Abstract
Bioinformatic analysis classifies the human protein encoded by immature colon carcinoma transcript-1 (ICT1) as one of a family of four putative mitochondrial translation release factors. However, this has not been supported by any experimental evidence. As only a single member of this family, mtRF1a, is required to terminate the synthesis of all 13 mitochondrially encoded polypeptides, the true physiological function of ICT1 was unclear. Here, we report that ICT1 is an essential mitochondrial protein, but unlike the other family members that are matrix-soluble, ICT1 has become an integral component of the human mitoribosome. Release-factor assays show that although ICT1 has retained its ribosome-dependent PTH activity, this is codon-independent; consistent with its loss of both domains that promote codon recognition in class-I release factors. Mutation of the GGQ domain common to ribosome-dependent PTHs causes a loss of activity in vitro and, crucially, a loss of cell viability, in vivo. We suggest that ICT1 may be essential for hydrolysis of prematurely terminated peptidyl-tRNA moieties in stalled mitoribosomes.
Keywords: mitoribosomes, peptidyl-tRNA hydrolase, translation release factor
Introduction
Human mitochondria are ubiquitous organelles that are essential for cell viability. Among many crucial functions, mitochondria couple the process of oxidative phosphorylation, where cellular respiration is harnessed to generate ATP. This demanding mechanism requires the synthesis and import of many nucleus-encoded proteins as well as the intramitochondrial production of 13 polypeptides that are encoded by the mitochondrial genome, mtDNA. Consequently, correct maintenance and expression of mtDNA is essential for cell viability. Although we are gradually learning more about the principal factors and mechanisms underlying the maintenance and transcription of mtDNA, the process of mitochondrial translation has proven extremely difficult to be characterised in detail. This is in part because isolated mitochondria lose their capacity to synthesise proteins after solubilisation of the inner membrane, consistent with loss of crucial membrane-associated factors. Furthermore, despite impressive efforts to reconstitute in vitro mitochondrial translation systems (Yasukawa et al, 2001; Takemoto et al, 2009), results have been limited. Invaluable contributions from the laboratories of Spremulli, Watanabe and O'Brien have identified or characterised constituents of both the bovine small 28S (mt-SSU) (Suzuki et al, 2001) and large 39S (mt-LSU) (Koc et al, 2001) mitoribosomal subunits and many proteins involved in translational initiation and elongation (Spremulli et al, 2004), but important factors remain to be unearthed.
To improve our understanding of this process, we have started to identify other principal components in mitochondrial protein synthesis. We have focused our efforts on a previous report where tagged mitochondrial ribosome recycling factor (mtRRF) was shown to immunoprecipitate mitoribosomes and associated proteins from mitochondrial lysates (Rorbach et al, 2008). Proteomic analysis uncovered a large number (73) of mitoribosomal proteins (MRPs). In addition, another 94 polypeptides were identified, a number of which have been tentatively identified as nucleoid proteins. Immature colon carcinoma transcript-1 (ICT1) was consistently associated with immunoprecipitation (IP), but it was neither known to be mitochondrial nor have an experimentally verified function, although it is predicted to be a member of the prokaryote/mitochondrial release factor family (uniprot Q14197). This family is intriguing. We have recently been able to show that of the four family members, only mtRF1a is necessary and sufficient to terminate the translation of all 13 mitochondrially encoded polypeptides (Soleimanpour-Lichaei et al, 2007; Temperley et al, 2010). What, therefore, is the function, if any, of the three remaining family members? We report here that a second member of this family, ICT1, is a component of the 39S mt-LSU. It has retained its ribosome-dependent peptidyl-tRNA hydrolase (PTH) activity that is essential for cell viability. Furthermore, this ribosome-dependent PTH activity is codon-non-specific. We speculate that this ribosome-associated activity may be involved in the hydrolysis of peptidyl-tRNAs that have been prematurely terminated and thus in the recycling of stalled mitoribosomes.
Results
ICT1 is an essential mitochondrial protein
To determine the subcellular location of ICT1, western blot analysis was performed using cell lysate and enriched mitochondria. As shown in Figure 1A, a 3- to 5-fold enrichment was seen for the mitochondrial matrix protein mtRF1a along with concomitant increase in ICT1. Crucially, ICT1 was resistant to addition of proteinase-K to intact mitochondria, but was lost on solubilisation of the organelle. To assess whether ICT1 is processed on import into mitochondria, full-length ICT1 (FL) or a truncated form lacking the N-terminal 29 residues (Δ29) were prepared. Both proteins were expressed in Escherichia coli as N-terminal glutathione-S-transferase (GST)-fusion proteins before cleavage and purification as detailed under Materials and methods. Migration of endogenous ICT1 in comparison with recombinant proteins (FL/Δ29) is consistent with cleavage on mitochondrial import, with the loss of ∼30 residues.
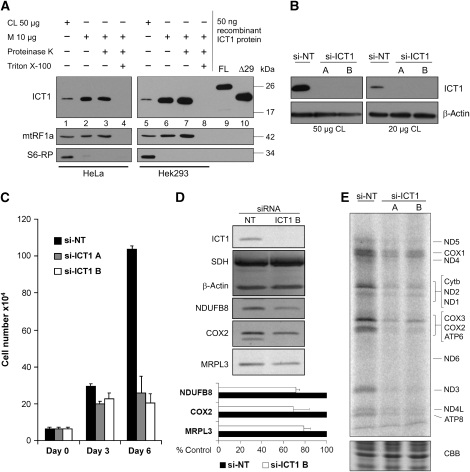
Figure 1.
ICT1 is an essential protein necessary for mitochondrial protein synthesis. (A) Human ICT1 is a mitochondrial protein. Cell lysate (CL 50 μg, lanes 1, 5) or mitochondria (10 μg, lanes 2–4, 6–8) were isolated from HeLa and HEK293T cells and subjected to western blot analysis either immediately (lanes 1, 2; 5, 6) or after treatment with proteinase-K (lanes 3, 7). Mitochondria were lysed with Triton X-100 to confirm the sensitivity of marker proteins to the protease (lanes 4, 8). Mitochondrial release factor-1a (mtRF1a) was used as a mitochondrial matrix marker and ribosomal protein-S6 (S6-RP) as a cytosolic marker. Purified FL (lane 9) and an ICT1 deleted of N-terminal 29 residues (Δ29, lane 10) are shown in comparison with the endogenous protein. (B–E) Depletion of ICT1 inhibits cell growth, impairs mitochondrial protein synthesis and decreases mitochondrial respiratory chain complexes. HeLa cells in standard glucose media were transfected with either of two siRNAs directed to ICT1 transcript (si-ICT1A or B) or a non-targeting control (si-NT), and cell numbers were counted at 3-day intervals. Standard errors were derived from three independent experiments (C). Cell lysates were isolated from non-targeting and ICT1-depleted cells (3 days) and subjected to western blotting for ICT1 (B) and various markers (D). The relative levels for the MRP MRPL3 and respiratory components NDUFB8 or COX2 were quantified (lower panel) with standard errors derived from three independent repeats. (E) After 3-day siRNA-mediated depletion, cells were subjected to metabolic labelling of mitochondrial proteins for 15 min after inhibition of cytosolic protein synthesis. Aliquots (50 μg) were separated by 15% SDS–PAGE and exposed to a PhosphorImager. Proteins are identified by comparison against those reported by Chomyn (1996). A section of the gel stained with Coomassie blue (CBB) following exposure is shown to indicate even loading of cell lysate.
To assess whether ICT1 serves an essential function in mitochondria, siRNAs were designed to target the transcript. Two siRNAs were highly efficient in depleting ICT1 from cells (Figure 1B), causing a morphological alteration and a reduction in cell number even when grown on standard, mainly glycolytic media (Figure 1C). To confirm that effects were specific and not off-target, cells lacking mtDNA (rho0 cells) were also transfected with ICT1-specific siRNA and control siRNAs. The rationale being, as rho0 cells lack mitochondrial gene expression and are still able to grow on glucose media supplemented with uridine and pyruvate, depletion of any protein involved solely in mitochondrial gene expression should have minimal effect. Accordingly, siRNA-mediated depletion of β-actin or HSP70 severely compromised the growth of rho0 cells, whereas growth was unaffected by depletion of ICT1 or the mitochondrial translation factor mtEF-Tu (Supplementary Figure S1). These data are strongly indicative that ICT1 functions in mitochondrial gene expression.
After only 3 days of ICT1 depletion, a decrease in the markers of the highly stable mitochondrial respiratory chain complex-I (NDUFB8) and IV (COX2) was confirmed by western blotting of HeLa lysates (Figure 1D). Interestingly, a similar decrease was also noted for the MRP MRPL3, indicating that levels of mitoribosome and possibly rates of mitochondrial protein synthesis may be compromised. Therefore, de novo synthesis of mitochondrial translation products was assessed by in vivo metabolic labelling. Figure 1E shows that in the ICT1-depleted cells 35S-met incorporation is indeed reduced.
ICT1 is a member of the large mitoribosomal subunit
Why does loss of ICT1 lead to reduction in mitochondrial protein synthesis? To investigate this question and to determine what components of the mitochondrial matrix associated with ICT1, a FLAG-tagged ICT1 was inducibly expressed in human HEK293T cells, facilitating IP. As shown in Figure 2A, silver staining uncovered a large number of proteins, similar to a previous profile where tagged mtRRF had immunoprecipitated the mitoribosome and associated proteins (Rorbach et al, 2008). Western blot analysis (Figure 2B) confirmed the presence of numerous MRPs and the predicted trace amounts of mtRRF. From proteomic data of the complete eluate, more than 200 mitochondrial proteins were identified (Supplementary Table S1), the MRPs being the most abundant (Supplementary Table S2); consistent with ICT1 interacting with entire mitoribosomes. As a second method to determine whether ICT1 was a component of the mitoribosome, complexes from untransfected cells were separated by isokinetic sucrose density gradients and fractions were subjected to blotting (Figure 2C). ICT1 co-sediments with MRPL3 and MRPL12, both components of the 39S mt-LSU. As has been found in other reports (Nolden et al, 2005; Williams et al, 2005), the mitochondrial monosome is not easily identified in cell or mitochondrial lysates by western blot after density-gradient centrifugation. This was also evidenced here by the lack of detectable small subunit marker DAP3 in the more dense fractions (Figure 2C, fractions 7–10). Therefore, to resolve this issue, we pre-concentrated samples by first immunoprecipitating mt-LSU and monosomes from mitochondrial lysates using FLAG-ICT1 and subjected the entire eluate to an identical gradient centrifugation. Fractions were then assessed by silver staining and western blotting. A similar distribution profile is still seen for ICT1 and MRPL3 (excluding free ICT1 caused by overexpression) (Figure 2D). Crucially, however, DAP3 is now visible in fractions 7–9, defining the monosomal fractions. Therefore, ICT1 behaves as an integral member of the 39S mt-LSU and a component of the intact 55S monosome.
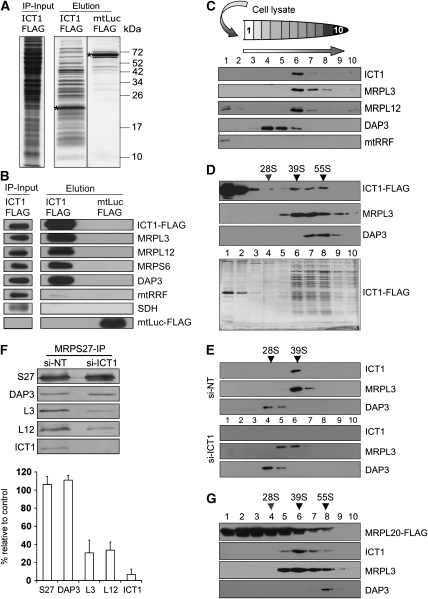
Figure 2.
ICT1 is an integral component of the mitoribosome. (A, B) FLAG-tagged ICT1 immunoprecipitates mitoribosomes. HEK293T cells expressing FLAG-tagged ICT1 or mitochondrially localised luciferase (mtLuc-FLAG) were induced for 3 days; mitochondria were isolated, lysed and subjected to IP as detailed. The eluate and mitochondrial lysate before IP (IP-input) were separated by 15% SDS–PAGE and visualised by silver staining. * designates the FLAG protein. (B) Aliquots of the eluates were also subjected to western blot analysis with the indicated antibodies: MRPL3, MRPL12, MRPS6 and DAP3 as mitoribosomal markers; mtRRF, mitoribosome recycling factor; SDH, 70-kDa component of complex-II. (C) ICT1 co-sediments with the large mitoribosomal subunit. HeLa cells were lysed (600 μg), separated through a 10–30% sucrose gradient and fractionated as detailed (HeLa and HEK293T lysates gave identical separations). Components of the 39S mt-LSU (MRPL3, MRPL12) and 28S mt-SSU (DAP3) mitoribosomal subunits were visualised by western blotting. On immediate lysis, mtRRF is used as a matrix-soluble marker. (D) ICT1 also co-sediments with the intact monosome. Mitochondria (3 mg) of ICT1-FLAG-expressing HEK293T cells were subjected to FLAG IP; the entire eluate was separated by isokinetic density gradients and fractions were blotted as detailed above or visualised by silver staining (lower panel). Mitochondrial SSU (DAP3) and mt-LSU (MRPL3) MRPs are visualised. The approximate indicators for 28S mt-SSU, 39S mt-LSU and 55S monosome are shown and were determined as described under Materials and methods. (E) ICT1 is an integral member of 39S mt-LSU. Cell lysates (600 μg) from ICT1-depleted (si-ICT1B) or non-targeted control cells (si-NT) were separated by isokinetic gradients and proteins were visualised in the fractions by western blotting as described. Sedimentation markers were identified as above. (F) Loss of ICT1 causes depletion of the monosome. Cells expressing MRPS27-FLAG were treated with si-NT or si-ICT1B, after which IP was performed. To assess monosome formation, levels of MRPL3 and MRPL12 were quantified by western blotting of three individual experiments (right panel; MRPL3 P=0.001, MRPL12 P<0.001, MRPS27 P=0.3). (G) ICT1's association with mitoribosomes is not FLAG-dependent. Mitochondria from cells expressing MRPL20-FLAG were subjected to FLAG IP and the eluate was analysed by western blotting after isokinetic density gradients as described in panel D.
Further support for ICT1 being an integral component of the 39S mt-LSU, was obtained from ICT1-depleted HeLa cells. Lysate was subjected to a similar isokinetic gradient, showing that the mt-LSU marker MRPL3, although present, was shifted into less dense fractions as compared with that in cells treated with non-targeting control siRNA (si-NT; Figure 2E). By contrast, the profile of the 28S mt-SSU protein DAP3 was unchanged, implying that only assembly of the mt-LSU and not the mt-SSU is affected on ICT1 depletion.
To confirm that the decrease in mt-LSU assembly observed in ICT1-depleted cells caused a concomitant decrease in the level of intact monosome, we used a cell line that expresses a FLAG-tagged component of the mt-SSU, MRPS27, that was able to IP both mt-SSU and entire monosome. After 3 days of ICT1 depletion and MRPS27-FLAG induction, FLAG-IP was performed and eluates were blotted (Figure 2F). Levels of DAP3 were unaffected by ICT1 depletion. However, anti-MRPL3 and MRPL12 antibodies showed a 60% reduction of each of these proteins in the ICT1-depleted cells, consistent with a decrease in the monosome formation, presumably due to decreased mt-LSU assembly. This was also consistent with the decreased 35S metabolic labelling of mitochondrial proteins as shown in Figure 1E. Finally, to show that mitoribosomal association of ICT1 was not simply mediated by the FLAG tag, similar isokinetic gradients were used to separate the immunoprecipitated eluate from cells expressing MRPL20-FLAG. When correctly assembled, this FLAG-tagged protein was able to IP mt-LSU, assembly intermediates and intact monosome. As show in Figure 2G, ICT1 was clearly associated with mt-LSU and monosome. These data confirm that ICT1 is an important component of the complete monosome.
ICT1 is a ribosome-bound, codon-independent PTH
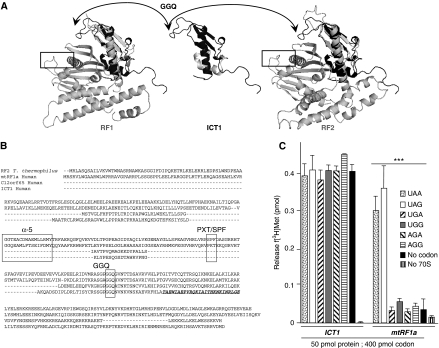
ICT1 is an integral component of the mitoribosome, but on the basis of homology it is predicted to be a ribosome-dependent PTH/translation release factor. This is surprising, as to our knowledge, no other PTH has been shown to be an integral ribosomal component. There are now four proteins classified as members of the mitochondrial release factor family, namely mtRF1a, mtRF1, ICT1 and another uncharacterised protein C12orf65 (Figure 3B). Curiously, both ICT1 and C12orf 65 have lost the two domains involved in codon recognition (α5 and PXT/SPF domain), potentially resulting in the dangerous situation of release factors that lack codon specificity. However, none of our previous IPs identified C12orf65 as a co-precipitant with mitoribosome components (data not shown). To determine whether ICT1 could promote hydrolysis of the ester bond between the growing peptide and the P-site tRNA, we used a well-established assay using isolated E. coli ribosomes, tritiated fmet-tRNAMet, synthetic codons and purified ICT1 (Caskey et al, 1971; Tate and Caskey, 1990; Soleimanpour-Lichaei et al, 2007). In comparison to the exquisite codon selectivity of mtRF1a, ICT1 promiscuously promoted the release of formylmethionine from its P-site tRNA irrespective of the codon sequence used to programme the assay and indeed even in the absence of codons in the A-site (Figure 3C). To determine whether ICT1 possessed a direct ribosome-independent PTH activity, or whether it functioned specifically to promote ribosome-dependent hydrolysis, purified ICT1 was incubated with f[3H]met tRNAmet in the absence of ribosomes before extraction and estimation of standard release factor activity. No significant increase in counts was noted over background, confirming that no significant hydrolysis of the substrate occurred in the absence of the ribosome (Figure 3C).
Figure 3.
ICT1 is a codon-independent PTH. (A) Structural comparisons between ICT1 and members of RF1 or RF2. Only limited structure is available for murine ICT1 (centre, PDB 1J26 unpublished structural genomics output), but this can be superimposed using Topmatch (Sippl and Wiederstein, 2008) onto either RF family (left T. maritima PDB 1RQ0 (Shin et al, 2004); right T. thermophilus PDB 2IHR (Zoldak et al, 2007)), making it unclear as to its evolutionary origin. The arrows show the ICT 1GGQ motif and the boxes enclose the codon recognition domains (Ito et al, 2000; Laurberg et al, 2008). (B) Primary sequence comparisons of ICT1 and translation release factor members. Representatives of the release factor families are shown; RF1 (human mtRF1a), RF2 (T. thermophilus) aligned with human sequences for ICT1 and a fourth member of the mitochondrial release factor family, C12orf65. Three regions are highlighted; the GGQ motif conferring PTH activity, and α-5/tripeptide domains that are implicated in codon recognition. The latter two domains are absent in ICT1 and C12orf65. There are two other families of PTHs, PTH1 and PTH2 (reviewed by Das and Varshney, 2006), which are predicted to be represented in the human mitochondrion by PTRH1 and PTRH2 (uniprot Q86Y79 and Q9Y3E5, respectively). These protein families function independently of the ribosome and on the basis of comparison of their three-dimensional structures (data not shown), are not homologous to the ribosome-dependent PTH family that contains ICT1. (C) ICT1 has codon-independent and ribosome-dependent translation release factor activity. E. coli ribosomes were programmed with tritiated P-site fmet-tRNAMet and A-site codons as indicated (detailed under Materials and methods). Activity was measured as hydrolysis of f[3H]met from its cognate tRNAMet and is represented as pmol f[3H]met released. Non-limiting amounts of protein (50 pmol) and RNA triplet (400 pmol) were used in the assay where required, with mtRF1a as a positive control. Activities are also evident where ribosomes were programmed with no codon or were absent from the assay, entirely. Reactions lacking 70S ribosomes contained the UAA triplet. Standard errors were calculated from a minimum of eight repeats; ***P<0.001.
A mutation of the GGQ domain of ICT1 causes loss of cell viability
ICT1 is an essential MRP with a ribosome-dependent yet codon-independent PTH activity, in vitro. Is it possible that ICT1 needs to maintain this activity when it is assembled into the mitoribosome, in vivo?
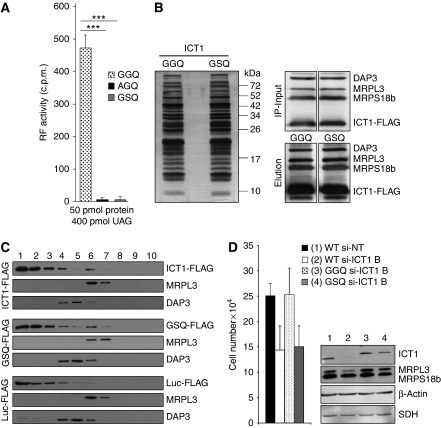
PTH activity of release factors is mediated by the domain containing the tripeptide motif GGQ (Frolova et al, 1999; Figure 3A and B). It has been shown that mutations in either of the two glycine residues completely abolishes in vitro PTH activity while retaining the structural integrity of the protein (Frolova et al, 1999). To determine whether the GGQ domain is critical for ICT1's function, we reproduced two of these GGQ mutants by site-directed mutagenesis. First, recombinant ICT1GSQ and ICT1AGQ were purified, monodispersion was confirmed by dynamic light scattering, and then they were subjected to the release assay described above (Figure 4A). Each retained ∼1% of peptide-release activity (GSQ 1.1%, AGQ 0.8% residual activity). Having confirmed the requirement of the GGQ domain for the PTH activity of ICT1, it was then possible to assess the importance of the PTH activity in vivo. If PTH activity is crucial then replacement of wild-type ICT1 with ICT1GSQ would be predicted to compromise mitochondrial gene expression, leading to reduced growth on galactose. Consequently, an HEK293T cell line was engineered to express a FLAG-tagged version of ICT1GSQ and compared with the wild-type ICT1-FLAG expressor. On induction, the ICT1 mutant was incorporated into mitoribosomes, which were assembled at levels similar to that in the wild-type transfected control (Figure 4B and C), but induction resulted in a substantial overexpression of ICT1GSQ such that the majority of the protein remained free and not mitoribosome-bound (Figure 4C). A similar overexpression was also noted on induction of wild-type ICT1-FLAG (Figure 4C, upper panels), which in itself produced a mild growth phenotype. To reduce the levels of the overexpressed protein to that of the endogenous untransfected controls and concomitantly deplete the endogenous ICT1, serial dilutions of si-ICT1B were used. A concentration of siRNA was achieved that resulted in levels of FLAG-tagged ICT1 equivalent to that of the untransfected controls (10 nM; Supplementary Figure S2). Using this strategy, growth rates of these various cell lines were compared, uncovering a slower doubling time in the ICT1GSQ mutant, confirming that ICT1 requires a functional GGQ domain to maintain cell viability (Figure 4D, 3 cf. 4).
Figure 4.
Mutations of the GGQ domain can affect cell viability. (A) GGQ-mutant derivatives of ICT1 have lost PTH activity. Wild type and mutant derivatives (AGQ, GSQ) of Δ29 ICT1 were expressed as GST-fusion proteins, cleaved and assayed for f[3H]met release as described in Figure 3. All assays were performed with UAG codons and purified proteins, all equally monodispersed as assessed by dynamic light scattering (data not shown); ***P<0.001. (B, C) Mutated ICT1 is assembled into the mitoribosome. (B) FLAG-tagged wild-type (GGQ) and mutant (GSQ) ICT1 were expressed in HEK293T cells and the eluate from FLAG IP was subjected to silver staining (left panel) or western blotting (right panels) after denaturing gel electrophoresis. Molecular weight markers are indicated. The western blots of mitochondrial lysates shown are those before (IP-input) and after (Elution) FLAG IP of the wild type and mutated ICT1 derivatives. (C) Cell lysates were subjected to isokinetic gradient analysis before fractionation and western blotting, as described. The upper three panels are from wild-type ICT1-FLAG, the middle panels from mutated GSQ ICT1-FLAG and the lower panels from control mtLuc-FLAG. (D) A mutation of the GGQ domain affects cell growth. Non-targeting si-RNA-treated cells served as negative control (1, WT si-NT). Cells with only endogenous ICT1 (WT), or overexpressing normal (GGQ) or mutated (GSQ) ICT1 were treated for 4 days with 10 nM si-ICT1B to deplete endogenous ICT1 (2–4), whereas lane-2 represented the fully depleted control (WT si-ICT1B). Growth rates were compared by counting populations after 3 days of siRNA treatment; 3 versus 4: P<0.01; 1 versus 4: P<0.001. Western blots of lysates (4 days of siRNA treatment) after interrogation with the indicated antibodies are shown to the right.
Discussion
Following stringent purification methods, isolation of bovine mt-SSU, mt-LSU and the 55S monosome has allowed characterisation of many constituents and a low-resolution structure (Sharma et al, 2003). These data are in agreement that the mammalian mitoribosome differs substantially from other ribosomes that have been characterised. In particular, the 30:70 (w:w) protein-to-RNA ratio is reversed, with only one reduced rRNA component for each subunit and an increased number of protein factors, as determined by initial LCMS, resulting in a larger, more porous structure (O'Brien, 2002). Were some MRPs missed in this original screen? Our data indicates that ICT1 is indeed one such protein.
ICT1 is classified bioinformatically as one of four members of the prokaryotic/mitochondrial release factor family. It is now known that all 13 mitochondrial encoded polypeptides terminate translation with the aid of mtRF1a (Soleimanpour-Lichaei et al, 2007; Temperley et al, 2010). A third member, mtRF1, has yet to be shown to have release-factor activity, but, similar to ICT1 and mtRF1a, is an essential mitochondrial protein. The final member, C12orf65 currently has an unknown function, although with a mitochondrial location (unpublished observation). ICT1 and C12orf65 are significantly smaller than the RF1 and RF2 types of class-I bacterial release factors. Alignments highlight that this loss occurs in three main regions: the N-terminus and the two sections (tripeptide motif and the tip of the α-5 helix) that come into close contact with the mRNA in the ribosomal A-site and determine the STOP codon specificity (Figure 3). Lack of these domains is consistent with the loss of codon specificity noted for ICT1 in this study. By contrast, there is striking sequence conservation of the GGQ motif. By superimposing the available structure of the mouse ICT1 on RF1 (Thermotoga maritima) and RF2 (Thermus thermophilus), it is clear that in all three proteins the GGQ motif reside in a flexible loop (Figure 3A). Furthermore, although there is reduced sequence identity in this domain, the structural features surrounding this motif are highly conserved and important in positioning the GGQ at the surface of the RF (Frolova et al, 1999). This suggests that maintaining the way in which the GGQ motif is presented to the substrate is important. Curiously, ICT1 contains a stretch of 25 residues that has no sequence similarity to C12orf65 or the other RFs (Figure 3B, bold, italics). In the structural superimpositions, this feature can also be distinguished, primarily in the unmatched horizontal helix in ICT1. As this feature is lacking in release factors that interact transiently with the ribosome and also from C12orf65 that does not IP with the mitoribosome, it is possible that this feature is involved in the integration of ICT1 into the mt-LSU.
What could be the function of a ribosome-bound, codon-independent PTH and how could its capacity for indiscriminate peptide release be controlled and positively harnessed? There are several possibilities. First, if mitoribosomes initiate translation on 3′ truncated or mutated mt-mRNA that lack termination codons, the terminus of the mRNA will eventually reach the P-site and be retained by the anchoring peptidyl-tRNA but will leave an A-site devoid of mRNA or release factors to trigger recycling. In the absence of a mitochondrial analogue to the bacterial tmRNA system that rescues stalled ribosomes (Keiler et al, 1996), PTH activity will be needed to liberate the peptide from the peptidyl-tRNA, forming a substrate that can be recycled. A similar activity would be necessary to help recycle mitoribosomes that have prematurely stalled. Alternatively, many genetic studies have highlighted the necessity of salvaging tRNA species from prematurely released peptidyl-tRNAs generated through abortive elongation events during protein synthesis, a function normally provided by a freely soluble PTH (reviewed by Das and Varshney, 2006). To date no human mitochondrial protein is known to show this activity (although candidates exist). Our data are consistent with mammalian mitoribosomes streamlining this process by incorporation of a codon-independent PTH activity that can recognise and hydrolyse peptidyl-tRNAs caught during their premature release from the mitoribosome (or even from the mt-LSU if the stalled ribosome dissociates, as has been suggested for E. coli ribosomes by Varshney and co-workers (Singh et al, 2008)).
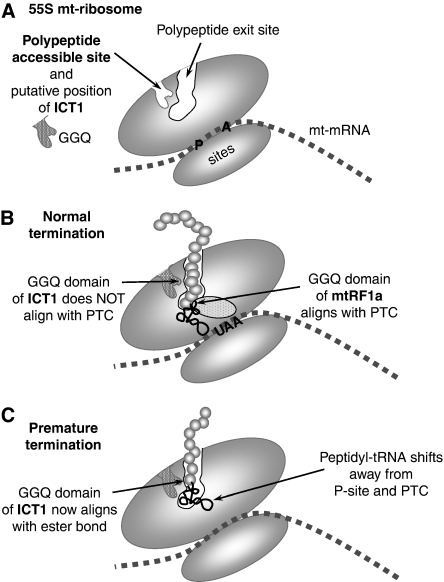
Cryo-EM data of mammalian mitoribosomes uncovers a feature absent in the bacterial ribosomes. This is a surface cavity in the mt-LSU near the exit tunnel, described as the polypeptide-accessible site (PAS) that prematurely exposes the nascent peptide to the solvent (Sharma et al, 2003; Figure 5A). It is created by loss of rRNA regions and proteins orthologous to bacterial components (O'Brien, 2002). Integration of ICT1 at this mitoribosomal site could allow access to peptidyl-tRNAs caught on release from the mitoribosome after aborted elongation (Figure 5C). Ribosomes use substrate-induced fit mechanisms to promote conformation changes that facilitate either peptide bond formation or hydrolysis (Schmeing et al, 2005). Premature release of a peptidyl-tRNA may cause ribosomal distortion, resulting in the juxtaposition of the GGQ motif and peptidyl-tRNA, promoting ester bond hydrolysis in a codon-independent manner. Crucially, this would release the tRNA for recharging. Clearly, to support any of these hypotheses, it would be of enormous benefit to generate high-resolution structural data to identify exactly where in the mitoribosome ICT1 is found. To date, there are only very limited structural data concerning the mammalian mitoribosome and producing such structural data is beyond the scope of this paper. Further experiments will be necessary to elucidate the exact role of this PTH activity; however, it is interesting to note that ICT1 is mitochondrial, has become incorporated into the mitoribosome, lost all stop codon specificity, but has acquired another function of equal importance to mitochondrial translation. Contrastingly, mtRF1a has retained its RF activity. It is tempting to speculate on what diverging fates may also have befallen the other two members of this family.
Figure 5.
A schematic representation of the putative position and function of ICT1 in the human mitochondrial ribosome. (A) A simplified cartoon of the 55S mitochondrial ribosome indicating the polypeptide exit tunnel and site (PES) and the PAS in the large subunit as defined by Sharma et al (2003). No orthologues of the proteins that would occupy the PAS have been found in mammalian mitochondria, and we postulate that is where ICT1 is positioned with the GGQ domain inserted deep into the pocket. Sites for the aminoacyl (A) and peptidyl (P) tRNAs are shown. The mt-mRNA is depicted between the large and small mt-ribosomal subunits. (B) Under conditions of normal termination, the ester bond of the peptidyl-tRNA is positioned close to the peptidyl-transferase centre (PTC); the release factor, mtRF1a, enters through the A-site, recognising the stop codon (UAA) and aligning the GGQ domain at the PTC to promote hydrolysis of the ester bond and release of the nascent peptide. (C) Where abortive elongation occurs, the peptidyl-tRNA may drop away from the P-site towards the PES, aligning the ester bond close to the GGQ domain of ICT1, promoting cleavage of the tRNA, which allows both mt-tRNA and truncated peptide to be released from the mitochondrial monosome (or potentially from dissociated 39S mt-LSU).
Materials and methods
Cell culture
Human HeLa cells were cultured (37°C, humidified 5% CO2) in Eagle's MEM (Sigma) supplemented with 10% (v/v) foetal calf serum (FCS), 1 × non-essential amino acids (NEAA) and 2 mM L-glutamine. 143B.206 rho0 osteosarcoma cells (provided by R Wiesner, University of Koeln) were propagated in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FCS, 50 μg/ml uridine and 1 × NEAA. Flp-InT-Rex-293 cells (HEK293T; Invitrogen) were grown in identical media supplemented with 10 μg/ml BlasticidinS and 100 μg/ml Zeocin (Invitrogen). Post-transfection selection was performed with HygromycinB (100 μg/ml). For growth on respiratory substrates, the medium contained glucose-free DMEM (Gibco), 0.9 mg/ml galactose, 1 mM sodium pyruvate, 10% (v/v) FCS, NEAA and 2 mM L-glutamine. For growth curve analyses, the galactose medium included 50 μg/ml uridine.
Subcellular localisation
Human mitochondria or cell lysates were prepared in a homogenisation buffer (10 mM Tris–HCl (pH 7.4), 0.6 M mannitol, 1 mM EGTA) by differential centrifugation. Aliquots of mitochondria were treated with proteinase-K (4 μg/100 μg protein) for 30 min at 4°C and either lysed (1% final v/v Triton X-100) or treated with 1 mM PMSF before separation through 15% PAG and transfer to a PVDF membrane. Western blots were performed and developed as described by Soleimanpour-Lichaei et al (2007).
siRNA constructs and transfection
Three sequences targeting ICT1 were tested for efficiency of protein depletion. The nucleotide positions are relative to the reference sequence NM_001545.1; sense strands were as follows:
si-ICT1A 5′ CUAGAUCGCUUGACAAUAU dTdT 3′ (nt 222–240);
si-ICT1B 5′ GCCGCUAUCAGUUCCGGAA dTdT 3′ (nt 421–439) and
si-ICT1C 5′GGGUCCCGAAUGGUGCAAA dTdT 3′ (nt 181–199).
Experiments were performed with si-ICT1B unless otherwise specified. si-ICT1C was found to be ineffective. Transfections were performed with 20% confluent HeLa cells using Oligofectamine (Invitrogen) in Optimem-I medium (Gibco) with 0.2 μM siRNA. Reverse transfections were performed as described by Ovcharenko et al (2005) using approximately 12 000 HEK293T cell per cm2 using Lipofectamine RNAiMAX (Invitrogen) in Optimem-I medium (10–33 nM siRNA). Custom and control non-targeting (NT; ref.: OR-0030-neg05) duplex siRNAs were purchased pre-annealed from Eurogentec.
In vivo mitochondrial protein synthesis
Mitochondrial protein synthesis in cultured cells was performed as described by Chomyn (1996) after addition of emetine and pulsed with [35S]-methionine for 15 min. Aliquots (50 μg) of total cell protein were separated by 15% (w/v) SDS–PAGE. The gels were exposed to PhosphorImage cassettes and visualised using the ImageQuant software.
Production of FLAG- and GST-fusion constructs, transfection, expression and purification
The original human ICT1 clone was obtained from MGC (MGC:21251; accession no. BC015335) and encodes the only isoform to be described. All constructs were prepared by PCR using the MGC clone as template. Constructs to facilitate inducible expression of C-terminal FLAG-tagged ICT1 were prepared by generating an amplicon from nt 2 to 620 using the following primers: 5′-CTTTCTTAAGCTTCCACCATGGCGGCCACCAGGTG-3′ and 5′-CTCTCCGATATCCTACTTATCGTCGTCATCCTTGTAATCGTCCATGTCGACCCTC-3′. The amplicon and pcDNA5/FRT/TO (Invitrogen) were digested with HindIII/EcoRV and ligated. C-terminal FLAG-tagged MRPS27 and MRPL20 were constructed by PCR amplification from MGC clones 6009616 and 3542715 (accession nos. BC064902 and BC009515), respectively, using primers 5′-CTTTCTTGGATCCCCACCATGGCTGCCTCCATAGTGCG-3′ and 5′-CTCTCTCTCGAGCTACTTATCGTCGTCATCCTTGTAATCGGCAGATGCCTTTGCTGCTT-3′ (MRPS27 Forward and Reverse) or 5-TACTATAAGCTTACCATGGTCTTCCTCACCG-3 and 5-ATACTACTCGAGCTACTTATCGTCGTCATCCTTGTAATCGTGGTACTGCACCACTC-3 (MRPL20 Forward and Reverse). The amplicons were digested with BamHI/XhoI or HindIII/XhoI, respectively, and ligated into similarly digested pcDNA5/FRT/TO. The GST-fusion constructs were made of the FL or N-terminal-deleted (Δ29) ICT1. These were generated by amplifying nt 2 or 90–623, respectively. The forward primers incorporated a BamHI site (FL Forward 5′-CTTTCTTGGATCCATGGCGGCCACCAGGTG-3′; Δ29 Forward 5′-CTTTCTTGGATCCCTGCACAAGCAGAAAGACG-3′) and the same reverse primer was used for both amplification reactions (Reverse 5′-CTCTCCCTCGAGTCAGTCCATGTCGACCCTC-3′ containing an XhoI site). The amplicons and vectors were digested to allow in-frame downstream fusion of ICT1-GST in pGEX-6P-1 (GE Healthcare). The constructs derived from pGEX-6P-1 or pcDNA5/FRT/TO were used for transfection of E. coli Rosetta pLysS (Merck Biosciences) or HEK293T cells, respectively. Bacteria were induced, and protein was purified and cleaved from GST using PreScission Protease exactly as previously described for mtRRF (Rorbach et al, 2008). Bacterial expression of both FL GST-ICT1-fusion protein and Δ29 was deleterious to growth, but was not lethal. Human HEK293T cells were transfected at ∼50% confluence with the vectors pOG44 and pcDNA5/FRT/TO containing sequences of the genes to be expressed (FLAG-tagged ICT1, mtLuc and derivatives) as previously described (Soleimanpour-Lichaei et al, 2007).
Dynamic light scattering
Dynamic light scattering measurements were performed using a commercial Zetasizer 1600 (Malvern Instruments) equipped with a He–Ne laser (633 nm, 5 mW). Aliquots (20 μl) of protein samples (concentrations of 0.1–1 mg/ml) were pipetted into low-volume glass cuvettes and equilibrated for 3 min in the apparatus before measurement. All samples were measured a minimum of five times at 25°C for 70 s.
Affinity purification and elution of FLAG peptides
Mitochondria were isolated from HEK293T cells expressing FLAG derivatives and treated with DNase-I (0.5 U/mg mitochondria; 15 min RT) and proteinase-K (5 μg/mg mitochondria; 30 min 4°C) followed by 1 mM PMSF inhibition. The pelleted mitochondria were washed in a homogenisation buffer, treated with digitonin to remove the outer membrane (400 μg/mg mitochondria), washed and resuspended in a lysis buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1% (v/v) TX-100, 1 × protease inhibitor cocktail (Roche), 1 mM PMSF). IP was performed using an anti-FLAG M2-agarose affinity gel following manufacturer's recommendations (Sigma Aldrich, St Louis, MO, USA). Elution was performed using 5 μg of 3 × FLAG peptide per 100 μl of an elution buffer.
LC MS/MS analysis of negative control and ICT1 IP
The immunopurification of ICT1 and negative control (IP eluate from the lysate of untransfected HEK293T cells subjected to the identical IP protocol with anti-FLAG M2-agarose) were separated using SDS–PAGE. Individual gel lanes were cut into three bands and were in-gel digested as described elsewhere (Wilm et al, 1996). The resulting peptide solutions were desalted and concentrated offline using STAGE tips (Rappsilber et al, 2003). Measurements were performed using an Agilent 1100 nanoflow system connected through a nano-electrospray ion source to a 7T linear ion-trap Fourier transform ion cyclotron resonance mass spectrometer (LTQ FT ULTRA; Thermo Scientific). Peptide separations were performed using a 15-cm fused silica emitter (New Objective, PicoTip Emitter; tip: 8±1 μm, ID: 100 μm, FS360-100-8-N-5-C15) packed with reversed-phase ReproSil-Pur C18AQ 3-μm resin purchased from Dr Maisch GmbH. Samples were loaded directly onto the analytical column at a flow of 600 nl/min eluent-A (3% acetonitrile, 0.5% acetic acid). Bound peptides were eluted from the column at a rate of 300 nl/min using an LC gradient of 10–40% eluent-B (80% v/v acetonitrile, 0.5% acetic acid) in 60 min, followed by a steep gradient of 40–100% eluent-B in 5 min. The mass spectrometer was programmed to select the top four most abundant ions from each survey scan in the ICR cell for subsequent fragmentation analysis using collision-induced dissociation (CID) in the linear ion trap. Only double and triple charged ions were selected for CID analysis, and dynamic exclusion with early expiration was enabled to prevent re-selection of previously fragmented ions.
MRP identification and data analysis
Precursor and fragment ions were extracted from raw data files using ExtractMSn (Thermo Scientific). Individual LC MS/MS data files of all three gel slices were combined for each sample before database search. Peptides were identified by automated database searches using Mascot (Matrix Science Mascot version 2.2) against the Refseq database (release 29) and a Refseq reverse database for automated validation using false-discovery rate calculations. Database searches were performed with a 20-p.p.m. precursor ion tolerance and 0.8-Da tolerance for MS/MS fragment ions using the following specifications: tryptic cleavage allowing one missed cleavage, carbamidomethylation (C) as fixed modification, and allowing deamidation (NQ) and oxidation (M) as variable modifications. Subsequent peptide identifications were blasted against the Refseq database and validated by false-discovery rate calculation using the PROVALT software (Weatherly et al, 2005). PROVALT was set to calculate peptide score cut-offs to achieve a calculated <1% false-discovery rate. Protein abundances were calculated as exponentially modified Protein Abundance Index (emPAI) values according to Ishihama et al (2005). MRPs were marked as enriched if (i) they were identified with at least three unique peptides and (ii) when emPAI values were at least twofold higher in the ICT1 IP sample as compared with that in the negative control. The table represents only identified human MRPs that appear in the Refseq database.
Isokinetic sucrose-gradient analysis of mitochondrial ribosomes
Total cell lysates (0.5–0.7 mg in lysis buffer) or eluted immunoprecipitates were loaded on a linear sucrose gradient (1 ml 10–30% (v/v)) along with 50 mM Tris–HCl (pH 7.2), 10 mM Mg(OAc)2, 40 mM NH4Cl, 0.1 M KCl, 1 mM PMSF, and 50 μg/ml chloramphenicol, and centrifuged for 2 h 15 min at 100 000 g at 4°C. Fractions (100 μl) were collected and 10-μl aliquots were analysed directly by western blotting or silver staining as follows: PAG were fixed in 50% methanol for 1 h, followed by 15-min incubation in staining solution (0.8% (w/v) AgNO3; 1.4% (v/v) NH4OH; 0.075% (w/v) NaOH), three washes of 5 min each in nanopure dH2O, and then developed in 0.055% (v/v) formaldehyde/0.005% (w/v) citric acid and fixed in 45% methanol/10% acetic acid.
Northern blot analysis for mitoribosomal subunit markers in gradient gel fractions
To identify the 28S mt-SSU and 39S mt-LSU in gradient fractions, the 12S and 16S mt-rRNA were visualised by northern blotting. RNA was prepared by phenol extraction from half of each gradient fraction. Northern blots were performed as described by Chrzanowska-Lightowlers et al (1994). Briefly, aliquots of RNA (5 μg) were electrophoresed through 1.2% agarose under denaturing conditions and transferred to GenescreenPlus membrane (NEN duPont) following the manufacturer's protocol. Probes were generated using random hexamers on PCR-generated templates corresponding to the internal regions of the 12S and 16S mt-rRNA. An estimate of 55S monosome was made from the position of the cytosolic 28S rRNA (representing the 60S cytosolic LSU) visualised on ethidium bromide staining of the agarose gel.
Mutagenesis of ICT1
Mutations in the GGQ tripeptide were introduced into the bacterial (AGQ, GSQ) and human (GSQ) ICT1 expression constructs following the QuikChange II site-directed mutagenesis protocol (Stratagene). The following primers and their complements were used; base changes from wild type are underlined: AGQ, 5′-GTAGTGGTCCTGCGGGGCAGAATG-3′; GSQ, 5′-GTGGTCCTGGGTCGCAGAATGTGAAC-3′.
In vitro release-factor assay
E. coli ribosomes were purified and assays were performed essentially as described by Tate and Caskey (1990) with modifications as detailed in reference Soleimanpour-Lichaei et al (2007). Briefly, an f[3H]met-tRNAmet substrate was made by combining 3.8 nmol L-[methyl-3H]-methionine (GE Healthcare) and cold methionine up to a concentration of 21.8 nmol, 3.5 mM leucovorin (Sigma) as the formyl donor, 20 μM amino acids (Promega), 0.3 mg N-formylmethionine-specific tRNA (Sigma), 1.2 mM ATP, 1 mM DTT, 10 mM MgCl2, in cacodylate buffer (100 mM, pH 6.8), and incubating for 30 min at 37°C. The ribosomal substrate (amounts given are per 50 μl of assay) was prepared by incubation of 70S ribosomes (5 pmol) with AUG (250 pmol) and f[3H]met-tRNAmet (2.5 pmol) in 20 mM Tris–HCl (pH 7.4), 10 mM Mg(OAc)2 and 150 mM NH4Cl at 30°C for 20 min. This ‘activated' ribosomal substrate was stored on ice before interrogation with release factors for activities with selected codons. Reactions that lacked the 70S were otherwise identical.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (grant number 074454/Z/04/Z), Biotechnology and Biological Sciences Research Council (grant number BB/F011520/1) and the Netherlands Genomics Initiative (Horizon Programme).
Footnotes
The authors declare that they have no conflict of interest.
References
- Caskey CT, Beaudet AL, Scolnick EM, Rosman M (1971) Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci USA 68: 3163–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyn A (1996) In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol 264: 197–211 [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Lightowlers ZM, Preiss T, Lightowlers RN (1994) Inhibition of mitochondrial protein synthesis promotes increased stability of nuclear-encoded respiratory gene transcripts. J Biol Chem 269: 27322–27328 [PubMed] [Google Scholar]
- Das G, Varshney U (2006) Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 152: 2191–2195 [DOI] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 4: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Ito K, Uno M, Nakamura Y (2000) A tripeptide ‘anticodon' deciphers stop codons in messenger RNA. Nature 403: 680–684 [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993 [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL (2001) The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276: 43958–43969 [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF (2008) Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123: 277–289 [DOI] [PubMed] [Google Scholar]
- O'Brien TW (2002) Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286: 73–79 [DOI] [PubMed] [Google Scholar]
- Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K, Brown D (2005) High-throughput RNAi screening in vitro: from cell lines to primary cells. RNA 11: 985–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM (2008) The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res 36: 5787–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA (2005) An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438: 520–524 [DOI] [PubMed] [Google Scholar]
- Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK (2003) Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115: 97–108 [DOI] [PubMed] [Google Scholar]
- Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH (2004) Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol 341: 227–239 [DOI] [PubMed] [Google Scholar]
- Singh NS, Ahmad R, Sangeetha R, Varshney U (2008) Recycling of ribosomal complexes stalled at the step of elongation in Escherichia coli. J Mol Biol 380: 451–464 [DOI] [PubMed] [Google Scholar]
- Sippl MJ, Wiederstein M (2008) A note on difficult structure alignment problems. Bioinformatics 24: 426–427 [DOI] [PubMed] [Google Scholar]
- Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R, Chrzanowska-Lightowlers Z (2007) mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol Cell 27: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spremulli LL, Coursey A, Navratil T, Hunter SE (2004) Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol 77: 211–261 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K (2001) Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28S small subunit. J Biol Chem 276: 33181–33195 [DOI] [PubMed] [Google Scholar]
- Takemoto C, Spremulli LL, Benkowski LA, Ueda T, Yokogawa T, Watanabe K (2009) Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res 37: 1616–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate WP, Caskey CT (1990) Termination of protein synthesis. In Ribosomes and Protein Synthesis: a Practical Approach, Spedding G (ed) Oxford: Oxford University Press, pp 81–100 [Google Scholar]
- Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZMA (2010) Hungry codons promote frameshifting in human mitoribosomes. Science 327: 301. [DOI] [PubMed] [Google Scholar]
- Weatherly DB, Atwood JA III, Minning TA, Cavola C, Tarleton RL, Orlando R (2005) A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteomics 4: 762–772 [DOI] [PubMed] [Google Scholar]
- Williams EH, Bsat N, Bonnefoy N, Butler CA, Fox TD (2005) Alteration of a novel dispensable mitochondrial ribosomal small-subunit protein, Rsm28p, allows translation of defective COX2 mRNAs. Eukaryot Cell 4: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M (1996) Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379: 466–469 [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K (2001) Wobble modification defect in tRNA disturbs codon–anticodon interaction in a mitochondrial disease. EMBO J 20: 4794–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoldak G, Redecke L, Svergun DI, Konarev PV, Voertler CS, Dobbek H, Sedlak E, Sprinzl M (2007) Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic Acids Res 35: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.