Abstract
Cellular signalling cascades regulate the activity of transcription factors that convert extracellular information into gene regulation. C/EBPβ is a ras/MAPkinase signal-sensitive transcription factor that regulates genes involved in metabolism, proliferation, differentiation, immunity, senescence, and tumourigenesis. The protein arginine methyltransferase 4 PRMT4/CARM1 interacts with C/EBPβ and dimethylates a conserved arginine residue (R3) in the C/EBPβ N-terminal transactivation domain, as identified by mass spectrometry of cell-derived C/EBPβ. Phosphorylation of the C/EBPβ regulatory domain by ras/MAPkinase signalling abrogates the interaction between C/EBPβ and PRMT4/CARM1. Differential proteomic screening, protein interaction studies, and mutational analysis revealed that methylation of R3 constraines interaction with SWI/SNF and Mediator complexes. Mutation of the R3 methylation site alters endogenous myeloid gene expression and adipogenic differentiation. Thus, phosphorylation of the transcription factor C/EBPβ couples ras signalling to arginine methylation and regulates the interaction of C/EBPβ with epigenetic gene regulatory protein complexes during cell differentiation.
Keywords: chromatin remodelling, differentiation, histone code, post-translational modification, signalling
Introduction
The transcription factor C/EBPβ is a member of the CCAAT/enhancer-binding protein family that is composed of C/EBPα, β, δ, γ, ɛ, and ζ. C/EBPβ is expressed in a variety of cell types and participates in tissue-specific gene expression, proliferation, and differentiation in a hormone, cytokine, and nutrient-dependent manner. C/EBPβ controls important functions in liver homeostasis, regeneration, acute phase response, female reproduction, innate and adopted immunity, senescence, and receptor tyrosine kinase/ras oncoprotein-mediated tumourigenesis (Roesler, 2001; Farmer, 2006; Sebastian and Johnson, 2006; Nerlov, 2007; Zahnow, 2009).
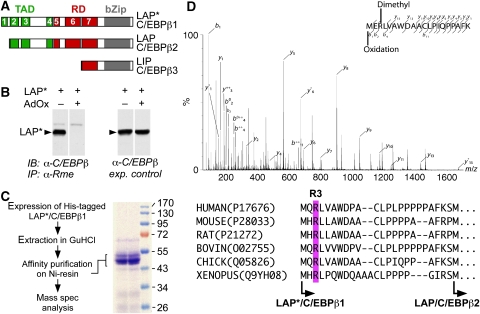
C/EBPβ carries at its N-terminus a modular, composite transactivation domain (TAD) that consists of four conserved regions (CR1–4), a composite regulatory domain (RD) consisting of CR5–7 at its centre that governs TAD functions, and a basic DNA binding and leucine dimerization domain (bZip) at the C-terminus (Kowenz-Leutz et al, 1994; Williams et al, 1995) (scheme in Figure 1A). Three C/EBPβ protein isoforms are expressed from a single, intronless transcript by signal-dependent alternative translation initiation from in-frame positioned start sites (Descombes and Schibler, 1991; Calkhoven et al, 2000). These N-terminally variant C/EBPβ isoforms of 38, 35, and 20 kDa are termed LAP*/C/EBPβ1, LAP/C/EBPβ2, and LIP/C/EBPβ3, respectively (Descombes and Schibler, 1991; Bundy and Sealy, 2003).
Figure 1.
C/EBPβ is post-translationally methylated on arginine residues. (A) Scheme of C/EBPβ isoforms that arise by alternative translation initiation from in-frame start codons termed as LAP*/C/EBPβ1, LAP/C/EBPβ2 and LIP/C/EBPβ3. (B) LAP*/C/EBPβ1 is precipitated by the ASYM24 anti-Rme2a antibody in the absence of AdOx. LAP*/C/EBPβ1-transfected fibroblasts were treated with adenosine dialdehyde (AdOx) for 12 h (as indicated) and immunoprecipitated with the ASYM24 antibody. Precipitated proteins were analysed by immunoblotting using an anti-C/EBPβ antibody. Right side shows expression controls in the absence and presence of AdOx. (C) Purification scheme of C/EBPβ. Briefly, His-tagged C/EBPβ was purified under denaturing conditions by affinity chromatography on a nickel chelating resin. Blot shows purified fractions of C/EBPβ. (D) Tandem mass spectrum of an R3me2 and oxidized N-terminal C/EBPβ peptide. b- and y-series of the LysC-generated peptides are indicated. Mass shifts indicate a 28 Da modification, corresponding to dimethylation at position R3. Underneath: alignment of conserved region 1 (CR1) of LAP*/C/EBPβ1 from various species shows conserved R3 (magenta box) and start sites (arrows) of the two long isoforms.
The C/EBPβ isoforms harbour distinct parts of the TAD and RD and display diverse or even opposite gene regulatory functions (Timchenko et al, 1999; Baer and Johnson, 2000; Calkhoven et al, 2000; Eaton et al, 2001). The two long isoforms (LAP*/C/EBPβ1 and LAP/C/EBPβ2) differ by a conserved sequence of 21–23 N-terminal amino acids in different species that represents CR1. CR1 functions as a gene regulatory module involved in the recruitment of the chromatin-remodelling SWI/SNF complex and the transcription regulatory Mediator complex (Kowenz-Leutz and Leutz, 1999; Pedersen et al, 2001; Mo et al, 2004). The short isoform (LIP/C/EBPβ3) lacks the TAD and part of the RD and represents a dominantly interfering protein that may neutralize both, transactivation and transrepression by all C/EBPs (Descombes and Schibler, 1991; Zahnow et al, 1997; Luedde et al, 2004).
The transcriptional activity of C/EBPβ is diversified by a multitude of interactions with other C/EBP family members, unrelated transcription factors, and transcription co-factors (Sebastian and Johnson, 2006; Nerlov, 2008; Zahnow, 2009). A major post-translational modification has been attributed to signalling through receptor tyrosine kinase–ras/mitogen-activated protein kinase (MAPK) pathway that phosphorylates an evolutionary conserved MAPK consensus site in the C/EBPβ RD (Thr-235 in human (Nakajima et al, 1993), Thr-188 in rat (Hanlon et al, 2001), Thr-220 in chicken (Kowenz-Leutz et al, 1994)). In its non-phosphorylated form, the RD of C/EBPβ is involved in masking the TAD through intra-molecular interaction (Kowenz-Leutz et al, 1994; Williams et al, 1995) and in binding a repressive or attenuated form of the Mediator complex. Phosphorylation of the MAPK site is accompanied by a conformational change in the protein, exchange of interacting Mediator components, and transcriptional activation (Mo et al, 2004).
Alignment and sequence comparison of vertebrate C/EBPβ revealed a number of highly conserved arginine residues in their N-termini that we considered to be involved in regulating C/EBPβ activity. Here, we show that the TAD of C/EBPβ interacts with the protein arginine methyltransferase PRMT4/CARM1 that methylates C/EBPβ at the highly conserved arginine residue at position 3 (R3) in CR1. Methylation of R3 is inversely correlated to phosphorylation of the MAPkinase site in the RD. The R3 methylation status determines the interaction with the SWI/SNF complex and with the Mediator complex to regulate endogenous target genes and cell differentiation in an MAPkinase signalling-dependent manner.
Results
C/EBPβ is methylated at arginine residues
Alignment and sequence comparison of vertebrate C/EBPβ family members revealed several conserved arginine residues in low complexity regions of the N-terminus. We considered the possibility that arginine side chains of C/EBPβ are post-translationally modified by methylation. As shown in Figure 1B, the LAP*/C/EBPβ1 isoform was immunoprecipitated by an antibody specific to methylated arginine (Rme). No Rme-specific immunoprecipitation was observed in the presence of adenosine dialdehyde (AdOx, a homocysteine hydrolase inhibitor that blocks the regeneration of the cellular methyl donor S-adenosyl methionine) (Bartel and Borchardt, 1984; Chen et al, 2004). The two long C/EBPβ isoforms reacted with antibodies that specifically recognize asymmetrically dimethylated arginine (ASYM24) or symmetrically dimethylated arginine (SYM10) side chains (Supplementary Figure 1). A small amount of the truncated LIP/C/EBPβ3 isoform was detected with the ASYM24 antibody, but not with SYM10 (Supplementary Figure 1B). Metabolic labelling with L-[methyl-3H]methionine in the presence of cycloheximide and chloramphenicol revealed tracer incorporation only in the long C/EBPβ isoforms (data not shown) (Pless et al, 2008). These results supported the idea that C/EBPβ is post-translationally modified by both, asymmetrical and symmetrical dimethylation on arginines. This notion was scrutinized by mass spectrometric analysis. A histidine-tagged LAP*/C/EBPβ1 construct was expressed in fibroblasts and enriched under denaturing conditions by nickel chelating affinity chromatography (Figure 1C). C/EBPβ fractions were digested to peptides, separated on a nano-HPLC system and analysed by MS/MS mass spectrometry, as shown in Figure 1D. The mass shift (28 m/z) of the modified R3 residue in the b-series of fragment ions identified a dimethylated arginine at position 3. Multiple-reaction monitoring (MRM) mass spectrometry from cell lysates approved the occurrence of arginine dimethylation on R3 of endogenous LAP*/C/EBPβ1 in cells, as shown in Supplementary Figure 2. As the N-terminally LAP*-specific peptide has a clearly defined modular function in chromatin remodelling and gene regulation, we focused on the molecular biology and functions of C/EBPβ R3 dimethylation; however, several other Rme/Rme2 residues were discovered and will be described elsewhere (data not shown). Taken together, the data showed that cellular C/EBPβ is modified by dimethylation of the conserved R3 (Figure 1D, lower part).
Proteomic screening reveals C/EBPβ R3 methylation-sensitive protein interactions
Protein interactions with non-modified and R3 asymmetrically dimethylated peptides (aa 1–41) were analysed by proteomic screening of a UNIPEX human cDNA expression library that covers approximately one third of the human proteome. Bacterially expressed His-tagged recombinant proteins, immobilized in duplicates on PVDF membrane were screened in replicate libraries with C-terminally biotinylated peptides that encompassed C/EBPβ CR1–2 with R3 either in its unmethylated or asymmetrically dimethylated form (R3me2a), followed by detection with streptavidine coupled to horseradish peroxidase, as shown in Figure 2A. Numerous interactions detected with methylated and unmethylated peptides fall into three distinct categories: protein interactions with no preference for either peptide, interactions that favoured R3 methylation, and interactions that favoured unmodified R3 (data not shown). Thus, the methylation status of C/EBPβ R3 directs the interaction between CR1 and other proteins.
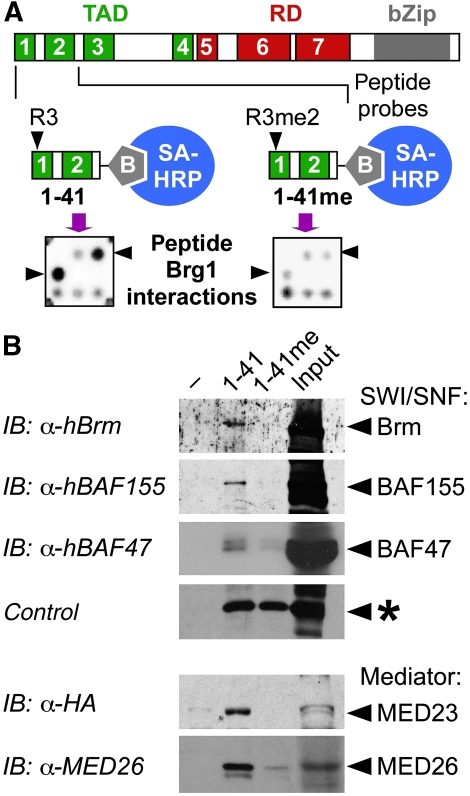
Figure 2.
Differential interaction of SWI/SNF and Mediator complexes with R3 methylated and unmethylated LAP*/C/EBPβ1 N-terminal peptides. (A) The N-terminal sequence of LAP*/C/EBPβ1 was synthesized with R3 in unmodified form (1–41) or in asymmetrically dimethylated form (1–41 Rme2a). Peptides with covalently attached C-terminal biotin moieties (grey pentagons) were applied to UNIPEX proteomic libraries and interactions were revealed by streptavidin-HRP (SA-HRP) and ECL. Positive clones were identified as characteristic duplicate pattern per square. Libraries contained 48x48 squares with four duplicate clones on two PVDF membranes. A single square containing the brama-related gene 1 (Brg1) expression clone interacts with unmethylated, but not with methylated peptide (arrow heads). (B) Unmethylated and methylated C/EBPβ peptides (1 μM), as shown in (A) were incubated with cell lysates and bound proteins separated by streptavidin Dynabeads. Cell lysates were prepared from Raji cells (top) or HEK-293 MED23-HA-transfected cell lysates (bottom). Western blots were incubated with anti-hBrm, anti-BAF155, anti-BAF47/Ini1, anti-HA, and anti-MED26 as indicated.
Of particular interest was the interaction with two core components of the SWI/SNF chromatin-remodelling complex, Brg1 (Figure 2A) and BAF47/Ini1 (data not shown). Both recombinant Brg1 and BAF47/Ini1 proteins interacted preferentially with non-modified peptide and weakly with the methylated peptide. Differential interaction between the SWI/SNF complex and unmethylated versus dimethylated peptide was confirmed in pull-down assays with cell lysates, as shown by retention of SWI/SNF subunits hBrm, BAF155, and BAF47/Ini1 (Figure 2B). In addition, Mediator components MED23/Sur2 and MED26 (not contained in the proteomic library), earlier shown to interact with CR1, likewise bound to unmodified LAP*/C/EBPβ1 N-terminal peptide (Figure 2B, bottom). These results are in accordance with the earlier genetic and biochemical results (Kowenz-Leutz and Leutz, 1999; Mo et al, 2004) and we conclude that methylation of R3 affects the interaction with SWI/SNF and Mediator complexes.
The effect of R3 modifications on protein complex binding was examined by substituting R3 with alanine (R3A) or leucine (R3L) to eliminate the methylation target or to mimic increased hydrophobicity after methylation, respectively. Immunoprecipitation and GST-pull-down assays with LAP*/C/EBPβ1 WT, R3A, and R3L mutants and SWI/SNF or Mediator components (Supplementary Figure 3A and B) showed preferential interaction of the R3A LAP*/C/EBPβ1 mutant with hBrm and with the Mediator component MED23, whereas R3L displayed decreased interactions with both complexes. Thus, the R3A and the R3L C/EBPβ1 mutant proteins reflect the binding specificity of the screening peptides and may serve as suitable tools for functional investigations.
Alterations in R3 specify gene regulation by LAP*/C/EBPβ1
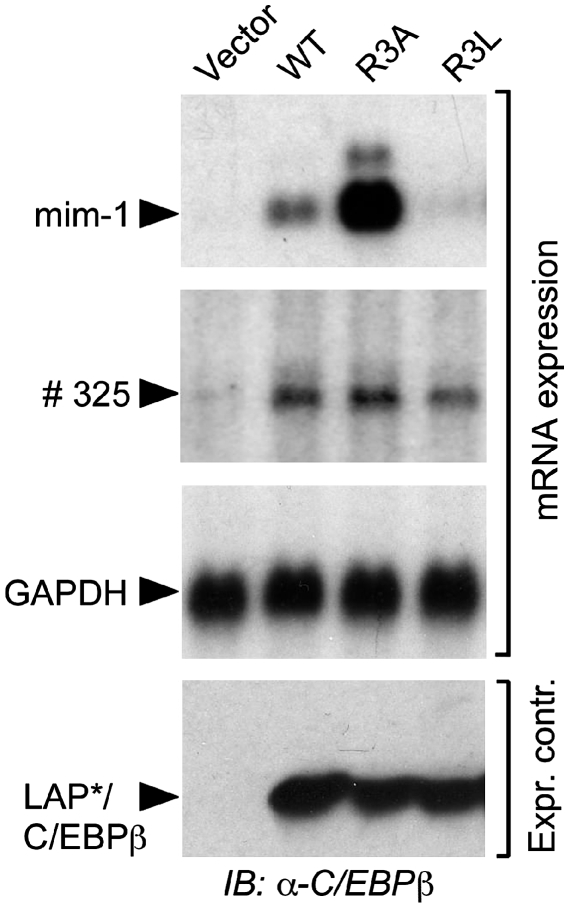
The interaction between CR1 and SWI/SNF is critical for the activation of a subset of C/EBPβ target genes, whereas other C/EBPβ target genes remain unaffected by CR1 (Kowenz-Leutz and Leutz, 1999). Therefore, the effects of R3 mutations were examined on the CR1-dependent endogenous myeloid target gene mim-1 and the CR1-independent goose-type lysozyme gene #325. As shown in Figure 3, the R3A LAP*/C/EBPβ1 mutant strongly activated endogenous mim-1 expression in comparison with WT LAP*/C/EBPβ1 that can be methylated at R3. In contrast, the R3L mutant was barely active. Importantly, WT and both mutants activated the #325 gene to similar extends. Thus, the gene regulatory activity of R3A and R3L mutants reflected CR1-specific co-factor interactions and CR1 R3-specific functions on chromatin-embedded target gene regulation.
Figure 3.
LAP*/C/EBPβ1 R3 mutations alter C/EBPβ target gene activation. LAP*/C/EBPβ1 constructs were transfected into QT6 fibroblasts as indicated. RNA was extracted 18 h post-transfection and subjected to serial northern hybridization to probes directed to the mim-1 gene, #325 gene, and GAPDH as a control.
PRMT4/CARM1 binds to and methylates C/EBPβ
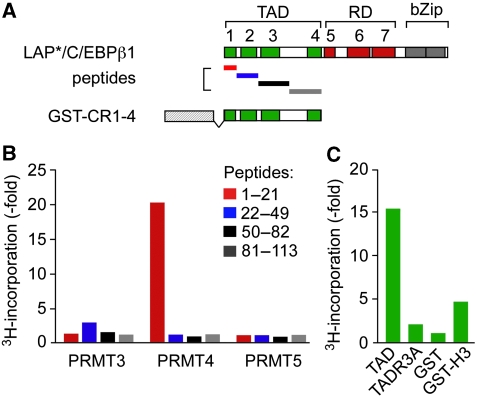
Lymphoid and myeloid cells that express C/EBPβ also highly express the protein arginine methyltransferases PRMT3, PRMT4/CARM1 (both: asymmetric arginine metylation), and PRMT5 (symmetric arginine methylation) (BioGPS). We, therefore, examined whether R3 of the LAP*/C/EBPβ1 isoform represents a target for one or more of these PRMTs. PRMT3, 4, and 5 were transiently expressed in HEK-293 cells, immunoaffinity purified from cell lysates, and incubated in the presence of S-adenosyl-L-[methyl-3H]methionine with peptides P1, P2, P3, and P4, tiling the TAD of the rat C/EBPβ N-terminus (Figure 4A; P1: aa 1–21; P2: aa 22–56; P3: aa 50–82; P4: aa 81–113). As shown in Figure 4B, radioactivity was specifically incorporated with PRMT4/CARM1 in P1, representing CR1 of LAP*/C/EBPβ1. Label incorporation was approximately 20-fold higher in P1 than in P2, P3, or P4 that also contained arginine and/or lysine residues. All three PRMTs were equally expressed (Supplementary Figure 4A) and functionally active, as determined by incorporation of 3H-methyl into recombinant GST-Histone H3 and H4 N-termini, but not into the GST moiety (Supplementary Figure 4B). These data show that PRMT4/CARM1 specifically methylates the CR1 N-terminus of LAP*/C/EBPβ1.
Figure 4.
PRMT4 methylates the N-terminus of C/EBPβ. (A) Scheme of C/EBPβ N-terminal peptides and C/EBPβ N-terminal GST-protein. (B) In vitro methylation of C/EBPβ N-terminal peptides by PRMT3, PRMT4/CARM1, and PRMT5. HA-tagged PRMT3, 4, and 5 were expressed in HEK-293 cells, purified from cell lysates by immunoprecipitation with anti-HA and incubated with 1 μM of the corresponding peptides as substrates in the presence of L-[methyl-3H]methionine as a 3H-methyl-donor. Bars represent relative incorporation of labelled S-adenosyl-L-[methyl-3H]methionine. (C) In vitro methylation of recombinant C/EBPβ N-terminal GST proteins by PRMT4/CARM1. Reactions were carried out as described in (B) with 3 μg GST protein as a substrate. Bars represent relative incorporation of labelled S-adenosyl-L-[methyl-3H]methionine. Histone H3 was used as a positive and the GST moiety as a negative control for PRMT4/CARM1-specific L-[methyl-3H]methionine labelling.
Methylation assays with the entire GST-C/EBPβ TAD (CR1–4; aa 1–113) showed that the C/EBPβ-WT, but not the R3A mutant TAD, was methylated by PRMT4/CARM1 (Figure 4C). The PRMT4/CARM1 enzyme activity and the C/EBPβ GST-protein expression were approved by auto-methylation and protein staining, respectively (Supplementary Figure 4C). In summary, these data confirm R3 of C/EBPβ as a PRMT4/CARM1 methylation target.
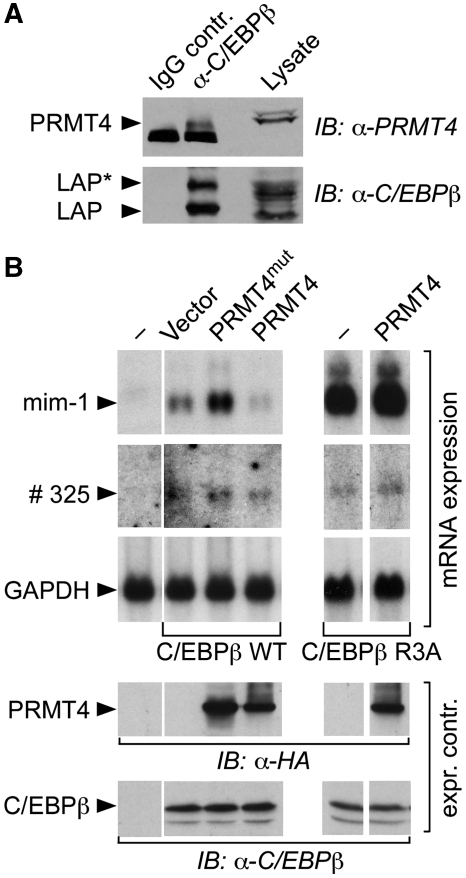
Immunoprecipitation of C/EBPβ from K562 cells revealed that endogenous PRMT4/CARM1 and C/EBPβ interacted in myeloid cells (Figure 5A). Co-immunoprecipitations of WT PRMT4/CARM1 or a catalytically inactive PRMT4/CARM1 mutant (amino-acids 189–191 VLD to AAA, dubbed PRMT4/CARM1mut) (Chen et al, 1999) with LAP*/C/EBPβ1 showed that interaction did not depend on the catalytic activity of the methyltransferase (Supplementary Figure 5A). Deletion mapping of C/EBPβ revealed PRMT4/CARM1 association with the C/EBPβ TAD (Supplementary Figure 5B) and GST-pull downs showed that PRMT4/CARM1 interacts preferentially with C/EBPβ TAD CR3 and CR4 (Supplementary Figure 5C).
Figure 5.
Physical and functional interaction between C/EBPβ and PRMT4/CARM1 in eukaryotic cells. (A) K562 cell lysates were incubated with antibodies (IgG, negative control) or anti-C/EBPβ and antigen–antibody complexes were immunoprecipitated. Proteins were analysed by immunoblotting with anti-PRMT4/CARM1 or anti-C/EBPβ as indicated. IB: immunoblot. (B) LAP*/C/EBPβ1 WT or R3A constructs, as indicated, were transfected in QT6 fibroblasts together with WT or PRMT4/CARM1mut constructs. RNA blots were subjected to serial hybridization to the mim-1, #325 and GAPDH gene probes.
Examination of the functional consequences of PRMT4/CARM1 interaction with C/EBPβ showed that the catalytically defective PRMT4/CARM1mut enhanced activation of the myeloid LAP*/C/EBPβ1 target gene mim-1, whereas WT PRMT4/CARM1 decreased activation of mim-1 (Figure 5B). Expression of the CR1-independent #325 myeloid gene remained indifferent to WT PRMT4/CARM1 or its mutant, excluding a general repressive effect of PRMT4/CARM1 on the transcriptional machinery or on C/EBPβ target genes. Importantly, co-expression of WT PRMT4/CARM1 did not suppress mim-1 activation by the non-methylatable C/EBPβ1 R3A mutant (Figure 5B, right), providing further evidence that methylation of R3 specifically interferes with the activation of LAP*/C/EBPβ1 CR1-dependent target genes. The results suggest that the methylation status of the R3 side chain has a decisive function in the regulation of SWI/SNF complex-dependent target genes.
C/EBPβ phosphorylation abrogates PRMT4/CARM1 interaction
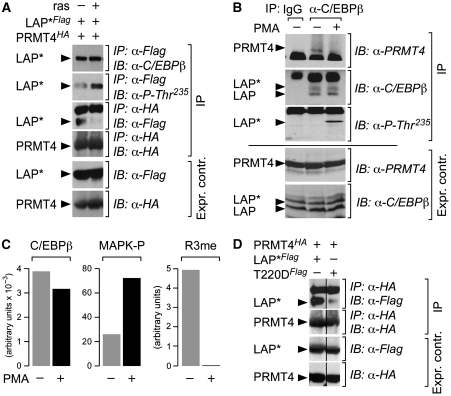
C/EBPβ is a repressed transcription factor that can be activated by EGF receptor tyrosine kinase and ras signalling through the MAP kinase Erk1/2 pathway (Nakajima et al, 1993; Kowenz-Leutz et al, 1994; Fan et al, 2009). Phosphorylation of the conserved MAP kinase site in the central RD of C/EBPβ is accompanied by a conformational change in C/EBPβ that alters interactions with protein complexes (Kowenz-Leutz et al, 1994; Williams et al, 1995; Mo et al, 2004). We, therefore, examined whether ras signalling also alters interaction between PRMT4/CARM1 and C/EBPβ. As shown in Figure 6A, co-expression of activated rasV12 with C/EBPβ increased phosphorylation at the MAPkinase site. Concomitantly, binding of PRMT4/CARM1 to C/EBPβ was diminished. Immunoprecipitation of endogenous C/EBPβ from K562 cells approved that PRMT4/CARM1 interacts with C/EBPβ in untreated, but not in phorbol ester (phorbol-12-myristate-13-acetate, PMA)-treated cells (Figure 6B). MRM of endogenous C/EBPβ from K562 cells showed inverse correlation between phosphorylation of the MAPK-site and dimethylation of R3 of LAP*/C/EBPβ1 and disappearance of R3 methylation after PMA treatment (Figure 6C).
Figure 6.
Effect of ras/MAPkinase signalling or a phospho-mimetic mutation on the interaction between C/EBPβ and PRMT4/CARM1. (A) FLAG-tagged LAP*/C/EBPβ1 was co-expressed with HA-tagged PRMT4/CARM1 in QT6 fibroblasts in the absence and presence of rasV12. Immunoprecipitation (IP) from cell lysates with anti-Flag or anti-HA antibody as indicated (top) and protein expression control (below, expr. control). The anti-P-Thr235 antibody specifically reveals C/EBPβ that is phosphorylated at the MAPkinase site. IP: immunoprecipitation. IB: immunoblot. (B) Lysates from K562 cells (with/without PMA treatment) were immunoprecipitated with anti-C/EBPβ or negative control IgG. Proteins were analysed by immunoblotting with anti-PRMT4/CARM1, anti-C/EBPβ, or anti-P-Thr235, as indicated. Protein expression control below (expr. contr.). IP: immunoprecipitation. IB: immunoblot. (C) Relative quantification of phosphorylation and R3me2 on LAP*/C/EBPβ1 by multiple-reaction monitoring (MRM). Left: three different tryptic peptides were used for quantification of C/EBPβ in total cell lysates from untreated and PMA-treated K562 cells, as indicated. Middle: quantification of the C/EBPβ Thr-235 phosphorylation MAPkinase site containing tryptic C/EBPβ peptide. Right: quantification of the R3-LAP*/C/EBPβ1 methylated tryptic peptide in total cell lysates by MRM. (D) FLAG-tagged LAP*/C/EBPβ1 or LAP*/C/EBPβ1 T220D constructs were co-expressed with HA-tagged PRMT4/CARM1 in QT6 fibroblasts. Cell lysates were immunoprecipitated with anti-HA and immunoblots (IP) were developed with anti-Flag or anti-HA, respectively (indicated on the right). IP: immunoprecipition. IB: immunoblot. Below: protein expression control (anti-Flag or anti-HA) as indicated. Vertical lines indicate assembly of two separated lanes on the same immunoblot.
Earlier, it was reported that EGF receptor tyrosine kinase signalling inhibits PRMT4/CARM1 methyltransferase activity and subsequent histone H3 methylation (Higashimoto et al, 2007). We, therefore, examined whether activated C/EBPβ, as represented by the LAP*/C/EBPβ1 phospho-mimetic MAPkinase site mutant T220D, interacts with PRMT4/CARM1. As shown in Figure 6D, interaction between PRMT4/CARM1 and C/EBPβ T220D was strongly diminished, as compared with WT C/EBPβ. In accordance, the C/EBPβ T220D mutant activated adipogenic target gene expression and fat cell differentiation, as compared with WT, whereas the C/EBPβ T220A displayed strongly diminished activity (Supplementary Figure 7). Taken together, these results suggested that phosphorylation of the C/EBPβ RD abolishes the interaction between the TAD of C/EBPβ and PRMT4/CARM1 and as a consequence leads to abrogation of methylation of R3 LAP*/C/EBPβ1.
Alteration of the LAP*/C/EBPβ1 R3 residue affects myeloid and adipogenic differentiation
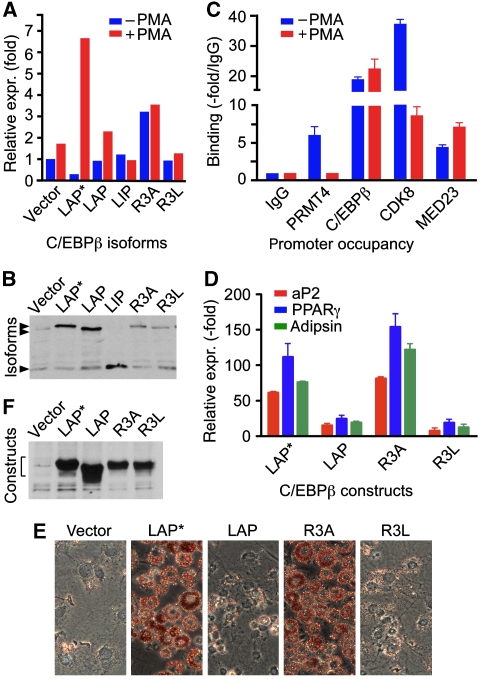
C/EBPβ associates with the human neutrophil elastase gene (hELA2) and activates hELA2 expression in an MAPkinase signal-dependent manner (Nuchprayoon et al, 1997; Lausen et al, 2006; Pless et al, 2008). Myeloid gene activation in heterologous cells and reprogramming by C/EBPβ are well established (Laiosa et al, 2006; Zahnow, 2009). Therefore, we examined whether the myeloid hELA2 gene can be activated by LAP*/C/EBPβ1 in fibroblasts. Expression of hELA2 in NIH 3T3 fibroblasts was strongly enhanced after PMA treatment with LAP*/C/EBPβ1, but not with LAP/C/EBPβ2 or LIP/C/EBPβ3. The LAP*/C/EBPβ1 R3A mutant was significantly more active than WT LAP*/C/EBPβ1, yet barely induced by PMA (Figure 7A, expression controls in 7B). These data are consistent with the rational that R3 in its unmethylated and methylated form is involved in activation and repression processes and that replacement of R3 (R3A) partially compromises both functions. The LAP*/C/EBPβ1 R3L mutant did not display significant activity with or without PMA, consistent with the notion that replacement of R3 with leucine stabilizes the inhibitory state of LAP*/C/EBPβ1. These data show that CR1 of LAP*/C/EBPβ1 has an important function in the activation of the endogenous hELA2 gene in fibroblasts.
Figure 7.
Myeloid and adipogenic gene regulation. (A) Expression of neutrophile elastase (hELA2) transcripts by C/EBPβ isoforms in NIH 3T3 cells before and after PMA treatment. NIH 3T3 cells were transfected with constructs (as indicated) and after 24 h stimulated with phorbol ester (PMA) for 1.5 h. Total mRNA and cDNA were prepared and hELA2 gene expression was analysed by RT–PCR and normalized to GAPDH expression. (B) Expression control of C/EBPβ isoforms detected by immunoblotting. (C) ChIP assay from either unstimulated or PMA-stimulated U937 cells. Antibodies were used as indicated. Quantitative PCR results are shown as fold binding compared with the IgG control. (D) Activation of PPARγ, aP2, and adipsin expression by LAP*/C/EBPβ1, LAP/C/EBPβ2 isoforms, and the LAP*/C/EBPβ1 R3A or R3L mutants in NIH 3T3 L1 cells in the absence of adipogenic differentiation hormone cocktail. NIH 3T3 L1 cells were transfected with vector, LAP*/C/EBPβ1, LAP/C/EBPβ2, LAP*/C/EBPβ1 R3A, and LAP*/C/EBPβ1 R3L and stable transfectants selected by puromycin. Cells were grown to confluency and total mRNA and cDNAs were prepared 10 days after confluency. The results were normalized to GAPDH expression. (E) Oil red O staining of stably transfected cells, ten days post-confluency, as shown in (D). (F) Protein expression control of cells as shown in (D).
Endogenous C/EBPβ in human U937 myeloid cells binds to a 117 bp fragment upstream of the hELA2 TATA-box (Nuchprayoon et al, 1997; Lausen et al, 2006) and PMA-induced phosphorylation of the C/EBPβ MAPkinase site upregulates hELA2 expression (Pless et al, 2008). As shown in Figure 7C, both C/EBPβ and PRMT4/CARM1 were found associated with the hELA2 promoter in U937 cells. Reciprocal re-immunoprecipitation of either C/EBPβ or PRMT4/CARM1 confirmed simultaneous occupancy of the hELA2 promoter (Supplementary Figure 6). PMA treatment abrogated association of PRMT4/CARM1 with the hELA2 promoter, whereas C/EBPβ occupancy persisted (Figure 7C). In addition, the Mediator complex component MED23/Sur2 was associated with the hELA2 promoter before PMA stimulation and increased after PMA treatment. In contrast, CDK8, a component of attenuated Mediator, was five-fold decreased after PMA treatment, probably reflecting the earlier described exchange between Mediator complex modules (Mo et al, 2004). Taken together, these data support the notion that C/EBPβ phosphorylation is accompanied by dissociation from PRMT4/CARM1 and loss of the Mediator CDK8 module during hELA2 gene activation.
C/EBPβ also regulates adipogenesis and controls early activation of a cascade of transcription factors that finally orchestrate adipogenic differentiation (Cao et al, 1991; Wu et al, 1995; Yeh et al, 1995; Mandrup and Lane, 1997; Birsoy et al, 2008). Recruitment of the SWI/SNF complex is an essential prerequisite for induced onset of the adipogenic program (Pedersen et al, 2001; Caramel et al, 2008). We, therefore, asked whether stable expression of WT and mutant LAP*/C/EBPβ1 isoforms in NIH 3T3 L1 cells alters adipogenesis. As shown in Figure 7E (expression controls Figure 7F), fat cell differentiation was strongly enhanced by the LAP*/C/EBPβ1 R3A mutant, as indicated by upregulation of the adipogenic genes PPARγ, aP2, and adipsin (Figure 7D), in the absence of the usually required hormone cocktail. In contrast, expression of the LAP/C/EBPβ2 isoform or the LAP*/C/EBPβ1 R3L mutant in NIH 3T3 L1 cells did not induce spontaneous differentiation. These results suggest that enhanced SWI/SNF recruitment by unmethylated R3 in LAP*/C/EBPβ1 CR1 predisposes mesenchymal precursor cells to adipogenic differentiation.
Discussion
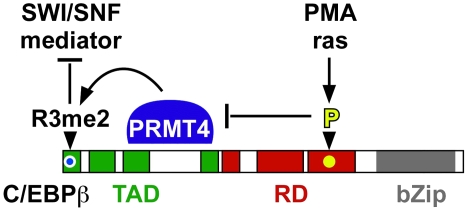
The transcription factor C/EBPβ functions at the interface of cell metabolism, stress, proliferation, and differentiation. Data presented here show that ras/MAPkinase signalling is coupled to R3 methylation and that the R3 methylation state specifies interactions between C/EBPβ, SWI/SNF and Mediator. Our data show how extracellular signals initially converted to kinase signalling connect to arginine methylation of a transcription factor that in turn determines interaction with the epigenetic and gene regulatory machinery in the nucleus to regulate gene transcription and differentiation. The data are summarized in the conceptual model shown in Figure 8.
Figure 8.
Model of crosstalk between phosphorylation and R3 methylation of C/EBPβ.
A distinctive feature of the intronless single C/EBPβ transcript is that alternative translation initiation at consecutive in-frame start sites may generate three protein isoforms with different N-termini that display activator and repressor functions (Descombes and Schibler, 1991; Calkhoven et al, 2000). The CR1 comprises 21–23 amino acids in different species and is contained in the largest LAP*/C/EBPβ1 isoform. LAP*/C/EBPβ1 regulates a subset of C/EBPβ target genes that are involved in mammary epithelial integrity, proliferation and cell differentiation, innate immunity, and potentially also fat metabolism (Eaton et al, 2001; Bundy and Sealy, 2003; Uematsu et al, 2007). Our data show that the conserved arginine at position 3 (R3) can be specifically methylated by PRMT4/CARM1 and that the methylation status of R3 determines the interaction between CR1 and other proteins. Proteins that interact with the unmethylated CR1 include components of the SWI/SNF complex and Mediator complex. Both multi-subunit protein complexes had been described earlier to mediate important functions of LAP*/C/EBPβ1 isoforms during chromatin remodelling and gene regulation.
Mass spectrometry combined with mutational analysis, biochemical, molecular genetic, and cell biological data establishes that PRMT4/CARM1 interacts with the TAD of C/EBPβ and dimethylates CR1 specifically at the evolutionary conserved R3 residue. Methylation of R3 impairs the interaction with SWI/SNF and Mediator complexes and, therefore, attenuates activation of myeloid and adipogenic genes. It has recently been shown that adipogenic induction requires activation of C/EBPβ expression and that the LAP* isoform was able to bypass an early regulatory network of pro-adipogenic transcription factors (Birsoy et al, 2008). At the same time, genes that are not affected by the presence of CR1 are exempt from R3 regulation. This interpretation is in agreement with a modular and appendicular function of N-terminal CR1 (Pedersen et al, 2001) and is supported by a recent knock in study that shows that LAP*/C/EBPβ1 may largely replace LAP/C/EBPβ2 (Uematsu et al, 2007).
Of particular attention is the finding of a functional crosstalk between ras/MAPkinase signalling, arginine methylation of C/EBPβ and recruitment of an epigenetic complex. Our data show that phosphorylation of a distal site and methylation of the N-terminal R3 has opposing functions on C/EBPβ-mediated gene activation. The interaction between PRMT4/CARM1 and C/EBPβ and their co-association on the hELA2 promoter is abolished when LAP*/C/EBPβ1 is phosphorylated. Concomitantly, the silent myeloid hELA2 C/EBPβ target gene was activated in fibroblasts, reflecting epigenetic activation of myeloid gene expression by LAP*/C/EBPβ1. The R3A mutation in C/EBPβ that removes the methylation target displayed enhanced co-factor interactions, myeloid gene activation, induction of adipogenesis, and, importantly, resists the repressive effect of PRMT4/CARM1. We conclude that in the absence of signals, association of PRMT4/CARM1 specifically restrains LAP*/C/EBPβ1 functions by R3 methylation, without repressing the overall C/EBPβ activity.
Earlier, we had shown that C/EBPβ is also methylated on a highly conserved lysine residue by G9a and that methylation at K39 compromised its transactivation (Pless et al, 2008). Here, we show that MAPkinase-phosphorylated C/EBPβ is undermethylated. Accordingly, our data support the idea of a negative crosstalk between C/EBPβ phosphorylation and methylation and raise the possibility that in the absence of an activating signal, C/EBPβ becomes methylated and remains transcriptional inactive or displays repressor functions, whereas MAPkinase signalling transiently abrogates C/EBPβ methylation and permits co-activator recruitment and gene activating functions.
Phosphorylation of the RD following receptor tyrosine kinase–ras/MAPK signalling has been shown to activate LAP*/C/EBPβ1 by inducing a structural change that affects interaction and composition of associated Mediator complex (Mo et al, 2004). Data presented here suggest that phosphorylation of the C/EBPβ RD also curtails interaction with PRMT4/CARM1 that is associated with inactive C/EBPβ. In this context, it is interesting to note that in studies of promoter-specific PRMT4/CARM1 functions, enhanced expression of the C/EBPβ target genes Il6 and COX2 (Akira et al, 1990; Poli, 1998; Gorgoni et al, 2001) was observed in CARM1-deficient cells (Covic et al, 2005), supporting an inhibitory function of CARM1 for distinct C/EBPβ functions.
PRMT4/CARM1 is primarily considered in gene transactivation; however, a repressive function on gene transcription has also been described for the histone acetyltransferase CREB-binding protein that when methylated at the KIX domain failed to interact with the transcription factor CREB. In addition, methylation of the steroid receptor co-activator 3 complex leads to co-activator disassembly and decreased steroid receptor-mediated transcription (Xu et al, 2001; Feng et al, 2006). Co-activator functions of PRMT4/CARM1 were described for nuclear hormone receptor-mediated target gene activation, including PPARγ-mediated adipogenesis, which is reduced by 40% in adipose tissue in PRMT4/CARM1 KOs (Yadav et al, 2008). It, therefore, seems that the activity of PRMT4/CARM1 is dispensable for the early activation of adipogenic co-factors (supported by data shown in Supplementary Figure 8), whereas PRMT4/CARM1 is required as a co-activator for PPARγ functions in terminal differentiation. In support of this notion is the observation that MAPkinase signalling and IBMX (enhances protein kinase-A signalling) supports early, but not late, adipogenic differentiation (Student et al, 1980; Kortum et al, 2005).
A function of PRMT4/CARM1 is the methylation of histone H3 at R17 (Ma et al, 2001; Bauer et al, 2002; Yadav et al, 2003). Although PRMT4/CARM1 is not the only H3R17-specific methyltransferase, it would have been informative to monitor the H3R17 methylation status in the vicinity of C/EBPβ-binding sites in the genome. Unfortunately, attempts to determine the H3R17 methylation status failed, as available R17 methylation-specific antibodies were found to cross-react with methylated C/EBPβ (data not shown). A more detailed analysis of chromatin structure and C/EBPβ R3 methylation must, therefore, await the availability of more specific reagents that discriminate between histone and C/EBPβ R3 modification states.
Interaction between the N-terminus of LAP*/C/EBPβ1 and the SWI/SNF complex was originally detected in chromatin-remodelling deficient yeast and confirmed biochemically and molecular genetically in vertebrate cells (Kowenz-Leutz and Leutz, 1999). Differential interaction of SWI/SNF complex proteins was now found again by proteomic screening with peptides that represent unmethylated and methylated derivatives of the LAP*/C/EBPβ1 N-terminus. These data confirm a modular function of CR1 in SWI/SNF recruitment and show the feasibility of proteomic expression screening approaches with transactivation modules that contain or lack post-translational modifications to uncover differential protein interactions.
Methylation of arginine side chains may either enhance interactions by increasing hydrophobicity or act as a hydrogen donor (for review see Dacwag et al, 2009; Lee and Stallcup, 2009), although more often abrogation of interactions between proteins has been observed. For example, it has been shown that methylation of RUNX1 by PRMT1 interferes with binding of the SIN3A complex, thus leading to gene activation by removal of an inhibitor (Zhao et al, 2008). However, our proteomic screening results support the notion that methylation of R3 not only abrogates interactions, but also enhances protein interactions. Whether or not such interactions through R3 contribute to gene silencing effects by LAP*/C/EBPβ1 is currently under investigation.
In addition to modifications at R3, C/EBPβ may carry a multitude of modifications including phosphorylation, acetylation, lysine, and arginine methylation at different sites, similar to the well-studied example of p53 (Yang, 2005; Olsson et al, 2007; Scoumanne and Chen, 2008). We, therefore, assume in analogy to a ‘histone code' (Turner, 1998, 2000; Jenuwein and Allis, 2001) a ‘transcription factor code' of modifications that tunes transregulatory functions (Sims and Reinberg, 2008). C/EBPβ might serve as a signal-dependent docking platform for epigenetic enzymes that once recruited may also alter the functionality of C/EBPβ by decoration with additional PTMs that, in further analogy to the histone code, may serve as docking/blocking/assembly sites for novel-interacting complexes. This notion has medical, pharmacological, and developmental implications, as ‘epigenetic drugs' might not only affect histone/chromatin modifications, but also transcription factor functions. The technical advance shown here, combining mass spectrometry with PTM-dependent proteomic screening, will help to establish a more general insight into the succession, interdependence, and functions of signal-dependent PTM patterns on C/EBPβ and interacting protein complexes.
Materials and methods
Peptides
C/EBPβ peptides covering the TAD were synthesized and are derived from the mouse sequence (Swiss-Prot P28033)—peptide 1: aa 1–21; peptide 2: aa 22–55; peptide 3: aa 49–81; peptide 4: aa 80–113. For protein macroarray screening and peptide pull-down analysis, two peptides were synthesized and HPLC purified covering aa 1–41 in a non-methylated and R3 asymetrically dimethylated form (PSL).
Protein macroarray screening (UNIPEX)
Two identical UNIPEX protein macroarrays (containing approximately 20 000 sequenced, annotated and verified cDNA clones; ImaGenes, Berlin) were overlayed with non-modified and R3 asymmetrically dimethylated N-terminal C/EBPβ peptides covering aa 1–41. In brief, protein macroarrays were rinsed in water, washed with TBS-T-T (500 mM NaCl, 20 mM Tris−HCl (pH 7.5), 0.05% Tween 20, 0.5% Triton X-100) and excess bacterial colony material was scraped off the macroarray. Subsequently, membranes were blocked with 3% skim milk in TBS-T (500 mM NaCl, 20 mM Tris−HCl (pH 7.5), 0.05% Tween 20) for 2 h. The libraries were incubated with 1 μM for both, unmodified and modified peptides, in blocking solution over night. Interaction partners were detected by using streptavidin-HRP and ECL. Positive scoring clones were localized, according to the manufacturer's instructions.
Peptide pull down
Streptavidin dynabeads were saturated with non-modified or R3 asymmetrically dimethylated N-terminal C/EBPβ peptides for 30 min in PBS (pH 7.4). Excess peptides were washed off with wash buffer (PBS (pH 7.4), 0.01% Tween, 0.1% BSA). Beads were then incubated with cell extracts prepared from Raji or HEK-293 cells for 1 h (lysis buffer: 20 mM Hepes (pH 7.6), 350 mM NaCl, 30 mM MgCl2, 1 mM EDTA (pH 8), 0.1 mM EGTA (pH 8), 20% glycerol, 0.5% NP40). After washing, bound proteins were eluted with SDS–PAGE loading buffer. Western blots were incubated with appropriate antibodies and revealed by ECL.
In vitro methylation
PCDNA3.1-HA PRMT3, 4, or 5 were transfected into HEK-293 cells; 48 h post transfection, cells were lysed (50 mM Tris pH 8, 150 mM NaCl, 0.1% NP40, 1 mM EDTA, 5 mM MgCl2 plus protease inhibitors) and immunoprecipitations were performed from lysates with anti-HA for 3 h. Immunocomplexes were coupled to protein G-coated Dynabeads (Invitrogen) for 1 h, washed with lysis buffer and used directly in in vitro methylation studies. Reactions were carried out in PBS, supplemented with 1 μCi of S-adenosyl-L-(methyl-3H)methionine as methyl donor and incubated with either 1 μM of corresponding peptides or 1–2 μg of C/EBPβ GST-fusion proteins for 3 h at 30°C. Three parts of the reaction was submitted to SDS–PAGE, methylated proteins were detected by fluorography, and one part of the reaction was spotted on phosphocellulose membrane to determine [3H]methyl incorporation by scintillation counting.
Mass spectrometry
pcDNA3-NFM-8xHis constructs were transiently transfected, expressed in QT6 quail fibroblasts, and extracted under denaturing conditions in lysis buffer (100 mM NaH2PO4 (pH 8.0); 10 mM Tris−HCl (pH 8.0); 6 M guanidine-HCl; 20 mM Imidazole; 1 mM Tris(2-carboxyethyl)phosphine; 1 mM Pefabloc; 40 μg/ml Bestatin; protease inhibitor cocktail (Roche)). Affinity purification was carried out on an ÄKTA purifier (GE Healthcare) equipped with a HisTrap HP Nickel column (GE Healthcare). Elution was performed with an Imidazole gradient (20–250 mM). For the identification of arginine methylation on C/EBPβ, peptides were generated using Lysyl-endopeptidase (LysC) as described (de Godoy et al, 2008). Peptides were separated by reverse phase chromatography on a nano-Acquity UPLC (Waters) and analysed by electrospray ionization on a Q-TOF premier mass spectrometer (Waters). Peptide spectra were assigned using the MASCOT software (Matrix Science) and refined by hand interpretation. For quantification of endogenous C/EBPβ R3 modification by MRM of untreated or PMA-treated K562 or U937, cell lysates were prepared in 6 M Urea, 2 M Thiourea, 10 mM HEPES (pH 8.0), 1 mM Pefablock, and protease inhibitor cocktail (Roche). Fragment ion spectra were obtained by fragmenting an arginine R3 dimethyl-modified hC/EBPβ N-terminal peptide. The peptide was injected into mass spectrometer using electrospray ionization. Fragment spectra were recorded in an MDS/Sciex AB 5500 Q-TRAP mass spectromenter and analysed with the Analyst 1.5 Software (Applied Biosystems). Signature fragments were selected for the creation of a MRM method; 5 μg of total cells extract were recorded with dwell times ranging from 5 to 100 ms. The intensities were integrated with the MRMPilot Software package (Applied Biosystems) and normalized to the actin measurement. Statistical analysis was performed using the R software package (V2.10).
Cell culture and transfection
C33A, HEKT-293, NIH 3T3, and NIH 3T3 L1, MEF −/+ PRMT4/CARM1 cells were grown in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 10% FCS (Invitrogen) and QT6 cells were grown in DMEM containing 8% FCS and 2% heat-inactivated chicken serum (Invitrogen). Raji, U937, and K562 cells were cultured in RPMI (Invitrogen) containing 10% FCS. All media were supplemented with 1% penicillin/streptomycin and cells were grown at 5% CO2, 37°C. Transient transfections were performed by calcium-phosphate precipitation or Metafectene (Biontex) according to the manufacturer's protocol.
Endogenous gene activation assay and northern blot analysis
Resident gene activation was determined after transient transfection of indicated constructs in QT6 fibroblasts as described earlier (Kowenz-Leutz et al, 1994). Briefly, total RNA was extracted by the guanidinium-isothiocyanate method, selected by magnetic oligo(dT) beads (Dynal), RNA was separated on 1.2% formaldehyde-agarose gel, transferred to nylon membrane (Hybond N+, Amersham), 32P-labelled probes to mim-1, #325 or GAPDH transcripts were hybridized in series to filters in Quik-Hyb hybridization solution (Stratagene) as indicated.
Plasmid constructs
Construction of C/EBPβ expression plasmids is based on the chicken sequence (accession number EMBL Z21646). The LAP*/C/EBPβ1 start site was altered by exchange of glutamine to glutamic acid at position 2 to generate an optimized Kozak consensus sequence. C/EBPβ-conserved regions and corresponding amino acids in C/EBPβ were published earlier (Kowenz-Leutz et al, 1994). Briefly, CR1 corresponds to AA 1–13; CR2 to AA 18–38; CR3 to AA 42–63; CR4 to AA 99–113; CR5 to AA 184–222; CR6 to AA 145–179; CR7 to AA 184–222, and CR8–9 (bZIP region) to AA 243–317. C/EBPβ mutations were introduced by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by sequencing.
GST constructs were cloned from PCR fragments derived from C/EBPβ or Histone H3 and H4 N-terminal tails as described earlier (Kowenz-Leutz and Leutz, 1999; Mo et al, 2004). All PRMT plasmids were generated by RT–PCR from HEK-293 or K562 cell cDNAs. PRMT3 was subcloned as BamHI/XhoI Fragment and PRMT4/CARM1 and PRMT5 expression plasmids were cloned as BamHI/EcoRI Fragments in pCDNA3.1-HA (Invitrogen).
PRMT3 primer: 5′-ATAGGATCCATGTGCTCGTTAGCGTCAGGC and 3′-TATCTCGAGTCACTGGAGACCATAAGTTTG. CARM1 primer: 5′-TGGGATCCCCGATGGCAGCGGCGGCAGCGGCG and 3′-CGGAATTCGCTCCCGTAGAGCATGGTGTTGGT. PRMT5 primer: 5′-CGCGGATCCGTGATTGGCTACTAGTATCAAGGAATCCCGGCGTGGACA and 3′-CGGGAATTCCTAGAGGCCAATGGTATATGA. CARM1VLD189−191 mutant was constructed as described (Chen et al, 1999).
GST-pull-down experiments
Bacterial expression and preparation of GST-fusion proteins was performed according to standard procedures. For pull-down assays, GST-fusion proteins were bound to Glutathione sepharose 4B and incubated with in vitro translated MED23 or PRMT4/CARM1 (TNT-Kit, Promega).
Immunoprecipitation and immunoblotting
Immunoprecipitations of C/EBPβ and hBrm, expressed in C33A cells, were performed as described (Kowenz-Leutz and Leutz, 1999). Briefly, cell lysates were prepared in lysis-buffer A (20 mM Hepes (pH 7.9), 350 mM NaCl, 30 mM MgCl2, 1 mM EDTA (pH 8), 0.1 mM EGTA (pH 8), 20% glycerol, 0.5% NP40, supplemented with protease inhibitor cocktail (Roche Applied Bioscience)). For IP, lysates were diluted with Buffer B (20 mM Hepes (pH 7.9), 30 mM MgCl2, 1 mM EDTA pH 8, 1 mM EGTA pH 8, 20% glycerol, 0.2% NP40). Immunoprecipitations of CARM1 and hBrm were performed in lysis buffer 50 mM Tris (pH 8), 150 mM NaCl, 0.5% NP40, 1 mM EDTA (pH 8) supplemented with protease inhibitor cocktail. Samples were immunoprecipitated with appropriate antibodies for 2–3 h at 4°C as indicated and immunoprecipitates were collected on protein A- or G-Sepharose beads. Immunoprecipitations of C/EBPβ constructs and PRMT4/CARM1 were performed after transient transfection of QT6 fibroblasts, HEK-293 cells, or K562 cells for endogenous protein interaction studies. Briefly, cells were lysed in buffer (50 mM Tris (pH 8), 150 mM NaCl, 0.5% NP40, 1 mM EDTA, and protease inhibitors). Samples were immunoprecipitated with appropriate antibodies for 2 h at 4°C as indicated and immunoprecipitates collected on protein A- or G-Sepharose beads. Antibodies: anti-C/EBPβ (Leutz lab), anti-C/EBPβ (Santa Cruz; C-19, H-7), anti-phospho-Thr235 C/EBPβ (Cell Signaling Technology; #3084), anti-FLAG (Sigma), anti-HA.11 (Covance), anti-ASYM24 (Upstate; 07-414), anti-SYM10 (Upstate; 07-412), anti-methyl Arginine (Abcam; ab412), anti-PRMT4/CARM1 (Cell Signaling Technology; #4438), anti-Brm (Biomol; A301-014A), anti-BAF155 (Biomol; A301-019A), anti-BAF47/Ini1 (Sigma; H9912), anti-MED23 (Santa Cruz; SC-12454), anti-MED26 (Santa Cruz; SC-9425), and anti-CDK8 (Santa Cruz; SC-1521). Immunoblots were visualized by ECL (GE Healthcare).
Chromatin immunoprecipitation
Chromatin immunoprecipitations with subsequent quantitative PCR of the hELA2 gene promoter and IgG control were performed as described (Pless et al, 2008). Re-ChIP analysis was performed using the ‘Re-ChIP-IT Magnetic Chromatin Re-immunoprecipitation kit' (Active Motif, #53016) according to the manufacturer's protocol.
Adipogenesis and RT–PCR analysis
Vector control, LAP*/C/EBPβ1, LAP/C/EBPβ2, LAP*/C/EBPβ1 R3A, or LAP*/C/EBPβ R3L were transfected by Metafectene according to the manufacturer's protocol (Biontex) in NIH 3T3 L1 fibroblasts and selected by 1.5 μg/ml puromycin. Stable transfectans were seeded in triplicates on tissue culture dishes and grown to confluency. Ten days past confluency, total RNA was isolated from one dish with High Pure RNA isolation Kit (Roche) and cDNA was prepared using SuperScript cDNA Kit (Invitrogen). Real-time PCR was performed using SYBR Green (Invitrogen) and a Light Cycler (Roche) according to the manufacturer's instructions. aP2 forward primer: CAAAATGTGTGATGCCTTTGTG; reverse primer: CTCTTCCTTTGGCTCATGCC; PPARγ forward primer: GCATGGTGCCTTCGCTGATGC; reverse primer: TACGTTTATCTGGTGTTTCAT; Adipsin forward primer: GCTATCCCAGAATGCCCTCGTT; reverse primer: CCACTTCTTTGTCCTCGTATTGC; PGC1a forward primer: TGCGTGTGTGTATGTGTGTGTG; reverse primer: CCTTGTTCGTTCTGTTCAGGTG. Alternatively, spontaneous NIH 3T3 L1 adipogenesis was visualized after fixation with 4% paraformaldehyd by oil red O staining. Protein expression of stable transfectans was determined by western blot.
Supplementary Material
Acknowledgments
We thank M Vermeulen and M Mann for initial help with mass spectrometric analysis, M Bedford for providing PRMT4/CARM1 KO MEF cell line and rescued MEFs, A Heuser for help with microscopy, U Bauer, and J Lausen for reagents and discussion and K Friedrich for excellent technical assistance. E Kowenz-Leutz, M Knoblich, and G Dittmar are supported by institutional funds of the Helmholtz Association and O Pless is supported by a grant of the Deutsche Forschungsgemeinschaft (DFG) to A Leutz (LE 770/4-1).Author contributions: EKL, OP, and AL designed, and EKL and OP performed experiments and analysed data. MK expressed and purified proteins, and GD performed mass spectrometric analysis. EKL, OP, and AL prepared the paper. AL supervised the work.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T (1990) A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J 9: 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer M, Johnson PF (2000) Generation of truncated C/EBPbeta isoforms by in vitro proteolysis. J Biol Chem 275: 26582–26590 [DOI] [PubMed] [Google Scholar]
- Bartel RL, Borchardt RT (1984) Effects of adenosine dialdehyde on S-adenosylhomocysteine hydrolase and S-adenosylmethionine-dependent transmethylations in mouse L929 cells. Mol Pharmacol 25: 418–424 [PubMed] [Google Scholar]
- Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T (2002) Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep 3: 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Chen Z, Friedman J (2008) Transcriptional regulation of adipogenesis by KLF4. Cell Metab 7: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy LM, Sealy L (2003) CCAAT/enhancer binding protein beta (C/EBPbeta)-2 transforms normal mammary epithelial cells and induces epithelial to mesenchymal transition in culture. Oncogene 22: 869–883 [DOI] [PubMed] [Google Scholar]
- Calkhoven CF, Muller C, Leutz A (2000) Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev 14: 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Umek RM, McKnight SL (1991) Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev 5: 1538–1552 [DOI] [PubMed] [Google Scholar]
- Caramel J, Medjkane S, Quignon F, Delattre O (2008) The requirement for SNF5/INI1 in adipocyte differentiation highlights new features of malignant rhabdoid tumors. Oncogene 27: 2035–2044 [DOI] [PubMed] [Google Scholar]
- Chen CS, Matsuoka R, Arai S, Momiyama Y, Murakami H, Kuno SY, Ishikawa K, Nakada K, Tawata M, Hayashi J (2004) Determination of normal ranges of mitochondrial respiratory activities by mtDNA transfer from 54 Human subjects to mtDNA-less HeLa cells for identification of the pathogenicities of mutated mtDNAs. J Biochem 135: 237–243 [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR (1999) Regulation of transcription by a protein methyltransferase. Science 284: 2174–2177 [DOI] [PubMed] [Google Scholar]
- Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO (2005) Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J 24: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacwag CS, Bedford MT, Sif S, Imbalzano AN (2009) Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol Cell Biol 29: 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- Descombes P, Schibler U (1991) A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell 67: 569–579 [DOI] [PubMed] [Google Scholar]
- Eaton EM, Hanlon M, Bundy L, Sealy L (2001) Characterization of C/EBPbeta isoforms in normal versus neoplastic mammary epithelial cells. J Cell Physiol 189: 91–105 [DOI] [PubMed] [Google Scholar]
- Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM, Richards JS (2009) MAPK3/1 (ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 324: 938–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR (2006) Transcriptional control of adipocyte formation. Cell Metab 4: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O'Malley BW (2006) Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 26: 7846–7857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Caivano M, Arizmendi C, Poli V (2001) The transcription factor C/EBPbeta is essential for inducible expression of the cox-2 gene in macrophages but not in fibroblasts. J Biol Chem 276: 40769–40777 [DOI] [PubMed] [Google Scholar]
- Hanlon M, Sturgill TW, Sealy L (2001) ERK2- and p90(Rsk2)-dependent pathways regulate the CCAAT/enhancer-binding protein-beta interaction with serum response factor. J Biol Chem 276: 38449–38456 [DOI] [PubMed] [Google Scholar]
- Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W (2007) Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci USA 104: 12318–12323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kortum RL, Costanzo DL, Haferbier J, Schreiner SJ, Razidlo GL, Wu MH, Volle DJ, Mori T, Sakaue H, Chaika NV, Chaika OV, Lewis RE (2005) The molecular scaffold kinase suppressor of Ras 1 (KSR1) regulates adipogenesis. Mol Cell Biol 25: 7592–7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Leutz A (1999) A C/EBP beta isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell 4: 735–743 [DOI] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A (1994) Novel mechanism of C/EBP beta (NF-M) transcriptional control: activation through derepression. Genes Dev 8: 2781–2791 [DOI] [PubMed] [Google Scholar]
- Laiosa CV, Stadtfeld M, Graf T (2006) Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol 24: 705–738 [DOI] [PubMed] [Google Scholar]
- Lausen J, Liu S, Fliegauf M, Lubbert M, Werner MH (2006) ELA2 is regulated by hematopoietic transcription factors, but not repressed by AML1-ETO. Oncogene 25: 1349–1357 [DOI] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR (2009) Minireview: protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol 23: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T, Duderstadt M, Streetz KL, Tacke F, Kubicka S, Manns MP, Trautwein C (2004) C/EBP beta isoforms LIP and LAP modulate progression of the cell cycle in the regenerating mouse liver. Hepatology 40: 356–365 [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR (2001) Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 11: 1981–1985 [DOI] [PubMed] [Google Scholar]
- Mandrup S, Lane MD (1997) Regulating adipogenesis. J Biol Chem 272: 5367–5370 [DOI] [PubMed] [Google Scholar]
- Mo X, Kowenz-Leutz E, Xu H, Leutz A (2004) Ras induces mediator complex exchange on C/EBP beta. Mol Cell 13: 241–250 [DOI] [PubMed] [Google Scholar]
- Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S (1993) Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci USA 90: 2207–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C (2007) The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol 17: 318–324 [DOI] [PubMed] [Google Scholar]
- Nerlov C (2008) C/EBPs: recipients of extracellular signals through proteome modulation. Curr Opin Cell Biol 20: 180–185 [DOI] [PubMed] [Google Scholar]
- Nuchprayoon I, Simkevich CP, Luo M, Friedman AD, Rosmarin AG (1997) GABP cooperates with c-Myb and C/EBP to activate the neutrophil elastase promoter. Blood 89: 4546–4554 [PubMed] [Google Scholar]
- Olsson A, Manzl C, Strasser A, Villunger A (2007) How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ 14: 1561–1575 [DOI] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C (2001) Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15: 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless O, Kowenz-Leutz E, Knoblich M, Lausen J, Beyermann M, Walsh MJ, Leutz A (2008) G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-{beta}. J Biol Chem 283: 26357–26363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli V (1998) The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem 273: 29279–29282 [DOI] [PubMed] [Google Scholar]
- Roesler WJ (2001) The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu Rev Nutr 21: 141–165 [DOI] [PubMed] [Google Scholar]
- Scoumanne A, Chen X (2008) Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol Histopathol 23: 1143–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian T, Johnson PF (2006) Stop and go: anti-proliferative and mitogenic functions of the transcription factor C/EBPbeta. Cell Cycle 5: 953–957 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Reinberg D (2008) Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol 9: 815–820 [DOI] [PubMed] [Google Scholar]
- Student AK, Hsu RY, Lane MD (1980) Induction of fatty acid synthetase synthesis in differentiating 3T3-L1 preadipocytes. J Biol Chem 255: 4745–4750 [PubMed] [Google Scholar]
- Timchenko NA, Welm AL, Lu X, Timchenko LT (1999) CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res 27: 4517–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (1998) Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci 54: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (2000) Histone acetylation and an epigenetic code. Bioessays 22: 836–845 [DOI] [PubMed] [Google Scholar]
- Uematsu S, Kaisho T, Tanaka T, Matsumoto M, Yamakami M, Omori H, Yamamoto M, Yoshimori T, Akira S (2007) The C/EBP beta isoform 34-kDa LAP is responsible for NF-IL-6-mediated gene induction in activated macrophages, but is not essential for intracellular bacteria killing. J Immunol 179: 5378–5386 [DOI] [PubMed] [Google Scholar]
- Williams SC, Baer M, Dillner AJ, Johnson PF (1995) CRP2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J 14: 3170–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Xie Y, Bucher NL, Farmer SR (1995) Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev 9: 2350–2363 [DOI] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM (2001) A transcriptional switch mediated by cofactor methylation. Science 294: 2507–2511 [DOI] [PubMed] [Google Scholar]
- Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT (2008) CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep 9: 193–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT (2003) Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA 100: 6464–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ (2005) Multisite protein modification and intramolecular signaling. Oncogene 24: 1653–1662 [DOI] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL (1995) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev 9: 168–181 [DOI] [PubMed] [Google Scholar]
- Zahnow CA (2009) CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med 11: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahnow CA, Younes P, Laucirica R, Rosen JM (1997) Overexpression of C/EBPbeta-LIP, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J Natl Cancer Inst 89: 1887–1891 [DOI] [PubMed] [Google Scholar]
- Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, Allis CD, Tempst P, Nimer SD (2008) Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev 22: 640–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.