Abstract
Caspases are cysteine-dependent proteases and are important components of animal apoptosis. They introduce specific breaks after aspartate residues in a number of cellular proteins mediating programmed cell death (PCD). Plants encode only distant homologues of caspases, the metacaspases that are involved in PCD, but do not possess caspase-specific proteolytic activity. Nevertheless, plants do display caspase-like activities indicating that enzymes structurally distinct from classical caspases may operate as caspase-like proteases. Here, we report the identification and characterisation of a novel PCD-related subtilisin-like protease from tobacco and rice named phytaspase (plant aspartate-specific protease) that possesses caspase specificity distinct from that of other known caspase-like proteases. We provide evidence that phytaspase is synthesised as a proenzyme, which is autocatalytically processed to generate the mature enzyme. Overexpression and silencing of the phytaspase gene showed that phytaspase is essential for PCD-related responses to tobacco mosaic virus and abiotic stresses. Phytaspase is constitutively secreted into the apoplast before PCD, but unexpectedly is re-imported into the cell during PCD providing insights into how phytaspase operates.
Keywords: abiotic stress, caspases, programmed cell death, subtilisin-like proteases, tobacco mosaic virus
Introduction
Programmed cell death (PCD, or apoptosis) is a basic process for elimination of redundant and damaged cells in multicellular organisms, which operates in the course of their development and in response to various stress-inducing stimuli. In animals, an important component of the PCD machinery is a family of apoptotic proteases termed as caspases (for cysteine-dependent aspartate-specific proteases). These proteolytic enzymes become activated in the course of apoptosis and introduce specific breaks after aspartate (D) residues in a limited number of cellular proteins thus mediating PCD (Thornberry and Lazebnik, 1998; Wolf and Green, 1999). Two types of caspases exist: initiator and effector caspases (Boatright and Salvesen, 2003). Initiator caspases (e.g. caspases-2, -8, -9 and -10) cleave inactive pro-forms of effector caspases, thereby activating them. Effector caspases (e.g. caspases-3, -6 and -7) in turn cleave other protein substrates within the cell, to trigger the apoptotic process. The initiation of this cascade reaction is regulated by caspase inhibitors (Ekert et al, 1999). Caspase inactivation is known to suppress cell death (Zheng et al, 1999).
PCD in the plant and animal kingdoms share a number of morphological and biochemical features, including condensation and shrinkage of the nucleus and cytoplasm, DNA laddering and cytochrome c release from mitochondria (Danon et al, 2000; Balk and Leaver, 2001; Lam et al, 2001; Hoeberichts and Woltering, 2003). However, direct structural homologues of animal caspases with an analogous cleavage specificity and function have not been shown to exist in plants (reviewed by Bonneau et al, 2008). Plants encode only distant homologues of caspases, metacaspases, that may be involved in PCD, but do not possess caspase-specific proteolytic activity (Bozhkov et al, 2005; Watanabe and Lam, 2005; Vercammen et al, 2006; He et al, 2007; Sundström et al, 2009). However, some specific peptide inhibitors of animal caspases have been shown to affect the development of plant PCD. One of the examples of PCD in plants is the hypersensitive response (HR) that results from incompatible plant–pathogen interactions, which serves to prevent the spread of pathogens from the infection site (Greenberg, 1997; Heath, 2000). For example, specific inhibitors of animal caspase-1 and -3 [N-acetyl-YVAD-chloromethylketone (Ac-YVAD-CMK) and Ac-DEVD-aldehyde (Ac-DEVD-CHO), respectively] could attenuate bacteria- and tobacco mosaic virus (TMV)-induced HR in tobacco leaves (del Pozo and Lam, 1998). The baculovirus anti-apoptotic proteins p35 and IAP (inhibitor of apoptosis), which are known to inhibit animal caspases, were also efficient in preventing plant PCD induced by bacterial, fungal and viral infections (Hansen, 2000; Dickman et al, 2001; del Pozo and Lam, 2003). Furthermore, caspase-like activities have been detected in plants using synthetic fluorogenic caspase substrates (reviewed by del Pozo and Lam, 1998; Bonneau et al, 2008) indicating that enzymes structurally distinct from classical caspases may operate as caspase-like proteases in plants.
Indeed, it has recently been shown that the plant protease vacuolar-processing enzyme (VPE) is required for PCD induced by TMV (Hatsugai et al, 2004) and fumonisin (Kuroyanagi et al, 2005) as well as for developmental cell death in seed integuments (Nakaune et al, 2005) and displays a caspase-1 (YVADase) activity (Hatsugai et al, 2004; Rojo et al, 2004). However, VPE is a legumain, a cysteine protease, that is structurally different from classical caspases. It has also been found that caspase inhibitors prevent ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) proteolysis during victorin-induced PCD, and the use of the pan-caspase substrate benzyloxycarbonyl-VAD-7-amino-4-trifluoromethylcoumarin (z-VAD-AFC) allowed the detection of caspase-like activity in extracts from victorin-treated Avena sativa leaves (Coffeen and Wolpert, 2004). The purified active protein contained amino-acid sequences homologous to plant subtilisin-like serine proteases and was named saspase (serine-dependent aspartate-specific protease). Saspase has also been shown to be indirectly involved in victorin-induced cleavage of Rubisco during PCD in oats (Coffeen and Wolpert, 2004). However, the function of saspase in PCD execution remains unknown.
Recently, we have identified a caspase-like activity in tobacco that seems to represent a functional analogue of animal caspases (Chichkova et al, 2004). This protease activity displayed an exceptional selectivity introducing a single break in the Agrobacterium tumefaciens VirD2 protein, after the D400 residue within the TATD motif, in a caspase-like manner. The tobacco enzyme, like animal caspases, was inactive in healthy tissues and became active in the course of TMV-induced HR in tobacco. Furthermore, suppression of enzyme activity by a peptide aldehyde matching its cleavage site inhibited PCD mediated by TMV in tobacco leaves (Chichkova et al, 2004).
Fragmentation of the A. tumefaciens VirD2 protein by tobacco caspase-like protease results in the detachment of the C-terminal nuclear localisation signal (NLS)-containing peptide of VirD2. The NLS in the VirD2 protein is essential for nuclear uptake of foreign DNA within the plant cell during bacterial infection and plant transformation (Steck et al, 1990; Shurvinton et al, 1992). Therefore, inactivation of VirD2 mediated by tobacco caspase-like protease may represent a protective mechanism aimed at limiting delivery and expression of foreign genes in plants. Indeed, substitution of the wild-type VirD2 protein with its mutant form, which is resistant to tobacco caspase-like protease, was shown to markedly improve A. tumefaciens-mediated foreign gene delivery (Reavy et al, 2007). Proteases with specificity and biochemical properties similar to those of the tobacco enzyme have been shown to be ubiquitous in mono- and dicotyledonous plants (Chichkova et al, 2008).
Here, we report the isolation and identification of this protease from tobacco and rice and propose to name it phytaspase (plant aspartate-specific protease). Mutational analysis of the recombinant enzyme showing serine dependence confirmed the designation of phytaspase as a subtilisin-like protease predicted from the amino-acid sequence. Being synthesised as a proenzyme, phytaspase is autocatalytically processed to generate a mature enzyme. Overexpression and silencing of the phytaspase gene showed that phytaspase is essential for PCD-related responses to biotic (TMV) and abiotic stresses. The substrate specificity of phytaspase is distinct from that of other known caspase-like proteases. Another intriguing peculiarity of phytaspase is that it is constitutively secreted into the apoplast before PCD, but unexpectedly is partially re-imported into the cell during PCD.
Results and discussion
Identification of phytaspase
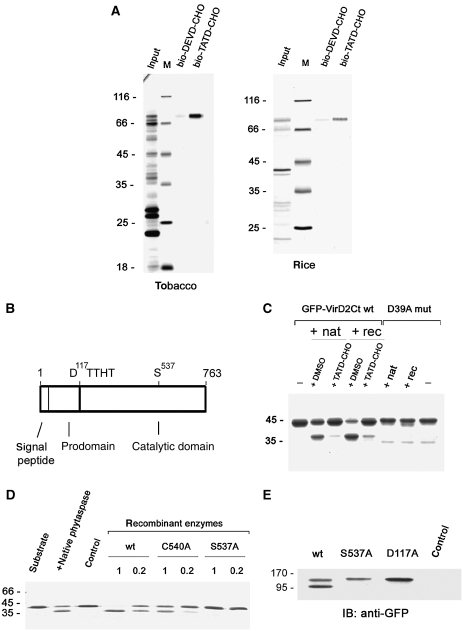
Using affinity chromatography with biotinylated TATD aldehyde (bio-TATD-CHO) on avidin resin, phytaspase was purified from Nicotiana tabacum leaves to homogeneity producing a single major ∼80 kDa protein (Figure 1A, left panel; Supplementary Figure S1). Mass spectrometric analysis of the protein allowed the identification of an N. tabacum EST encoding a central portion of the enzyme and by amplification of a cDNA, the complete ORF of the protein was obtained and sequenced (these sequence data have been submitted to the GenBank databases under accession No. GQ249168). The deduced complete amino-acid sequence of tobacco phytaspase indicated that it corresponds to a putative subtilisin-like protease of the S8 family predicted to possess a 24 amino-acid N-terminal signal peptide (for extracellular targeting) and a prodomain (Figure 1B; Supplementary Figure S2). Indeed, the mature tobacco phytaspase was found to lack both the signal sequence and the prodomain (Supplementary Figure S2). A rice phytaspase was similarly identified (accession GI 32977156) and it displayed 53% amino-acid sequence identity with the tobacco phytaspase (Figure 1A, right panel; Supplementary Figure S2).
Figure 1.
Identification and molecular characterisation of tobacco phytaspase. (A) Affinity chromatography purification of tobacco (left panel) and rice (right panel) phytaspases with their inhibitor, bio-TATD-CHO on avidin resin; bio-DEVD-CHO, which does not inhibit phytaspase, was used as a control. Protein samples were fractionated by SDS–gel electrophoresis and stained with Coomassie Blue or zinc-imidazole. Positions of MW protein markers (M) are indicated on the left. (B) Schematic representation of phytaspase domains. (C) Recombinant tobacco phytaspase (rec) displays the same ability to cleave GFP-VirD2Ct protein as natural tobacco enzyme (nat). The bio-TATD-CHO was used at 100 μM for enzyme inhibition. Coomassie Blue-stained gel is shown. The D39A mutation (D39A mut) (Chichkova et al, 2004) preventing cleavage in the GFP-VirD2Ct protein corresponds to the D400 residue in full-length VirD2. (D) The S537A mutation prevents phytaspase-mediated cleavage of GFP-VirD2Ct in vitro (Coomassie Blue staining). C540A, a mutation of a nearby residue, only marginally affects proteolytic activity. Recombinant enzymes were produced as in (C). 1 and 0.2 indicate relative amounts of recombinant enzymes taken to assess their hydrolytic activity, as verified by western blotting with anti-GST antibody. Control sample (control) obtained in an identical manner from vector-only agroinfiltrated leaves. (E) Phytaspase mutants with impaired processing. Western blot analysis of the crude extracts from leaves agroinfiltrated with constructs expressing wt and mutated phytaspase forms fused to GFP with anti-GFP antibody. In contrast to the purified enzyme (A) showing only one band (mature enzyme), extracts containing wt phytaspase display two bands (proenzyme and mature enzyme) apparently because in the latter case, we used immediate denaturation of samples (boiling in SDS). Both the S537A (catalytic residue) and D117A (the prodomain/catalytic domain junction) mutations impair proenzyme processing in N. benthamiana leaves. Control, vector-only agroinfiltrated leaves.
The tobacco phytaspase gene was expressed in N. benthamiana leaves as phytaspase-glutathione-S-transferase (GST) fusion using an Agrobacterium-mediated transient expression system and purified by glutathione Sepharose chromatography. The recombinant phytaspase displayed the same ability to cleave a substrate protein GFP-VirD2Ct (Chichkova et al, 2004) at the TATD motif in vitro, as the native enzyme (Figure 1C). S8 subtilisin-like proteases are serine proteases. Accordingly, mutation of the predicted catalytic S537 residue (Rawlings et al, 2008; Rawlings and Barrett, 2009) of tobacco phytaspase abolished its proteolytic activity (Figure 1D).
To determine whether processing of the prodomain occurs autocatalytically, green fluorescent protein (GFP) was fused to the C-terminus of the full-length tobacco phytaspase. Western blots of total protein extracted from these leaves were reacted with anti-GFP antibody, and two protein bands (Figure 1E) were detected corresponding to the partially processed enzyme-containing QSETYVIHM at its N-terminus (proenzyme, lacking the predicted 24-amino-acid signal peptide) and completely processed protein with the TTHTSQFL as the N-terminal amino-acid sequence (lacking the signal peptide and prodomain), respectively, as was shown by Edman degradation sequencing (Supplementary Figure S2). Of note, the inactive S537A mutant displayed only one protein band with an MW similar to the wild-type proenzyme (Figure 1E) and its N-terminal sequence determined by Edman degradation sequencing as QSETYVIHM corresponded to the amino-terminus of the proenzyme lacking the signal peptide (Supplementary Figure S2). This indicates that no cleavage of the prodomain had occurred in the catalytically inactive mutant and shows that phytaspase seems to be a self-processing enzyme—a property shared by subtilases of various origin including plant subtilisin-like proteases (Cedzich et al, 2009). Consistent with this conclusion, the D residues in both tobacco and rice phytaspase precursors immediately precede the TTHT motif, which is the N-terminus of the mature enzyme (Figure 1B; Supplementary Figure S2). Accordingly, mutation of the D117 to A in tobacco phytaspase impaired proenzyme processing (Figure 1E).
Substrate specificity of phytaspase
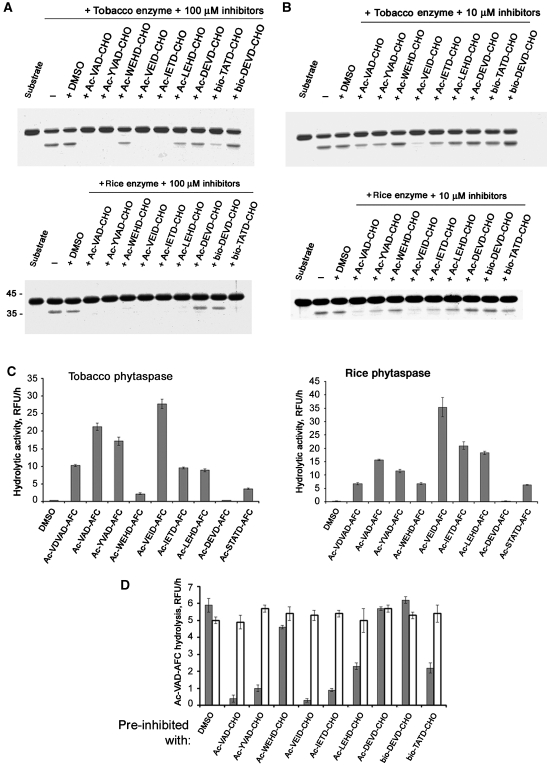
To determine the substrate specificities of the tobacco and rice phytaspases, we used a panel of peptide aldehyde inhibitors of animal caspases in the in vitro GFP-VirD2Ct cleavage assay. Their inhibitory potential was markedly distinct, with Ac-VEID-CHO showing the strongest inhibition, whereas DEVD-CHO produced no effect (Figure 2A and B). Employment of a range of peptide-based fluorogenic substrates of animal caspases produced complementary results: Ac-VEID-AFC (7-amino-4-trifluoromethylcoumarin) was found to be the optimal substrate of phytaspases, whereas Ac-DEVD-AFC remained uncleaved (Figure 2C). Phytaspase inactivation by peptide aldehyde inhibitors was found to be reversible (Figure 2D). These results describing VEID as a preferred motif for phytaspase cleavage differentiate tobacco and rice phytaspases from the saspase obtained from A. sativa, which does not cleave VEID (nor VDVAD or WEHD) at all (Coffeen and Wolpert, 2004). By the same substrate specificity criterion, phytaspase is more similar to animal caspase-6 and to an unidentified protease involved in PCD during embryogenesis in Norway spruce (Bozhkov et al, 2004).
Figure 2.
Substrate specificities of tobacco and rice phytaspases. (A) Peptide aldehyde inhibitors of animal caspases at 100 μM, with the exception of DEVD-CHO, impair tobacco (upper panel) and rice (lower panel) phytaspase-mediated fragmentation of GFP-VirD2Ct substrate protein. (B) The use of the same set of inhibitors at 10 μM shows their markedly distinct inhibitory potential and minor differences between tobacco and rice phytaspase specificities. Note that in both cases, Ac-VEID-CHO is the most potent phytaspase inhibitor. (C) Fluorogenic peptide substrates of animal caspases are hydrolysed by tobacco (left panel) and rice (right panel) phytaspases. Relative rates of peptide-AFC (20 μM) hydrolysis expressed as relative fluorescence units per hour (RFU/h, mean values from three samples) are slightly different for rice and tobacco enzymes. Note that the naturally occurring phytaspase cleavage site, STATD, in the VirD2 protein is one of the poorest cleavage sites at the peptide level. (D) Inhibition of phytaspase with peptide aldehyde inhibitors is reversible. Phytaspase was pre-incubated with the indicated inhibitors (at 100 μM), samples were diluted five-fold and fluorogenic substrate Ac-VAD-AFC was added up to 20 μM directly to the mixture to determine relative rates of substrate hydrolysis in the presence of the inhibitors (grey bars). Alternatively, before addition of the fluorogenic substrate, free inhibitors were eliminated by spin gel filtration followed by 1 h incubation at room temperature to allow possible reversion of inhibition (open bars). Data (means from three experiments) are given for tobacco phytaspase. Rice phytaspase behaves similarly (data not shown).
Overexpression and silencing of phytaspase gene in tobacco transgenic plants
To address the possible function of phytaspase in the HR, we generated transgenic N. tabacum Samsun NN plants, which either overproduced tobacco phytaspase or possessed markedly decreased levels of phytaspase activity because of RNAi silencing induced by transgenic expression of three independent hairpin RNA constructs designed to avoid ‘off-target' silencing. Using the siRNA scan website (http://bioinfo2.noble.org/RNAiScan.htm) (Xu et al, 2006), sequences of these three independent fragments of the phytaspase ORF were screened against datasets from tobacco, tomato and Arabidopsis to seek 21 nt stretches of homology with other genes and thus the potential for ‘off-target' silencing. For fragment 1 (nt 7–237), no matches were identified to any sequences in all the datasets searched. For the other two fragments, fragment 2 (nt 1096–1446) and fragment 3 (nt 1888–2295) matches were made to sequences of only one tomato gene (SGN-U329832) annotated as a gene for subtilisin-like protease, which may or may not be a homologue of tobacco phytaspase. However, taking into account the unavailability of full-genome sequence of tobacco, we performed a functional complementation assay to ascertain that the phenotypic effects observed on RNAi were due to specific silencing of the phytaspase gene, as described below.
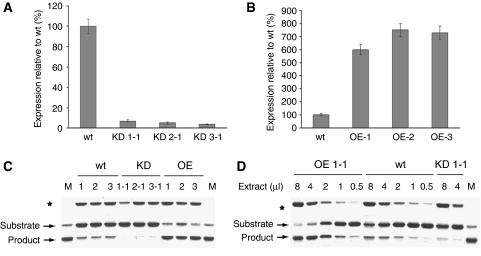
Tests using qRT–PCR revealed >90% reduction in transcript level in leaf tissues of T1 seedlings in each of five to seven lines of phytaspase-silenced (knocked down, KD) tobacco plants generated with each of the hairpin RNAi constructs. Typical results characteristic of all the KD transgenic lines for each construct are shown in Figure 3A. In all the phytaspase overexpressing (OE) lines, exemplified by three of them in Figure 3B, phytaspase gene expression was increased about seven- to eight-fold compared with wild-type tobacco. Measurements of phytaspase enzymatic activity confirmed the significant increase (approximately four-fold) and reduction (approximately eight-fold) of phytaspase-specific activity in overproducing and silenced transgenic plants, respectively (Figure 3C and D). In spite of these differences, neither overproduction of phytaspase nor its silencing showed any discernible altered phenotype. It is possible that phytaspase overexpression could be neutralised by export of the enzyme out of the cell (see below), whereas (as suggested by the silencing experiments) phytaspase is possibly not involved in plant growth and development (at least under the optimal glasshouse conditions). Alternatively, functional redundancy may exist and another enzyme may compensate for the low level of phytaspase during plant growth and development. The results presented below were typical of all transgenic lines; most tests were performed using silenced line KD1-1 generated with the RNAi fragment 1 and OE line OE-1.
Figure 3.
Relative expression of phytaspase ORF in transgenic N. tabacum Samsun NN plants with silenced (knocked down, KD) or overexpressed (OE) phytaspase ORF. (A, B) Phytaspase ORF expression was measured by qRT–PCR in wild-type (wt) plants and in three independent phytaspase-silenced transgenic lines KD1-1, KD2-1 and KD3-1 generated using different RNAi constructs (A) and three independent lines overexpressing phytaspase (B, OE-1, OE-2 and OE-3). The results presented in (A) and (B) were typical of all transgenic lines produced. Data are means±s.d. from six independent plants. The levels of ubiquitin mRNA used as a constitutively expressed internal control were similar in all the samples analysed above (data not shown). (C, D) Transgenic tobacco plants with overexpressed (OE) and silenced (KD) phytaspase display altered phytaspase enzymatic activity relative to wild-type (wt) plants. (C) Crude leaf extracts from three independent OE and KD lines and from wt plants were tested for in vitro cleavage of GFP-VirD2Ct substrate protein. Reaction mixtures were fractionated by 12% SDS–gel electrophoresis. An unknown protein band marked with an asterisk comes from extracts and indicates similar sample loading. Lane M, substrate protein cleaved with purified tobacco phytaspase. (D) Serial dilutions of arbitrary chosen extracts of each type allow quantification of phytaspase enzymatic activity: approximately four-fold increase in OE plants and approximately eight-fold decrease in KD plants, relative to wt plants.
Phytaspase is involved in the hypersensitive cell death response and resistance to TMV
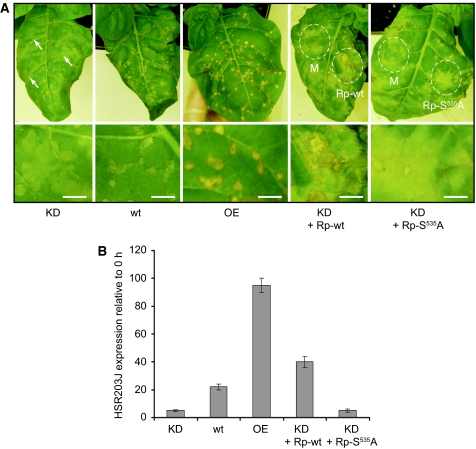
N. tabacum plants carrying the N gene (e.g. Samsun NN) are resistant to TMV and exhibit an HR at temperatures <27°C: TMV is localised to the vicinity of the necrotic lesions formed at the late stages of the HR (Kassanis, 1952). At higher temperatures (30°C), the resistance response is inoperative and TMV is able to multiply and spread, but after decreasing the temperature, the HR is synchronously induced in all of the infected cells (Mittler et al, 1995). To assess a potential function of phytaspase in the TMV-mediated HR, transgenic plants overproducing or silencing phytaspase were infected with TMV at 30°C and examined after induction of the HR by temperature shift to 24°C. TMV-mediated HR was suppressed in phytaspase-silenced (KD) plants, resulting in the formation of less severe amorphous lesions some of which continued to grow reaching up to 1 cm in size compared with wild-type plants, which under these conditions formed necrotic lesions of ∼2–3 mm in diameter (Figure 4A). In contrast, the HR in plants overproducing phytaspase (OE lines) resulted in the formation of more severe and sharply defined necrotic lesions compared with those in wild-type plants, although their sizes did not differ significantly (2–3 mm) (Figure 4A). These data indicate that deficiency of phytaspase markedly suppressed TMV-mediated HR, whereas its overproduction facilitated HR. The same conclusion was drawn from evaluation of expression of a plant gene, HSR203J that is an early marker of HR (Pontier et al, 1994) (Figure 4B). To ascertain whether the phenotypic effects observed on RNAi were due to specific silencing of the phytaspase gene, we performed a functional complementation assay: Agrobacterium-mediated expression of the heterologous wild-type rice phytaspase gene in the RNAi transgenic leaves 24 h before induction of HR by temperature shift (30–24°C) of leaves pre-infected with TMV at 30°C led to remarkable strengthening of HR (necrotisation of lesions and HSR203J expression) (Figure 4A and B). In contrast, expression of the inactive rice phytaspase (S535A mutant; Supplementary Figure 2) did not affect the HR phenotype of phytaspase-silenced plants, indicating specificity of RNA silencing (Figure 4A and B). Of note, no 21 nt regions of homology were found between any of the three RNAi constructs used for generation of transgenic plants and the rice phytaspase ORF.
Figure 4.
Effects of transgenic phytaspase deficiency or overproduction on PCD in tobacco (Samsun NN) plants induced by TMV. (A) TMV-mediated HR is suppressed in phytaspase-silenced (KD; line KD1-1) plants compared with control (wt) plants, resulting in the formation of less severe amorphous lesions some of which (shown by arrows) continue to grow. The HR in plants overproducing (OE; OE-1 line) phytaspase results in the formation of more severe and sharply defined necrotic lesions. Infected plants were incubated at 30°C for 36 h and then transferred to 24°C. The photographs were taken 12 h after the temperature shift. Heterologous Agrobacterium-mediated expression of the wild-type rice phytaspase gene in the phytaspase-silenced transgenic tobacco leaves (KD + Rp-wt), 24 h before temperature shift led to strengthening of necrotisation, whereas S535 A mutant of phytaspase (KD + Rp-S535A) did not affect the HR phenotype. Areas of agroinfiltration with empty vector (mock), Rp-wt or Rp-S535A constructs are indicated by dashed lines. Lower panels show higher magnification images. Bars are 5 mm. (B) Expression of the HR-specific marker gene HSR203J induced by TMV after the temperature shift is suppressed in phytaspase-silenced (KD; KD1-1 line) plants and is enhanced in phytaspase overproducing (OE; OE-1 line) plants compared with control (wt) plants as shown by qRT–PCR 5 h after the temperature shift. Heterologous expression of the wild-type rice phytaspase gene in the phytaspase-silenced transgenic tobacco leaves (KD + Rp-wt), 24 h before stress treatments led to increase in expression of HSR203J, whereas S535A mutant of phytaspase (KD + Rp-S535A) did not affect HSR203J levels. Data are means±s.d. from three experiments with three independent replicates in each. The levels of ubiquitin mRNA used as an internal control were similar in all of the samples. Data (including photographs) were typical of all generated KD and OE transgenic lines, respectively, tested in, at least, three experiments with three replicates in each.
Consistent with these observations, rates of TMV accumulation in phytaspase-silenced plants were markedly higher after the temperature shift compared with non-transgenic control plants, whereas in plants overproducing phytaspase, accumulation of TMV under these conditions was reduced (Table 1). When plants were continuously maintained at 30°C (in the absence of HR), no differences in TMV accumulation were found in plants expressing different levels of phytaspase (Table 1). Collectively, these data indicate that phytaspase is involved in the HR and N gene-mediated resistance to TMV.
Table 1. Effect of the phytaspase overproduction or deficiency on TMV accumulation in tobacco Samsun NN plants.
| Plant | Amount of TMV antigen (μg/g leaf) | |
|---|---|---|
| 30°C → 24°C | 30°C | |
| wt | 30±6 | 115±8 |
| OE | 6±2 | 110±7 |
| KD | 95±8 | 120±8 |
| Accumulation of TMV in wild-type (wt) and transgenic N. tabacum Samsun NN plants overexpressing (OE; OE-1 line) or silencing (KD; KD1-1 line) tobacco phytaspase, was determined by ELISA using serial dilutions of TMV preparation as concentration standards. TMV-infected tobacco plants were incubated at 30°C for 24 h and then transferred to 24°C. The samples were taken 24 h after the temperature shift. Control plants were incubated at 30°C for 48 h. Data shown are means±s.d. of three independent experiments with three replicates each. Data were typical of all generated KD and OE transgenic lines, respectively. | ||
Phytaspase is involved in the cell death induced by abiotic stress
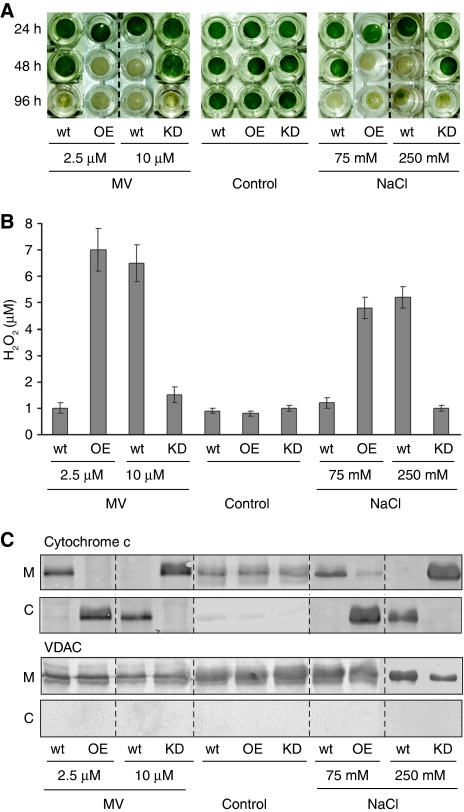
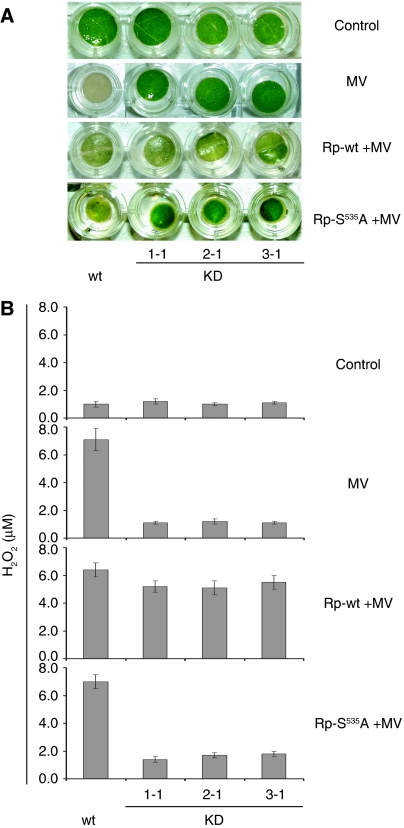
To assess whether phytaspase is also involved in the PCD-related response to abiotic stresses, we used methyl viologen (MV), a redox cycling oxidative stress inducer (Bassham, 2007) and NaCl, which causes osmotic stress, imbalance in the cellular ion concentration and oxidative stress (Affenzeller et al, 2009). Treatment of tobacco leaf discs with each chemical resulted in the loss of their viability (bleaching) (Figure 5A). These effects were associated with important hallmarks of PCD, accumulation of ROS (Skopelitis et al, 2006) expressed as H2O2 production (Figure 5B) and release of cytochrome c to the cytosol (Kim et al, 2003) (Figure 5C), which occurred before visible physiological changes. Overproduction of phytaspase in the OE transgenic lines significantly accelerated all of these processes induced by 2.5 μM MV and by 75 mM NaCl compared with wild-type plants (Figure 5A–C). To discriminate between the wild type and KD lines, higher concentrations of stress inducers were required. At 10 μM MV and at 250 mM NaCl, phytaspase silencing in KD transgenic lines considerably enhanced viability (suppressed bleaching) of leaf discs and impaired H2O2 accumulation and release of cytochrome c to the cytosol compared with wild-type plants (Figure 5A–C). Heterologous expression of the active rice phytaspase in the RNAi KD transgenic leaves before MV treatment led to restoration of the wild-type phenotype (including bleaching and H2O2 levels), whereas expression of the mutated (S535A) enzymatically inactive rice enzyme did not, indicating specificity of RNA silencing (Figure 6).
Figure 5.
Effects of transgenic phytaspase deficiency or overproduction on PCD in tobacco (Samsun NN) plants induced by abiotic stresses. (A) Loss of viability (bleaching) of leaf discs, (B) generation of H2O2 and (C) cytochrome c release from mitochondria (M) to cytosol induced by MV or NaCl (versus untreated controls) are facilitated in leaf discs overexpressing phytaspase (OE, OE-1 line) and are suppressed in phytaspase-silenced leaf discs (KD, KD1-1 line) compared with wild type (wt). (A) Leaf discs were immersed in the aqueous solutions of MV or NaCl, or control solution (lacking MV or NaCl) and photographed 24, 48 and 96 h after treatment. (B) H2O2 production was determined 24 h after treatment; data are means±s.d. from three experiments with three independent replicates in each. (C) Western blot analysis of mitochondrial (M) and cytosolic (C) fractions was performed with the cytochrome c monoclonal antibody 24 h after treatment. As a control for cell fractionation, western blots were probed with an anti-VDAC monoclonal antibody. VDAC is localised in the outer membrane of mitochondria (Tsujimoto and Shimizu, 2002). Concentrations of MV and NaCl are indicated. Data (including photographs) were typical of all generated KD and OE transgenic lines, respectively, tested in, at least, three experiments with three replicates in each.
Figure 6.
Functional complementation of phytaspase deficiency in methyl viologen (MV)-treated tobacco leaves. (A) Heterologous expression of the wild-type rice phytaspase gene (Rp-wt) in the phytaspase-silenced transgenic tobacco leaves, 24 h before stress treatments led to restoration of the wild-type PCD phenotype as exemplified for MV stress, whereas S535A mutant of phytaspase (KD + Rp-S535A) did not affect viability of leaf discs. Data are shown for three independent RNAi silencing lines (KD1-1, KD2-1 and KD3-1). MV at the concentration of 10 μM induced PCD in wild type (wt), but not in phytaspase RNAi (KD) plants. Photographs were taken 48 h after treatment. (B) Accumulation of H2O2 induced by MV (versus untreated control) was suppressed in phytaspase-silenced leaf discs compared with wild-type leaves and was restored by Agrobacterium-mediated transient expression of wild-type rice phytaspase (Rp-wt), whereas it was unaffected by S535A mutant of phytaspase (Rp-S535A). H2O2 production was determined 24 h after treatment. Data are means±s.d. from three independent experiments with three replicates in each. Data (including photographs) were typical of all generated KD and OE transgenic lines, respectively, tested in, at least, three experiments with three replicates in each.
Collectively, all these data indicate that phytaspase is also involved in pathways related to PCD induced by abiotic stresses operating upstream of ROS accumulation and cytochrome c release.
Localisation and processing of phytaspase
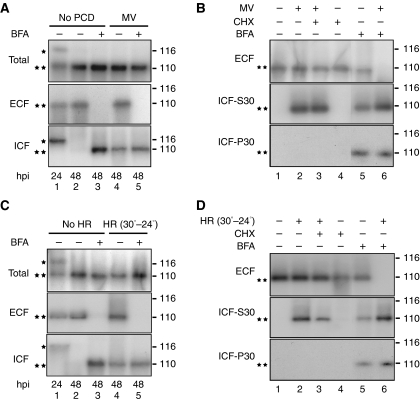
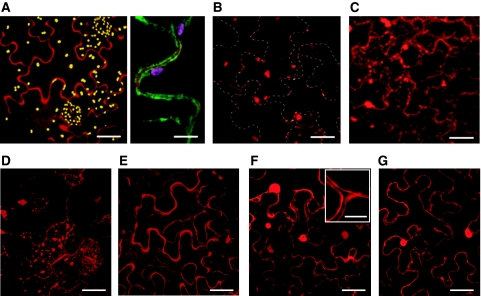
To determine whether the signal peptide targets phytaspase to the apoplast, monomeric red fluorescent protein (mRFP) was fused to the C-terminus of the full-length tobacco phytaspase and the construct was expressed in N. tabacum leaves by agroinfiltration. Leaf discs prepared from these plants were treated as described below. Western blots of total protein extracted from the untreated leaf discs (before PCD) were reacted with an mRFP antibody, and two protein bands of ∼120 and 110 kDa (Figure 7A, lane 1) were detected at 24 h post-infiltration (hpi), corresponding to the partially processed (proenzyme, lacking the predicted 24-amino-acid signal peptide) and completely processed (lacking the signal peptide and prodomain) proteins, respectively, as was shown by Edman degradation sequencing (Supplementary Figure S2). Fractionation of plant tissues at this stage showed that the mature phytaspase was almost completely localised in extracellular (apoplastic) fluid (ECF), whereas the proenzyme was exclusively associated with the intracellular fraction (ICF) (Figure 7A, lane 1). Interestingly, the proenzyme was specifically cleaved over time converting into the processed mature enzyme and secreted into the apoplast (48 hpi; Figure 7A, lane 2). Apoplastic localisation of the enzyme fused to mRFP was also confirmed by confocal microscopy (Figure 8A); measurement of optical density as described in Materials and methods in the images of 50 cells showed that ∼95% of fluorescence is associated with the apoplast.
Figure 7.
Localisation (subcellular fractionation) and processing of tobacco phytaspase. (A, B) Subcellular fractionation of phytaspase-mRFP before and after PCD induced by MV. PCD was induced in leaf discs prepared from N. tabacum Samsun NN leaves agroinfiltrated with phytaspase-mRFP construct by immersing in MV solution (10 μM) 42 hpi. (A) BFA was added to discs 24 hpi. (B) BFA and CHX were applied 42 hpi. Plant tissues were fractionated into apoplastic (ECF) and intracellular fractions (ICF, ICF-S30 and ICF-P30) as described in Materials and methods section 24 hpi (A) or 48 hpi (A, B). Concentrations of BFA and CHX were 10 and 100 μg/ml, respectively. Proteins presented in each fraction were analysed by western blot analysis using anti-mRFP antibody. *—and **—protein bands correspond to partially processed (proenzyme lacking the signal peptide, *) and completely processed (mature enzyme lacking the signal peptide and prodomain, **) proteins, as confirmed by Edman degradation sequencing (Supplementary Figure S2). Positions of MW markers are shown on the right. (C, D) Subcellular fractionation of phytaspase-mRFP before and after PCD induced by TMV (HR). N. tabacum Samsun NN leaves simultaneously agroinfiltrated with phytaspase-mRFP and infected with TMV were incubated at 30°C. PCD (HR) in leaf discs prepared from these leaves was induced by temperature shift to 24°C (30−24°C) 42 hpi. (C) BFA was added to discs 24 hpi. (D) BFA and CHX were applied 42 hpi. Plant tissues were fractionated into apoplastic (ECF) and intracellular fractions (ICF, ICF-S30 and ICF-P30) 24 hpi (C) or 48 hpi (C, D) as indicated. For other details see (A) and (B). Data were reproducible over three independent experiments.
Figure 8.
Localisation of phytaspase-mRFP in N. tabacum leaves (confocal microscopy). (A) Apoplastic localisation of phytaspase-mRFP. Agrobacterium-mediated expression of phytaspase-mRFP (48 hpi) results in largely apoplastic fluorescence (left panel) as confirmed by co-expression of this construct with the plasma membrane marker EGFP-LT16b, with clear mRFP fluorescence detected outside the cell membrane (right panel). Yellow (left panel) and purple (right panel) granules are chloroplasts showing natural autofluorescence. (B) Inhibition of phytaspase-mRFP secretion by BFA. BFA (10 μg/ml) was infiltrated 24 h after agroinfiltration of the phytaspase-mRFP construct and fluorescence was monitored 24 h later. Phytaspase-mRFP forms aggregates within the cell treated with BFA. Dashed lines show cell borders. (C) Phytaspase-mRFP is partially redistributed into cytoplasm during TMV-mediated HR. N. tabacum plants were simultaneously agroinfiltrated with phytaspase-mRFP construct and infected with TMV at 30°C. HR was induced by temperature shift to 24°C 24 hpi, and fluorescence was monitored 24 h later. (D) Redistribution of phytaspase-mRFP during stress induced by MV (similar redistribution was observed during NaCl-induced PCD). MV (10 μM) was applied 24 hpi, and fluorescence was monitored 24 h later (E) Apoplastic localisation of cathepsin B (Gilroy et al, 2007) is retained after stress treatment exemplified here with MV. Cathepsin B-mRFP fusion construct was agroinfiltrated into N. tabacum leaves and MV was applied 24 hpi. Fluorescence was monitored 24 h later. (F, G) Intracellular (presumably cytoplasmic and nuclear) localisation of free mRFP (used as an additional control) expressed from Agrobacterium in untreated N. tabacum leaves (F) was not changed during MV-induced PCD (G). Insert in (F) clearly shows unlabelled (by free mRFP) apoplastic space close to cell junctions. Of note, intracellular localisation of free mRFP (F, G) was different from that of phytaspase–mRFP under stress conditions (C, D). Scale bars are 15 μm [A-left panel, B, C, D, E, F (apart from insert) and G], 50 μm (A-right panel and insert (in F).
Treatment with brefeldin A (BFA), an inhibitor of secretion (Nebenführ et al, 2002), applied 24 hpi resulted in retention of phytaspase-mRFP within the cell during the next 24 h (48 hpi; Figures 7A and C, lane 3; Figure 8B), but did not affect processing of phytaspase (Figures 7A and C, lane 3), suggesting that secretion is not required for this process. Under these conditions, phytaspase-mRFP (produced and pre-imported into the apoplast before BFA treatment) was not detected in ECF presumably because of degradation within the apoplast (see Figure 7A, lane 3). During shorter treatments with BFA (as shown below for 6 h), some phytaspase-mRFP was still present in ECF (Figure 7B and D, lane 5).
PCD was induced in leaf discs at 42 hpi by treatment with MV, or by temperature shift (30–24°C) of leaves pre-infected with TMV at 30°C as described above, and leaf discs were analysed 6 h later. Processing of phytaspase leading to formation of the mature enzyme was not affected by these treatments (Figure 7A and C, lane 4 versus lane 2 and lane 5 versus lane 3). However, under these conditions mature phytaspase-mRFP was partially redistributed from ECF to ICF (Figure 7A and C, lane 4 versus lane 2). Accordingly, fluorescence of the fusion protein was also relocalised from the apoplast to inside the cell (Figure 8C and D); ∼40–70% of fluorescence was detected within the cells by measurements of optical density in images of 50 cells. Another protease, cathepsin B fused to mRFP known to be constitutively present in the apoplast (Gilroy et al, 2007), was not relocalised into the cell after induction of PCD by either MV or TMV (as exemplified by MV treatment in Figure 8E) indicating specificity of phytaspase redistribution.
Further fractionation of the ICF to separate soluble, mainly cytosolic (S30) and membranous (P30) proteins (as described in Materials and methods), showed that phytaspase-mRFP redistributed during MV- or TMV-induced PCD from the apoplast to inside the cell primarily localises to the soluble S30 fraction (Figure 7B and D, lane 2 versus lane 1). Treatment of leaf discs with BFA applied 42 hpi for 6 h led to retention of phytaspase-mRFP in both soluble S30 and membrane-enriched P30 fractions. Under non-stress conditions, phytaspase-mRFP was also detected in the ECF (Figure 7B and D, lane 5). In contrast, under stress conditions, phytaspase-mRFP was not detected in the ECF (Figure 7B and D, lane 6). Collectively, these data suggest that phytaspase, which had accumulated in the apoplast before BFA treatment, can be re-imported into the cell.
To verify whether PCD-induced redistribution of phytaspase-mRFP occurs through a retrograde transport of the enzyme from the apoplast, but is not due to cessation of phytaspase secretion under PCD conditions, the effect of the protein synthesis inhibitor, cycloheximide (CHX), on the redistribution was studied. Although CHX inhibited protein synthesis (including synthesis of phytaspase-mRFP) in the tobacco leaf discs (Supplementary Figure 3), CHX treatment did not affect the redistribution of phytaspase-mRFP from ECF to the intracellular (primarily soluble S30) fraction, induced by MV- or TMV-mediated PCD (Figure 7B and D, lanes 2, 3 and 4). It is thus conceivable that the enzyme under these PCD conditions is physically relocalised from the apoplast to inside the cell (possibly to the cytoplasm) rather than internalised within the cell after translation (which was blocked by CHX). The more detailed intracellular localisation of the mature phytaspase during PCD will be a major task for the future.
Intracellular accumulation of phytaspase upon stress is in full agreement with our earlier data, which showed the cleavage of the intracellularly located VirD2 protein during TMV-mediated HR, but not in the absence of HR (Chichkova et al, 2004). TMV infection in a non-HR host or at higher temperatures (in the absence of HR) did not lead to relocalisation of tobacco phytaspase into the cell (data not shown), suggesting a functional link between such a relocalisation and PCD.
Concluding remarks
As the first caspase-like activity was reported in plants (del Pozo and Lam, 1998), a growing body of information has accumulated on various plant caspase-like activities detected using synthetic tetrapeptide substrates and inhibitors of animal caspases (see for review, Bonneau et al, 2008). However, plants encode only distant homologues of caspases, metacaspases, which though do not possess caspase-specific proteolytic activity (Bozhkov et al, 2005; Watanabe and Lam, 2005; Vercammen et al, 2006; He et al, 2007; Sundström et al, 2009), may be involved in PCD. Instead, it has been shown that some plant proteases such as VPE (Hatsugai et al, 2004; Rojo et al, 2004; Kuroyanagi et al, 2005; Nakaune et al, 2005; Yamada et al, 2005) and the unidentified subtilisin-like serine protease, saspase (Coffeen and Wolpert, 2004), structurally different from classical caspases, display caspase-like substrate cleavage specificity. In this work, we report the identification and characterisation of a novel subtilisin-like protease from tobacco and rice named phytaspase that possesses caspase specificity distinct from that of other known caspase-like proteases. For example, VEID is a preferred motif for phytaspase cleavage that differentiates it from saspase, which does not cleave VEID at all (Coffeen and Wolpert, 2004). Another remarkable peculiarity of phytaspase is its specific localisation.
We suggest a model whereby after translation, phytaspase is activated (self-processed) and secreted into the apoplast in which it may be sequestered before PCD and/or perhaps fulfils a guarding function. In response to a variety of death-inducing stimuli, phytaspase is partially relocalised from the apoplast to inside the cell. Thus, in contrast to other known caspase-like proteases, which operate either in the apoplast (saspase) (Coffeen and Wolpert, 2004) or the vacuole (VPE; Hatsugai et al, 2004), phytaspase may function in both apoplast and intracellular compartments. It is also possible that cell exclusion might represent a way to prevent unintended phytaspase-mediated proteolysis of intracellular targets in the absence of PCD. This could be viewed as an alternative or additional plant-specific mechanism to the mammalian mechanism using caspase inhibiting proteins. Owing to its capability to hydrolyse a wide range of fluorogenic peptide-based caspase substrates (Figure 2C), phytaspase alone could account for the majority of caspase-like activities implicated in diverse plant PCD systems (Bozhkov et al, 2004; Bonneau et al, 2008). For example, it may operate at early stages of PCD (upstream of ROS generation and cytochrome c release) or may be involved in direct cleavage of final target proteins such as the VirD2 protein (Chichkova et al, 2004) and hence seems to be multifunctional. The challenge for the future is to elucidate how apoptotic proteases from animals (caspases) and plants (phytaspases), being structurally completely unrelated, manage to possess similar substrate cleavage specificity and function. The relative functions and interactions of other different caspase-like proteases and metacaspases in various subcellular locations during PCD also remain to be elucidated.
Materials and methods
Isolation and identification of phytaspase
Phytaspases were purified from N. tabacum ‘Xanthi nc' and Oryza sativa cv. ‘Leader' leaves as described (Chichkova et al, 2008). Phytaspase samples were then preincubated with 0.4 mM bio-TATD-CHO to obtain an adduct with high yield and subjected to affinity chromatography on a SoftLink Soft Release Avidin resin (Promega). Biotinylated proteins eluted with 5 mM biotin were analysed by SDS–PAGE and stained either with zinc-imidazole (for mass spectrometry) or with Coomassie Blue followed by blotting onto PVDF membrane (for N-terminal protein sequencing). Edman degradation sequencing was performed on a Protein Sequencing System 491cLC (Applied Biosystems).
Mass spectrometric protein identification
Proteins were subjected to in-gel reduction, alkylation and trypsin digestion (Chichkova et al, 2004). The resulting peptide mixtures were analysed by liquid chromatography multistage mass spectrometry on a Thermo Electron LTQ Fourier transform ion cyclotron resonance mass spectrometer equipped with an Eksigent nanoLC system. Data were analysed using the GPM software with tobacco EST and rice genomic databases from NCBI and TIGR.
Cloning, mutagenesis and production of recombinant tobacco phytaspase
cDNA was synthesised by the SMART method (Zhu et al, 2001) from total RNA isolated from N. tabacum meristematic tissues and used for 5′ and 3′ step-out RACE (Evrogen, Russia) as described (Matz et al, 1999). Oligonucleotide primers were designed on the basis of the EST GI 76871138 sequence. PCR products encoding the missing 5′ and 3′ portions of tobacco phytaspase ORF were cloned and sequenced. cDNA encoding the complete ORF was then generated by PCR using primers designed against the obtained sequence data, and SMART-amplified cDNA as template. The full-length PCR product (∼2500 bp) was cloned and sequenced. A full-length cDNA clone (AK067138) corresponding to rice phytaspase was obtained from the Rice Genome Resource Center of the National Institute of Agrobiological sciences. Mutations were introduced into cDNA using QuikChange site-directed mutagenesis kit (Stratagene). For production of recombinant tobacco phytaspase-GST, -GFP and -mRFP fusion proteins, PCR-amplified GST, GFP or mRFP cDNAs were in-frame ligated upstream of the termination codon of phytaspase ORF. The resultant cDNA was inserted into pLH7000 binary vector and transformed into A. tumefaciens. Leaves of N. benthamiana plants were agroinfiltrated and recombinant phytaspase-GST was purified using Glutathione Sepharose (GE Healthcare) chromatography.
In vitro assays of phytaspase proteolytic activity
Phytaspase activity of purified enzymes and crude extracts was determined using a protein substrate GFP-VirD2Ct (Chichkova et al, 2004, 2008). Peptide aldehyde inhibitors (Bachem; Calbiochem) were used at the concentrations indicated in the figures and figure legends. Control samples were supplied with anequivalent amount of DMSO. Fluorogenic peptide substrates (Anaspec; Bachem; Calbiochem) were tested at the concentration of 20 μM. Kinetic measurements of fluorescence intensities were performed at 37°C for 3 h in triplicate using FLUOstar OPTIMA reader equipped with 405 nm excitation and 520 nm emission filters (BMG Labtech).
Transgenic plants
For transgenic overexpression, the full-length tobacco phytaspase cDNA was cloned into pDONR207 to give the entry clone compatible with Gateway recombination system. The generated clone was inserted into the expression vector pROK2attR, which had the Gateway cassette (Invitrogen) cloned into pROK2. The resultant plasmid was electroporated into A. tumefaciens to transform N. tabacum Samsun NN as described (Taliansky et al, 2004). For transgenic silencing, the cDNA fragments described in Results and discussion section were cloned into the pDONR vector and transferred to a Gateway version of binary vector pFGC5941 containing Gateway cassettes flanking the chalcone synthase intron. This resulted in the cloned RNAi fragments being in opposite orientations leading to the formation of a hairpin loop. Plant transformation was performed as above. Expression of phytaspase gene in transgenic plants was analysed by qRT–PCR (Lacomme et al, 2003). For SYBR green-based real-time PCR (QuantiTest SYBR Green PCR kit, Qiagen), primer pairs were designed outside the regions of cDNA targeted for silencing using PRIMER EXPRESS software supplied with ABI PRISM 7700 Sequence Detection System (Applied Biosystems).
HSR203J gene expression analysis
This was performed essentially as described above for phytaspase mRNA.
Analysis of TMV multiplication
ELISA analysis of TMV multiplication was performed as described (Chichkova et al, 2004).
Abiotic stress treatments
Leaf discs were immersed in aqueous solutions of MV or NaCl (at concentrations indicated in Figure 5), vacuum infiltrated and incubated for up to 96 h in Petri dishes in continuous light.
Chemiluminescence assay for H2O2
Generation of ROS in leaf discs was assayed by measuring the H2O2-dependent luminescence of luminol as described (Gomez-Gomez et al, 1999) using a scintillation spectrometer LS 6000SE (Beckman).
Detection of cytochrome c release
Cytosolic and mitochondrial fractions were obtained as described by Kim et al (2003). Proteins were analysed by SDS–PAGE and probed with the antibodies against cytochrome c (Pharmingen) and human voltage-dependent anion channel (VDAC)/porin (Calbiochem).
Subcellular fractionation of phytaspase-mRFP fusion
Proteins in tobacco leaf apoplastic fluid (ECF) were isolated by vacuum-infiltrating leaf discs with a solution of 500 mM NaCl and 0.01% Silwet L-77 and centrifuging at 1350 g for 10 min at 4°C (Lohaus et al, 2001). It should be noted that the most abundant intracellular leaf protein, Rubisco, was not detected in any of the tested ECF fractions by western blot analysis using an anti-Rubisco antibody (data not shown), suggesting no detectable contamination of the apoplastic fluid with intracellular proteins. The remaining leaf material was macerated in 0.2 M Tris–HCl buffer (pH 9.0) containing 0.2 M KCl, 30 mM MgCl2, 0.2 M sucrose and 10 mM 2-mercaptoethanol. After filtering, the extract was centrifuged for 10 min at 1000 g to yield a low-speed pellet fraction (P1) enriched with nuclei and plastids (Mas et al, 2000). The P1 fraction contained phytaspase neither before nor after stress treatments (data not shown) and, therefore, the resulting supernatant was used as ICF. For further fractionation, ICF supernatant was centrifuged at 30 000 g for 30 min to produce a membrane-enriched high-speed pellet fraction (P30) and a supernatant (mainly cytosolic) soluble fraction (S30) (Mas et al, 2000). The S30 fraction was dialysed and concentrated using polyethylene glycol. Protein amounts in each fraction were quantified by using a Coomassie blue dye binding assay (Ghosh et al, 1988) and equal amounts were analysed by western blot assay.
Western blot analysis
For western blot analysis of phytaspase-GST, phytaspase-GFP and phytaspase-mRFP, proteins were extracted from plant tissues or subcellular fractions with SDS-loading buffer and immediately boiled. The immunoblots were probed with anti-GST, -GFP and -mRFP antibodies. Immunoreactions were detected using the ECL western blotting system (Amersham Pharmacia Biotech).
Confocal laser scanning microscopy
Localisation of phytaspase-mRFP was monitored using a Leica TCS-SP2 AOBS confocal microscope 48 h after agroinfiltration. mRFP was imaged using 561 nm excitation with emissions between 600 and 630 nm. GFP was imaged using 488 nm excitation with emissions between 500–530 nm. Quantification of the levels of accumulation of mRFP in the apoplast versus intracellular space was carried out using ImageJ software (Abramoff et al, 2004). For measurements of intensity of fluorescence, the images were converted to 8-bit greyscale and calibrated against a standardised optical density step tablet to give a mean optical density value for the apoplastic and intercellular areas separately. BFA was infiltrated 24 h after agroinfiltration of the phytaspase-mRFP construct and fluorescence was monitored 24 h later.
Immunoprecipitation of phytaspase-mRFP and its N-terminal sequencing
Protein extracts from plants expressing phytaspase-GFP or phytaspase-mRFP were fractionated by SDS–PAGE. Gel bands corresponding to phytaspase-GFP or phytaspase-mRFP detected by parallel western blot analysis were excised and subjected to immunoprecipitation as described (Kirstein et al, 2005) using anti-GFP or anti-mRFP antibodies, respectively. The bands separated by electrophoresis after immunoprecipitation were transferred to PVDF membranes and subjected to N-terminal sequencing by Edman degradation using Applied Biosystems 494 Procise Protein sequencing system.
Supplementary Material
Acknowledgments
This work was supported by grants from Russian Foundation for Basic Research (ABV, NVC), the Royal Society (MT and ABV), Rural Development Administration, Republic of Korea (BioGreen 21 Program, SHK) and by Scottish Government Rural and Environment Research and Analysis Directorate (MT, LT, JS, GD) and by Institute of Mitoengineering, Moscow State University. We thank P Birch (SCRI) for providing the cathepsin B-mRFP construct and T Ovchinnikova (Institute of Bioorganic Chemistry) for protein sequencing.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Internat 11: 36–42 [Google Scholar]
- Affenzeller MJ, Darehshouri A, Andosch A, Lütz C, Lütz-Meindl U (2009) Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J Exp Bot 60: 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC (2007) Plant autophagy-more than a starvation response. Curr Opin Plant Biol 10: 587–593 [DOI] [PubMed] [Google Scholar]
- Boatright KM, Salvesen GS (2003) Mechanisms of caspase activation. Curr Opin Cell Biol 15: 725–731 [DOI] [PubMed] [Google Scholar]
- Bonneau L, Ge Y, Drury G, Gallois P (2008) What happened to plant caspases? J Exp Bot 59: 491–499 [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Filonova LH, Suarez MF, Helmersson A., Smertenko AP, Zhivotovsky B, Von Arnold S (2004) VEIDase is a principal caspase-like activity involved in plant programmed cell death during plant embriogenesis. Cell Death Differ 11: 175–182 [DOI] [PubMed] [Google Scholar]
- Bozhkov PV, Suarez MF, Filonova LH, Daniel G, Zamyatnin AA Jr, Rodriguez-Nieto S, Zhivotovsky B, Smertenko A (2005) Cysteine protease mcII-Pa executes programmed cell death during plant embryogenesis. Proc Natl Acad Sci USA 102: 14463–14468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedzich A, Huttenlocher F, Kuhn BM, Pfannstiel J, Gabler L, Stintzi A, Schaller A (2009) The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SISBT3). J Biol Chem 284: 14068–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova NV, Kim SH, Titova ES, Kalkum M, Morozov VS, Rubtsov YP, Kalinina NO, Taliansky ME, Vartapetian AB (2004) A plant caspase-like protease activated during the hypersensitive response. Plant Cell 16: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkova NV, Galiullina RA, Taliansky ME, Vartapetian AB (2008) Tissue disruption activates a plant caspase-like protease with TATD cleavage specificity. Plant Stress 2: 89–95 [Google Scholar]
- Coffeen WS, Wolpert TJ (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16: 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Delorme V, Mailhac N, Gallois P (2000) Plant programmed cell death: a common way to die. Plant Physiol Biochem 38: 647–655 [Google Scholar]
- del Pozo O, Lam E (1998) Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr Biol 8: 1129–1132 [DOI] [PubMed] [Google Scholar]
- del Pozo O, Lam E (2003) Expression of the baculovirus p35 protein in tobacco affects cell death progression and compromises N gene-mediated disease resistance response to tobacco mosaic virus. Mol Plant Microbe Interact 16: 485–494 [DOI] [PubMed] [Google Scholar]
- Dickman MB, Park YK, Oltersdorf T, Li W, Clemente T, French R (2001) Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc Natl Acad Sci USA 98: 6957–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekert PG, Silke J, Vaux, DL (1999) Caspase inhibitors. Cell Death Differ 6: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Geptsein S, Heikkila JJ, Dumbroff EB (1988) Use of a scanning densitometer or an ELISA plate reader for measurement of nanogram amounts of protein in crude extracts from biological tissues. Anal Biochem 169: 227–233 [DOI] [PubMed] [Google Scholar]
- Gilroy E, Hein I, van der Hoorn R, Boevink PC, Venter E, McLellan H, Kaffarnik F, Hrubrikova K, Shaw J, Holeva M, Lopez EC, Borras-Hidalgo O, Pritchard L, Loake GJ, Lacomme C, Birch PRJ (2007) Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J 52: 1–13 [DOI] [PubMed] [Google Scholar]
- Greenberg JT (1997) Programmed cell death in plant-pathogen interactions. Annu Rev Plant Physiol Plant Mol Biol 48: 525–545 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Hansen G (2000) Evidence for Agrobacterium-induced apoptosis in maize cells. Mol Plant Microbe Interact 13: 649–657 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- He R, Drury GE, Rotari VI, Gordon A, Willer M, Farzaneh T, Woltering EJ, Gallois P (2007) Metacaspase-8 modulates programmed cell death induced by UV and H2O2 in Arabidopsis. J Biol Chem 283: 774–783 [DOI] [PubMed] [Google Scholar]
- Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Hoeberichts FA, Woltering EJ (2003) Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays 25: 47–57 [DOI] [PubMed] [Google Scholar]
- Kassanis B (1952) Some effect of high temperature on susceptibility of plants to infection with viruses. Ann Appl Biol 26: 358–369 [Google Scholar]
- Kim M, Ahn J-W, Jin U-H, Choi D, Paek K-H, Pai H-S (2003) Activation of programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem 278: 19406–19415 [DOI] [PubMed] [Google Scholar]
- Kirstein J, Zuhlke D, Gerth U, Turgay K, Hecker M (2005) A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J 24: 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi M, Yamada K, Hatsugai N, Kondo M, Nishimura M, Hara-Nishimura I (2005) VPE is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J Biol Chem 280: 32914–32920 [DOI] [PubMed] [Google Scholar]
- Lacomme C, Hrubikova K, Hein I (2003) Enhancement of virus-induced gene silencing through viral-based production of inverted-repeats. Plant J 34: 543–553 [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411: 848–853 [DOI] [PubMed] [Google Scholar]
- Lohaus G, Pennewiss K, Sattelmacher B, Hussmann M, Hermann Muehling K (2001) Is the infiltration-centrifugation technique appropriate for the isolation of apoplastic fluid? A critical evaluation with different plant species. Physiol Plant 111: 457–465 [DOI] [PubMed] [Google Scholar]
- Mas P, Sanchez-Pina MA, Balsalobre JM, Pallas V (2000) Subcellular localisation of Cherry leaf roll virus coat protein and genomic RNAs in tobacco leaves. Plant Sci 153: 113–124 [DOI] [PubMed] [Google Scholar]
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A (1999) Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res 27: 1558–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Shulaev V, Lam E (1995) Coordinated activation of programmed cell death and defense mechanisms in transgenic tobacco plants expressing a bacterial proton pump. Plant Cell 7: 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M, Hara-Nishimura I (2005) A vacuolar processing enzyme, δVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell 17: 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, Ritzenthaler C, Robinson DG (2002) Brefeldin A: deciphering an enigmatic inhibitor of secretion. Plant Physiol 130: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Godiard L, Marco Y, Roby D (1994) hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant pathogen interactions. Plant J 5: 507–521 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ (2008) MEROPS: the peptidase database. Nucleic Acids Res 36: D320–D325I [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ (2009) MEROPS, the peptidase database, http://merops.sanger.ac.uk/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reavy B, Bagirova S, Chichkova NV, Fedoseeva SV, Kim SH, Vartapetian AB, Taliansky ME (2007) Caspase-resistant VirD2 protein provides enhanced gene delivery and expression in plants. Plant Cell Rep 26: 1215–1219 [DOI] [PubMed] [Google Scholar]
- Rojo E, Martin R, Carter C, Zouhar J, pan S, Plotnikova J, Jin H, Paneque M, Sanchez-Serrano JJ, Baker B (2004) VPE [gamma] exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Shurvinton CE, Hodges L, Ream W (1992) A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc Natl Acad Sci USA 89: 11837–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Paschalidis KA, Pliakonis ED, Delis ID, Yakoumakis DI, Kouvarakis A, Papadakis AK, Stephanou EG, Roubelakis-Angelakis KA (2006) Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18: 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck TR, Lin TS, Kado CI (1990) VirD2 gene product from the nopaline plasmid pTiC58 has at least two activities required for virulence. Nucleic Acids Res 18: 6953–6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA Jr, Välineva T, Saarikettu J, Frilander MJ, Suarez MF, Zavialov A, Ståhl1 U, Hussey PJ, Silvennoinen O, Sundberg E, Zhivotovsky B, Bozhkov PV (2009) Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 11: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Taliansky M, Kim SH, Mayo MA, Kalinina NO, Fraser G, McGeachy KD, Barker H (2004) Escape of a plant virus from amplicon-mediated RNA silencing in associated with biotic or abiotic stresses. Plant J 39: 194–205 [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y (1998) Caspases: enemies within. Science 281: 1312–1316 [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Shimizu S (2002) The voltage-dependent anion channel: an essential player in apoptosis. Biochimie 84: 187–193 [DOI] [PubMed] [Google Scholar]
- Vercammen D, Belenghi B, van de Cotte B, Beunens T, Gavigan J-A, De Rycke R, Brackenier A, Inze D, Harris JL, Van Breusegem F (2006) Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J Mol Biol 179: 625–636 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E (2005) Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J Biol Chem 280: 14691–14699 [DOI] [PubMed] [Google Scholar]
- Wolf BB, Green DR (1999) Suicidal tendencies: apoptotic cell death by caspase family proteinases. J Biol Chem 274: 20049–20052 [DOI] [PubMed] [Google Scholar]
- Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS (2006) Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Shimada T, Nishimura M, Hara-Nishimura I (2005) A VPE family supporting various vacuolar functions in plants. Physiol Plant 123: 369–375 [Google Scholar]
- Zheng TS, Hunot S, Kuida K, Flavell RA (1999) Caspase knockouts: matters of life and death. Cell Death Differ 6: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Zhu YY, Machleder EM, Chenchik A, Li R, Siebert PD (2001) Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30: 892–897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.