Abstract
The discovery of the DNA double helix structure half a century ago immediately suggested a mechanism for its duplication by semi-conservative copying of the nucleotide sequence into two DNA daughter strands. Shortly after, a second fundamental step toward the elucidation of the mechanism of DNA replication was taken with the isolation of the first enzyme able to polymerize DNA from a template. In the subsequent years, the basic mechanism of DNA replication and its enzymatic machinery components were elucidated, mostly through genetic approaches and in vitro biochemistry. Most recently, the spatial and temporal organization of the DNA replication process in vivo within the context of chromatin and inside the intact cell are finally beginning to be elucidated. On the one hand, recent advances in genome-wide high throughput techniques are providing a new wave of information on the progression of genome replication at high spatial resolution. On the other hand, novel super-resolution microscopy techniques are just starting to give us the first glimpses of how DNA replication is organized within the context of single intact cells with high spatial resolution. The integration of these data with time lapse microscopy analysis will give us the ability to film and dissect the replication of the genome in situ and in real time.
Integration of genomic studies and super-resolution microscopy indicates that, in vivo, DNA replication is self-propagating and increases in efficiency through S phase.
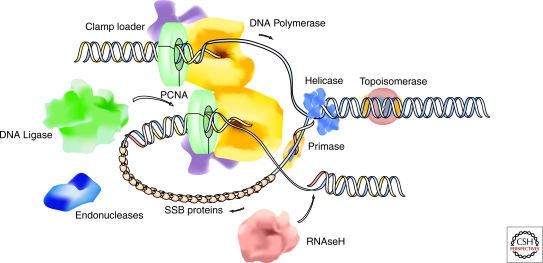
Following the elucidation of the DNA double helix structure by Watson and Crick (Watson and Crick 1953) and the isolation of the first DNA polymerase by Kornberg (Kornberg 1960), a fundamental biological question has been how genomes are duplicated prior to cell division. In the 50 years since these seminal discoveries, the basic mechanisms of DNA replication have been established by a powerful combination of genetics and in vitro biochemistry. From this body of work, it is clear that the fundamental features and components of DNA replication are well conserved throughout evolution from bacteria to mammals. A comprehensive list of proteins that are involved in the process and their activities has been compiled, and it is now possible to provide a relatively detailed description of the DNA replication machinery on a molecular level (Fig. 1) (Perumal et al. 2009). However, genomic DNA in eukaryotic cells is hierarchically packed within the nucleus and genome duplication requires the concerted effort of many thousands of individual replication units. As such, an equally important question is how DNA replication is coordinated in space and time across the entire genome within the living cell.
Figure 1.
Schematic outline of the DNA replication fork and molecular components and enzymatic activities of the replisome. PCNA, proliferating cell nuclear antigen (also termed DNA polymerase clamp or processivity factor). SSB, single strand DNA binding proteins.
DNA REPLICATION: THE BASICS
Every cycling cell needs to duplicate its genome before it divides. This entails not only duplicating precisely and completely its genetic information, which is the focus of this article, but also restoring its epigenetic information to build up the differentiated chromatin structures.
After cells divide, the so-called prereplication complexes including the DNA helicase complex (MCM 2-7) assemble onto the DNA throughout the G1 phase (Blow and Dutta 2005). At the end of G1, input from the cell cycle machinery via the cell-cycle dependent kinases triggers initiation of DNA replication at discrete sites, known as origins of replication, scattered throughout the genome. Each origin fires once per cell cycle and their spacing must ensure that the entire genome is replicated during the S phase. Early results from cell fusion experiments indicated that replicated DNA differed from unreplicated DNA because it was not permissive for replication unless it passed through one mitotic division (Rao and Johnson 1970). Subsequent experiments using cell-free replication assays further refined this idea and a model was proposed whereby replication origins were “licensed” to replicate at late mitosis and G1, and the “license” was removed as the DNA was replicated in S phase (Blow and Laskey 1988). Detailed biochemical evidence is now available and indicates that preventing rebinding of the MCM complex to DNA is the key to avoid rereplication (Blow and Dutta 2005).
In bacteria and lower eukaryotes, replication origins are defined by specific DNA sequences; whereas in metazoans the defining characteristics of these sites remain less clear (Robinson and Bell 2005; Aladjem 2007; Hamlin et al. 2008). At each origin, replication proteins are assembled, which then duplicate one segment (replicon) of the genome in a processive manner (Jacob and Brenner 1963; Jacob 1993). The active units of DNA replication, consisting of the replication machinery (also called replisome), the existing DNA template, and the nascent DNA strand, are referred to as replication forks. Following initiation of DNA synthesis, replication forks proceed bidirectionally from the origin, unwinding the genomic DNA as they traverse the chromosome (Huberman and Riggs 1968).
At the molecular level, replication of a genomic DNA template involves at least two different DNA polymerases, a DNA primase, a DNA helicase, a single strand DNA binding (SSB) protein complex (replication protein A, RPA), DNA topoisomerases, a clamp loading complex (replication factor C, RFC), and a DNA polymerase clamp or processivity factor (proliferating cell nuclear antigen, PCNA). Furthermore, the inherent polarity of the DNA synthesis reaction (5′ to 3′ direction) necessitates discontinuous duplication of the lagging strand in addition to the continuous leading-strand synthesis. This process involves the synthesis of short (180–200 bp in eukaryotic cells) DNA fragments known as Okazaki fragments (Okazaki et al. 1968) that are subsequently processed and ligated together by a number of additional enzymes, including the flap endonucleases (FEN-1) and DNA ligase I (Hubscher and Seo 2001). To explain how the synthesis of both strands is coordinated, an asymmetric dimer of DNA polymerases and associated factors has been proposed first for prokaryotic DNA replication (McHenry 1988) and subsequently extended for eukaryotes (Tsurimoto and Stillman 1989). In order that the dimeric polymerases can extend the two antiparallel strands at the same rate and in the same direction, looping back of the lagging strand into the replisome (“trombone” model) has been postulated, with recycling of the lagging-strand polymerase from the end of one Okazaki fragment to the next RNA primer forming a priming loop (Sinha et al. 1980; Pandey et al. 2009). This situation suggests that the replication machinery at a given replication fork likely consists of at least two functional (sub)modules, one responsible for leading-strand synthesis, and the other for lagging-strand synthesis (Fig. 1). Biochemical evidence points to the existence of large preformed multiprotein replication complexes that contain all activities (Noguchi et al. 1983; Tom et al. 1996). However, evidence from live-cell microscopy analysis points to short time interactions between the individual components, suggesting highly dynamic complexes (Sporbert et al. 2002; Sporbert et al. 2005; Schermelleh et al. 2007; Gorisch et al. 2008).
DNA REPLICATION: IN VIVO
Investigating how the distinct activities required for DNA synthesis are organized within the cell nucleus relates to the larger issue of understanding how DNA replication is regulated on a cellular level. A cell must duplicate its entire genome once and only once every time it divides. Therefore, in the broadest sense, investigating the global regulation of DNA replication can be distilled into the question of how the activity of individual replication units is coordinated throughout a cell cycle. This rather daunting problem can be broken down into more specific questions. How is replication propagated along the chromosomes? How does the cell ensure that the whole genome gets replicated? Where along the chromosome does replication begin? These questions were originally addressed in bacteria, where the replication program proceeds in a rather straightforward fashion with genetically well defined replication origins, which fire once per cell cycle (Mott and Berger 2007).
However, the much larger size and complexity of eukaryotic genomes impose additional difficulties on the organization of DNA replication. Hence, eukaryotic DNA replication represents a more complex situation that remains poorly understood. The confusion centers around two seemingly contradictory observations. First, a number of studies have clearly shown that DNA replication follows a defined, temporal progression (Sparvoli et al. 1994; Cardoso et al. 1997; Jackson and Pombo 1998; Ma et al. 1998; Dimitrova and Gilbert 2000; Leonhardt et al. 2000; Sadoni et al. 2004; Easwaran et al. 2005). Actively transcribed, euchromatic regions of the genome tend to be duplicated early in S phase, whereas heterochromatin, which is more condensed and often transcriptionally silent, replicates late in S phase (Fig. 2 and Movie 1 online at http://cshperspectives.cshlp.org/). This phenomenon was originally described by observing replication along Giemsa stained chromosomes and correlating DNA synthesis with banding patterns (Drouin et al. 1990). Second, eukaryotic replication origins fire in a stochastic fashion throughout S phase (Dijkwel et al. 2002; Patel et al. 2006). Therefore, the distribution of active origins, and thus replication initiation, changes between each cell cycle. Given the random nature of replication origin firing, it is hard to understand how a cell can maintain the temporal progression of replication. Reconciling these data acquired at very different spatial resolution levels (from hundreds of base pairs in stretched DNA fibers and by two-dimensional gel electrophoresis analysis to megabase chromatin domains in whole cells in situ) presents a significant hurdle to our understanding of how replication proceeds in eukaryotic cells.
Figure 2.
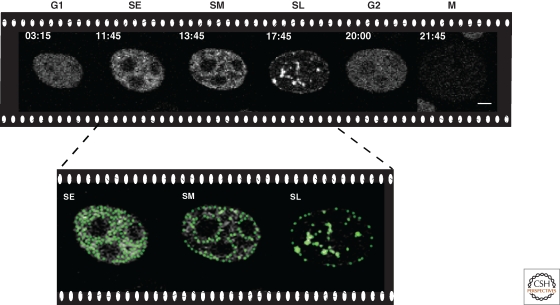
Temporal progression of genome replication. Snapshots of a time-lapse confocal microscopy movie of DNA replication throughout the cell cycle in human HeLa cells stably expressing GFP-tagged PCNA. Times are indicated in hours:minutes and cell cycle phases as G1/SE (S early)/SM (S mid)/SL (S late)/G2/M. Scale bar, 5 µm. During early S phase, small foci distributed throughout the nucleus and mostly corresponding to euchromatic genomic regions are duplicated. Subsequently, the DNA replication machinery loads at perinuclear and perinucleolar heterochromatin regions followed at later times by large constitutive heterochromatic chromosomal regions. This temporal replication program is recapitulated at each cell cycle. See also Movie 1 online at http://cshperspectives.cshlp.org/.
It should be noted that the budding yeast Saccharomyces cerevisiae represents an exception to the standard eukaryotic strategy for genome duplication. Similar to bacteria, S. cerevisiae possess well-defined replication origin sequences that can fire at a very efficient rate during S phase, leading to a very homogenous pattern of DNA replication (Fangman and Brewer 1991; Gilbert 2001). Furthermore, genome-wide analysis of replication initiation indicates no bias for sites of active transcription and no observable delay for any distinct regions of the genome (Raghuraman et al. 2001). For these reasons, it is perhaps misleading to generalize conclusions obtained with this system when contemplating eukaryotic DNA replication timing. Therefore, this article focuses on how DNA replication is regulated within the nucleus of metazoan systems.
GENOMIC APPROACHES TO STUDYING REPLICATION TIMING
The recent development of genomic assays has for the first time permitted a genome-wide examination of replication timing in populations of eukaryotic cells. For example, microarray analysis has been successfully used in a variety of systems to generate genome-wide profiles of replication timing. First performed in S. cerevisiae, and more recently in S. pombe, Drosophila melanogaster, and Homo sapien cells (Raghuraman et al. 2001; Schubeler et al. 2002; Watanabe et al. 2002; White et al. 2004; Woodfine et al. 2004; Jeon et al. 2005; Eshaghi et al. 2007; Hiratani et al. 2008; Watanabe et al. 2008), this technique usually requires the synchronization of a population of cells at the G1/S boundary or their flow cytometric sorting based on increasing DNA content through S phase. Following release into S phase, the accumulation of newly synthesized DNA over time is measured by hybridization to DNA arrays. Alternatively, isolated nascent DNA can be directly sequenced using novel high-throughput deep sequencing techniques. A summary of these studies is compiled in Table 1.
Table 1.
Genomic studies of DNA replication.
| Genome duplication characteristic | Species | Genome segment investigated | Reference |
|---|---|---|---|
| Timing of replication | |||
| Chromosome bands/ isochores; GC/AT content | Human | Chromosome 22 | (White et al. 2004) |
| Human | MN1/PITPNB region of Chromosome 22 | (Schmegner et al. 2007) | |
| Human | Whole genome (at 1 Mbp resolution), Chromosomes 22q and 6 |
(Woodfine et al. 2004; Woodfine et al. 2005) | |
| Mouse | Whole genome | (Farkash-Amar et al. 2008) | |
| Human | ENCODE regions | (Karnani et al. 2007) | |
| Human | Chromosomes 21q, 11q and 6 | (Costantini and Bernardi 2008) | |
| Alu/LINE repeats content | Human | Whole genome (at 1 Mbp resolution), Chromosomes 22q and 6 |
(Woodfine et al. 2004; Woodfine et al. 2005) |
| Transcription activity; Coding sequences |
Human | Chromosome 22 | (White et al. 2004) |
| Human | Whole genome (at 1 Mbp resolution), Chromosomes 22q and 6 |
(Woodfine et al. 2004; Woodfine et al. 2005) | |
| Human | 1,589 genes in Chromosomes 21 and 22 | (Jeon et al. 2005) | |
| Human | ENCODE regions | (Karnani et al. 2007) | |
| Mouse embryonic | Whole genome | (Hiratani et al. 2008) | |
| House keeping/Tissue specific genes | Mouse | Whole genome | (Farkash-Amar et al. 2008) |
| H3K4 and H3K9 methylation; H3K9 and H3K14 acetylation; H3K4 and H3K36 trimethylation | Human | ENCODE regions | (Karnani et al. 2007) |
| Human | MYC, LMNB2, HBB genes, FRAXA region, 1075 kbp of Chromosome 22 | (Lucas et al. 2007) | |
| Mouse embryonic | Whole genome | (Hiratani et al. 2008) | |
| Human | ENCODE regions | (Cadoret et al. 2008; Necsulea et al. 2009) | |
| Mode of replication | |||
| Asynchronous replication of 9–60% of the sequences throughout S-phase | Human | 1,589 genes in Chromosomes 21 and 22 | (Jeon et al. 2005) |
| Human | ENCODE regions | (Karnani et al. 2007) | |
| Mouse | Whole genome | (Farkash-Amar et al. 2008) | |
| Acute transition between individual replication timing zones | Human | ENCODE regions | (Karnani et al. 2007) |
| Human | MN1/PITPNB region of Chromosome 22 | (Schmegner et al. 2007) | |
| Gradient transition between individual replication timing zones with large replicons locating at the boundaries of replication timing zones | Human | Chromosomes11q and 21q | (Watanabe et al. 2008; Watanabe et al. 2009) |
| Mouse embryonic | Whole genome | (Hiratani et al. 2008) | |
| Ori/Replicon distribution | |||
| X inactivation independent CpG-associated Ori activity | Mouse | X chromosome | (Gomez and Brockdorff 2004) |
| Correlation of Ori activity with promoter regions and gene density | Mouse embryonic | 10 Mbp on chromosome X and 3 | (Sequeira-Mendes et al. 2009) |
| Replicon clusters with interorigin distance ≤100 kbp | Mouse embryonic | 10 Mbp on chromosome X and 3 | (Sequeira-Mendes et al. 2009) |
| Human | MYC, LMNB2, HBB genes, FRAXA region, 1075 kbp of Chromosome 22 | (Lucas et al. 2007) | |
| Human | Chromosomes 21q, 11q and 6 | (Costantini and Bernardi 2008) | |
| Single large replicons (≥300 kbp) | Human | MYC, LMNB2, HBB genes, FRAXA region, 1075 kbp of Chromosome 22 | (Lucas et al. 2007) |
| Human | ENCODE regions | (Cadoret et al. 2008) | |
| Mouse | Whole genome | (Farkash-Amar et al. 2008) | |
| Human | Chromosomes 21q, 11q and 6 | (Costantini and Bernardi 2008) | |
| Human | Chromosomes11q and 21q | (Watanabe et al. 2008; Watanabe et al. 2009) | |
ENCODE – ENCyclopedia Of DNA Elements represent regions covering ∼1% (∼30Mb) of human genome.
DNA combing is another technique that has recently been used to systematically investigate replication initiation and elongation at the level of single DNA fibers. Long, individual DNA molecules are stripped of proteins, uniformly stretched across a glass surface, and examined by standard fluorescence microscopy (Bensimon et al. 1994). By pulse-labeling cells with nucleotides prior to this treatment, it is possible to directly examine sites of DNA synthesis (Pasero et al. 2002; Anglana et al. 2003). Originally used with radioactively labeled nucleotides to accurately measure the rate of replication fork progression (Huberman and Riggs 1968), more recent studies have used this approach to compare the efficiency of origin firing between early and late replicating regions (Patel et al. 2006).
The application of these methods has, for the first time, permitted the examination of how DNA replication proceeds at the genomic level. The availability of several complete genome sequences has further allowed in silico evaluation of the correlation of DNA replication with structural and functional genomic features. For example, whereas a relationship between transcriptional activity and replication timing had been suggested by earlier reports analyzing single genes (Gilbert 2002), microarray studies investigating the timing of DNA replication along D. melanogaster and human chromosomes convincingly linked these processes on a genome-wide level (Schubeler et al. 2002; Woodfine et al. 2004; Jeon et al. 2005). Specifically, early replicating regions displayed a strong correlation with gene rich areas that possessed a high GC content and contained actively transcribed genes. In fact, a high-resolution human genome analysis revealed that boundaries between regions with different GC content (the so-called isochores) correlated with borders between DNA replication timing zones (Costantini and Bernardi 2008). In addition, recent studies also showed that a substantial portion of the genome (approx. 60%) does not replicate until much later in S phase (Eshaghi et al. 2007).
The dynamics of origin firing has also been examined by genome-wide analysis. Consistent with earlier reports, this process was found to be stochastic, leading to a random distribution of replication initiation sites across the chromosomes (Patel et al. 2006). It was also shown that early S phase origins are quite inefficient at initiating replication. In contrast, the relative firing efficiency of late S phase origins, although still occurring in a random manner, was significantly higher (Eshaghi et al. 2007). This could reflect the fact that less DNA is “licensed” to replicate at later S phase stages leading to a seemingly higher firing efficiency. Alternatively, assuming recycling of a factor limiting replication initiation—firing propensity redistribution—has also allowed reasonable modeling of S phase based on stochastic firing of individual origins (Lygeros et al. 2008). Cyclin-dependent kinases were suggested as factors limiting initiation. These factors are recycled to late-firing origins, thus increasing the probability of their activation.
Together, these results have led to the “increasing efficiency model,” which has the potential to explain many of the questions surrounding the regulation of DNA replication. The idea centers around the hypothesis that as a cell progresses through S phase, the overall efficiency with which the remaining fraction of available origins initiates replication increases (Lucas et al. 2000; Hyrien et al. 2003). An appealing aspect of this model is that it accounts for how cells can avoid the problem of gaps in DNA replication. Furthermore, by assuming that different regions of the genome possess variable efficiencies of origin firing, it can also explain how a cell could maintain stochastic firing of replication origins and still replicate its DNA in an ordered fashion (Rhind 2006).
In general, this proposal provides a satisfactory explanation for how replication timing is modulated in eukaryotic nuclei. However, two very important questions remain. First, how does the firing efficiency of replication origins increase over the course of S phase? Second, what determines the inherent replication efficiency of a given genomic region? Regarding the first question, one model centers on the concept of polymerase recycling, whereby a fixed number of DNA polymerase complexes are available to the cell. At the onset of S phase, only the most efficient and/or accessible origins have a chance of initiating replication. As the genome becomes duplicated, the number of potential origins decreases, thereby increasing the probability that they will fire. The answer to the second question remains less clear, but alterations in chromatin structure are likely to play a central role.
The likelihood of a connection between chromatin structure and replication timing has been well established (Donaldson 2005). This discussion is most often framed by the correlation between the open chromatin structure present at transcriptionally active regions and the fact that these sites are often replicated early in S phase. In the simplest view, a relaxed chromatin structure results in more accessible genomic DNA, which leads to more efficient binding by both transcription and replication factors. There has been some discussion as to whether replication or transcription plays a causal role in this relationship (i.e., whether early replicating sites “define” regions of transcription or vice versa) but no conclusive results have been obtained. One shortcoming of the genomic methods described earlier is that whereas replication timing profiles certainly reflect the influence of chromatin structure, it is difficult to backtrack and directly examine the nature of this chromatin. This is due in part to the ensemble/pooled nature of genomic analysis, as well as the fact that neither assay permits a particularly detailed examination of nuclear structure.
CELLULAR ORGANIZATION OF REPLICATION
Chromosomes and chromosomal domains are nonrandomly organized within eukaryotic nuclei and their topology is thought to have functional significance (Cremer and Cremer 2001; Kumaran et al. 2008; Takizawa et al. 2008). Microscopic inspection of nuclei is a powerful approach to investigate the spatio-temporal organization of replication within the context of nuclear architecture. Ongoing DNA synthesis provides a way to directly detect the nuclear sites of DNA replication after introducing labeled nucleotides into the cells. Initially, radioactive thymidine was used (Milner 1969) and later, with the development of antibodies specifically detecting halogenated thymidine analogs (Gratzner 1982; Aten et al. 1992), immunofluorescence analysis of nuclear replication structures became reality (Nakamura et al. 1986; Nakayasu and Berezney 1989; O’Keefe et al. 1992). More recently, live-cell microscopy analysis of replication progression was made possible by introducing fluorescently conjugated nucleotides (Schermelleh et al. 2001) or by expression of fluorescent replication factors (Cardoso et al. 1997; Leonhardt et al. 2000).
With these approaches, DNA replication was found to occur at subnuclear sites called replication foci, which accumulate numerous DNA replication factors and cell cycle proteins (Cardoso et al. 1993; Cardoso et al. 1997). These foci show distinct patterns of localization over the course of S phase; as such, studying their composition and dynamics enables an examination of how DNA replication is regulated on a cellular level. Time-lapse microscopy of living mammalian cells over the course of an entire cell cycle (Fig. 2 and Movie 1 online at http://cshperspectives.cshlp.org/) has shown that early in S phase, immediately following the onset of DNA replication, a multitude of small replication foci are distributed throughout the nucleus. During mid S phase, the replication foci are uniformly larger, and are distributed around the periphery of the nucleoli and nuclear envelope. Finally, at the end of S phase, the sites of replication have been consolidated into a small number of very large foci. In addition to reflecting the stage of S phase progression, the pattern of replication foci also correlates with the nature and topology of the chromatin that is being replicated. For example, early replication patterns represent actively transcribed euchromatin, whereas late replication patterns are associated with heterochromatic regions. Thus, this type of microscopic analysis permits the simultaneous visualization of both replication dynamics and chromatin structure in a single cell basis.
The role of chromatin modifications and structural rearrangements in replication organization is yet to be established. Some experimental evidence suggests a role of chromatin remodeling and assembly factors in facilitating replication through heterochromatin domains (Collins et al. 2002; Quivy et al. 2008). However, the impact of histone modifications is less clear. For example, disruption of histone H3 lysine 9 trimethylation, the typical epigenetic mark for heterochromatin, does not significantly affect the late replication of mouse chromocenters (Wu et al. 2006). Other histone modifications, such as phosphorylation of the linker histone H1 by Cdk2, have been proposed to play a role in the large-scale decondensation of chromatin associated with replication (Alexandrow and Hamlin 2005).
Examining the dynamics of the replication machinery during S phase has in addition provided a detailed view of how DNA replication proceeds on a cellular level. The combination of time-lapse microscopy with fluorescence photobleaching/activation indicated that the processivity of the replication machinery is built on transient interactions of various replication enzymes with a stable core consisting of the processivity factor PCNA (Sporbert et al. 2002; Sporbert et al. 2005; Schermelleh et al. 2007; Gorisch et al. 2008). The latter stayed associated with the DNA and upon photobleaching no recovery was measured for periods of over 10 minutes. When assembly of PCNA was measured, it was found to occur at sites adjacent to the ones previously labeled. This indicated that once replication is completed at a given site, a new replication focus assembles de novo at a neighboring site (Sporbert et al. 2002; Sadoni et al. 2004). Consistent data has been reported using double nucleotide pulse-chase-pulse experiments, whereby sequentially replicated DNA is labeled by two consecutive pulses of modified nucleotides (Manders et al. 1996; Jackson and Pombo 1998). Thus, replication of a specific genomic region facilitates subsequent loading of new replication factors at neighboring sites. One interpretation of this result is that the act of replication induces local changes in chromatin condensation, which in turn promotes the access/recruitment of replication factors and the initiation of additional replication cycles. Replication would begin at sites with an “open” chromatin conformation, similar to what has previously been proposed. As a result of these initial replication events, the chromatin at adjacent regions, which normally would not support replication initiation, would begin to decondense, leading to an increased probability that origins present at these sites would fire. It is tempting to suggest that the activity of replication helicases such as the MCM proteins would promote local chromatin decondensation. However, it is equally feasible that the mere act of DNA polymerization is sufficient to induce such changes. This propagating chromatin fiber decondensation can be visualized as analogous to pulling on a shoelace.
On a genome-wide scale, this model, which we refer to as the domino model, leads to a simple, self-propagating mode by which the entire chromosomes become fully duplicated by simply spreading the replication process using the “nearest neighbor” principle (Fig. 3). This hypothesis also dovetails nicely with the increasing efficiency model, in that it provides a physical basis for why origin efficiency would increase over the course of S phase. The increased efficiency of late origins is not because of polymerase recycling; rather it can be accounted for by the fact that as a cell progresses through S phase, it becomes more and more likely that any given region of the genome is proximal to a site of DNA replication. Therefore, even origins contained in hyper-condensed, late replicating regions such as the heterochromatin become accessible as more and more of the surrounding chromatin undergoes replication.
Figure 3.
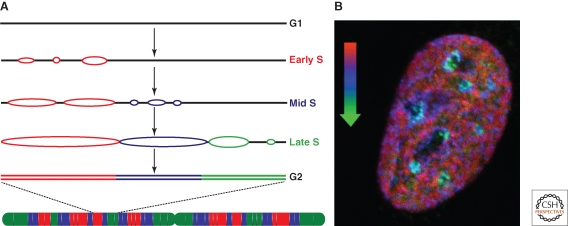
Model for the progression of genome replication—domino model. (A) One replicon cluster (red) initiates replication at early S phase and DNA synthesis proceeds bidirectionally (replication bubbles). This activates initiation of neighboring replicon clusters (blue), which in turn activate replication at later replicon clusters (green) until the whole chromosome (see scheme below) is fully duplicated in G2 phase. (B) Mouse primary fibroblast cell was pulse labeled for 30 minutes with the nucleotide analogue IdU (early S, red) followed by 3 hours chase, another 30 minutes pulse labeling with CldU (mid S, blue) and 3 hours chase, fixation and detection of replication sites with an antibody to DNA ligase I (late S, green). The micrograph represents a mid optical section of the late S phase fibroblast cell depicting the overlay of the three sequential replication labels and the spatiotemporal progression of genome replication.
FUTURE DIRECTIONS
Despite significant advances in the characterization of the process of DNA replication, several basic questions remain unanswered. For example, it is still not fully evident how propagation of DNA replication over chromatin is coordinated with other nuclear processes. In other words, how is duplication of the (epi)genome once and only once per cell cycle achieved with high precision in a highly variable environment including parallel transcription, repair, and other DNA metabolic activities, and on such a differentiated template as chromatin? How are chromatin epistates maintained at every cell cycle? Is transcriptional activity or chromatin structure determining replication timing or are they rather determined by the time they are replicated during S phase?
Interdependency of all these nuclear processes is also influenced by the fact that multiple molecular components acting on DNA metabolism are shared. On the one hand, the fact that, e.g., many DNA replication factors are shared with DNA repair pathways (the same is also the case between repair and transcription factors) is far more economic for the cell when dealing with similar tasks. On the other hand, this can be a dangerous strategy as mutations on single factors will have pleiotropic effects and factors can get exhausted and become rate limiting.
Finally, understanding the DNA replication process will require the ability to connect data from genomic studies with data from single cells in a unified coherent model. Closing this (temporal and spatial) gap is becoming reality with the advent of novel super-resolution nanoscopy techniques. In fact, recent analysis of DNA replication in intact cells provided much increased resolution and consequently much higher numbers of replication foci throughout S phase (Baddeley et al. 2009). In the near future, a major goal of the field will be to visualize and to characterize in full detail single replicons in intact cells, which were previously only identified in stretched DNA fibers (Fig. 4). The integration of these data with time lapse analysis will give us the ability to film and dissect the replication of the genome in situ and in real time.
Figure 4.
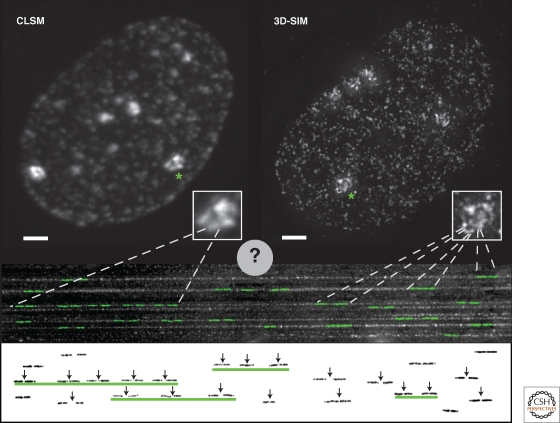
Connecting replicon analysis on DNA fibers to whole cell in situ replication foci. Human HeLa cells expressing GFP-tagged PCNA were imaged using either conventional confocal laser scanning microscopy (CLSM) or super-resolution 3D-structured illumination microscopy (3D-SIM). Shown are maximal intensity projections of the full 3D image stacks illustrating the spatial organization of replication foci in late S phase. The inset shows a twofold magnification of the areas containing replicating heterochromatic regions marked by an asterisk in the image. Scale bar, 2 µm. Even in these condensed heterochromatin domains, the much larger number of individual replicating sites revealed by the increased resolution of the 3D-SIM technique is apparent. Below the cell images, combed DNA fibers are shown with a cartoon representation of nucleotide labeled replicon units overlayed in green. The same replicons are shown underneath with the vertical arrows indicating the origin of replication of each replicon. Several of the replicons are closed together in clusters (underlined by green lines), which initiate replication coordinately. By comparing labeling of replicons in combed DNA fibers and the numbers of replication foci counted by conventional light microscopy, it was concluded that one replication focus corresponds to a spatially organized cluster of replicons, as indicated by the dashed lines. Using 3D-SIM and other super-resolution nanoscopy techniques, much higher numbers of replication foci are resolved. This will soon allow us to measure in detail the characteristics of individual replicons that were formerly visualized only on stretched DNA fibers.
ACKNOWLEDGMENTS
We are indebted to Robert M. Martin for the artwork in Figure 1 and to Corella Casas Delucchi for the DNA fibers in Figure 4. We also thank all the past and present members of our laboratory for their many contributions along the years. Last but not least, we thank our many collaborators, which have made our work possible and enjoyable. Our research has been supported by grants of the Deutsche Forschungsgemeinschaft and the Volkswagen foundation. We regret that because of space constraints, we had to eliminate many important and relevant citations.
Footnotes
Editors: David Spector and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Aladjem MI 2007. Replication in context: Dynamic shownion of DNA replication patterns in metazoans. Nat Rev Genet 8:588–600 [DOI] [PubMed] [Google Scholar]
- Alexandrow MG, Hamlin JL 2005. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J Cell Biol 168:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M 2003. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385–394 [DOI] [PubMed] [Google Scholar]
- Aten JA, Bakker PJ, Stap J, Boschman GA, Veenhof CH 1992. DNA double labelling with IdUrd and CldUrd for spatial and temporal analysis of cell proliferation and DNA replication. Histochem J 24:251–259 [DOI] [PubMed] [Google Scholar]
- Baddeley D, Chagin VO, Schermelleh L, Martin S, Pombo A, Carlton PM, Gahl A, Domaing P, Birk U, Leonhardt H, et al. 2009. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res doi: 10.1093/nar/gkp901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D 1994. Alignment and sensitive detection of DNA by a moving interface. Science 265:2096–2098 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A 2005. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA 1988. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 332:546–548 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105:15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MC, Joseph C, Rahn HP, Reusch R, Nadal-Ginard B, Leonhardt H 1997. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J Cell Biol 139:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H, Nadal-Ginard B 1993. Reversal of terminal differentiation and control of DNA replication: Cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74:979–992 [DOI] [PubMed] [Google Scholar]
- Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet 32:627–632 [DOI] [PubMed] [Google Scholar]
- Costantini M, Bernardi G 2008. Replication timing, chromosomal bands, and isochores. Proc Natl Acad Sci 105:3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292–301 [DOI] [PubMed] [Google Scholar]
- Dijkwel PA, Wang S, Hamlin JL 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol Cell Biol 22:3053–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM 2000. Temporally coordinated assembly and disassembly of replication factories in the absence of DNA synthesis. Nat Cell Biol 2:686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD 2005. Shaping time: Chromatin structure and the DNA replication programme. Trends Genet 21:444–449 [DOI] [PubMed] [Google Scholar]
- Drouin R, Lemieux N, Richer CL 1990. Analysis of DNA replication during S-phase by means of dynamic chromosome banding at high resolution. Chromosoma 99:273–280 [DOI] [PubMed] [Google Scholar]
- Easwaran HP, Leonhardt H, Cardoso MC 2005. Cell cycle markers for live cell analyses. Cell Cycle 4:453–455 [DOI] [PubMed] [Google Scholar]
- Eshaghi M, Karuturi RK, Li J, Chu Z, Liu ET, Liu J 2007. Global profiling of DNA replication timing and efficiency reveals that efficient replication/firing occurs late during S-phase in S. pombe. PLoS ONE 2:e722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman WL, Brewer BJ 1991. Activation of replication origins within yeast chromosomes. Annu Rev Cell Biol 7:375–402 [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I 2008. Global organization of replication time zones of the mouse genome. Genome Res 18:1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2001. Making sense of eukaryotic DNA replication origins. Science 294:96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2002. Replication timing and transcriptional control: Beyond cause and effect. Curr Opin Cell Biol 14:377–383 [DOI] [PubMed] [Google Scholar]
- Gomez M, Brockdorff N 2004. Heterochromatin on the inactive X chromosome delays replication timing without affecting origin usage. Proc Natl Acad Sci 101:6923–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorisch SM, Sporbert A, Stear JH, Grunewald I, Nowak D, Warbrick E, Leonhardt H, Cardoso MC 2008. Uncoupling the replication machinery: Replication fork progression in the absence of processive DNA synthesis. Cell Cycle 7:1983–1990 [DOI] [PubMed] [Google Scholar]
- Gratzner HG 1982. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 218:474–475 [DOI] [PubMed] [Google Scholar]
- Hamlin JL, Mesner LD, Lar O, Torres R, Chodaparambil SV, Wang L 2008. A revisionist replicon model for higher eukaryotic genomes. J Cell Biochem 105:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6:e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman JA, Riggs AD 1968. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol 32:327–341 [DOI] [PubMed] [Google Scholar]
- Hubscher U, Seo YS 2001. Replication of the lagging strand: A concert of at least 23 polypeptides. Mol Cells 12:149–157 [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A 2003. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25:116–125 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A 1998. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140:1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F 1993. The replicon: Thirty years later. Cold Spring Harb Symp Quant Biol 58:383–387 [DOI] [PubMed] [Google Scholar]
- Jacob F, Brenner S 1963. On the regulation of DNA synthesis in bacteria: The hypothesis of the replicon. C R Hebd Seances Acad Sci 256:298–300 [PubMed] [Google Scholar]
- Jeon Y, Bekiranov S, Karnani N, Kapranov P, Ghosh S, MacAlpine D, Lee C, Hwang DS, Gingeras TR, Dutta A 2005. Temporal profile of replication of human chromosomes. Proc Natl Acad Sci 102:6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor C, Malhotra A, Dutta A 2007. Pan-S replication patterns and chromosomal domains defined by genome-tiling arrays of ENCODE genomic areas. Genome Res 17:865–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A 1960. Biologic synthesis of deoxyribonucleic acid. Science 131:1503–1508 [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Thakar R, Spector DL 2008. Chromatin dynamics and gene positioning. Cell 132:929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC 2000. Dynamics of DNA replication factories in living cells. J Cell Biol 149:271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas I, Chevrier-Miller M, Sogo JM, Hyrien O 2000. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J Mol Biol 296:769–786 [DOI] [PubMed] [Google Scholar]
- Lucas I, Palakodeti A, Jiang Y, Young DJ, Jiang N, Fernald AA, Le Beau MM 2007. High-throughput mapping of origins of replication in human cells. EMBO Rep 8:770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygeros J, Koutroumpas K, Dimopoulos S, Legouras I, Kouretas P, Heichinger C, Nurse P, Lygerou Z 2008. Stochastic hybrid modeling of DNA replication across a complete genome. Proc Natl Acad Sci 105:12295–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng P, Meng C, Berezney R 1998. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143:1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EM, Stap J, Strackee J, van Driel R, Aten JA 1996. Dynamic behavior of DNA replication domains. Exp Cell Res 226:328–335 [DOI] [PubMed] [Google Scholar]
- McHenry CS 1988. DNA polymerase III holoenzyme of Escherichia coli. Annu Rev Biochem 57:519–550 [DOI] [PubMed] [Google Scholar]
- Milner GR 1969. Changes in chromatin structure during interphase in human normoblasts. Nature 221:71–72 [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol 5:343–354 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C 1986. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res 165:291–297 [DOI] [PubMed] [Google Scholar]
- Nakayasu H, Berezney R 1989. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol 108:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Guillet C, Cadoret JC, Prioleau MN, Duret L 2009. The relationship between DNA replication and human genome organization. Mol Biol Evol 26:729–741 [DOI] [PubMed] [Google Scholar]
- Noguchi H, Prem veer Reddy G, Pardee AB 1983. Rapid incorporation of label from ribonucleoside disphosphates into DNA by a cell-free high molecular weight fraction from animal cell nuclei. Cell 32:443–451 [DOI] [PubMed] [Google Scholar]
- O’Keefe RT, Henderson SC, Spector DL 1992. Dynamic organization of DNA replication in mammalian cell nuclei: Spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol 116:1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A 1968. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci 59:598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS 2009. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature 462:940–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16:2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N 2006. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell 17:308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal SK, Yue H, Hu Z, Spiering MM, Benkovic SJ 2009. Single-molecule studies of DNA replisome function. Biochim Biophys Acta doi: 10.1016/j.bbapap.2009.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Gérard A, Cook AJL, Roche D, Almouzni G 2008. The HP1–p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Structural Mol Biol 15:972–979 [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294:115–121 [DOI] [PubMed] [Google Scholar]
- Rao PN, Johnson RT 1970. Mammalian cell fusion: Studies on the regulation of DNA synthesis and mitosis. Nature 225:159–164 [DOI] [PubMed] [Google Scholar]
- Rhind N 2006. DNA replication timing: Random thoughts about origin firing. Nat Cell Biol 8:1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Bell SD 2005. Origins of DNA replication in the three domains of life. FEBS J 272:3757–3766 [DOI] [PubMed] [Google Scholar]
- Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D 2004. Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci 117:5353–5365 [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H 2007. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res 35:4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Solovei I, Zink D, Cremer T 2001. Two-color fluorescence labeling of early and mid-to-late replicating chromatin in living cells. Chromosome Res 9:77–80 [DOI] [PubMed] [Google Scholar]
- Schmegner C, Hameister H, Vogel W, Assum G 2007. Isochores and replication time zones: A perfect match. Cytogenet Genome Res 116:167–172 [DOI] [PubMed] [Google Scholar]
- Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32:438–442 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5:e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NK, Morris CF, Alberts BM 1980. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J Biol Chem 255:4290–4293 [PubMed] [Google Scholar]
- Sparvoli E, Levi M, Rossi E 1994. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J Cell Sci 107:3097–3103 [DOI] [PubMed] [Google Scholar]
- Sporbert A, Domaing P, Leonhardt H, Cardoso MC 2005. PCNA acts as a stationary loading platform for transiently interacting Okazaki fragment maturation proteins. Nucleic Acids Res 33:3521–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell 10:1355–1365 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T 2008. The meaning of gene positioning. Cell 135:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom TD, Malkas LH, Hickey RJ 1996. Identification of multiprotein complexes containing DNA replication factors by native immunoblotting of HeLa cell protein preparations with T-antigen-dependent SV40 DNA replication activity. J Cell Biochem 63:259–267 [DOI] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B 1989. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, α and δ. Embo J 8:3883–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Abe T, Ikemura T, Maekawa M 2009. Relationships between replication timing and GC content of cancer-related genes on human chromosomes 11q and 21q. Gene 433:26–31 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Fujiyama A, Ichiba Y, Hattori M, Yada T, Sakaki Y, Ikemura T 2002. Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: Disease-related genes in timing-switch regions. Human Mol Gen 11:13–21 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Shibata K, Ikemura T, Maekawa M 2008. Replication timing of extremely large genes on human chromosomes 11q and 21q. Gene 421:74–80 [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH 1953. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171:737–738 [DOI] [PubMed] [Google Scholar]
- White EJ, Emanuelsson O, Scalzo D, Royce T, Kosak S, Oakeley EJ, Weissman S, Gerstein M, Groudine M, Snyder M, Schubeler D 2004. DNA replication-timing analysis of human chromosome 22 at high resolution and different developmental states. Proc Natl Acad Sci 101:17771–17776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Beare DM, Ichimura K, Debernardi S, Mungall AJ, Fiegler H, Collins VP, Carter NP, Dunham I 2005. Replication timing of human chromosome 6. Cell Cycle 4:172–176 [DOI] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP 2004. Replication timing of the human genome. Hum Mol Genet 13:191–202 [DOI] [PubMed] [Google Scholar]
- Wu R, Singh PB, Gilbert DM 2006. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol 174:185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]