Abstract
Actin participates in several essential processes in the cell nucleus. Even though the presence of actin in the nucleus was proposed more than 30 years ago, nuclear processes that require actin have been only recently identified. Actin is part of chromatin remodeling complexes; it is associated with the transcription machineries; it becomes incorporated into newly synthesized ribonucleoproteins; and it influences long-range chromatin organization. As in the cytoplasm, nuclear actin works in conjunction with different types of actin-binding proteins that regulate actin function and bridge interactions between actin and other nuclear components.
Actin functions in the nucleus as part of chromatin-remodeling complexes and cooperates with a nuclear myosin motor to regulate the transcriptional machinery.
Actin is a highly conserved protein of approximately 42 kDa found in all eukaryotic cells. It is a major component of the cytoskeleton and plays fundamental roles in essential biological processes such as determining cell shape, cell migration, and intracellular trafficking. The functions of actin in the cytoplasm are intrinsically coupled to the dynamics of actin polymerization, which is a tightly regulated process that responds to extracellular signals (reviewed by Moustakas and Heldin 2008; Papakonstanti and Stournaras 2008).
Early studies raised the possibility that actin is also present in the cell nucleus and is implicated in the expression of protein-coding genes (Scheer et al. 1984; Egly et al. 1984). However, the existence of the so-called “nuclear actin” was initially met with massive skepticism (reviewed by Pederson and Aebi 2002). Biochemists could not rule out contamination artifacts in nuclear preparations because of the high abundance of actin in the cytoplasm, and microscopists could not visualize in the cell nucleus the conspicuous actin filaments that are commonly observed in the cytoplasm. Nevertheless, research performed in the last 10 years has provided convincing evidence for the existence of actin in the cell nucleus and for the involvement of actin in fundamental nuclear processes. Actin is part of the chromatin remodeling complex; it is associated with the transcription machineries; it associates with newly synthesized ribonucleoproteins; and it influences long-range chromatin organization.
ACTIN AND CHROMATIN REMODELING
Actin participates in gene expression as a component of chromatin-modifying complexes. Early findings by Crabtree and coworkers revealed that actin interacts with Brg1, the ATPase subunit of the BAF (Brg or Brm Associated Factors) SWI/SNF-like chromatin remodeling complex (Zhao et al. 1998). Since then, β-actin and a considerable number of actin-related proteins (ARPs) have been identified as components of different types of chromatin remodeling and histone acetyltransferase (HAT) complexes in a wide range of organisms from yeast to man (reviewed by Olave et al. 2002; Chen and Shen 2007; Farrants 2008). A central question that has not been fully answered concerns the mechanism(s) by which actin and ARPs contribute to chromatin remodeling.
Not all chromatin remodeling complexes contain actin or ARPs, which indicates that these proteins are not essential for chromatin remodeling per se. The fact that actin and ARPs often bind to the ATPase subunit of the chromatin remodeling complexes suggests that actin and ARPs work as allosteric regulators of chromatin remodeling (Blessing et al. 2004). In agreement with this possibility, Zhao et al. (1998) showed that actin is needed for the full ATPase activity of the BAF complex. Actin also mediates or facilitates the binding of BAF to chromatin (Zhao et al. 1998). The ability of Brg1 to bind actin filaments in vitro (Rando et al. 2002) and the association of actin with the RNA polymerase (see the following discussion) suggest that actin recruits BAF to transcribed genes by bridging the interaction between the chromatin remodeling complex and the transcription machinery. In other cases, the binding of ARPs to core histones might bridge the interaction (reviewed by Blessing et al. 2004).
Independently of its function in chromatin remodeling, actin participates in the regulation of a subset of genes that code for components of the actin cytoskeleton and that are activated by the serum response factor (SRF) (reviewed by Farrants 2008; Vartiainen 2008). This function of actin is mediated by MAL, a transcriptional coactivator of SRF that can sense variations in the cellular concentration of G-actin (Miralles et al. 2003). MAL is an actin-binding protein that shuttles continuously between the nucleus and the cytoplasm, and only the actin-bound form of MAL is efficiently exported from the nucleus. Actin regulates both the nucleocytoplasmic transport of MAL and the ability of the MAL-SRF complex to activate SRF target genes. Serum stimulation reduces the rate of MAL nuclear export. Under these conditions, MAL and SRF associate with the SRF target promoters, and the SRF target genes become activated upon disruption of the MAL-actin interaction (Vartiainen et al. 2007).
ACTIN IN ASSOCIATION WITH THE TRANSCRIPTION MACHINERY
Actin is associated with all three RNA polymerases, Pol I, II, and III (Fomproix and Percipalle 2004; Philimonenko et al. 2004; Hofmann et al. 2004; Hu et al. 2004; Kukalev et al. 2005). The actin–polymerase interaction has not been characterized at the structural level. Three Pol III subunits have been detected in association with actin in coimmunoprecipitation experiments (Hu et al. 2004). It has been proposed that two of them, RPABC2 and RPABC3, build the actin-binding site, because these subunits are common to all three polymerases and are located close to each other at the surface of the polymerase (Cramer et al. 2001; Hu et al. 2004). Coimmunoprecipitation experiments also showed that actin can interact with the largest Pol I subunit and with the carboxy-terminal domain (CTD) of the largest Pol II subunit (Fomproix and Percipalle 2004; Kukalev et al. 2005), which provides additional actin-binding sites specific for these polymerases.
The interaction of actin with the RNA polymerases is functionally relevant. Anti-actin antibodies inhibit transcription by Pol I and Pol II in vivo and in vitro (Hofmann et al. 2004; Philimonenko et al. 2004), and actin restores the full activity of partially purified, inactive Pol III preparations in vitro (Hu et al. 2004). These observations indicate that actin associates with RNA polymerases that are engaged in transcription.
The inhibitory effect of anti-actin antibodies on Pol I and II, as well as the ability of actin to activate Pol III, have been observed in vitro on naked DNA (Hofmann et al. 2004; Philimonenko et al. 2004; Hu et al. 2004), which suggests that actin is involved in basal transcription independently of its role in chromatin regulation. These observations have led to the proposal that actin is required for the activity of the basal transcription machinery. However, we are far from understanding how actin contributes to the transcription process and whether all three RNA polymerases use actin in the same manner.
A relevant question is whether actin is required for transcription initiation or elongation. Actin occupancy at promoters of Pol I and Pol II genes and copurification of actin with preinitiation complexes (PIC) suggest that actin is required for the assembly of transcription-competent polymerases (Hofmann et al. 2004; Philimonenko et al. 2004; Percipalle et al. 2006; Obrdlik et al. 2008; Louvet and Percipalle 2009). However, in vitro rDNA transcription assays that discriminate between transcription initiation (defined operationally by the synthesis of the initial trinucleotide) and elongation show that actin and nuclear myosin 1 (NM1) are needed for Pol I transcription elongation (Philimonenko et al. 2004; Percipalle et al. 2006).
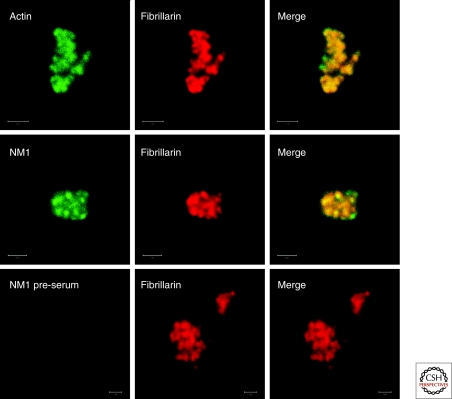
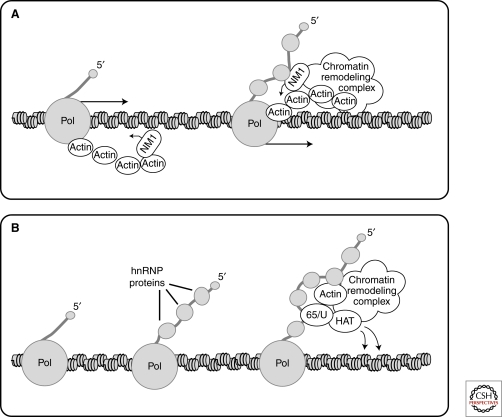
An important contribution toward understanding the role of actin in association with the transcription machinery was provided by the finding that NM1 is also intimately associated with Pol I and Pol II (Pestic-Dragovich et al. 2000; Fomproix and Percipalle 2004; Hofmann et al. 2006a). NM1 is a short-tailed monomeric myosin that acts as an actin-dependent ATPase. NM1 was found on active nucleolar transcription sites of both interphase and mitotic HeLa cells (Fig. 1) (Fomproix and Percipalle 2004), and microinjection of anti-NM1 antibodies into living cells inhibited rRNA synthesis (Philimonenko et al. 2004). Furthermore, depletion of either actin or NM1 from transcription extracts repressed the ability of the extracts to support transcription in vitro (Hofmann et al. 2004; Philimonenko et al. 2004). The facts that endogenous actin and NM1 associate with promoter and coding regions of rRNA genes, that actin and NM1 are associated with the transcription machinery, and that impairment of the myosin ATPase activity down-regulates Pol I transcription led to the proposal that an actomyosin activity is required for Pol I transcription (Fomproix and Percipalle 2004; Philimonenko et al. 2004; Percipalle et al. 2006). A recent study by Ye and coworkers (2008) provided further experimental support to this hypothesis. Actin mutants that are deficient in polymerization failed to associate with Pol I and did not support Pol I transcription. NM1 mutants that were defective in ATP-binding, actin-binding, or calmodulin-binding failed to associate with the rDNA and with Pol I and finally, the association of actin and NM1 with Pol I was regulated by ATP hydrolysis (Ye et al. 2008). These observations have enabled a model to be proposed in which the actin-NM1 complex acts as a molecular motor that helps the transcription machinery slide along the rDNA (Ye et al. 2008). Such a motor should interact with the transcription machinery itself and with the DNA, directly or indirectly (Fig. 2A). The actin-Pol I interaction is well-documented. The direct interaction between NM1 and DNA in vitro (Hofmann et al. 2006b) might explain how the actin-NM1 complex fastens to the DNA.
Figure 1.
Actin and NM1 localize to mammalian nucleoli. Homogeneous preparations of nucleoli from HeLa cells were analyzed by immunofluorescence labeling and confocal microscopy using rabbit polyclonal antibodies to actin (scale bar, 2 µm) and NM1 (scale bar, 2 µm) or corresponding preimmune sera (scale bar, 1 µm). All the specimens were coimmunostained with a mouse monoclonal antibody to fibrillarin as marker for nucleoli.
Figure 2.
Two possible ways by which actin contributes to transcription elongation. (A) An actomyosin motor helps the transcription machinery slide along the DNA. The architecture of the motor is not well understood but we envisage that the actin-NM1 complex makes contact with both the transcription machinery and the DNA/chromatin. Two possible configurations are depicted in the figure. (B) Actin participates in the recruitment of histone modifiers to protein-coding genes. Actin binds to hnRNP proteins and becomes incorporated into the nascent pre-mRNPs. Actin forms a complex with specific hnRNP adaptor proteins such as Hrp65 in Chironomus tentans and hnRNP U in mammals, and recruits HATs that acetylate histones and facilitate transcription elongation. The HATs might be components of larger chromatin remodeling complexes that might also establish direct contacts with actin.
ACTIN IN NASCENT TRANSCRIPTS
Actin not only interacts with the transcription machineries, it also associates with the nascent transcripts (Percipalle et al. 2001; Percipalle et al. 2002). Immunohistochemistry experiments performed in the dipteran C. tentans provided initial insights into the cotranscriptional binding of actin to nascent transcripts, because antibodies to actin labeled many loci in polytene chromosome preparations but failed to label after treatment of the chromosomes with RNase A (Percipalle et al. 2001). Immunoelectron microscopy experiments on ultrathin sections of C. tentans salivary glands showed that actin was preferentially associated with the distal region of the active transcription unit, away from the promoter (Percipalle et al. 2001). Later on, anti-actin antibodies coprecipitated coding regions of Pol I and Pol II genes in chromatin immunoprecipitation experiments (Hofmann et al. 2004; Philimonenko et al. 2004; Obrdlik et al. 2008), emphasizing the presence of actin along active genes. The connection between actin and RNA was further shown at the molecular level by chromatin RNA immunoprecipitation assays, in which it was possible to analyze the cotranscriptional association of protein factors with nascent RNA (Obrdlik and Percipalle 2009; Obrdlik et al. 2008).
Specific heterogeneous nuclear ribonucleoproteins (hnRNPs) bind actin and mediate its association with RNA. Actin-associated hnRNP proteins were discovered mainly by resolving nuclear extracts or RNP preparations with DNase I affinity chromatography. In both C. tentans and mammals, a significant fraction of actin-associated hnRNPs belong to the A/B type family. The hnRNP A/B proteins contain two conserved RNA recognition motifs (RRMs) flanked at the amino terminus by an acidic domain and at the carboxyl terminus by a divergent module for protein–protein interactions, termed the “auxiliary domain” (Krecic and Swanson 1999; Dreyfuss et al. 2002). In C. tentans, actin is associated with the shuttling mRNA-binding protein hrp36 (Visa et al. 1996), whereas in mammals, in which the repertoire of A/B-type hnRNPs is somewhat larger (Krecic and Swanson 1999; Dreyfuss et al. 2002), actin associates with several proteins, including hnRNP A2, hnRNP A3, and the transport mediator CArG-box binding factor (CBF-A) (Percipalle et al. 2002; Raju et al. 2008).
Another actin-associated hnRNP protein of C. tentans, hrp65, shares strong homology with the human proteins PSF (Patton et al. 1993), p54nrb/NonO (Dong et al. 1993), PSP1 (Fox et al. 2002), and with the Drosophila NonA/BJ6 (Jones and Rubin 1990; von Besser et al. 1990). These proteins do not belong to the A/B-type and they are characterized by a central domain of about 320 amino acids termed “DBHS” (Drosophila behavior and human splicing). This domain is evolutionarily conserved. The DBHS domain comprises two RRMs and an additional carboxy-terminal stretch of approximately 100 amino acids that mediates dimerization or oligomerization among DBHS proteins (Kiesler et al. 2003). The association of DBHS proteins with actin is conserved in mammals; the human PSF and NonO proteins have been identified as components of a large nuclear protein complex together with actin, Pol II, and N-WASP, a key regulator of cytoplasmic microfilaments that is also present in the cell nucleus (Wu et al. 2006).
Actin associates with hnRNP U/SAF-A (Scaffold attachment factor A), a large 120-kDa tripartite protein with ATP-binding activity in mammals. The amino terminus of this protein displays DNA-binding activity, the carboxyl terminus contains two RRMs for RNA binding, and the central domain displays Pol II-binding activity (Kukalev et al. 2005).
Some of the actin-associated hnRNP proteins identified so far interact directly with actin, whereas others are part of actin-containing complexes and are not in direct contact with actin. The C. tentans hrp65-2 (hrp65 isoform 2) and the human hnRNP U proteins interact with actin through a conserved motif (Percipalle et al. 2003; Kukalev et al. 2005). Remarkably, inhibition of the actin-hrp65-2 or actin-hnRNP U interactions in living cells inhibits transcription elongation by Pol II as monitored by incorporation of labeled nucleotide precursors in run-on assays (Percipalle et al. 2003; Kukalev et al. 2005). Based on these results, we proposed a regulatory mechanism in which actin facilitates the cross talk between chromatin, elongating polymerase, and nascent RNA (Louvet and Percipalle 2009).
How do actin-hnRNP complexes facilitate transcription elongation? Experiments in insects and mammals suggest that specific actin-hnRNP complexes facilitate the elongation phase by recruiting transcriptional coactivators to remodel the nucleosome barrier imposed by chromatin (Percipalle and Visa 2006; Miralles and Visa 2006; Louvet and Percipalle 2009). Actin-hrp65-2 and actin-hnRNP U complexes mediate the recruitment of the HATs p2D10 and PCAF, respectively, to transcribed genes (Fig. 2B) (Sjölinder et al. 2005; Obrdlik et al. 2008). The disruption of these actin-hnRNP interactions in vivo reduced the level of histone acetylation in chromatin, blocked Pol II transcription, and affected HAT occupancy along active genes (Sjölinder et al. 2005; Kukalev et al. 2005; Obrdlik et al. 2008). These results indicate that the integrity of specific actin-hnRNP interactions is required for coactivator recruitment to active genes. The topology of the actin complex is not known. However, an antibody specific for the actin-binding site on hnRNP U coprecipitates endogenous hnRNP U and PCAF but fails to precipitate actin in mammals (Obrdlik et al. 2008). This leads us to favor the idea that hnRNP U is able to interact simultaneously with both PCAF and actin. There is also recent evidence that the HAT PCAF and BRG1, the ATPase subunit of the SWI/SNF chromatin remodeling complex, are sequentially recruited to the promoter of the myogenin gene and are both involved in gene activation (Li et al. 2007). Actin is also a component of the SWI/SNF complex (reviewed in Farrants 2008). We speculate, therefore, that during elongation of nascent pre-mRNA, the actin-RNP complexes work as modules that facilitate recruitment of HATs and chromatin remodeling complexes to active genes (see Fig. 2B).
Studies by Obrdlik and coworkers (2008) revealed that the interplay between actin and the nascent RNP also involves interaction with the RNA polymerase in some cases. During elongation, the carboxy-terminal domain (CTD) of the largest subunit of Pol II is subjected to a series of covalent modifications that modulate recruitment of transcriptional coactivators. The amino acid residues Ser2 and Ser5 within the conserved CTD heptapeptide repeats become combinatorially phosphorylated and the different phosphorylation states represent hallmarks for individual transcription steps (Meinhart et al. 2005). Therefore, if specific actin-hnRNP complexes are required for efficient elongation, it is likely that they are cotranscriptionally associated with specific CTD phosphorylation states. Recent evidence shows that this is the case. Actin together with hnRNP U associates only with Ser2-phosphorylated heptapeptide repeats or with heptapeptide repeats that are phosphorylated on both Ser5 and Ser2, and fails to interact with unphosphorylated heptapeptide repeats or with those repeats that are phosphorylated on Ser5 only (Obrdlik et al. 2008). These findings support the idea that specific actin-hnRNP complexes physically associate with the elongating Pol II, a view that is corroborated by evidence that PSF and p54nrb, mammalian homologs of the C. tentans hrp65 protein, also interact with actin and with the CTD (Emili et al. 2002; Wu et al. 2006).
As discussed earlier, actin and NM1 are required for transcription elongation by Pol I, and NM1 also appears to mediate the association of specific chromatin-modifying components to the rDNA genes. NM1 is a core component of the B-WICH chromatin remodeling complex together with the Williams syndrome transcription factor (WSTF) and the ATPase SNF2 h (Percipalle et al. 2006), and this complex turned out to also contain precursor rRNA (Cavellan et al. 2006). Remarkably, inhibition of endogenous NM1 or WSTF in abortive initiation assays did not affect the formation of initial pre-rRNA transcripts but affected the synthesis of the entire transcript when chromatin templates were used in run-on transcription assays (Philimonenko et al. 2004; Percipalle et al. 2006). These results suggest that the core components of the B-WICH complex, including NM1, are required after transcription initiation, and a model has been proposed in which the dynamic interaction between actin and NM1 is required for local recruitment of the B-WICH complex to active genes (Percipalle and Östlund Farrants 2006; Louvet and Percipalle 2009).
The previous considerations suggest that actin modulates the elongation of nascent pre-mRNAs and pre-rRNAs by controlling the cotranscriptional association of chromatin-modifying components with the active genes. This view is of particular interest in the case of Pol I transcription, where active rRNA genes are believed to be devoid of nucleosomes. For protein-coding genes, it is intriguing that actin together with hnRNP U and the HAT PCAF associate with the CTD in a phosphorylated Ser2-dependent manner (Obrdlik et al. 2008), an observation that strengthens the hypothesis that the coupling of actin to the elongating Pol II facilitates the modifications of chromatin structure.
ACTIN BEYOND TRANSCRIPTION
The studies performed in C. tentans showed that actin is cotranscriptionally added to nascent pre-mRNA, remains incorporated in mature RNPs released into the nucleoplasmic milieu, and accompanies the mRNA all the way to polysomes (Percipalle et al. 2001). There is no evidence of nuclear actin-based motors promoting directional movement of RNP particles or even preribosomal subunits (Singh et al. 1999; Politz et al. 2003; Siebrasse et al. 2008). However, actin is associated with mature mRNPs and with pre-60S subunits, it decorates pore-linked filaments that project inward into the nucleoplasm from the nuclear pore basket, and it has been implicated in controlling the export of viral mRNA transcripts (Percipalle et al. 2001, 2002; Hofmann et al. 2001; Kiseleva et al. 2004; Oeffinger et al. 2007). Also, NM1 is loaded into preribosomes and involved in the export-competent pre-60S subunits (Obrdlik et al. 2009). In addition, NM1 accompanies preribosomal subunits in transit to the nuclear envelope (Cisterna et al. 2007; Obrdlik et al. 2009), is present at the nuclear envelope (Holaska et al. 2007), and has been recently incorporated in the export of small ribosomal subunits (Cisterna et al. 2009). Finally, there is recent evidence that a complex containing rRNA and NM1 is associated with the RNA-binding protein Nup153 at the nuclear pore (Obrdlik et al. 2009), which suggests that NM1 accompanies the rRNA from the gene to the nuclear pore complex. These observations suggest that the synergy between actin and NM1 plays a more general role, and is not limited to the gene. We favor the idea that actin and NM1 contribute to the maturation and assembly of export-competent mRNPs and rRNPs by promoting the remodeling of RNP structures through dynamic protein–protein interactions (reviewed in Percipalle et al. 2009).
REGULATION OF ACTIN POLYMERIZATION
The presence of actin in the cell nucleus raises the possibility that there are nuclear-based mechanisms regulating actin polymerization (Pederson 2008; Louvet and Percipalle 2009). It has been suggested that nuclear actin coexists in monomeric (G) or short oligomeric forms and in a polymeric form (Louvet and Percipalle 2009; Gieni & Hendzel 2009). It is unclear how transitions between these forms occur, but dynamic alterations in actin polymerization may be instrumental in modulating the function of actin in RNA biogenesis.
Canonical actin filaments, which are commonly seen in the cytoplasm by immunofluorescence methods, have not been convincingly visualized in the cell nucleus. However, many of the factors that are known to control cytoplasmic actin polymerization and stabilize filamentous F-actin structures have been detected in the cell nucleus. N-WASP and the ARP2/3 complexes cooperate in the nucleation and branching of new actin filaments. These proteins are present in the cell nucleus, where they are implicated in the elongation of nascent mRNA transcripts (Wu et al. 2006; Yoo et al. 2007). More recently, other proteins that facilitate cytoplasmic actin nucleation independently of N-WASP and ARP2/3 have been discovered in the nucleus, including the formin-like mDia proteins and JMY (Miki et al. 2008; Zuchero et al. 2009). These findings indeed suggest the presence of polymeric actin in the nucleus, and it is likely that there are several independent mechanisms that regulate nuclear actin polymerization, as is the case in the cytoplasm. It is possible that the need to have several mechanisms for actin polymerization reflects the multiple roles that actin plays in the cell nucleus. This may or may not be the case, but it is clear that nuclear actin assembled into higher order structures is rather dynamic with a high turnover (McDonald et al. 2006), which indicates that a pool of monomeric actin is also present in the nucleus. This pool is probably necessary to feed dynamic polymeric actin structures.
The existence of monomeric actin was indirectly suggested by the discovery of G-actin binding proteins such as cofilin, profilin, β-thymosin, and a gelsolin-like protein in the cell nucleus (Prendergast et al. 1991; Pendleton et al. 2003; Skare et al. 2003; Huff et al. 2004). Actin can be copurified with hnRNP U and the Pol II machinery using DNase I affinity chromatography (Kukalev et al. 2005; Obrdlik et al. 2008), which has higher affinity for monomeric than for filamentous actin (Zechel 1980). In addition, monoclonal antibodies designed to recognize epitopes exclusively available in monomeric actin or actin dimers revealed significant immunostaining of the cell nucleus (reviewed in Jokusch et al. 2006). There is also evidence that monomeric or short oligomeric actin is directly involved in nuclear functions such as the role of monomeric actin in the regulation of SRF-dependent transcription in complex with the coactivator MAL (Vartiainen et al. 2007).
The G-actin binding protein profilin affects transcription of rRNA genes, and drugs that inhibit actin polymerization, such as cytochalasin D and latrunculin B, inhibit the transcription of rDNA genes in vivo and in vitro, whereas drugs that favor actin assembly do not influence rDNA transcription (Ye et al. 2008). Furthermore, a recent study indicated that actin polymerization takes place in retinoic acid (RA)-induced HoxB transcription (Ferrai et al. 2009). These observations suggest that polymeric actin is functional in the cell nucleus. Because N-WASP and ARP2/3 are important during pre-mRNA elongation, and an actin-NM1 complex is clearly implicated in transcription elongation by Pol I, we favour the idea that both monomeric and polymeric actin are present along active genes, and that cotranscriptional transitions in the actin polymerization states result in the existence of different actin forms associated with different parts of the gene.
What is the polymerization state of actin in RNPs? Ultrastructural analysis has not revealed canonical filamentous actin structures attached to mature RNPs (reviewed in Percipalle 2009), which hints at an intrinsic mechanism that keeps actin from polymerizing as soon as it is assembled into RNPs. An equally intriguing possibility is that actin undergoes covalent modifications that do not favor the establishment of polymers. Some of these ideas are speculative, but they emphasize the importance of regulating actin polymerization for nuclear function.
LONG-RANGE MOVEMENT
Most of our current knowledge of nuclear actin originates from research in the field of chromatin regulation and transcription. However, recent studies point to the possibility that actin plays a role also in the large-scale organization of the genome. The eukaryotic nucleus is highly compartmentalized and, in spite of the lack of membrane-bound organelles, most nuclear components show specific distributions restricted to certain nuclear domains (reviewed by Lamond and Spector 2003; Schneider and Grosschedl 2007). The functional compartmentalization of the nucleus is reflected in the fact that different types of sequences occupy different positions in the nucleus. For example, gene-rich chromosome regions tend to occupy internal positions, whereas gene-poor sequences are associated with the nuclear periphery, and repressed genes are often associated with heterochromatic regions, whereas active genes are usually located near nuclear speckles (reviewed by Spector 2003). What is most interesting is that the position of a gene locus can change in response to transcriptional activation (Parada and Misteli 2002) and that the transcriptional activity of a gene is affected by its position in the nucleus (Finlan et al. 2008). Chuang and coworkers (2006) visualized the dynamics of gene repositioning by constructing cell lines that carry tandem gene arrays under the control of an inducible promoter. When the expression of the array was activated, the entire locus relocated from the cell periphery to the nuclear interior. Unidirectional trajectories were recorded over distances of 1–5 µm, which suggests that an active, directed mechanism for long-range chromatin movement is present. Interestingly, the expression of mutant actin or NM1 with defects in polymerization and actin-binding inhibited the relocation of the array on transcriptional activation (Chuang et al. 2006). Dundr and coworkers (2007) used a similar experimental design based on the use of synthetic inducible arrays to study the association of Cajal bodies (CBs) with U2 snRNA genes in human cells. After transcription had been induced, the U2 snRNA gene array moved over long distances inside the nucleus (2–3 µm) and became stably associated with CBs. Also in this case, the expression of a nonpolymerizable actin mutant inhibited the movement of the U2snRNA locus (Dundr et al. 2007). In a third study, Hu and coworkers identified long-range, estrogen-induced interactions among genes located in different chromosomes. The interactions were lost after treatment of the cells with drugs that either blocked actin polymerization or inhibited actin depolymerization. Depletion of NM1 by RNAi or nuclear injection of antibodies against NMI also blocked the estrogen-induced interactions (Hu et al. 2008). Moreover, the inhibitory effect of the anti-NM1 antibodies could be reversed by the expression of wild-type NM1, but not by the expression of NM1 mutants that were defective in actin binding or lacked ATPase activity (Hu et al. 2008).
The results reveal that the dynamics of actin in the cell nucleus affect gene positioning and large-scale chromatin organization, in a direct or indirect manner. The molecular mechanisms underlying these long-range chromatin movements are still unclear. The type of movement observed could be explained by active transport mechanisms based on actomyosin motors. This would imply the existence of mechanisms for the regulation of actin dynamics at micrometer scale inside the nucleus. If this is the case, further work must characterize such an actin large-scale regulatory network and elucidate the mechanisms that impose directionality inside the nucleus. Alternatively, and considering the fact that the observed relocations take place after transcription activation, the actin dependence could be at the transcriptional level and the chromatin movements could be a consequence of chromatin decondensation. This model, however, does not provide a satisfactory explanation of the fact that the U2 snRNA genes are targeted to relatively stable positions in the nucleus, nor does it provide any basis for the directionality of the movements. Further work is needed to understand the molecular mechanisms of large-scale chromatin relocation and to answer the questions raised by the discovery of gene relocation events. We anticipate that the answers to these questions will provide a fundamental advance in our understanding of the dynamics of the eukaryotic genome.
CONCLUDING REMARKS
There is an increasing body of evidence that supports the idea that actin plays a role in controlling multiple phases of gene transcription as a component of chromatin-remodeling complexes and RNP particles, and that it is closely associated with all RNA polymerases. Two possible models by which actin may regulate transcription elongation are illustrated in Figure 2. One model proposes the existence of an actin-NM1 motor, whereas the other one emphasizes the role of actin-RNP complexes in recruiting chromatin modifiers to transcribed genes. Both models are based on solid experimental data and are not mutually exclusive. We do not know whether both mechanisms act in all types of genes. Support for the existence of an actin-NM1 motor comes mostly from studies of Pol I transcription, whereas the recruitment of HATs mediated by RNPs has been mostly documented for protein-coding genes. It is possible that the way in which actin acts in transcription is different for different types of genes.
After many years of debate about the existence of actin in the nucleus, it is now clear that actin is an abundant nuclear protein and many independent studies support the idea that it is involved in gene expression. An interesting question is whether the nuclear functions of actin are linked to—or regulated by—the dynamics of actin in the cytoplasm. The actin cytoskeleton is highly dynamic and reacts to a variety of extracellular signals (for recent reviews, see Moustakas and Heldin 2008; Papakonstanti and Stournaras 2008), and there is evidence that the cytoplasmic and actin pools are not independent of each other (Vartiainen et al. 2007). It is therefore tempting to suggest that actin acts as a sensor of extracellular signals with the ability to transduce the signals to the genome and modulate gene expression. The question remains whether the overall activity of actin in gene expression is also sensitive to extracellular stimuli.
ACKNOWLEDGMENTS
We thank Ann-Kristin Östlund Farrants for critical reading of the manuscript and George Farrants for language editing. Our work is supported by grants from the Swedish Research Council (Vetenskapsrådet), the Swedish Cancer Society (Cancerfonden), and the European Science Foundation (Eurocores Programme RNAQuality).
Footnotes
Editors: David Spector and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Blessing CA, Ugrinova GT, Goodson HV 2004. Actin and ARPs: Action in the nucleus. Trends Cell Biol 14:435–442 [DOI] [PubMed] [Google Scholar]
- Cavellan E, Asp P, Percipalle P, Östlund Farrants AK 2006. The chromatin remodelling complex WSTF-SNF2 h interacts with several nuclear proteins in transcription. J Biol Chem 281:16264–16271 [DOI] [PubMed] [Google Scholar]
- Chen M, Shen X 2007. Nuclear actin and actin-related proteins in chromatin dynamics. Curr Opin Cell Biol 19:326–330 [DOI] [PubMed] [Google Scholar]
- Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS 2006. Long-range directional movement of an interphase chromosome site. Curr Biol 16:825–831 [DOI] [PubMed] [Google Scholar]
- Cisterna B, Malatesta M, Dieker J, Muller S, Prosperi E, Biggiogera M 2009. An active mechanism flanks and modulates the export of the small ribosomal subunits. Histochem Cell Biol 131:743–753 [DOI] [PubMed] [Google Scholar]
- Cisterna B, Necchi D, Prosperi E, Biggiogera M 2006. Small ribosomal subunits associate with nuclear myosin and actin in transit to the nuclear pores. FASEB J 20:1901–1903 [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863–1876 [DOI] [PubMed] [Google Scholar]
- Dong B, Horowitz DS, Kobayashi R, Krainer AR 1993. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucl Acids Res 21:4085–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N 2002. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3:195–205 [DOI] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung M-H, John S, Upender M, Reid T, Hager GL, Matera AG 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo, J. Cell Biol 179:1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egly JM, Miyamoto NG, Moncollin V, Chambon P 1984. Is actin a transcription initiation factor for RNA polymerase B? EMBO J 3:2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, Blencowe BJ, Ingles CJ 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrants AK 2008. Chromatin remodelling and actin organisation. FEBS Lett 582:2041–2050 [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4:e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomproix N, Percipalle P 2004. An actin-myosin complex on actively transcribing genes. Exp Cell Res 294:140–148 [DOI] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI 2002. Paraspeckles: A novel nuclear domain. Curr Biol 12:13–25 [DOI] [PubMed] [Google Scholar]
- Ferrai C, Naum-Onganía G, Longobardi E, Palazzolo M, Disanza A, Diaz VM, Crippa MP, Scita G, Blasi F 2009. Induction of HoxB transcription by retinoic acid requires actin polymerization. Mol Biol Cell May 28. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieni RS, Hendzel MJ 2009. Actin dynamics and functions in the interphase nucleus: Moving toward an understanding of nuclear polymeric actin. Biochem Cell Biol 87:283–306 [DOI] [PubMed] [Google Scholar]
- Harata M, Oma Y, Mizuno S, Jiang YW, Stillman DJ, Wintersberger U 1999. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol Biol Cell 10:2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Johnson T, Klapczynski M, Fan J-L, de Lanerolle P 2006b. From transcription to transport: Emerging roles for nuclear myosin I. Biochem Cell Biol 84:418–426 [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Reichart B, Ewald A, Muller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, et al. 2001. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol 152:895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol 6:1094–1101 [DOI] [PubMed] [Google Scholar]
- Hofmann WA, Vargas GM, Ramchandran R, Stojiljkovic L, Goodrich JA, de Lanerolle P 2006a. Nuclear myosin I is necessary for the formation of the first phosphodiester bond during transcription initiation by RNA polymerase II. J Cell Biochem 99:1001–1009 [DOI] [PubMed] [Google Scholar]
- Holaska JM, Wilson KL 2007. An emerin “proteome”: Purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry 46:8897–8908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Hernandez N 2004. A role for β-actin in RNA polymerase III transcription. Genes Dev 18:3010–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. 2008. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci 105:19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff T, Rosorius O, Otto AM, Muller CS, Ballweber E, Hannappel E, Mannherz HG 2004. Nuclear localisation of the G-actin sequestering peptide thymosin β4. J Cell Sci 117:5333–5341 [DOI] [PubMed] [Google Scholar]
- Jockusch BM, Schoenenberger CA, Stetefeld J, Aebi U 2006. Tracking down the different forms of nuclear actin. TRENDS Cell Biol 16:391–396 [DOI] [PubMed] [Google Scholar]
- Jones KR, Rubin GM 1990. Molecular analysis of no-on-transient A, a gene required for normal vision in Drosophila. Neuron 4:711–723 [DOI] [PubMed] [Google Scholar]
- Kiesler E, Miralles F, Ostlund Farrants AK, Visa N 2003. The Hrp65 self-interaction is mediated by an evolutionarily conserved domain and is required for nuclear import of Hrp65 isoforms that lack a nuclear localization signal. J Cell Sci 116:3949–3956 [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Drummond SP, Goldberg MW, Rutherford SA, Allen TD, Wilson KL 2004. Actin- and protein-4.1-containing filaments link nuclear pore complexes to subnuclear organelles in Xenopus oocyte nuclei. J Cell Sci 117:2481–2490 [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS 1999. hnRNP complexes: Composition, structure, and function. Curr Opin Cell Biol 11:363–371 [DOI] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P 2005. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol 12:238–244 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL 2003. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol 4:605–612 [DOI] [PubMed] [Google Scholar]
- Li ZY, Yang J, Gao X, Lu JY, Zhang Y, Wang K, Cheng MB, Wu NH, Zhang Y, Wu Z, et al. 2007. Sequential recruitment of PCAF and BRG1 contributes to myogenin activation in 12-O-tetradecanoylphorbol-13-acetate-induced early differentiation of rhabdomyosarcoma-derived cells. J Biol Chem 282:18872–18878 [DOI] [PubMed] [Google Scholar]
- Louvet E, Percipalle P 2009. Actin and myosin in gene transcription. Int Rev Cell Mol Biol 272:107–147 [DOI] [PubMed] [Google Scholar]
- McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ 2006. Nucleoplasmic β-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 172:541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P 2005. A structural perspective of CTD function. Genes Dev 19:1401–1415 [DOI] [PubMed] [Google Scholar]
- Miki T, Okawa K, Sekimoto T, Yoneda Y, Watanabe S, Ishizaki T, Narumiya S 2008. mDia2 shuttles between the nucleus and the cytoplasm through the importin-{α}/{β}- and CRM1-mediated nuclear transport mechanism. J Biol Chem 284:5753–5762 [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329–342 [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH 2008. Dynamic control of TGF-β signaling and its links to the cytoskeleton. FEBS Lett 582:2051–2065 [DOI] [PubMed] [Google Scholar]
- Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P 2008. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol 28:6342–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik A, Louvet E, Kukalev A, Naschekin D, Kiseleva E, Fahrenkrog B, Percipalle P 2009. Nuclear myosin 1 is in complex with mature rRNA transcripts and associates with the nuclear pore complex. FASEB J, 2009 Sept 3 [Epub ahead of print] PMID: 19729515 [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4:951–956 [DOI] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR 2002. Nuclear actin and actin-related proteins in chromatin remodelling. Annu Rev Biochem 71:755–781 [DOI] [PubMed] [Google Scholar]
- Papakonstanti EA, Stournaras C 2008. Cell responses regulated by early reorganization of actin cytoskeleton. FEBS Lett 582:2120–2127 [DOI] [PubMed] [Google Scholar]
- Parada L, Misteli T 2002. Chromosome positioning in the interphase nucleus. Trends Cell Biol 12:425–432 [DOI] [PubMed] [Google Scholar]
- Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev 7:393–406 [DOI] [PubMed] [Google Scholar]
- Pederson T 2008. As functional nuclear actin comes into view, is it globular, filamentous, or both? J Cell Biol 180:1061–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Aebi U 2002. Actin in the nucleus: what form and what for? J Struct Biol 140:3–9 [DOI] [PubMed] [Google Scholar]
- Pendleton A, Pope B, Weeds A, Koffer A 2003. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem 278:14394–14400 [DOI] [PubMed] [Google Scholar]
- Percipalle P 2009. The long journey of actin and actin-associated proteins from gene to polysomes. Cell Mol Life Sci 66:2151–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Obrdlik A 2009. Analysis of nascent RNA transcripts by chromatin RNA immunoprecipitation. Meth Mol Biol 567:215–235 [DOI] [PubMed] [Google Scholar]
- Percipalle P, Visa N 2006. Molecular functions of nuclear actin in transcription. J Cell Biol 172:967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Östlund Farrants AK 2006. Chromatin remodelling and transcription: Be-WICHed by nuclear myosin 1. Curr Opin Cell Biol 18:267–274 [DOI] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Cavellán E, Voit R, Reimer G, Krüger T, Thyberg J, Scheer U, Grummt I, Farrants AK 2006. The chromatin remodelling complex WSTF-SNFh interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep 7:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, Visa N 2003. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci 100:6475–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Jonsson A, Nashchekin D, Karlsson C, Bergman T, Guialis A, Daneholt B 2002. Nuclear actin is associated with a specific subset of hnRNP A/B-type proteins. Nucleic Acids Res 30:1725–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Zhao J, Pope B, Weeds A, Lindberg U, Daneholt B 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J Cell Biol 153:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, Shabanowitz J, Hunt DF, Hozak P, de Lanerolle P 2000. A myosin I isoform in the nucleus. Science 290:337–341 [DOI] [PubMed] [Google Scholar]
- Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, et al. 2004. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 6:1165–1171 [DOI] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Pederson T 2003. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell 14:4805–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast GC, Ziff EB 1991. Mbh 1: a novel gelsolin/severin-related protein which binds actin in vitro and exhibits nuclear localization in vivo. EMBO J 10:757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju CS, Göritz C, Nord Y, Hermanson O, Lopez-Iglesias C, Visa N, Castelo-Branco G, Percipalle P 2008. In cultured oligodendrocytes the A/B-type hnRNP CBF-A accompanies MBP mRNA bound to mRNA trafficking sequences. Mol Biol Cell 19:3008–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Zhao K, Janmey P, Crabtree GR 2002. Phospatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodelling complex. Proc Natl Acad Sci 99:2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hinssen H, Franke WW, Jockusch BM 1984. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell 39:111–122 [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R 2007. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21:3027–3043 [DOI] [PubMed] [Google Scholar]
- Siebrasse JP, Veith R, Dobay A, Leonhardt H, Daneholt B, Kubitscheck U 2008. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc Natl Acad Sci 105:20291–20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh OP, Björkroth B, Masich S, Wieslander L, Daneholt B 1999. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp Cell Res 251:135–146 [DOI] [PubMed] [Google Scholar]
- Sjölinder M, Björk P, Soderberg E, Sabri N, Östlund Farrants AK, Visa N 2005. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev 19:1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare P, Kreivi JP, Bergström A, Karlsson R 2003. Profilin I colocalizes with speckles and Cajal bodies: A possible role in pre-mRNA splicing. Exp Cell Res 286:12–21 [DOI] [PubMed] [Google Scholar]
- Spector DL 2003. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72:573–608 [DOI] [PubMed] [Google Scholar]
- Vartiainen MK 2008. Nuclear actin dynamics–from form to function. FEBS Lett 582:2033–2040 [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316:1749–1752 [DOI] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Björkroth B, Wurtz T, Daneholt B 1996. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell 84:253–264 [DOI] [PubMed] [Google Scholar]
- von Besser H, Schnabel P, Wieland C, Fritz E, Stanewsky R, Saumweber H 1990. The puff-specific Drosophila protein Bj6, encoded by the gene no-on transient A, shows homology to RNA-binding proteins. Chromosoma 100:37–47 [DOI] [PubMed] [Google Scholar]
- Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL 2006. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol 8:756–763 [DOI] [PubMed] [Google Scholar]
- Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I 2008. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev 22:322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y, Wu X, Guan JL 2007. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem 282:7616–7623 [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95:625–636 [DOI] [PubMed] [Google Scholar]
- Zechel K 1980. Isolation of polymerization competent cytoplasmic actin by affinity chromatography on immobilized DNase I using formamide as eluant. Eur J Biochem 110:343–348 [DOI] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD 2009. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol 11:451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]