 homodimers have been proposed to repress transcription, because they lack transcription activation do-mains. Additionally, DNA-bound p50 homodimers have been shown to recruit histone deacetylases to promoters to epigenetically inactivate transcriptionally permissive chromatin state (Zhong et al. 2002; Williams et al. 2006). Because the NF-κB1 gene, which encodes the p105 precursor to p50, is thought to be an NF-κB target gene, this route of gene suppression is yet another form of feedback inhibition mediated by induced NF-κB.
Post-transcriptional Regulation of NF-κB Target Genes
The variety of ways by which NF-κB is down-regulated from the nucleus indicates that long-term NF-κB activation is detrimental. To effectively terminate NF-κB-dependent gene expression, however, post-transcriptional regulatory mechanisms are also in place. For example, if NF-κB target gene mRNA persists significantly after termination of transcription, the effect of NF-κB down-regulation will not be obvious. Similarly, NF-κB transcription factor down-regulation will have little impact if proteins expressed from NF-κB target genes remain functionally active in cells. One of the earliest characterized examples of the functional consequences of dysregulating NF-κB target genes at the post-transcriptional level is that of TNFα. The 3′ untranslated region of the TNFα mRNA contains AU-rich elements (AREs) that determine mRNA stability and translational control (Kontoyiannis et al. 1999; Hitti et al. 2006). Germline deletion of the ARE results in increased TNFα production and development of chronic inflammatory pathologies (Kontoyiannis et al. 1999).
Recently, Hao and Baltimore reported the first systematic analysis of the role of mRNA stability in regulating NF-κB-dependent gene expression (Hao and Baltimore 2009). They found that the expression patterns of TNFα-inducible genes could be grouped on the basis of mRNA stability. Specifically, mRNAs that were rapidly induced and rapidly down-regulated had short half-lives (t1/2), mRNAs that were rapidly induced and then maintained had intermediate t1/2, and those that were induced gradually over the time course of their studies had long t1/2. The t1/2s were determined by the 3′ UTRs of these genes, with shorter t1/2 correlating with higher numbers of AREs within the 3′ UTR. Interestingly, genes within each category were functionally related with regard to their roles during inflammation. Thus, post-transcriptional mechanisms constitute a critical component of NF-κB-dependent responses.
CONCLUDING REMARKS
NF-κB dimers can be induced in virtually all cell types by a remarkably broad range of stimuli, yet the set of target genes induced in a cell type by a given stimulus is highly variable. Much of this selectivity can be attributed to differences in the set of signaling pathways induced by each stimulus, which lead to the induction of multiple transcription factors that act in concert with NF-κB dimers through combinatorial mechanisms to activate specific target genes. Although considerable effort has been devoted to the elucidation of signaling pathways, and associated transcription factors, induced by NF-κB inducing stimuli, there exists a considerable gap in our mechanistic understanding of how these factors cooperate with NF-κB to coordinate cellular responses. Given the growing importance of NF-κB dysregulation in human disease, it is likely that such combinatorial specificities will provide additional targets for therapeutic intervention.
In this article, we attempted to minimize documenting the considerable advances that have been made in NF-κB-dependent transcriptional control. Instead, we highlight a subset of regulatory strategies that reveal important goals for the future. Amongst these are the elaboration of different functions of closely related NF-κB family members as homo- or heterodimeric species, and the mechanism by which NF-κB complexes work with other transcription factors to activate transcription. Similarly, although the duration of NF-κB response is now known to be tightly regulated and contributes to selective gene activation, much remains to be learned about the coordination between various levels of regulation that impact NF-κB-dependent transcription. Additionally, it is likely that NF-κB regulation itself, as well as its transcriptional cross talk with other factors, will vary considerably between tissue types. Further studies in this area will be particularly interesting. Finally, the control regions associated with each NF-κB target gene must, by necessity, contain all of the information required to regulate cell type and stimulus-specific activation, yet we lack a complete understanding of how genes are selected to become NF-κB responsive. The availability of complete genome sequences and of an increasing repertoire of molecular and bioinformatic techniques for analyzing mechanisms of selective gene transcription will undoubtedly lead to rapid progress.
ACKNOWLEDGMENTS
Research in the NF-κB field performed in the authors’ laboratories was supported by the Intramural Research Program of the National Institute of Aging (R.S.) and by NIH grant R01 AI073868 (S.T.S.).
Footnotes
Editors: Louis M. Staudt and Michael Karin
Additional Perspectives on NF-κB available at www.cshperspectives.org
REFERENCES
- Banerjee D, Liou HC, Sen R 2005. c-Rel-dependent priming of naïve T cells by inflammatory cytokines. Immunity 4:445–458 [DOI] [PubMed] [Google Scholar]
- Bunting K, Rao S, Hardy K, Woltring D, Denyer GS, Wang J, Gerondakis S, Shannon MF 2007. Genome-wide analysis of gene expression in T cells to identify targets of the NF-κ B transcription factor c-Rel. J Immunol 178:7097–7109 [DOI] [PubMed] [Google Scholar]
- Carmody RJ, Ruan Q, Liou HC, Chen YH 2007. Essential roles of c-Rel in TLR-induced IL-23 19 gene expression in dendritic cells. J Immunol 178:186–191 [DOI] [PubMed] [Google Scholar]
- Chen FE, Ghosh G 1999. Regulation of DNA binding by Rel/NF-κB transcription factors: Structural views. Oncogene 18:6845–6852 [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J 21:6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LF, Fischle W, Verdin E, Greene WC 2001. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293:1653–1657 [DOI] [PubMed] [Google Scholar]
- Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S 2008. Repression of gene expression by unphosphorylated NF-κB 65 through epigenetic mechanisms. Genes Dev 22:1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Xing L, Poljak L, Shores EW, Brown KD, Leonardi A, Tran T, Boyce BF, Siebenlist U 1997. Requirement for NF-κB in osteoclast and B-cell development. Genes Dev 11:3482–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondakis S, Grossmann M, Nakamura Y, Pohl T, Grumont R 1999. Genetic approaches in mice to understand Rel/NF-κB and IκB function: Transgenics and knockouts. Oncogene 18:6888–6895 [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A 2006. Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene 25:6781–6799 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Hayden MS 2008. New regulators of NF-κB in inflammation. Nat Rev Immunol 8:837–848 [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB 1998. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol 16:225–260 [DOI] [PubMed] [Google Scholar]
- Grumont R, Lock P, Mollinari M, Shannon FM, Moore A, Gerondakis S 2004. The mitogen-induced increase in T cell size involves PKC and NFAT activation of Rel/NF-κB-dependent c-mye expression. Immunity 21:19–30 [DOI] [PubMed] [Google Scholar]
- Hao S, Baltimore D 2009. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nature Immunology 10:281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj EW, Sun SC 1999. Regulation of RelA subcellular localization by a putative nuclear export signal and 50. Mol Cell Biol 19:7088–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S 2004. Signaling to NF-κB. Genes Dev 18:2195–2224 [DOI] [PubMed] [Google Scholar]
- Himes SR, Coles LS, Reeves R, Shannon MF 1996. High mobility group protein I(Y) is required for function and for c-Rel binding to CD28 response elements within the Gm-CSF and IL-2 promoters. Immunity 5:479–489 [DOI] [PubMed] [Google Scholar]
- Hitti E, Iakovieve T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A, Gaestel M 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol 26:2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G 2006. Transcriptional regulation via the NF-κB signaling module. Oncogene 25:6706–6716 [DOI] [PubMed] [Google Scholar]
- Huang TT, Miyamoto S 2001. Postrepression activation of NF-κB requires the amino-terminal nuclear export signal specific to IκBα. Mol Cell Biol 21:4737–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Van Antwerp D, Hope TJ 1999. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J 18:6682–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A 2006. IκBε provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J Biol Chem 173:659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M 2003. Post-activation turn-off of NF-κ B-dependent transcription is regulated by acetylaltion of 65. J Biol Chem 278:2758–2766 [DOI] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity 10:387–398 [DOI] [PubMed] [Google Scholar]
- Kunsch C, Ruben SM, Rosen CA 1992. Selection of optimal κ B/Rel DNA-binding motifs: Interaction of both subunits of NF-κ B with DNA is required for transcriptional activation. Mol Cell Biol 12:4412–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Bebien M, Liu GY, Nizet V, Karin M 2005. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 434:1138–1143 [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D 2004. One nucleotide in a κB site can determine cofactor specificity for NF-κB dimers. Cell 20:453–464 [DOI] [PubMed] [Google Scholar]
- Liss AS, Bose HR Jr 2002. Mutational analysis of the v-Rel dimerization interface reveals a critical role for v-Rel homodimers in transformation. J Virol 76:4928–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GN, Mao X, Komarck CM, Burstein E 2007. COMMD1 promotes the ubiquitination of NF-κB subunits through a cullin-containing ubiquitin ligase. EMBO J 26:436–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Gluck N, Li D, Maine GN, Li H, Zaidi IW, Repaka A, Mayo MW, Burstein E 2009. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-κB/RelA. Genes Dev 23:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I 2005. Interactions of NF-κB with chromatin: The art of being at the right place at the right time. Nat Immunol 6:439–445 [DOI] [PubMed] [Google Scholar]
- Natoli G 2009. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol 1:a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Schulze-Luehrman J, Hayden MS, Hashimoto N, Ogawa W, Kasuga M, Ghosh S 2009. The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-κB and activate T cells. Nat Immunol 10:158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CM, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138:114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20:282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Gerondakis S, Woltring D, Shannon MF 2003. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol 170:3724–3731 [DOI] [PubMed] [Google Scholar]
- Rupec RA, Poujol D, Grosgeorge J, Carle GF, Livolsi A, Peyron JF, Schmid RM, Baeuerle PA, Messer G 1999. Structural analysis, expression, and chromosomal localization of themouse ikba gene. Immunogenetics 49:395–403 [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2002. 38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat Immunol 3:69–75 [DOI] [PubMed] [Google Scholar]
- Saccani S, Marazzi I, Beg AA, Natoli G 2004. Degradation of promoter-bound 65/RelA is essential for the prompt termination of the nuclear factor in κB response. J Exp Med 200:107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST 2000. Selective requirement for c-Rel during IL-12 40 gene induction in macrophages. Proc Natl Acad Sci USA 97:12705–12710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S, Williams KJ, Saccani S, Zhou L, Hoffmann A, Ghosh G, Gerondakis S, Natoli G, Smale ST 2005. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev 19:2138–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, Baltimore D 1986. Inducibility of κ immunoglobulin enhancer-binding protein Nf-κ B by a posttranslational mechanism. Cell 47:921–928 [DOI] [PubMed] [Google Scholar]
- Shapiro VS, Truitt KE, Imboden JB, Weiss A 1997. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol Cell Biol 17:4051–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyang H, Phillips R, Douglas I, Ghosh S 1996. Role of unphosphorylated newly synthesized I κ B β in persistent activation of NF-κB. Mol Cell Biol 16:5444–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WF, Sen R 2001. IκB family members function by different mechanisms. J Biol Chem 276:7701–7704 [DOI] [PubMed] [Google Scholar]
- Tam WF, Wang W, Sen R 2001. Cell-specific association and shuttling of IκBα provides a mechanism for nuclear NF-κB in B lymphocytes. Mol Cell Biol 21:4837–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam WF, Lee LH, Davis L, Sen R 2000. Cytoplasmic sequestration of rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol 20:2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Grusby MJ, Kaisho T 2007. PDLIM2-mediated termination of transcription factor NF-κB activation by intranuclear sequestration and degradation of the 65 subunit. Nat Immunol 8:584–591 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M 2009. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol 27:693–733 [DOI] [PubMed] [Google Scholar]
- Venkataraman L, Wang W, Sen R 1996. Differential regulation of c-Rel translocation in activated B and T cells. J Immunol 157:1149–1155 [PubMed] [Google Scholar]
- Wang J, Wang X, Hussain S, Zheng Y, Sanjabi S, Ouaaz F, Beg AA 2007. Distinct roles of different NF-κ B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J Immunol 178:6777–6788 [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309:1857–1861 [DOI] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC 2006. NF-κB 50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25:139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF 2009. Negative regulation of NF-κ B action by Set9-mediated lysine methylation of the RelA subunit. EMBO J 28:1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Baeuerle PA 1990. Purified human I κ B can rapidly dissociate the complex of the NF-κ B transcription factor with its cognate DNA. Cell 61:255–265 [DOI] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S 1998. Phosphorylation of NF-κ B 65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell 1:661–671 [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S 2002. The phosphorylation status of nuclear NF-κ B determines its association with CBP/p300 or HDAC-1. Mol Cell 9:625–636 [DOI] [PubMed] [Google Scholar]