Abstract
ATMTel1 and ATRRad3 checkpoint kinases phosphorylate the C-terminus of histone H2AX (H2A in yeasts) in chromatin flanking DNA damage, establishing a recruitment platform for checkpoint and repair proteins. Phospho-H2A/X (γH2A/X)-binding proteins at double-strand breaks (DSBs) have been characterized, but those required for replication stress responses are unknown. Here, we present genetic, biochemical, small angle X-ray scattering (SAXS), and X-ray structural studies of the Schizosaccharomyces pombe Brc1, a 6-BRCT-domain protein that is structurally related to Saccharomyces cerevisiae Rtt107 and mammalian PTIP. Brc1 binds γH2A to form spontaneous and DNA damage-induced nuclear foci. Spontaneous Brc1 foci colocalize with ribosomal DNA repeats, a region prone to fork pausing and genomic instability, whereas DNA damage-induced Brc1 foci colocalize with DSB response factors. γH2A binding is critical for Brc1 function. The 1.45 Å resolution crystal structure of Brc1–γH2A complex shows how variable BRCT insertion loops sculpt tandem-BRCT phosphoprotein-binding pockets to facilitate unique phosphoprotein-interaction specificities, and unveils an acidic DNA-mimicking Brc1 surface. From these results, Brc1 docking to γH2A emerges as a critical chromatin-specific response to replication-associated DNA damage.
Keywords: chromatin, DNA replication, genome stability, Schizosaccharomyces pombe, X-ray crystal structure
Introduction
DNA damage responses for maintaining genome integrity require that DNA repair and checkpoint proteins work in concert with factors that bind or modify chromatin at DNA lesions (Stucki and Jackson, 2006; Harper and Elledge, 2007). Central to this integration of chromatin metabolism and DNA repair, the checkpoint kinases ATMTel1 and ATRRad3/Mec1 phosphorylate the carboxyl terminus of histone H2AX (or H2A in yeasts) in multi-kilobase regions of chromatin surrounding DNA double-strand breaks (DSBs), creating a phospho-H2A/X (γH2A/X) recruitment platform for checkpoint and DSB repair factors (Rogakou et al, 1998; Celeste et al, 2003; Stucki and Jackson, 2006; Williams et al, 2007). Although functions of γH2A/X at DSBs have been extensively investigated, its functions in cellular responses to spontaneous and DNA replication-associated DNA damage have received comparatively little attention and remain a mystery. As cells are constantly exposed to endogenous (i.e. metabolic) sources of DNA damage that stall or collapse replication forks, and the most widely used anti-cancer drugs cause S-phase-specific DNA damage, such unknown effectors of ATM/ATR-dependent γH2A/X signalling during replication are arguably the most crucial.
Evidence for such unknown S-phase γH2A effectors comes from genetic and microscopic studies of the Schizosaccharomyces pombe checkpoint mediator protein Crb2. In response to DSB producing ionizing radiation (IR), the Crb2 tandem-BRCT (BRCA1 carboxy terminus) domains directly bind γH2A in a phosphorylation-dependent manner (Nakamura et al, 2004; Du et al, 2006; Kilkenny et al, 2008). γH2A is required for Crb2 IR-induced focus formation and for amplification and maintenance of DNA damage checkpoint signals. However, H2A phosphorylation is also critical for fission yeast responses to hydroxyurea (HU), which stalls replication forks without creating DNA double breaks, and for survival of exposure to camptothecin (CPT) (Redon et al, 2003; Nakamura et al, 2004), a prototypical anti-cancer drug that collapses replication forks by poisoning topoisomerase I (Pommier, 2006). Yet, ablation of Crb2 and γH2A uncovers non-epistatic genetic interactions (Du et al, 2006), suggesting the existence of additional important γH2A-interacting factors.
Candidate S-phase γH2A effectors include S. pombe Brc1 and budding yeast Rtt107 (Esc4), which are members of a poorly understood family of 6-BRCT-domain proteins that appear to be conserved in fungi. Brc1 and Rtt107 are required for survival of CPT treatment, but are dispensable for survival of IR (Verkade et al, 1999; Rouse, 2004; Sheedy et al, 2005; Roberts et al, 2006; Zappulla et al, 2006). Rtt107 is required for stabilization of stalled replication forks and reestablishment of collapsed forks (Rouse, 2004; Chin et al, 2006; Roberts et al, 2008), a function, which may involve localization to sites of DNA replication-associated damage through uncharacterized protein–protein interactions. However, the mechanism of DNA damage recruitment of these 6-BRCT genome maintenance factors and how they enable specific repair of replication-associated damage remains unknown. Brc1 and Rtt107 are structurally related to mammalian PTIP proteins that have 6-BRCT domains and participate in DNA damage responses (Muñoz and Rouse, 2009). Although it is unclear as to what extent these proteins are functionally related, a detailed structure defining the functional interactions for any member of this group would provide a critical framework for improved understanding of their structural and functional relationships.
Here, we provide integrated microscopic, genetic, biochemical and structural results showing that the C-terminal pair of Brc1 BRCT domains (BRCT5–BRCT6) dock to γH2A and act in S-phase damage responses. Abrogating Brc1–γH2A interactions confers sensitivity to replication fork-damaging agents. A high-resolution X-ray structure of the BRCT5–BRCT6:γH2A phosphoprotein complex reveals stereo-specific recognition of pSer phosphates by BRCT repeats, and a probable general mechanism for generating alternate phosphoprotein interaction specificities in the large BRCT-containing superfamily of replication and DNA repair proteins. Together, these results define Brc1 as a major effector of Tel1/Rad3 and γH2A signalling in mechanisms that protect genome integrity from replication-associated DNA damage.
Results
Brc1 protects the genome from endogenous DNA damage
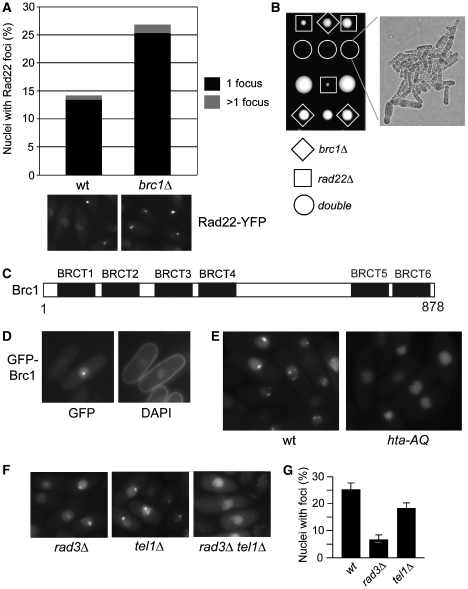
Brc1 is required for survival of genotoxins that interfere with DNA replication (Verkade et al, 1999; Sheedy et al, 2005; Dovey and Russell, 2007), but we suspected it might also protect genome integrity during an unperturbed cell cycle. We tested this by monitoring nuclear foci of Rad22Rad52, a critical HR protein that mediates assembly of Rad51 onto ssDNA (New et al, 1998). Live-cell analysis revealed that spontaneous Rad22-YFP foci were increased ∼2-fold in brc1Δ cells (Figure 1A). Interestingly, a similar effect is seen in hta-AQ cells that lack the C-terminal SQ phosphorylation motif required to form γH2A (Nakamura et al, 2004).
Figure 1.
Brc1 is required for genome stability and forms γH2A-dependent spontaneous nuclear foci. (A) Deletion of brc1+ causes an increase in Rad22-YFP foci. Cells were grown to mid-log phase in YES medium and imaged. Foci numbers from >1000 cells for each strain were quantified. (B) Tetrad analysis of genetic crosses between brc1Δ and rad22Δ reveals a severe synergistic growth in the double mutants. Representative spores from three asci are shown. (C) Schematic of the Brc1 BRCT domain organization. BRCT5 and BRCT6 form a tandem pair containing a consensus phosphoprotein-binding motif. (D) Cells were grown to mid-log phase in selective medium and fixed for 10 min in 70% ethanol at room temperature. DAPI staining was performed to visualize the nuclei and cell morphology. (E) Brc1 foci formation is dependent on phosphorylation of γH2A. Live cell microscopy of ectopically expressed GFP-Brc1 driven by the nmt41 promoter in wild-type (wt) or hta-AQ cells. (F) Brc1 foci formation is abolished in the rad3Δtel1Δ mutant. (G) Brc1 foci formation is reduced in the rad3Δ and tel1Δ mutants. Live cell microscopy of GFP-Brc1 was examined in the indicated strain backgrounds.
To explore whether brc1Δ cells require HR repair for survival, we tested for genetic interactions between brc1Δ and rad22Δ. Tetrad analysis uncovered a severe synergistic growth defect in brc1Δ rad22Δ cells (Figure 1B). Double mutants were either inviable or formed microcolonies containing highly elongated cells indicative of a prolonged DNA damage checkpoint arrest (Figure 1B). As even one broken replication fork is lethal in rad22Δ cells (Roseaulin et al, 2008), brc1Δ cells are likely suffering increased rates of replication fork collapse or rearrangement that are repaired by Rad22-dependent HR.
Brc1 forms γH2A-dependent nuclear foci
Brc1 has 4 BRCT domains (BRCT1–BRCT2–BRCT3–BRCT4) near its N-terminus followed by an acidic linker region and a tandem pair of BRCTs (BRCT5–BRCT6) at its extreme C-terminus (Figure 1C). We postulated that as a BRCT domain-containing protein that is required for genome stability, Brc1 might localize into nuclear foci. Expression of functional, C-terminally tagged GFP-Brc1 from its genomic locus was insufficient for microscopic detection. However, when GFP-Brc1 was expressed from the moderate strength nmt41 promoter, it formed distinct nuclear foci in ∼15–25% of cells in an asynchronous population (Figure 1D).
As brc1Δ and hta-AQ mutations confer sensitivity to an overlapping spectrum of S-phase DNA-damaging agents (Verkade et al, 1999; Nakamura et al, 2004; Sheedy et al, 2005), we hypothesized that Brc1 foci formation might require γH2A. Indeed, spontaneous nuclear GFP-Brc1 foci were absent in hta-AQ cells (Figure 1E).
Rad3 and Tel1 create γH2A (Nakamura et al, 2004). Rad3-Rad26ATRIP/Ddc2 complex associates with DNA lesions by binding the single-stranded (ss) DNA-binding protein replication protein A (Zou and Elledge, 2003), whereas Tel1 associates with DSBs by binding Nbs1 (Falck et al, 2005; You et al, 2005). To investigate which types of interactions create spontaneous Brc1 foci, we expressed GFP-Brc1 in rad3Δ or tel1Δ strains. GFP-Brc1 foci were detected in both mutant backgrounds (Figure 1F), although they were reduced in number (Figure 1G), indicating that Rad3 and Tel1 both regulate Brc1 localization. As predicted, GFP-Brc1 foci were abolished in a rad3Δtel1Δ strain (Figure 1F).
Brc1 BRCT5–BRCT6 domains bind γH2A
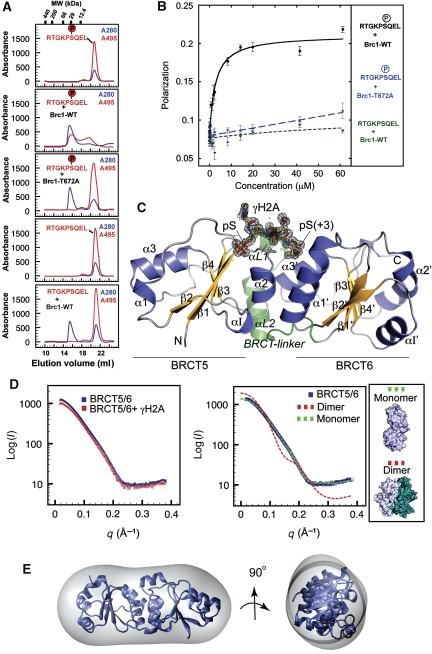
As tandem-BRCT domains in MDC1 and Crb2 bind γH2A/X (Stewart et al, 2003; Stucki et al, 2005; Lou et al, 2006; Kilkenny et al, 2008), and Brc1 foci formation is independent of Crb2 (Supplementary Figure S1), we hypothesized that a pair of BRCT domains in Brc1 may interact directly with γH2A. Sequence analysis of the six Brc1 BRCT domains revealed that only BRCT5 has phosphoserine-binding consensus motifs (Williams et al, 2004), referred to here as C1: (S/T)G, and C2: (T/S)XK (X is any residue). To test whether a Brc1:γH2A interaction is mediated via direct association, we analysed binding of recombinant Brc1 BRCT5–BRCT6 to N-terminal fluorescein-conjugated γH2A peptides by analytical gel filtration and fluorescence polarization (FP) (Figure 2A and B). In isolation, an FITC-γH2A peptide migrates as an ∼4 kDa peak in gel filtration, but coelutes at ∼25 kDa in the presence of purified BRCT5–BRCT6, consistent with a direct Brc1–γH2A interaction. FP-binding analysis shows Brc1 BRCT5–BRCT6 binds the γH2A tail with a 2.6-μM affinity, similar to other BRCT–phosphoprotein interactions (Figure 2A and B; Manke et al, 2003; Stucki et al, 2005). A Brc1 C1 motif mutant (T672A) does not bind, and interactions with an unphosphorylated H2A peptide are not detected by either method, indicating the interaction is phosphorylation dependent (Figure 2A and B). The hMDC1 BRCT domains bind human γH2AX C-terminal tail (Stucki et al, 2005), but unlike the Brc1–γH2A interaction, hMDC1 specifically recognizes the human ‘SQEY-coo(−)' γH2AX C-terminus, and will not bind the yeast ‘SQEL-coo(−)' carboxy terminus (Stucki et al, 2005). Thus, critical differences in these γH2A interactions are dictated by molecular recognition of the C-terminal H2A residue at the pSer +3 position.
Figure 2.
Brc1 BRCT5–BRCT6 domains interact directly with the phosphorylated γH2A C-terminal tail. (A) Analytical gel filtration binding analysis of Brc1–γH2A protein–peptide interactions. BRCT5–BRCT6–γH2A binding was monitored by following coelution of FITC-labelled γH2A peptides that absorb at 495 nm (red trace) with recombinant BRCT5–BRCT6 (blue trace). All peptides are FITC conjugated on the N-terminal amide and bear carboxylated C-termini. γH2A peptide coelution with wild-type BRCT5–BRCT6 is observed only for the phosphorylated γH2A peptide and is disrupted by pSer binding site mutation T672A. (B) FP-binding analysis of the Brc1–γH2A.1 interaction. Brc1 binds the γH2A tail with an apparent affinity of 2.6 μM, whereas the Brc1–T672A mutant or unphosphorylated peptide display no detectable binding. (C) X-ray crystal structure of the Brc1–γH2A complex. The γH2A peptide (gold with grey electron density) binds at the interface between Brc1 BRCT5 and BRCT6 domains (yellow β-sheets and blue α-helices). Sigma-A weighted 1.45 Å Fo–Fc density contoured at 3 σ was calculated before building the peptide chain. (D) Brc1 BRCT5/6 and the BRCT5/6–γH2A complex were analysed by SAXS (left). No significant change in Brc1 oligomeric state occurs on peptide binding. SAXS scattering for apo BRCT5/6 fits well to a monomeric Brc1 model (χ2=4.9), but not to the dimeric NCS-related dimer model (χ2=54.1) found in the apo-Brc1 crystal structure (see inset for NCS dimer structure). (E) Low resolution averaged ab initio GASBOR shape reconstruction for apo BRCT5/6 closely matches the monomeric structure.
Architecture of the Brc1–γH2A complex
To examine the structural basis for specificity differences between mammalian and yeast γH2A–BRCT interactions, and provide general insights into the structural determinants of phosphoprotein interaction specificities in the BRCT-containing superfamily of proteins, we solved high-resolution X-ray structures for unliganded Brc1 BRCT5–BRCT6 and the BRCT5–BRCT6:γH2A complex (Figure 2C, see Materials and methods). Three key observations from the Brc1:γH2A structure extend our understanding of BRCT domain-phosphoprotein recognition and specificity. First, the 1.45 Å structure of the BRCT5-BRCT6:γH2A complex provides an unprecedented high-resolution visualization of the BRCT–phosphoprotein interaction, revealing critical conserved determinants of phosphoserine recognition and binding specificity. Second, comparative analysis of the second primary BRCT peptide-binding determinant, the pSer +3 binding pocket, shows how variable sequences and structures at the BRCT–BRCT interface mold pSer +3 pockets to accommodate different peptide target specificities. Third, structural conservation of an exposed acidic Brc1 and MDC1 surface outside of the γH2A/X tail binding cleft suggests additional histone core particle interactions augment phosphorylated histone tail binding in histone-interacting BRCT proteins.
Each Brc1 C-terminal BRCT domain folds as an α–β sandwich with a central four-stranded parallel β-sheet that is flanked on one side by two helices (α1 and α3), and on the another by helix α2. BRCT5 and BRCT6 also contain unique helical elements (α I1 and α I1′) inserted between strands β2 and β3. Packing of an α2–α1′–α3′ three-helix bundle forms a 40 × 30 × 70 Å elongated dual repeat BRCT–BRCT interface resembling other family members (Williams et al, 2001; Glover et al, 2004). γH2A binds in the groove between the BRCT domains. Conformational change of helix α1, and repositioning of the BRCT5 and BRCT6 repeats relative to one another accompany peptide binding. α1 conformational change seems appropriate to relay BRCT peptide-binding state to neighbouring regions in BRCT-containing proteins, or to other binding partners.
As Brc1 BRCT5–BRCT6 associates as a non-crystallographic dimer with an extensive interface (1590 Å2 buried solvent-accessible surface) in the unliganded crystal form, but is monomeric in the peptide-bound structure, we hypothesized monomer-dimer transitions might occur with γH2A binding. To test this, we evaluated the solution oligomeric state of BRCT5–BRCT6 and the BRCT5–BRCT6/γH2A complex in solution with small angle X-ray scattering (SAXS), which can provide accurate shape and assembly information in solution (Putnam et al, 2007; Hura et al, 2009). X-ray solution scattering obtained from purified BRCT5–BRCT6 in either the presence or absence of peptide reveals a monomeric assembly state in solution and is not consistent with dimeric models (Figure 2D). Also, low-resolution ab initio solution structures for apo- and peptide-liganded states closely match the elongated 40 × 40 × 70 Å architectures of monomeric BRCT5–BRCT6 (Figure 2E). Gel filtration also shows both unbound and peptide-bound BRCT5–BRCT6 migrate with an apparent molecular weight of ∼25 kDa, consistent with the monomeric form and a 1:1 protein:peptide-binding stoichiometry (Figure 2A). Together, SAXS and gel filtration analyses show that Brc1 BRCT5–BRCT6 is monomeric in solution and interacts with the γH2A.1 tail with 1:1 heterodimer stoichiometry.
Brc1–γH2A tail recognition
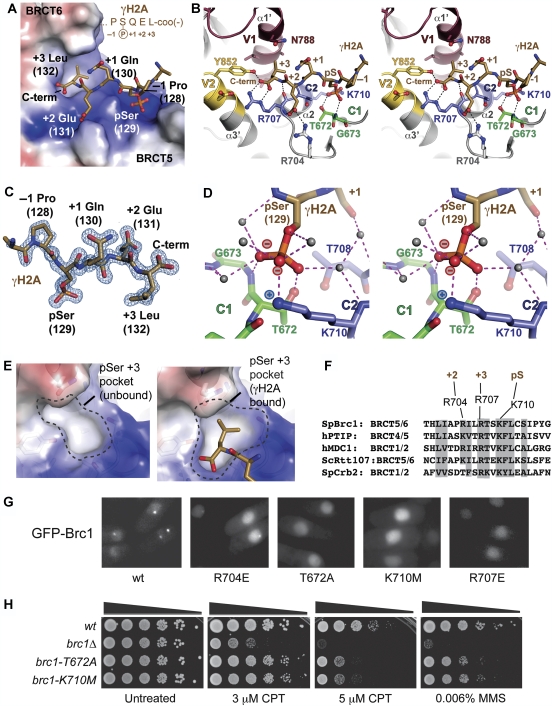
Three surface pockets embrace the pSer, pSer +2 Glu and pSer +3 Leu residues of the γH2A tail (Figure 3A–E) and a 604 Å2 solvent-accessible surface is buried at the Brc1–γH2A interface. Residues N-terminal to γH2A pSer129 (Lys127 and Pro128) do not make direct protein contacts. γH2A binding is likely a conserved function as residues lining the walls of the interaction pockets are conserved in fungal Brc1 orthologs (Figure 3F). Interestingly, these residues are also conserved in mammalian PTIP, which requires γH2AX for the formation of IR-induced foci, although this is thought to involve an indirect mechanism requiring MDC1 and RNF8 (Stucki et al, 2005; Gong et al, 2009). The binding pockets exhibit precise shape and electrostatic complementarity with the bound peptide, and seem well suited for γH2A binding (Figure 3A). Comparisons of our high-resolution X-ray structures of the γH2A-free and γH2A-bound Brc1 show how flexure and rearrangements at the BRCT5–BRCT6 interface accommodate γH2A binding (Figure 3E). Strikingly, the partially formed pSer +3 (Leu) interaction pocket in the apo structure changes to mold around γH2A Leu132, revealing induced-fit γH2A binding to a structurally plastic Brc1 pocket (Figure 3E).
Figure 3.
Brc1–γH2A interactions. (A) The γH2A-binding pocket of Brc1 BRCT5–BRCT6 harbours two positively charged grooves (blue electrostatic potential) for the pSer and pSer +2 (Glu) residues, and a hydrophobic pocket for the pSer +3 (Leu). The γH2A peptide (gold) is displayed overlaid on an electrostatic surface of the Brc1 phosphoprotein-binding surface. (B) γH2A is bound by constant BRCT pSer binding motifs C1 (green) and C2 (blue) and variable regions V1 (maroon) and V2 (yellow). A stereoview is displayed. (C) Final Sigma-A weighted 2Fo–Fc electron density (blue) at 1.45 Å is contoured at 1.5 σ and displayed for the peptide (gold). (D) Molecular interactions and water hydration shell for the γH2A phosphoserine. Stereospecific coordination of the pSer phosphate moiety maximizes charge–charge interactions with C2 motif K710 epsilon amino group. (E) Structural plasticity of the Brc1 pSer +3 Leu binding pocket. A partially formed +3 pocket in the apo-Brc1 structure rearranges to interact with γH2A Leu132 on phosphoprotein binding (right). (F) Sequence conservation of the Brc1 peptide-binding groove. (G) Mutation of conserved residues in the γH2A-binding pocket of Brc1 ablates spontaneous foci formation for mutants T672A, K710M and R707E. Expression of the GFP-Brc1 proteins was induced in a wt strain for 18 h at 30°C and live fluorescence microscopy was performed. (H) Genetic analysis of the Brc1 γH2A-binding mutants. Ten-fold serial dilutions of cells were exposed to the indicated DNA-damaging agent and incubated at 30°C for 3 days.
Thr672 (motif C1) and Lys710 (motif C2) (Figure 3B and D) from BRCT5 and an associated network of protein–solvent–phosphate interactions mediate the γH2A pSer129 interaction. An analysis of phosphoserine oxygen-coordinating atoms indicates the phosphate moiety is likely bound as a polarized anion with two negatively charged oxygen atoms tetrahedrally coordinated by protein and solvent, and a third neutral, doubly bonded oxygen with linear coordination. Consistent with oxygen coordination, refinement of the complex to 1.45 Å (Figure 3C) with relaxed geometric restraints on the phosphoserine yields bond lengths appropriate for the two equidistant P–O bonds (1.55 Å), and a third shorter P=O (1.48 Å) bond (Figure 3D). We therefore propose that stereo-specificity of the pSer interaction is a critical conserved determinant of BRCT-peptide binding, which maximizes coulombic attraction between the charged phosphate oxygen and the conserved C2 motif lysine (Brc1 K710). Selection for a polarized phosphoserine (versus resonance binding) probably contributes to moderate (high nM to low mM) binding affinities for four-residue BRCT molecular recognition sequences, despite the modest size of the BRCT protein–phosphoprotein interfaces. This phosphoserine interaction geometry may also ensure specific BRCT–phosphoprotein recognition of phosphoserine-phosphorylated targets over phospho-mimicking sequences containing Asp or Glu.
γH2A-pSer and pSer +3 contacts are critical for Brc1 foci formation and survival of S-phase damage
On the basis of our Brc1–γH2A structure, we used mutational analysis to directly assess the functions for γH2A-pSer, pSer +2 and pSer +3 contact residues in mediating spontaneous Brc1 nuclear focus formation. A salt bridge between Brc1 Arg704 and γH2A Glu131 mediates the pSer +2 interaction. An R704E mutant localized to nuclear foci, albeit at a reduced frequency compared with WT, suggesting that the Brc1–γH2A pSer +2 interaction is important, but not essential for γH2A binding in cells (Figure 3G). In contrast, despite their nuclear localization, Brc1 foci formation was ablated for the T672A (pSer binding), K710M (pSer binding) and R707E (pSer +3 carboxylate binding) mutants. Thus, structural and in vivo observations suggest that the critical determinants of Brc1–γH2A binding are phosphoserine binding and recognition of the pSer +3 position.
To assess the significance of the Brc1 BRCT5–BRCT6–γH2A interaction and Brc1 foci formation, we introduced the T672A and K710M mutations into the genomic brc1+ locus and challenged these mutants with chronic exposure to S-phase genotoxins. Both mutants were sensitive to CPT and the DNA alkylating agent methyl methanesulphonate (MMS) (Figure 3H), which also interferes with DNA replication, indicating that γH2A binding and foci formation are critical for Brc1 function. Interestingly, comparison to a brc1Δ mutant revealed that T672A and K710M are hypomorphic mutations (Figure 3H), indicating that Brc1 may retain submicroscopic interactions with DNA lesions when it cannot bind γH2A.
To evaluate the genetic epistasis relationships between Brc1 and γH2A, we introduced brc1Δ, brc1-T672A or brc1-K710M mutations into the hta-AQ background and tested genotoxin sensitivity relative to the parent strains. All of the brc1 mutants were sensitive to 4 mM HU, as was the hta-AQ parent, and this sensitivity was not further enhanced by combining the mutations (Supplementary Figure S2). These data suggest that in response to replication fork arrest caused by HU, the function of γH2A can be largely if not entirely explained by its recruitment of Brc1. The results were largely the same in cells treated with 2 μM CPT (Supplementary Figure S2). However, when tested in media containing 5 μM CPT, the brc1-T672A hta-AQ or brc1-K710M hta-AQ strains were more sensitive than the parent strains. There are several possible explanations for these data, with our favoured model being that γH2A has both Brc1 dependent and independent functions in recovery from collapsed replication forks.
Sculpting of BRCT pSer +3 binding pockets
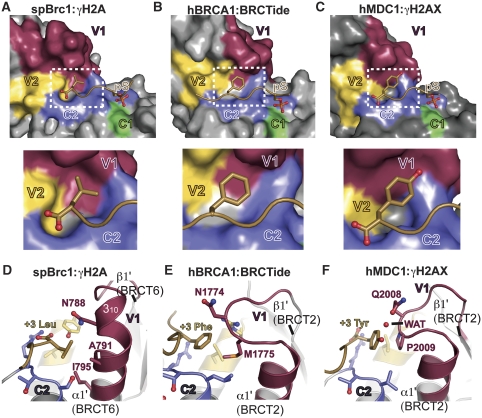
To understand Brc1 pSer +3 binding and specificity determinants, we compared the Brc1 pSer +3 (Leu) recognition pocket with structures of the MDC1–γH2AX complex (Stucki et al, 2005) and the BRCA1–BRCtide complex (Williams et al, 2004) (Figure 4). In all three structures, two variable surface regions (V1 and V2), and one constant motif (C2) converge to form contiguous pSer +3 binding pockets (Figure 4A–C). Whereas V2 and C2 are structurally similar for all interactions, the V1 regions (comprising the β1′–α1′ connecting loop and α1′ of the second BRCT of the BRCT pair) show marked structural heterogeneity, and are a primary pSer +3 pocket sculpting and specificity determinant.
Figure 4.
BRCT V-loop structural variability molds the pSer +3 binding pocket and defines peptide-binding specificity. (A) V-loop pSer +3 binding pocket molding in the Brc1–γH2A complex. A surface representation of Brc1 is displayed for BRCT phosphoprotein interactions with constant regions (C1—green and C2—blue), and variable BRCT peptide-binding loops (V1—maroon and V2—yellow). Inset: The Brc1 pSer +3 binding pocket tightly contours around the γH2A Leu132 carboxyl terminus. (B) hBRCA1–BRCTide complex constant and variable loops. Inset: BRCA1 phosphoprotein-binding pocket (RCSB 1T2V) is larger than Brc1, and binds Phe at γH2AX pSer +3. (C) hMDC1–γH2AX complex constant and variable loops. Inset: the mammalian γH2AX-binding pocket is larger than the Brc1 pocket, and binds Tyr, but not Leu or Phe at the γH2AX pSer +3 position. (D) Side view of the Brc1 pSer +3 binding pocket. V1 of Brc1 forms an extended loop and 310 helix that surrounds γH2A pSer +3 Leu on three sides. (E, F) The BRCA1 and hMDC1 V1 loops do not form 310 helical structures like Brc1, but adopt extended loops contributing two residues to γH2AX pSer +3 binding pocket.
In Brc1, the V1 α1′ is capped by a 310 helix. Three α1′ residues (Asn788, Ala791 and Ile795) form the base of the hydrophobic γH2A pSer +3 (Leu132) side-chain-binding pocket. Two residues (Tyr852 from V2 and Arg707 from C2) directly bind the γH2A C-terminal carboxylate. Carboxyl terminus positioning by Tyr852 and Arg707, and precise pocket contouring around γH2A Leu132 Cδ1 and Cδ2, suggests that Brc1 binding to larger aromatic side chains (Phe or Tyr) would require major distortions of the binding pocket. Thus, Brc1 seems evolved to specifically bind Leu at pSer +3. In contrast to Brc1, two residues in extended β1′–α1′ loops of BRCA1 and MDC1 make the pSer +3 side-chain contacts. These fundamentally different V1 pocket configurations form larger pockets adapted for binding Phe/Tyr (BRCA1) (Williams et al, 2001; Manke et al, 2003) or Tyr (MDC1) (Stucki et al, 2005), but not Leu at the pSer +3 position as seen here for Brc1 (Figure 4). Thus, our current structural data show how sequence and structural variability in the BRCT V1 loops gives rise to markedly different pSer +3 binding pocket topologies and binding specificities.
Spontaneous Brc1 foci do not colocalize with Crb2 or Rad22
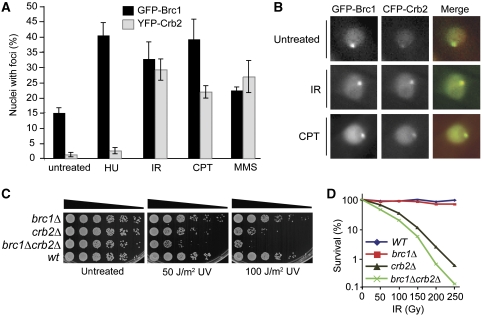
Crb2 and Brc1 both bind γH2A through C-terminal BRCT repeats; yet they have different functions: Crb2 is required for Chk1 activation and efficient DSB repair, whereas Brc1 is required for recovery from replication fork collapse. To investigate whether Crb2 and Brc1 localize into distinct structures, we first compared the foci patterns of GFP-Brc1 or YFP-Crb2 in unperturbed cells. This analysis revealed a large difference in the percentage of cells with GFP-Brc1 foci (∼15%) or YFP-Crb2 foci (∼2%) (Figure 5A). Analysis of a strain that expressed both fluorescently tagged proteins confirmed that most GFP-Brc1 foci do not overlap with CFP-Crb2 foci in untreated cells (Figure 5B). This analysis was extended by examining a strain that coexpressed GFP-Brc1 and Rad22-RFP, in which only ∼5% of the GFP-Brc1 foci overlapped with Rad22-RFP foci (Figure 6A). This pattern contrasts with the nearly total overlap of YFP-Crb2 and Rad22-CFP foci in IR-treated cells (Du et al, 2003).
Figure 5.
Specificity of DNA damage-induced foci formation by Brc1 and Crb2. (A) Foci formed by Crb2 and Brc1 increase in response to IR, CPT and MMS, whereas spontaneous and HU-induced Brc1 foci occur more frequently than Crb2. Cells expressing either GFP-Brc1 or 2YFP-Crb2 were grown for 17 h at 25°C in minimal medium before being split. Treatment was with 12 mM HU for 4 h, 30 μM CPT for 4 h, 0.03% MMS for 5 h or 120 Gy IR plus 2 h recovery. (B) Brc1 and Crb2 foci colocalize following exogenous DNA damage. Cells expressing GFP-Brc1 (green) and CFP-Crb2 (red) were grown for 17 h at 25°C in minimal medium and left untreated or subjected to treatment with 30 μM CPT for 5 h or 120 Gy IR plus 2 h recovery before imaging. The weak focus in the untreated CFP-Crb2 image is GFP bleed through into the red channel. (C) Synthetic additivity of brc1 and crb2 mutations. Five-fold serial dilutions of cells were exposed to the indicated doses of UV and incubated at 30°C for 2 days. (D) The brc1Δcrb2Δ double mutant shows enhanced sensitivity to IR.
Figure 6.
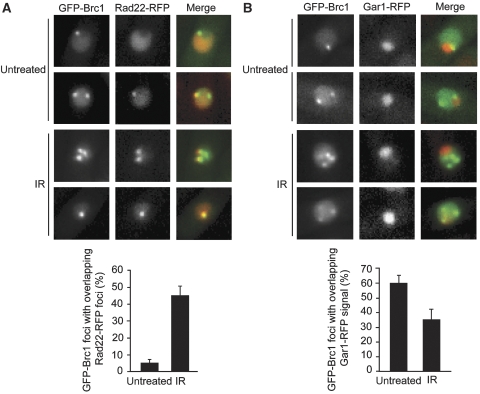
Brc1 foci localize to sites of genome instability. (A) IR-induced Brc1 foci frequently colocalize with the Rad22 DNA repair protein. Live cells expressing GFP-Brc1 (green) and Rad22-RFP (red) were photographed. Images were merged in the right panel. Samples were left untreated or exposed to 120 Gy IR followed by a 1 h recovery. Only ∼5% of spontaneous GFP-Brc1 and Rad22-RFP foci colocalize under normal conditions, whereas an increased overlap in signal is observed following induction of DSBs. For each sample, foci were scored in at least 300 cells and mean values of colocalized foci per field were plotted with error bars representing the standard deviation of the mean. (B) Live cells expressing GFP-Brc1 (green) and nucleolar Gar1-RFP (red) were photographed. Images were merged in the right panel. Samples were left untreated or exposed to 120 Gy IR followed by a 1 h recovery. Quantification is shown on the right. For each sample, foci were scored in at least 300 cells and the mean values of colocalized foci per field were plotted with error bars representing the standard deviation of the mean.
From these data, we hypothesized that most of the spontaneous Brc1 foci occur at sites of replication fork arrest, which do not activate the Crb2-dependent DNA damage checkpoint and are not substrates for Rad22-dependent HR repair. To test this hypothesis, we compared GFP-Brc1 and YFP-Crb2 foci in cells treated with HU, a compound that causes replication fork arrest by depleting dNTPs. Consistent with earlier results (Du et al, 2006), HU treatment did not significantly increase YFP-Crb2 foci (Figure 5A). In contrast, HU treatment caused a large increase in cells with GFP-Brc1 foci (Figure 5A). As observed for spontaneous GFP-Brc1 foci, HU-induced GFP-Brc1 foci were abolished in hta-AQ cells (Supplementary Figure S3).
Spontaneous Brc1 foci colocalize with rDNA
As HU-induced replication fork arrest triggers Brc1 foci formation, we inquired whether spontaneous Brc1 foci occur in genomic regions prone to fork arrest and genomic instability. The most obvious candidate was the ribosomal DNA (rDNA) repeats where polar replication fork barriers (RFBs) are required to maintain the integrity of the rDNA locus (Krings and Bastia, 2004; Sanchez-Gorostiaga et al, 2004). These repetitive rDNA arrays are located on the outer arms of chromosome III and protrude into the nucleolus (Uzawa and Yanagida, 1992). We therefore coexpressed GFP-Brc1 with RFP-tagged Gar1, a protein that associates the small nucleolar RNA. Approximately 60% of GFP-Brc1 foci were perinucleolar in untreated cells (Figure 6B), indicating that the majority of spontaneous Brc1 foci occur in the rDNA loci.
DNA damage-induced Brc1 foci colocalize with Rad22 and Crb2
As brc1Δ cells are sensitive to genotoxins such as CPT and MMS that cause DNA damage during S-phase, we hypothesized that exposure to these clastogens might induce Brc1 foci. Indeed, these agents caused a large increase in GFP-Brc1 foci (Figure 5A). IR had a similar effect, which was unexpected because brc1Δ cells are not IR sensitive (Figure 5D; Dovey and Russell, 2007). In all these treatments, GFP-Brc1 foci formation required γH2A (Supplementary Figure S3). These observations suggested that besides forming foci in response to replication arrest, Brc1 also forms foci at DSBs. Indeed, we found that IR caused a large increase in the percentage of GFP-Brc1 foci that overlap with Rad22-YFP (Figure 6A). Moreover, the percentage of GFP-Brc1 foci that were perinucleolar decreased in response to IR (Figure 6B), suggesting that Brc1 is recruited to IR-induced DSBs occurring throughout the genome.
CPT, MMS and IR induction of GFP-Brc1 foci were paralleled by YFP-Crb2 foci formation (Figure 5A). To test whether Brc1 and Crb2 colocalize at DSBs, cells coexpressing GFP-Brc1 and CFP-Crb2 were exposed to IR or CPT. Both treatments caused extensive colocalization of CFP-Crb2 and GFP-Brc1 foci (Figure 5B).
The IR induction of GFP-Brc1 foci suggested that Brc1 might have an unappreciated function in DSB repair. We examined this possibility by carrying out genetic epistasis analysis with brc1Δ and crb2Δ. As observed earlier, brc1Δ cells were not sensitive to IR or UV. However, the brc1Δ crb2Δ double mutant displayed additive sensitivity to these genotoxins (Figure 5C and D). This result shows that Brc1 and Crb2 share a DSB repair function, or a unique function of Brc1 can be bypassed through a Crb2-dependent mechanism.
Discussion
Brc1 is a critical S-phase γH2A effector
Although MDC1–γH2AX and Crb2–γH2A interactions are critical for responding to IR-induced DSBs (Stewart et al, 2003; Nakamura et al, 2004; Stucki et al, 2005; Du et al, 2006; Lou et al, 2006; Kilkenny et al, 2008), the effectors of γH2A signalling in response to replication stress or S-phase DNA damage have been elusive. Here, we identify Brc1 as a critical γH2A-docking factor required for recovery from replication-associated DNA damage in fission yeast. With a high-resolution visualization of the Brc1–γH2A binding interface, our results reveal how key conserved determinants of phosphate recognition combined with evolutionary sculpting of BRCT pSer +3 binding pockets achieve maximum binding affinity with minimalist four-residue phosphorylated target recognition sequences. Disruption of this interface ablates Brc1 foci formation and confers sensitivity to genotoxins that disturb DNA replication. Taken together, these data show that Tel1/Rad3-regulated binding of Brc1 to γH2A is a critical chromatin-specific response to DNA damage in S-phase.
Distinct types of γH2A-dependent Brc1 foci
ATM and ATR checkpoint kinases regulate cellular responses to many forms of genotoxic stress, which vary with respect to the damage type, chromosomal context and the replicative state of the genome. These kinases also have many substrates, few of which are understood in detail. Chromatin-specific responses largely flow through γH2A formation, raising the question of how these responses are tailored to specific circumstances of DNA damage. From the results presented here, it emerges that there are at least two γH2A-docking proteins in S. pombe, Crb2 and Brc1, which both use C-terminal BRCT domains to bind γH2A.
Notably, there are at least two types of Brc1 foci important for tailoring chromatin-specific responses to varying types of genotoxic stress: those that colocalize with Crb2 and Rad22, and those that do not. Both types require γH2A. The former class is induced by clastogens that cause DSBs such as CPT or IR, whereas the latter class arises spontaneously and largely in the rDNA locus, or in response to HU-induced fork arrest. As Rad22 foci increase and Rad22 is required for viability in brc1Δ cells, Brc1 foci likely mark stalled replication forks. Failure to stabilize these forks leads to their collapse and creates a requirement for Rad22-mediated HR repair. Once a replication fork collapses, as when CPT poisons topoisomerase I, both Brc1 and Crb2 are required for efficient recovery along with Rad22-mediated HR repair. These Brc1 and Crb2 functions are substantially enhanced through γH2A interactions.
What accounts for the differential localization of Brc1 and Crb2 when both proteins bind γH2A? Part of the explanation must involve Crb2's requirement for two types of histone modifications to form IR-induced foci (Du et al, 2004, 2006; Sanders et al, 2004). Proximal to its γH2A-binding BRCT domains, Crb2 has Tudor domains that bind Set9-methylated histone H4 (H4K20Me) (Sanders et al, 2004; Botuyan et al, 2006; Du et al, 2006). Mutations abolishing γH2A or H4K20Me binding are epistatic. As bulk H4K20Me is constitutive in fission yeast, DNA damage may alter chromatin structure and expose H4K20Me to control Crb2 localization (Du et al, 2006). So H4K20Me may be demethylated at stalled replication forks, or chromatin structure around stalled forks may not expose H4K20Me for productive engagement by the Crb2 Tudor domains. By relying on a histone code, checkpoint kinases can differentially regulate Crb2 and Brc1 localization by a shared histone modification.
γH2A's dual function in recruiting both Brc1 and Crb2 to chromatin at CPT-induced DSBs can explain why hta-AQ mutants are preferentially sensitive to genotoxins that collapse replication forks (Nakamura et al, 2004). As hta-AQ mutants are defective in recruiting both Brc1 and Crb2, while set9Δ mutants are defective only in recruiting Crb2, γH2A recruitment of two factors explains the heretofore perplexing observation that the CPT sensitivity of hta-AQ mutants exceeds that of set9Δ mutants (Du et al, 2006).
Dual modes of recruiting Brc1 to damaged DNA?
The Brc1 γH2A-binding mutants are hypomorphic, which implies that Brc1 has both γH2A-dependent and γH2A-independent mechanisms of interacting with DNA lesions. As this situation resembles that for Crb2, it may be a common feature of proteins that interact with γH2A (Du et al, 2006). The γH2A-independent functions of Brc1 may involve the BRCT1−4 domains binding to other DNA or chromatin-associated ligands, although these are unlikely mediated by direct Brc1–phosphoprotein interactions. Our results suggest three specific possibilities. First, Brc1 may interact with Cut5TopBP1, as this protein acts in DNA replication as well as DNA damage checkpoint signalling (Garcia et al, 2005), and it is required for the γH2A-independent mechanism of Crb2 function (Du et al, 2006). Second, Brc1 may bind ssDNA exposed at DNA lesions. Third, Brc1 may interact with nucleosomes at sites of DNA damage independent of post-translational modifications. However, expression of Brc1 BRCT5–BRCT6 domains is both necessary and sufficient for nuclear foci formation (Supplementary Figure S4), suggesting that binding to γH2A is critical for recruitment to sites of genome instability that can be detected microscopically.
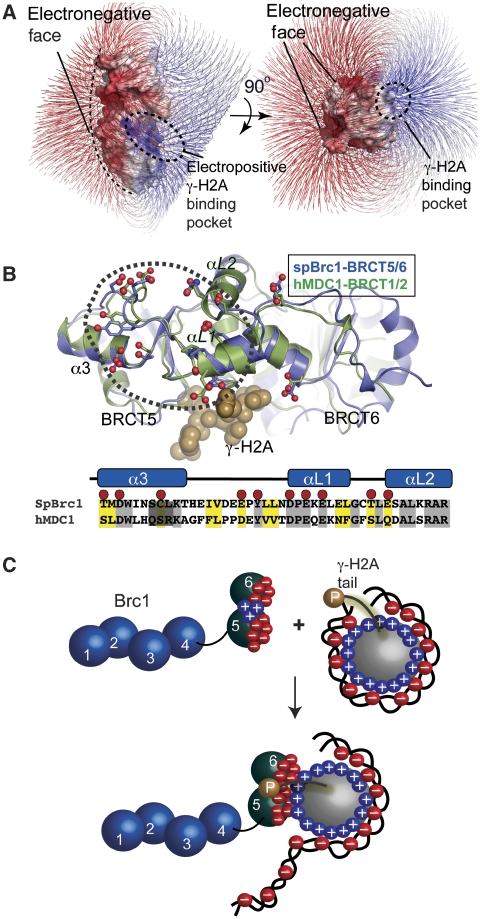
Interestingly, the Brc1 BRCT5–BRCT6 interdomain linker is formed by two helices (αL1 and αL2) closely resembling the MDC1 helix-loop-helix linker structure (Stucki et al, 2005). This was unexpected because BRCT linker structures are the most structurally variable regions in tandem-BRCT repeat domains (Glover et al, 2004). Moreover, a conserved contiguous surface patch comprising 12 acidic (Asp, Glu) and polar residues (Tyr, Thr and Ser) populate the BRCT linker and neighbouring helix α3 of Brc1–BRCT5, or the equivalent region of MDC1–BRCT1 (Figure 7A and B). Direct tethering of this electronegative BRCT surface (Figure 7A) adjacent to the histone octamer suggests that additional interactions with the positively charged histone-fold core particle surfaces may occur.
Figure 7.
A conserved BRCT electronegative surface on γH2A and γH2AX-interacting proteins. (A) An electrostatic surface potential (displayed with field lines) for Brc1 shows a highly polarized bar magnet surface with an electropositive γH2A interaction cleft (blue), and an extensive electronegative face (red). (B) A structural overlay of the mammalian γH2AX-binding MDC1 tandem-BRCT domains with the yeast Brc1–γH2A complex reveals an extensive conserved electronegative surface. (C) Schematic for a proposed two-site nucleosome interaction involving phosphorylation-dependant γH2A tail binding and nucleosome-Brc1 charge–charge interactions. Docking to the electronegative surface may augment γH2A histone tail docking through direct interactions with the positively charged DNA-binding nucleosome histone core fold.
We hypothesize that the conserved electronegative patch of MDC1 and Brc1 γH2A-interacting BRCT proteins seems adapted for recognition of histone core particle surfaces exposed on disruption of nucleosome architecture at DNA damage (Figure 7C). Charge–charge Brc1–histone interactions could facilitate γH2A histone exchange and/or localized remodelling of chromatin structure proximal to damaged DNA through an interaction predicted to compete with histone-DNA binding (Figure 7C). Consistent with this hypothesis, the Brc1 acidic surface exhibits the hallmark characteristics of a DNA mimic (Putnam et al, 1999; Putnam and Tainer, 2005).
Spontaneous Brc1 foci at rDNA loci
Brc1 foci are formed more frequently than Crb2 foci in untreated cells, suggesting an important function for Brc1 localization to chromatin during an unperturbed cell cycle. Spontaneous Brc1 foci often colocalize to the nucleolar periphery with the rDNA repeats (Figure 6B), so Brc1 may have specific functions in maintaining rDNA integrity. Eukaryotic cells have genetically programmed sites of replication pausing, the most abundant being polar RFBs in the rDNA loci (Krings and Bastia, 2004; Sanchez-Gorostiaga et al, 2004). RFBs may reduce replisome collisions with the transcription machinery (Takeuchi et al, 2003; Tsang and Carr, 2008), and defects in RFB function lead to genomic instability. Brc1 may therefore facilitate RFB formation or maintenance. An additional ∼40% of spontaneous Brc1 foci are not perinucleolar (Figure 6B). Non-rDNA foci may be sites of Brc1 accumulation serving general functions in promoting the stability of genomic regions prone to fork pausing, such as non-specific sites of endogenous DNA damage.
BRCT–phosphoprotein recognition in the DNA damage response
Tandem-BRCT domains are key molecular readout modules for the assembly of phosphorylation-regulated DNA damage response effector and signalling complexes (Manke et al, 2003, Glover et al, 2004). In mammalian cells, ATM and ATR phosphorylate over 700 proteins in response to genotoxic insult (Matsuoka et al, 2007). With over 2400 curated BRCT domain sequences in the PFAM database (http://pfam.sanger.ac.uk/family?acc=PF00533), it is likely that many other critical phosphoprotein–BRCT interaction combinations have evolved with unique interaction specificities. The molding of BRCT phosphoprotein-binding specificity pockets through evolution of variable BRCT insertion loops, as observed here for the Brc1–γH2A complex, provides a probable general mechanism for generating the variability required for assembly of specific and distinct BRCT–phosphoprotein complexes in vivo.
Materials and methods
Strains, media and genetic analysis
Strain genotypes are listed in Supplementary Table S1. Growth media and methods for S. pombe were performed as described (Moreno et al, 1991). Ectopic expression of pRep41-N-GFP-brc1+ for microscopy was under control of the thiamine-repressible nmt41 promoter. Induction of plasmid expression in selective medium lacking thiamine was for 18–21 h. Spot assays were performed by plating 5- or 10-fold serial dilutions of exponentially growing cells onto YES in the absence or presence of the indicated DNA-damaging agents. Plates were incubated at 30°C and scanned after 2–3 days of growth. The survival assay was performed by counting cells plated in triplicate onto rich medium after exposure to the indicated IR doses. Normalization was to the untreated sample.
Microscopy
Cells were photographed using a Nikon Eclipse E800 microscope equipped with a Photometrics Quantix CCD camera. Cells were treated with 12 mM HU (Sigma), 30 μM CPT (Sigma) or 0.03% MMS (Sigma) and grown for 4–5 h at 30°C or 5–6 h at 25°C. For IR treatment, mid-log phase cells were exposed to the indicated dose and allowed time for recovery at 30°C for 1–4 h. Quantitation was performed by scoring 500 or more nuclei from at least two independent experiments. For DAPI-staining experiments, cells were fixed for 10 min at RT in 70% EtOH and washed with H2O before treatment with 5 μl of DAPI (500 μg/ml).
Protein expression, purification and crystallization
Brc1–BRCT5–BRCT6 (residues 659–878) was expressed in Escherichia coli Rosetta2 (DE3) (Novagen) at 20°C in Terrific Broth from pET15b as N-terminal 6-histidine tagged protein. After Ni-NTA affinity chromatography and elution with an on-resin thrombin cleavage that removed the 6his-tag, BRCT5–BRCT6 was purified with Superdex 75 gel filtration chromatography (GE-Amersham) in BRCT buffer (200 mM NaCl, 20 mM Tris–HCl pH 7.5, 0.1% β-mercaptoethanol). The T672A variant was introduced using the Quikchange (Stratagene).
Crystallization data collection
Crystals of selenomethionine-derivitized BRCT5–BRCT6 were grown at 22°C by mixing 2 μl of 15 mg/ml BRCT5–BRCT6 in BRCT buffer with 2 μl of well solution 1 (20–22% w/v PEG3350 100 mM NaCl, 20 mM ammonium phosphate, 0.1 M sodium citrate pH 5.0). Single crystals were obtained with streak seeding from clustered monoclinic rods that grew within 1–2 days. Native orthorhombic crystals of apo-BRCT5–BRCT6 used for refinement were grown using well solution 2 (20–22% w/v PEG 3350, 100 mM NaCl, 100 mM K2HPO4/KH2PO4 pH 5.0).
All peptides used in this study were synthesized at the Scripps Research Institute core facility. The BRCT5–BRCT6–γH2A complex was preformed before crystallization by mixing BRCT5–BRCT6 at 10 mg/ml in BRCT buffer with γH2A phosphopeptide (Ac-KPpSQEL-COO-) at a 1.5:1 phosphopeptide:protein ratio. In all, 30 × 30 × 500 μM orthorhombic needles of the complex grew at 22°C within 2–3 days after mixing 1 μl of complex with 1 μl well solution 3 (17–20% PEG3350, 200 mM ammonium chloride). For data collection, the crystals were flash frozen in liquid nitrogen after transfer to cryo-protectant solutions comprised of the appropriate mother liquor supplemented with 26% ethylene glycol.
X-ray diffraction data collection and processing
All single crystal X-ray diffraction data were collected at the SIBYLS beamline (BL12.3.1) at the Advanced Light Source, Lawrence Berkeley National Laboratory. X-ray data reduction and scaling was performed with the HKL2000 suite (Otwinowski and Minor, 1997).
X-ray structure determination and refinement
All four selenium positions in the crystallographic asymmetric unit of the monoclinic apo-BRCT5–BRCT6 space group (2 selenomethionines per 220 residue BRCT5–BRCT6 polypeptide) were located and refined with SOLVE (Terwilliger and Berendzen, 1999) and a 3-wavelength Se-MET monoclinic (P21) MAD data set (Supplementary Table S2). Crystallographic phases and experimental electron maps were improved by density modification in RESOLVE (Terwilliger, 2000). Automated model building in RESOLVE and manual fitting in O (v10.0) (Jones et al, 1991) produced a model that was preliminarily refined to 1.8 Å using REFMAC (Murshudov et al, 1997; Winn et al, 2001) and the MAD data set remote wavelength (λ3). This model was used for molecular replacement search with MOLREP (Vagin and Teplyakov, 2000) to determine the orientation of the structure in the related native P212121 crystal form. Subsequent cycles of positional and anisotropic temperature factor refinement against the 1.55 Å orthorhombic data set.
Phases for the BRCT5–BRCT6–γH2A complex were obtained by molecular replacement in MOLREP and the unbound BRCT5–BRCT6 structure. The BRCT5–BRCT6–γH2A complex crystal form contains one complex in the asymmetric unit. The phosphorylated γH2A peptide chain was traced into unambiguous sigma-A weighted 1.45 Å Fo–Fc and 2Fo–Fc difference electron density maps, and the model was refined as for the apo-BRCT5–BRCT6 structure. The refined models of apo-BRCT5–BRCT6 at 1.55 Å (R/Rfree=17.9/22.5) and the BRCT5–BRCT6–γH2A complex at 1.45 Å (R/Rfree=13.1/18.8) exhibit excellent geometric parameters (Supplementary Table S2).
Accession codes
Coordinates and structure factors for BRCT5–BRCT6 (RCSB accession code, 3L40) and the BRCT5–BRCT6–γH2A complex (RCSB accession code, 3L41) are in the Protein Data Bank.
Peptide binding studies
FP equilibrium binding measurements with amino-terminal fluorescein (FITC)-conjugated peptides were taken on a Fluoromax-3 (Horiba/Yobin-Yvon) with constant wavelength excitation at 490 nm and emission monitored at 520 nM. Titrations used a constant concentration (10 nM) of phosphorylated (FITC-KRTGKPpSQEL) or unphosphorylated (FITC-KRTGKPSQEL) H2A peptide, and increasing amounts (25 nM–80 μM) of WT-BRCT5–BRCT6 or T672A–BRCT5–BRCT6. A measure of 400 μl binding reactions were equilibrated for 10 min at 20°C in binding buffer (40 mM NaCl, 20 mM Tris–HCl pH 7.5, 5% glycerol, 0.1 mg/ml bovine serum albumin, 0.1% β-mercaptoethanol) before FP measurement. Equilibrium binding constants were calculated with KaleidaGraph (v4.03).
Small angle X-ray scattering
SAXS scattering data for samples of apo- for apo-BRCT5–BRCT6 (at 2.5, 5.0 and 10.0 mg/ml), and for the BRCT5–BRCT6–γH2A complex (at 5.0 and 10.0 mg/ml) in the momentum transfer range 0.015 < S <0.388 Å−1 was collected at beamline 12.3.1 (SIBYLS) at the Advanced Light Source.
SAXS data analysis
No evidence of aggregation for samples was evident as judged by linearity of Guinier plots. Radius of gyration (Rg) and pair distance distribution functions P(r) were evaluated using GNOM (Svergun, 1992). Theoretical scattering was generated with CRYSOL (Svergun et al, 1995). Ab initio solution envelopes were obtained by averaging 10 independent dummy residue models from reciprocal space minimization in GASBOR (Svergun et al, 2001).
Supplementary Material
Acknowledgments
We thank Oliver Limbo for plasmid construction, Charly Chahwan and Eishi Noguchi for helpful suggestions, Li-Lin Du for strains, and the Russell and Tainer laboratories members plus the Scripps Cell Cycle Group for discussions. RSW is supported by postdoctoral fellowships from the Canadian Institutes of Health Research (CIHR), the Alberta Heritage Foundation for Medical Research (AHFMR), and the Skaggs Institute for Chemical Biology. Work on Brc1 in the laboratories is supported in part by National Cancer Institute grants CA117638, GM59447 and CA77325. Support for SAXS at the SIBYLS beamline of the Advanced Light Source came from the program Integrated Diffraction Analysis Technologies (IDAT) under DOE contract DE-AC02-05CH11231.
Footnotes
The authors declare that they have no conflict of interest.
References
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chin JK, Bashkirov VI, Heyer WD, Romesberg FE (2006) Esc4/Rtt107 and the control of recombination during replication. DNA Repair (Amst) 5: 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey CL, Russell P (2007) Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe. Genetics 177: 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Moser BA, Russell P (2004) Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J Biol Chem 279: 38409–38414 [DOI] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Moser BA, Russell P (2003) Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol Cell Biol 23: 6150–6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P (2006) Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP (2005) Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434: 605–611 [DOI] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM (2005) Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS (2004) Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci 29: 579–585 [DOI] [PubMed] [Google Scholar]
- Gong Z, Cho YW, Kim JE, Ge K, Chen J (2009) Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem 284: 7284–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL II, Tsutakawa SE, Jenney FE Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nature Methods 6: 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH (2008) Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev 22: 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D (2004) swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci USA 101: 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J (2006) MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21: 187–200 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Muñoz IM, Rouse J (2009) Control of histone methylation and genome stability by PTIP. EMBO Rep 10: 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P (2004) Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24: 6215–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391: 407–410 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Carter CW, Sweets RM (eds) pp 307–326. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Pommier Y (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nature Rev Cancer 6: 789–802 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA (1999) Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J Mol Biol 287: 331–346 [DOI] [PubMed] [Google Scholar]
- Putnam CD, Tainer JA (2005) Protein mimicry of DNA and pathway regulation. DNA Repair (Amst) 4: 1410–1420 [DOI] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Rogakou EP, Orr AH, Lowndes NF, Bonner WM (2003) Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep 4: 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA M II, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW (2006) Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol Biol Cell 17: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Zaidi IW, Vaisica JA, Peter M, Brown GW (2008) Regulation of rtt107 recruitment to stalled DNA replication forks by the cullin rtt101 and the rtt109 acetyltransferase. Mol Biol Cell 19: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B (2008) Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J 27: 1378–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J (2004) Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J 23: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gorostiaga A, Lopez-Estrano C, Krimer DB, Schvartzman JB, Hernandez P (2004) Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol 24: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Sheedy DM, Dimitrova D, Rankin JK, Bass KL, Lee KM, Tapia-Alveal C, Harvey SH, Murray JM, O'Connell MJ (2005) Brc1-mediated DNA repair and damage tolerance. Genetics 171: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP (2006) gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 5: 534–543 [DOI] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25: 495–503 [Google Scholar]
- Svergun DI, Barberato C, Koch MHJ (1995) Crysol—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr 28: 768–773 [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang E, Carr AM (2008) Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair (Amst) 7: 1613–1623 [DOI] [PubMed] [Google Scholar]
- Uzawa S, Yanagida M (1992) Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J Cell Sci 101: 267–275 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (2000) An approach to multi-copy search in molecular replacement. Acta Crystallogr D Biol Crystallogr 56: 1622–1624 [DOI] [PubMed] [Google Scholar]
- Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ (1999) Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell 10: 2905–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Green R, Glover JN (2001) Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nature Struct Biol 8: 838–842 [DOI] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN (2004) Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nature Struct Mol Biol 11: 519–525 [DOI] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA (2007) Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol 85: 509–520 [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57: 122–133 [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P (2005) ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Maharaj AS, Connelly JJ, Jockusch RA, Sternglanz R (2006) Rtt107/Esc4 binds silent chromatin and DNA repair proteins using different BRCT motifs. BMC Mol Biol 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.