Abstract
VHS (Vps27, Hrs, and STAM) domains occur in ESCRT-0 subunits Hrs and STAM, GGA adapters, and other trafficking proteins. The structure of the STAM VHS domain–ubiquitin complex was solved at 2.6 Å resolution, revealing that determinants for ubiquitin recognition are conserved in nearly all VHS domains. VHS domains from all classes of VHS-domain containing proteins in yeast and humans, including both subunits of ESCRT-0, bound ubiquitin in vitro. ESCRTs have been implicated in the sorting of Lys63-linked polyubiquitinated cargo. Intact human ESCRT-0 binds Lys63-linked tetraubiquitin 50-fold more tightly than monoubiquitin, though only 2-fold more tightly than Lys48-linked tetraubiquitin. The gain in affinity is attributed to the cooperation of flexibly connected VHS and UIM motifs of ESCRT-0 in avid binding to the polyubiquitin chain. Mutational analysis of all the five ubiquitin-binding sites in yeast ESCRT-0 shows that cooperation between them is required for the sorting of the Lys63-linked polyubiquitinated cargo Cps1 to the vacuole.
Keywords: Cps1, crystal structure, protein structure, vacuole, yeast genetics
Introduction
The covalent addition of ubiquitin (Ub) is one of the most widespread of all regulatory post-translational modifications of proteins (Hershko et al, 2000; Kerscher et al, 2006).
Ubiquitination is a major signal for the endocytosis of receptors and other plasma membrane proteins (Hicke and Dunn, 2003; Kirkin and Dikic, 2007), as well as for a subset of cargo that transit from the trans-golgi to endosomes. A distinctive property of Ub, in comparison to many other modifying moieties, is its ability to form covalent chains. Ub contains seven Lys residues, any one of which can participate in chain formation through a covalent bond with the C-terminal carboxylate of the subsequent Ub moiety. Monoubiquitination alone is, in some cases, a competent endocytic signal (Hicke and Dunn, 2003), but endocytosed receptors are also abundantly modified by Lys63-linked polyUb (K63-Ub) and other forms of multiubiquitination (Huang et al, 2006; Kirkin and Dikic, 2007). Some endosomal cargo is recycled, whereas other cargo is directed via the ESCRT pathway to the lysosome (or vacuole in yeast) for degradation. K63 ubiquitination is required for the degradative sorting of some, and perhaps most, cargo, such as the yeast permease Gap1 and the hydrolase Cps1, into the lysosomal/vacuolar branch of the pathway (Lauwers et al, 2009). In mammalian cells, polyubiquitination promotes the degradative sorting of the EGF receptor via ESCRT-0 (Umebayashi et al, 2008). The ESCRT-0, -I, and -II complexes all contain Ub-binding domains (UBDs) (Raiborg and Stenmark, 2009). Indeed the role of the ESCRT system in sorting ubiquitinated cargo was discovered based on the presence of a predicted UBD in the ESCRT-I subunit Vps23 (Katzmann et al, 2001).Therefore, a key question is whether any of the ESCRT complexes bind preferentially to polyUb, and to K63-Ub in particular. Here we show that ESCRT-0 binds with high avidity to polyUb chains.
As the furthest upstream component of the ESCRT machinery, ESCRT-0 acts at a branchpoint in endosomal traffic to bind to a certain cargo and commit it to the lysosomal pathway (Raiborg et al, 2008). In the absence of an interaction with ESCRT-0, these cargo will typically be recycled instead of being degraded (Raiborg et al, 2008). ESCRT-0 consists of two subunits, known as Hrs and STAM in humans, and Vps27 and Hse1 in yeast. Both subunits contain UBDs that engage cargo (Bilodeau et al, 2002; Raiborg et al, 2002; Shih et al, 2002; Bache et al, 2003; Mizuno et al, 2003). Hrs binds monoUb at two sites on its DUIM motif (Hirano et al, 2006), whereas Vps27 also binds two monoUb moieties through its tandem UIM1 and UIM2 motifs (Bilodeau et al, 2002; Shih et al, 2002; Fisher et al, 2003; Swanson et al, 2003). STAM and Hse1 each contain a single Ub-binding UIM motif. In addition to UIM and other motifs, Hrs, STAM, and their yeast orthologs contain N-terminal VHS domains.
VHS domains (Lohi and Lehto, 1998) are octahelical bundles (Mao et al, 2000; Misra et al, 2000), whose functions have only been defined in a few cases. The VHS domains of the human GGA trafficking adapters bind to the C-terminal tails of mannose 6-phosphase receptors (MPRs) via helices 6 and 8 (Misra et al, 2002; Shiba et al, 2002), but the MPR-binding site is not conserved in other VHS domains. The VHS domain of STAM binds to Ub (Mizuno et al, 2003; Hong et al, 2009; Ren et al, 2009) through the canonical Ile44 patch (Hong et al, 2009). Here we report the crystal structure of the STAM1 VHS domain–Ub complex, shows that the Ub binding motif in STAM is also present in the large majority of other VHS domains, and directly confirm that Ub binding is a widespread, conserved property of VHS domains.
VHS domains are present in the sequences of ESCRT-0 subunits, the GGA trafficking adapters (Bonifacino, 2004), and the GGA-like Tom1 and Tom1-like family of proteins. All of these proteins have in common a role in endosomal sorting, and all of them contain at least one other UBD in addition to the VHS domain. In addition to the UIM and DUIM motifs of ESCRT-0 subunits, the GGA and Tom1 families of proteins contain Ub-binding GAT domains. We sought to understand the significance of the co-occurrence of the VHS domain with other UBDs using ESCRT-0 as a model system. We show that the cooperation of VHS domains with UIMs is essential for function in yeast, and contributes to avid binding to polyUb chains in vitro. These observations explain the conservation of VHS domains and suggest a molecular mechanism for the recognition and sorting of polyubiquitinated proteins into the lysosomal degradation pathway by the ESCRT system.
Results
The STAM1 VHS domain–Ub complex
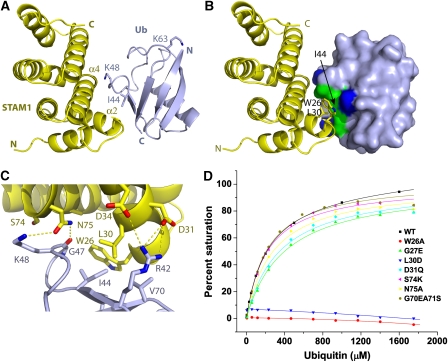
MonoUb was co-crystallized with the human STAM1 VHS domain, and the structure determined to 2.6 Å resolution (Table 1). The asymmetric unit contains three copies of the VHS–Ub complex, and one copy of an uncomplexed VHS domain. The three copies of the complex are essentially identical in conformation, with r.m.s.d. values of 1.0 Å for all Cα atoms in the complex when both the VHS domain and Ub are superimposed simultaneously. The Ub-binding site of the uncomplexed VHS domain is blocked by a lattice contact, and shows small structural differences in side-chains of residues 23–28. The three crystallographically independent VHS–Ub complexes provide a highly consistent picture of the interaction. The interface covers 360 Å2 of accessible surface area, a value typical of the low-affinity complexes of trafficking domains with Ub (Hicke et al, 2005; Hurley et al, 2006). On Ub, the interface centers on Ile44 (Figure 1A–C). Both Ile44 and Gly47 are completely buried in the interface, whereas Arg42, Lys48, Gln49, His68, and Val70 are partially buried. On the VHS domain, helices α2 and α4 form the binding site for Ub (Figure 1B). Trp26 and Leu30 together contribute more than half the total interaction area on the VHS domain. Gly27, Asp31, Asp34, Ala71, Ser74, and Asn75 are the other major interactors on the VHS domain. These interacting residues are consistent with those identified in solution using NMR chemical shift perturbation analysis (Hong et al, 2009). Hydrogen bonds are observed between the side chains of STAM1 Asp31and Asp43 and Ub Arg42, and between the side-chain of STAM1 Asn75 and Ub Gly47 carbonyl oxygen and Lys48 ɛ-amino group (Figure 1C).
Table 1. Crystallographic data collection and refinement.
| Native | |
|---|---|
| Constructs | STAM1 (2–143) |
| Ubiquitin (1–76) | |
| Space group | P21 |
| Unit cell | a=66.61 Å, b=112.13 Å, c=68.66 Å, β=99.43 |
| X-ray source | APS 22-ID |
| Wavelength (Å) | 1.0000 |
| Resolutiona (Å) | 2.60 (2.69–2.60) |
| No. of unique reflections | 30 656 |
| I/σ(I)a | 16.9 (2.9) |
| Rsyma (%) | 18 (37) |
| Completenessa (%) | 93.4 (59.0) |
| R factor (%) | 22.9 |
| Free R factorb (%) | 23.6 |
| RMS bond length dev. (Å) | 0.009 |
| RMS bond angle dev. (deg) | 1.11 |
| Average B-factorc (Å2) | 74.9 |
| Number of atoms | 6105 |
| aValues in parentheses refer to the highest-resolution shell. | |
| bRfree is calculated for a randomly chosen 5% subset of reflections omitted from refinement. | |
| cAverage B value of all atoms in an asymmetric unit. | |
Figure 1.
The STAM VHS (Vps27, Hrs, and STAM) domain–ubiquitin (Ub) complex. (A) Overall structure of the complex with the STAM1 VHS domain shown in yellow and Ub in light blue. The side-chains of the key Ub residues Ile44, Lys48, and Lys63, are shown. (B) The STAM1 VHS domain binds to the hydrophobic patch (colored green) consisting of Ub Ile44, Gly47, and Val70. Basic residues Arg42, Lys48, and His68 involved in binding are colored dark blue, whereas Gln49 is colored white. The remainder of the non-interacting residues on the Ub surface are colored light blue. (C) Interactions in the complex. Hydrogen bonds are shown with dashed lines. (D) Binding of wild-type and mutant VHS domains to Ub. Binding curves for wild-type and mutant VHS domains are identified in the inset.
The STAM1 VHS-domain binds to Ub with Kd=220 μM as judged by surface plasmon resonance (SPR). In order to quantitate the roles of STAM1 residues in binding, the structural interaction residues were mutated and binding was measured by SPR. The residues that comprise the largest portion of the interface, Trp26 and Leu30, are essential for the interaction, as judged by the complete abolition of binding in the W26A and L30D mutants (Figure 1D). Mutations in the periphery of the interface leads to at most a two-fold increase in Kd (Figure 1D, Table 1).
Ub binding is a conserved property of VHS domains
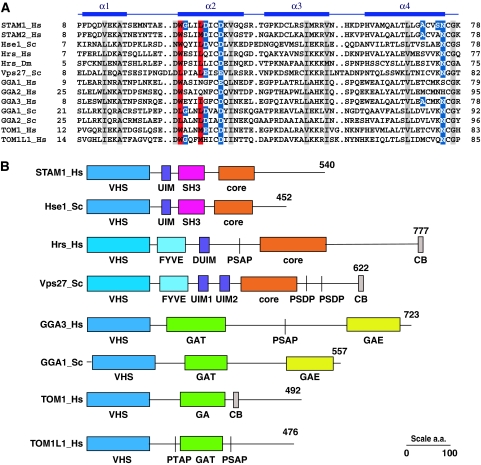
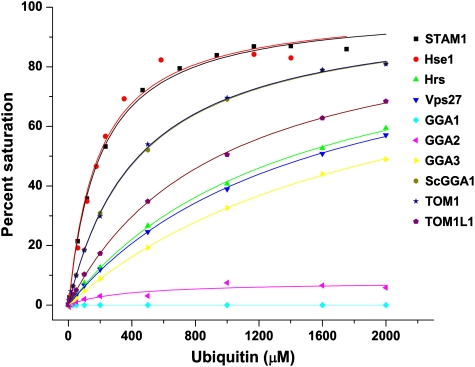
Diverse aligned sequences of VHS domains reveal that the key Ub-binding Trp26 and Leu30 residues of STAM1 are conserved in most other VHS domains (Figure 2A, Supplementary Figure S1). In some cases, Trp26 is replaced by another large hydrophobic residue, Leu. Some peripheral residues, such as Asp31, Asp34, and Asn75, are also well-conserved. This suggested to us that Ub binding might be a conserved property of VHS domains. The VHS domains of the yeast and human ESCRT-0 subunits, GGAs, and human Tom1 and Tom1L1 were purified and their Ub binding was measured by SPR (Figure 3). All VHS domains were bound to Ub with affinities in the 100 μM–2 mM range, with the exception of human GGA1 and GGA2. In the GGA1 and 2 sequences, the residue corresponding to STAM1 Leu30 is either Asn or Gln, which probably explains the lack of binding. The weak binding to the Hrs VHS domain was surprising, in that the canonical Trp and Leu residues are preserved. Mutations in peripheral residues were chosen in part on the basis of the corresponding residues in Hrs, for example, G27E, D31Q, and S74K (Figure 1D). Two of these mutations decrease binding by approximately two-fold each, and it seems likely that the cumulative effect of these and other peripheral changes not tested must account for the weak binding to the Hrs VHS domain. Yeast Vps27, Gga1, and Gga2 VHS domains have a Leu residue in place of STAM1 Trp26, a replacement that is apparently well-tolerated.
Figure 2.
The structural ubiquitin (Ub)-binding motif is conserved in nearly all VHS (Vps27, Hrs, and STAM) domains. (A) Aligned sequences of VHS domain-containing proteins in the region corresponding to the N-terminal half of the domain. The secondary structure of the human STAM1 VHS domain is shown with solid bars to indicate α-helices. Residues shown to be critical for VHS domain–Ub binding are shaded in red, and other residues involved in, but not essential for, the interaction are shaded in blue. Identically conserved residues that are not directly involved in binding are shaded gray. Species abbreviations: Hs, Homo sapiens; Dm, Drosophila melanogaster; and Sc, Saccharomyces cerevisiae. (B) Schematic of the domain organization of selected VHS domain-containing proteins. Of the sequences shown in (A), the domain structure of STAM2_Hs is like that of STAM1_Hs; Hrs_Dm like that of Hrs_Hs, GGA1_Hs, and GGA2_Hs like GGA3_Hs; and GGA2_Sc like GGA1_Sc. SH3, Src homology-3 domain; core, ESCRT-0 core GAT and coiled coil region responsible for heterodimerization; PSAP, PTAP, PSDP are ESCRT-I binding motifs; CB, clathrin binding; and GAE, γ-adaptin ear domain.
Figure 3.
Ubiquitin binds to nearly all VHS (Vps27, Hrs, and STAM) domains. Surface plasmon resonance (SPR)-binding curves between ubiquitin (Ub )and the indicated VHS domains are identified by the key in the inset.
UBDs of ESCRT-0 cooperate in polyubiquitin-chain binding
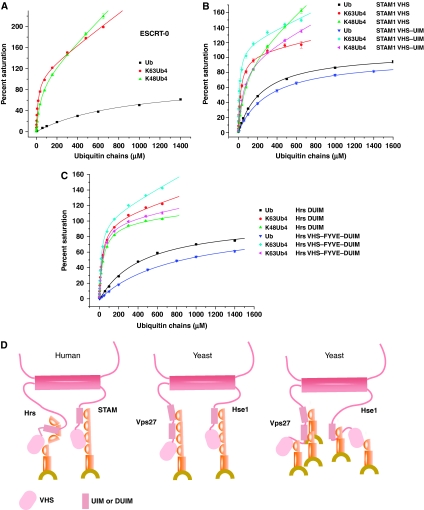
Trials for the mono- and polyUb binding of the complete ESCRT-0 complex and UBD-containing fragments thereof were carried out by SPR to determine whether polyUb binding could account for the biological function of ESCRT-0 in sorting polyubiquitinated substrates. Recombinant full-length human ESCRT-0 was purified from insect cells (Ren et al, 2009) and immobilized for SPR analysis. In spite of the presence of multiple UBDs, binding to monoUb could be fit to a single-site model allowing for a non-saturable non-specific component, yielding Kd=920 μM (Table 2, Figure 4A, Supplementary Figure S2). The low affinity reflects the average behavior of higher- (STAM VHS, UIM, and Hrs DUIM) and lower- (Hrs VHS domain)affinity sites. K48-Ub and linear head-to-tail (NC-Ub) chains are not known to have a role in endosomal sorting, but they were studied here as controls to assess the chain specificity of ESCRT-0 and its fragments. Binding to ESCRT-0 to Ub2 chains was significantly enhanced compared with monoUb, with three-fold increases for K48-Ub2 and NC-Ub2, and a nine-fold increase for K63-Ub2. Binding of ESCRT-0 to K48- and K63-Ub4 was tighter than to either monoUb or diUb, as expected. The increase in affinity for NC-Ub4 was 7-fold, with larger increases of 21-fold for K48-Ub4 and 50-fold for K63-Ub4. The reason for the lack of more avid binding to NC-Ub4 is not clear. In terms of chain specificity, the main conclusion from these experiments is that ESCRT-0 has a modest 2.4-fold preference for K63-Ub in the context of a Ub4 chain, and a somewhat greater 3.7-fold preference in the context of a Ub2 chain.
Table 2. Dissociation constants (Kd) of ESCRT-0 UBDs binding to monoUb or polyUb as obtained from SPR experiments.
| Kd for Ub (μM) | |||||||
|---|---|---|---|---|---|---|---|
| STAM1–VHS wild type | 220±3.2 | ||||||
| STAM1–VHS W26A mutant | NB | ||||||
| G27E | 400±29 | ||||||
| L30D | NB | ||||||
| D31Q | 350±36 | ||||||
| S74K | 220±18 | ||||||
| N75A | 280±32 | ||||||
| G70E/A71S | 220±12 | ||||||
| Sc Hse1–VHS | 190±28 | ||||||
| Hrs–VHS | 1400±68 | ||||||
| Sc Vps27–VHS | 1500±56 | ||||||
| GGA1–VHS | NB | ||||||
| GGA2–VHS | NB | ||||||
| GGA3–VHS | 2100±53 | ||||||
| Sc GGA1–VHS | 450±8.0 | ||||||
| TOM1–VHS | 440±16 | ||||||
| TOM1L1–VHS | 950±40 | ||||||
| STAM1–UIM | 430±22 | ||||||
| Sc Vps27–UIM1 | 600±17 | ||||||
| Sc Vps27–UIM1 (A266Q/S270D) | NB | ||||||
| Sc Vps27–UIM2 | 360±4 | ||||||
| Sc Hse1–UIM | 490±5 | ||||||
| Sc Hse1–UIM (A170Q/S174D) | NB | ||||||
| Kd for: (μM) | MonoUb | K63-Ub2 | K48-Ub2 | NC-Ub2 | K63-Ub4 | K48-Ub4 | NC-Ub4 |
| Full ESCRT-0 | 920±90.5 | 110±2.8 | 370±52 | 350±23 | 18±2.3 | 43±2.2 | 140±11 |
| STAM VHS | 220±3.2 | 110±7.8 | 380±71 | 100±9.1 | 33±3.7 | 76±4.7 | 48±4.1 |
| STAM VHS–UIM | 320±5.0 | 76±7.3 | 310±16 | 90±11 | 17±3.3 | 55±3.8 | 80±6.4 |
| Hrs DUIM | 450±50.8 | 190±9.3 | 820±53 | 180±16 | 31±3.7 | 41±2.5 | 80±5.3 |
| Hrs VHS–FYVE–DUIM | 830±52.8 | 206±13.4 | 978±50.9 | 165±7.5 | 20±2.2 | 32±1.9 | 36±5.5 |
| Vps27-tandem UIMs | 350±1.2 | 70±6.0 | 180±12 | 65±7.4 | 16±2.6 | 32±3.0 | 50±9.1 |
| NB, no binding could be detected; SPR, surface plasmon resonance; Ub, ubiquitin; UBD, ubiquitin-binding domain; VHS, Vps27, Hrs, and STAM. | |||||||
| These data are the averages of three independent experiments. Values are shown to two significant figures. | |||||||
Figure 4.
ESCRT-0 and VHS (Vps27, Hrs, and STAM) domain-containing fragments of STAM1 bind polyubiquitin chains avidly. (A) Surface plasmon resonance (SPR) curves for full length ESCRT-0. (B) SPR curves for STAM VHS domain and VHS–UIM construct. (C) SPR curves for Hrs DUIM and VHS–FYVE–DUIM construct. SPR binding curves between the constructs noted at top and the ubiquitin (Ub) chains identified by the key in the inset. Binding curves are normalized to 100% for the saturable component. The total binding includes a non-saturable, non-specific component in some cases, leading to values higher than 100%. (D) Model for mono and polyubiquitinated cargo recognition by human and yeast ESCRT-0.
The STAM VHS domain showed increased affinity for K63-Ub4 over monoUb, with a slightly smaller increase for K48-Ub4 (Table 2, Figure 4B). This VHS domain and the other single and multi-domain fragments tested consistently showed a 3–5-fold preference for K63-Ub2 over K48-Ub2 (Table 2, Supplementary Figure S2), compared with a 1.5- to 2.5-fold preference for K63-Ub4 over K48-Ub4 (Table 2, Figure 4B). The only exception was that the Vps27 UIM1-2 construct displayed a two-fold preference for K63-Ub that was independent of chain length (Table 2, Supplementary Figure S2). A STAM fragment comprising only the VHS and UIM domains was investigated to determine if this domain combination bound chains cooperatively. A 19-fold increase was observed for binding to K63-Ub4 over Ub (Table 2, Figure 4B). A somewhat smaller six-fold increase was observed for K48-Ub4 relative to Ub (Table 2, Figure 4B). The 19- and 6-fold increases in binding to the two types of chains account for a significant fraction of the corresponding 50-fold and 21-increases seen in the complete ESCRT-0 complex. Even more strikingly, the Kd values for the two types of chains are nearly identical in the full-length ESCRT-0 complex and the tandem UBD fragment of STAM.
In order to complete the characterization of all known Ub-binding regions of ESCRT-0, the N-terminal region of Hrs with and without the VHS domain was characterized. The Hrs DUIM domain had similar 30–40 μM affinities for both K63 and K48-Ub4 (Table 2, Figure 4C). The Hrs DUIM had strikingly poor binding to K48-Ub2 as opposed to other Ub2 chains, but this selectivity disappeared at the level of Ub4 chains. The larger fragment, including both the VHS domain and the FYVE domain and linker regions between the VHS and DUIM, bound somewhat more tightly to both types of Ub chain. As seen for the dual UBD construct for STAM, the VHS–FYVE–DUIM construct of Hrs bound Ub chains with affinities that are close to those of the intact ESCRT-0 complex. From these analyses, we conclude that ESCRT-0 binds avidly, but without significant linkage selectivity, to long polyubiquitin chains. A moderate degree of specificity for K63>K48 is seen only in the context of diubiquitin chains. The similar polyubiquitin-binding properties of the N-terminal multiple UBD-containing fragments of Hrs and STAM can account for the behavior of the full complex.
VHS and UIM domains of ESCRT-0 cooperate in sorting Cps1
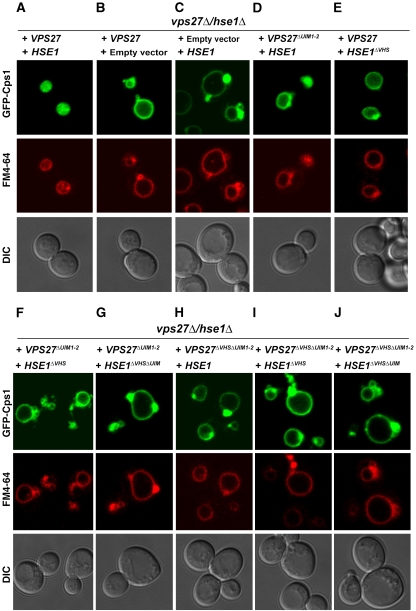
Tetra-K63-Ub bound ESCRT-0 with sufficiently high affinity that it seemed likely to us to be involved in the physiological sorting function of the complex. We proceeded to test the hypothesis that ESCRT-0 UBDs cooperate to avidly bind Ub chains. ESCRT-0 is a heterodimer (Prag et al, 2007; Ren et al, 2009) which, including the newly identified Ub-binding activities of the Hrs, Vps27, and Hse1 VHS domains, contains a total of five known Ub-binding sites in both yeast and human complexes. Yeast Vps27 contains two UIMs, Hse1 contains one, and all three are competent to bind Ub (Fisher et al, 2003) (Table 2). In order to determine whether the two VHS domains of yeast ESCRT-0 subunits cooperate with the three UIMs in sorting polyubiquitinated cargo, we adapted the approach previously used to study the cooperativity of UBDs in cargo sorting by ESCRT-I and II (Shields et al, 2009). Cps1 is a vacuolar hydrolase that is K63-ubiquitinated and is normally sorted to the lumen of the vacuole by the ESCRT pathway. Disruption of the ESCRT pathway can be diagnosed by the mislocalization of Cps1 to the limiting membrane of the vacuole and to aberrant structures that appear as puncta in light microscopy and are known as ‘class E compartments'.
The VHS domains of the yeast ESCRT-0 subunits Vps27 and Hse1 were mutationally inactivated alone and in combination with UIMs. The sorting of the Lys63-polyubiquitination-dependent cargo Cps1 (Lauwers et al, 2009) to MVBs was monitored in vps27Δ hse1Δ cells complemented with rescue plasmids bearing various combinations of UIM and VHS domain mutants of VPS27 and HSE1. The VHS domains of Vps27 and Hse1 were inactivated by the mutations W28A–L32D and W25A–L29D, respectively. UIM domains were mutationally inactivated as follows: Vps27–UIM1, A266Q–S270D; Vps27–UIM2, A309Q–S313D; and Hse1–UIM, A170Q–S174D. These mutations are all on the exterior of the protein domain structures and are therefore expected to have no effects on the stability of the Vps27 and Hse1 proteins. Rescue of GFP-Cps1 sorting by expression of VPS27 alone was partial (Figure 5A and B) consistent with the weak class E phenotype of hse1Δ (Bilodeau et al, 2002). Expression of HSE1 alone did not rescue GFP-Cps1 sorting at all and manifested large class E puncta (Figure 5C), consistent with the strong class E phenotype of vps27Δ (Piper et al, 1995). Expression of VPS27ΔUIM1–2 with HSE1-manifested GFP fluorescence localized predominantly to the limiting membrane of the vacuole, but with some diffuse lumenal GFP fluorescence, indicated that a portion of the GFP-Cps1 was delivered to the vacuole (Figure 5D). The VPS27ΔUIM1–2 HSE1-expressing cells showed a few class E puncta, but they were markedly smaller and fewer in number than expected for a full block in ESCRT sorting. This is interpreted as a weak class E phenotype, and is consistent with reduced Cps1 sorting seen in a similar VPS27 double UIM mutant (Shih et al, 2002).
Figure 5.
Cps1 sorting in yeast depends on the cooperative action of ESCRT-0 Ub binding domains. The indicated plasmids were transformed into vps27Δhse1Δ yeast cells and imaged for GFP-Cps1 (cargo) and FM4-64 (membrane) fluorescence as shown individually in panels (A–J).
Expression of VPS27 and HSE1ΔVHS led to a weak class E phenotype (Figure 5E) similar to that of the VPS27 double UIM mutant, indicating that Ub binding by the Hse1 VHS domain is involved in, but not critical for, Cps1 sorting. In contrast, the triple Ub-binding-site knockout, VPS27ΔUIM1–2 HSE1ΔVHS showed almost no lumenal GFP-Cps1 (Figure 5F), hence its phenotype was significantly stronger than that of either VPS27ΔUIM1–2 HSE1 or HSE1ΔVHS. As might be expected, given these results, the quadruple knockout VPS27ΔUIM1–2HSE1ΔUIMΔVHS (Figure 5G) had a strong class E phenotype, indicating that in the context of a triple UIM defect, the additional loss of Hse1 VHS domain-binding results in a complete loss of Cps1 sorting.
Because the Vps27 VHS domain bound Ub 10-fold more weakly than the Hse1 VHS domain, VPS27ΔVHS was not expected to have a phenotype and was not tested by itself. In combination with the Vps27 UIM double knockout, VPS27ΔUIM1–2ΔVHS HSE1 yielded a strong class E phenotype (Figure 5H). This demonstrated that the Ub-binding site on the Vps27 VHS domain does have a role in Cps1 sorting despite its low affinity for monoUb in vitro. Not surprisingly, VPS27ΔUIM1–2ΔVHS HSE1ΔVHS (Figure 5I) and VPS27ΔUIM1–2ΔVHS HSE1ΔUIMΔVHS (Figure 5J) have the same strong phenotype. The threshold for partial loss of function thus appears to occur after mutation of one to two UBDs. Once three UBDs are mutated, the loss of function is nearly complete, so there is little further change when four or five UBDs are mutated. These data show that while the Ub-binding activities of the VHS domains of the Vps27 and Hse1 subunits are not essential on their own, their cooperation with the UIMs is required for sorting.
Discussion
The VHS domain was identified as a conserved motif in trafficking proteins more than a decade ago (Lohi and Lehto, 1998). Shortly after the identification of the VHS domain, the VHS domains of the human GGA proteins were shown to bind to mannose 6-phosphate receptors and sortilin, and to sort them from the TGN to the endolysosomal pathway (Nielsen et al, 2001; Puertollano et al, 2001; Takatsu et al, 2001; Zhu et al, 2001). The key residues on helices α6 and α8 that form the MPR/sortilin-binding site (Misra et al, 2002; Shiba et al, 2002) were not conserved in GGAs of simple eukaryotes or in other VHS domain proteins. The Ub-binding property of the STAM VHS domain has been known for some time (Mizuno et al, 2003), yet its generalization to other VHS domains was not clear. Here we have shown that the VHS domain binds Ub through a conserved interface whose size and interaction strength is typical for UBDs. Ub binding was found in nearly every VHS domain tested, spanning most known VHS domain proteins in yeast and humans. The two notable exceptions are human GGA1 and GGA2, which may have gained the specialized ability to bind MPRs during evolution while losing their Ub-binding site. In the case of GGA3, whose VHS domain binds both MPRs and Ub, the two sites are at opposite ends of the domain structure and there is nothing to preclude their simultaneous binding to both types of ligand. Based on the results obtained here, Ub binding is a conserved function of the VHS domain, so at least one function can now be assigned to all known VHS domains in yeast and humans.
The human GGAs (Puertollano and Bonifacino, 2004), yeast Ggas (Scott et al, 2004; Deng et al, 2009), and Tom1–Tom1L1-family proteins (Katoh et al, 2004; Puertollano, 2005) all have roles in the sorting of ubiquitinated cargo. All of these proteins also contain Ub-binding GAT domains, and it has generally been accepted that the GAT domain was sufficient to recognize cargo. The identification of a second UBD in these proteins adds new complexity to the picture. In the case of yeast ESCRT-0, we found that the trafficking of the ubiquitination-dependent cargo Cps1 is robust to the inactivation of single UBDs but sensitive to inactivation of multiple UBDs. Sensitivity to multiple, but not single, UBD inactivation was previously shown for the yeast ESCRT-I–II supercomplex (Shields et al, 2009). This pattern of cooperative binding to polyubiquitinated proteins could be a common feature of the many multi-UBD proteins and complexes involved in trafficking, and likely extends to the recognition of multiubiquitinated proteins as well.
The presence of multiple VHS domains and UIMs within ESCRT-0 gives the complex the ability to bind avidly to polyubiquitin chains. The enhanced binding of polyubiquitin to ESCRT-0 explains how polyubiquitination could potentially serve as an effective signal to direct the cargo Cps1 into the MVB pathway. The level of specificity for K63- over K48-Ub is moderate, 3.7- or 2.4-fold depending on whether di- or tetraubiquitin chains, respectively, are examined. At present, the minimum length of Ub chains required for sorting of polyubiquitin-dependent ESCRT cargoes, such as Cps1, remains to be established. Indeed, polyubiquitination does not appear to be required for all ESCRT cargoes. A counterexample of monoubiquitin-dependent sorting into the MVB pathway is provided by Fth1. Fth1 is not normally an ESCRT cargo, but can be directed into the ESCRT pathway by gene fusion with a monoubiquitin mutant designed to block chain elongation through either Lys48 or Lys63 (Bilodeau et al, 2002). The presence of multiple UBDs in ESCRT-0 may provide an effective mechanism for binding both monoubiquitinated and polyubiquitinated proteins (Figure 4D). Consistent with this view, ESCRT-0 can cluster a model monoubiquitinated cargo present in a bulk concentration of just 65 nM, provided that the cargo is tethered to a membrane containing the ESCRT-0-binding lipid phosphatidylinositol 3-phosphate (Wollert and Hurley, 2010). The presence of multiple Ub moieties in a polyubiquitin chain may be more important in some cases than others, such as for substrates present at low concentrations in the cell. We have found that polyubiquitination can enhance ESCRT-0 binding relative to monoubiquitination by nearly two orders of magnitude. Our other observations suggest that membrane tethering of both Ub and ESCRT-0 can enhance affinity by more than three orders of magnitude (Wollert and Hurley, 2010), increasing the affinity for even monoUb to the point where physiological levels of cargo are efficiently bound. Thus, the multiplicity of UBDs in ESCRT-0 appears to confer multiple functions in mono- and polyubiquitinated cargo sorting. Here we have shown that the VHS domain is a conserved UBD and has a key role in the ability of ESCRT-0 to avidly bind Ub chains. We have shown Ub recognition by VHS domains is important in sorting Cps1. Other functions will be interesting to explore in the future.
Materials and methods
Protein expression and purification
DNA coding for human STAM1 residues 2–143 was subcloned into the pGST2 vector (Sheffield et al, 1999), providing for an N-terminal glutathione S-transferase (GST) tag followed by a TEV protease cleavage site. Plasmids were transformed into Escherichia coli BL21 (DE3) cells and overexpressed in ZYM-5052 autoinduction medium (Studier, 2005). Pellets were resuspended in 1 × PBS buffer (pH 7.4), 5 mM β-ME, and protease inhibitors (Sigma), and lysed by sonication. The clarified supernatant was purified using a glutathione–sepharose column (GE Healthcare). Samples for crystallized were incubated with His6-tagged TEV protease to cleave the histidine tag, followed by passage through Ni-NTA resin. The protein was concentrated and purified on a Superdex 75 column (GE Healthcare) in 20 mM Tris pH 7.4, 50 mM NaCl, 5 mM DTT. Bovine Ub (Sigma) was dissolved in 20 mM Tris pH 7.4, and then further purified on a Superdex 75 column before use in crystallization experiments. Site-directed mutants of STAM1 (1–143) were generated using the Quick change kit (Stratagene). For SPR studies of VHS domains of various proteins and fragments of various ESCRT-0 subunits, the appropriate DNA fragments (Supplementary Table S1) were PCR amplified and subcloned into pGST1 (for Hse1, the pGST2 vector was used). All constructs were verified by DNA sequencing. Plasmids were transformed into BL21 (DE3) cells, and then overexpressed in LB medium. Cells were induced with 0.2 mM IPTG and grown at 18°C overnight. The lysate was purified using glutathione–sepharose column, and then applied to Superdex 200 column in 10 mM HEPES pH 7.0, 150 mM NaCl. The insect cell expression and purification of the full-length human Hrs–STAM1 complex was described previously (Ren et al, 2009). The masses of all recombinant proteins were confirmed by electrospray mass spectrometry.
Crystallization, data collection, and structure determination
Ubiquitin and STAM1 VHS domain were mixed at a 1:1 molar ratio and concentrated. Crystals of the complex were obtained at 35 mg ml−1 protein and equilibrated against 0.2 M Na thiocyanate, 20% PEG3350, and 0.1 M imidazole pH 8.0 in hanging drops. Crystals were cryoprotected in reservoir solution supplemented with 25% glycerol and frozen in liquid nitrogen. Data were collected at 2.6 Å resolution from a single frozen crystal using a MAR CCD detector at beamline 22-ID, Advanced Photon Source. A molecular replacement solution was found using STAM2 VHS domain structure (PDB: 1X5B) as a search model with Phaser (McCoy et al, 2007) in the space group P21. Model building and refinement was using coot (Emsley and Cowtan, 2004), CNS (Brunger et al, 1998) and AutoBuster (Blanc et al, 2004) (Table 1). Structural figures were generated in PyMol (W Delano, http://pymol.sourceforge.net/).
Polyubiquitin synthesis
K48- and K63-Ub chains were synthesized following the procedure of (Pickart and Raasi, 2005). Briefly, His-tagged mouse Ub-activating enzyme E1 was purified from baculovirus-infected Hi5 insect cells. Ub-K48C and Ub-D77, human E2-25K (E2) were used to make the K48-linked chain. Ub-K63R, Ub-D77, yeast Ubc13-Mms2 complex (E2), and Yuh1 hydrolase for the K63-linked chain.
Surface plasmon resonance
The binding of Ub to different VHS fragments and mutants was measured using a Biacore T100 instrument. Anti-GST (GE Healthcare) was immobilized on a CM5 chip by amine coupling following the manufacturer's protocol. GST-fusion proteins were captured on this surface to a density of 700–1000 U; the monoUb or polyUb analyte was injected at a flow rate of 20 μl min−1 in 10 mM HEPES pH 7.0, 150 mM NaCl, and 0.005% P20 at 25°C. A total of 10 mM glycine–HCl pH 2.0 was used for surface regeneration. Each protein was chromatographed by gel filtration immediately before SPR experiments in order to avoid immobilizing any aggregated material. The data were processed using BioEvaluation software (Biacore), and curve fitting was carried out with Origin software (OriginLab). A single-site-binding model is used to fit monoUb binding, but this model did not provide acceptable fits for most of the polyUb binding curves. A non-specific binding model incorporated a linear term was used to fit polyUb binding as follows:
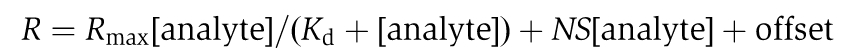 |
For plotting, data were scaled such that Rmax=100. As a control to determine whether the immobilization for SPR or the fitting procedure had introduced systematic errors into the Kd values for polyUb binding, the tandem UIMs of Vps27 were purified and polyUb binding was assessed (Supplementary Figure S2, Table 2). The Kd values obtained by fitting to the above equation were 16 and 32 μM, respectively, for K63-Ub4 and K48-Ub4. These are similar to the corresponding values of 29 and 31 μM obtained from fluorescence anisotropy-binding measurements in solution (Sims and Cohen, 2009).
Plasmid construction and yeast strains
The yeast strain vps27Δ∷KanR was purchased from Open Biosystems. The vps27Δ hse1Δ strain was prepared by replacing HSE1 with a nourseothricin-resistance gene from the hse1Δ strain by homologous recombination. The complete expression cassettes of VPS27 and HSE1 were amplified from yeast genomic DNA (Invitrogen) and cloned into the single-copy plasmids Ycplac111 and Ycplac22, respectively. Plasmids used in this study are listed in Supplementary Table S2. The Ub-binding inactivation mutants were generated by site-directed Quickchange mutagenesis (Stratagene), and confirmed by DNA sequencing. Plasmids bearing wild-type and mutant VPS27and HSE1, and GFP-CPS1 (Odorizzi et al, 1998) were co-transformed into vps27Δ hse1Δ. The strains were used in this study were: BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), BY4741 vps27Δ∷KanR, BY4741 vps27Δ∷KanR hse1Δ∷NATR.
Microscopy
Yeast strains expressing the appropriate alleles were harvested at an A600 of 0.3–0.5 and labeled with FM4-64 for vacuolar membrane staining (Vida and Emr, 1995). Uptake of FM4-64 by live cells was performed at 30°C for 1 h, after which cells were resuspended in selection media and incubated for 30 min at 30°C. Visualization of cells was performed on an LSM510 fluorescence microscope (Carl Zeiss MicroImaging) equipped with fluorescein isothiocyanate (FITC) and rhodamine filters and captured with a digital camera.
Supplementary Material
Acknowledgments
We thank D Kloer for assistance with crystallographic analysis, B Baibokov and Y Im for assistance with microscopy, L Saidi for cell culture support, J Dean for microscope access, K Iwai, C Wolberger, S Misra, Y Ye, and A Weissman for reagents and advice on polyUb chain synthesis, C Philpott, J Bonifacino, and R Puertollano for cDNAs, and W Prinz for yeast strains. Use of the APS was supported by the US DOE, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. This work was supported by the NIDDK and IATAP programs of the NIH intramural research program. Crystallographic coordinates have been deposited in the Protein Data Bank with the accession code 3LDZ.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bache KG, Raiborg C, Mehlum A, Stenmark H (2003) STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem 278: 12513–12521 [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC (2002) The Vps27p-Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol 4: 534–539 [DOI] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr Sect D 60: 2210–2221 [DOI] [PubMed] [Google Scholar]
- Bonifacino JS (2004) The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol 5: 23–32 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Deng Y, Guo Y, Watson H, Au WC, Shakoury-Elizeh M, Basrai MA, Bonifacino JS, Philpott CC (2009) Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-golgi network to the vacuole. J Biol Chem 284: 23830–23841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP (2003) Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem 278: 28976–28984 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A (2000) The ubiquitin system. Nat Med 6: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S (2006) Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol 13: 272–277 [DOI] [PubMed] [Google Scholar]
- Hong YH, Ahn HC, Lim J, Kim HM, Ji HY, Lee S, Kim JH, Park EY, Song HK, Lee BJ (2009) Identification of a novel ubiquitin binding site of STAM1 VHS domain by NMR spectroscopy. FEBS Lett 583: 287–292 [DOI] [PubMed] [Google Scholar]
- Huang FT, Kirkpatrick D, Jiang XJ, Gygi S, Sorkin A (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell 21: 737–748 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Shiba Y, Mitsuhashi H, Yanagida Y, Takatsu H, Nakayama K (2004) Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J Biol Chem 279: 24435–24443 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Devel Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kirkin V, Dikic I (2007) Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr Opin Cell Biol 19: 199–205 [DOI] [PubMed] [Google Scholar]
- Lauwers E, Jacob C, Andre B (2009) K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol 185: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi O, Lehto VP (1998) VHS domain marks a group of proteins involved in endocytosis and vesicular trafficking. FEBS Lett 440: 255–257 [DOI] [PubMed] [Google Scholar]
- Mao YX, Nickitenko A, Duan XQ, Lloyd TE, Wu MN, Bellen H, Quiocho FA (2000) Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100: 447–456 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Beach BM, Hurley JH (2000) Structure of the VHS domain of human Tom1 (target of myb 1): Insights into interactions with proteins and membranes. Biochemistry 39: 11282–11290 [DOI] [PubMed] [Google Scholar]
- Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH (2002) Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415: 933–937 [DOI] [PubMed] [Google Scholar]
- Mizuno E, Kawahata K, Kato M, Kitamura N, Komada M (2003) STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol Biol Cell 14: 3675–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Madsen P, Christensen EI, Nykjaer A, Gliemann J, Kasper D, Pohlmann R, Petersen CM (2001) The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J 20: 2180–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95: 847–858 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Raasi S (2005) Controlled synthesis of polyubiquitin chains. Methods Enzymol 399: 21–36 [DOI] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH (1995) Vps27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol 131: 603–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G, Watson H, Kim YC, Beach BM, Ghirlando R, Hummer G, Bonifacino JS, Hurley JH (2007) The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev Cell 12: 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R (2005) Interactions of Tom1L1 with the multivesicular body sorting machinery. J Biol Chem 280: 9258–9264 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Aguilar RC, Gorshkova I, Crouch RJ, Bonifacino JS (2001) Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS (2004) Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol 6: 244–251 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol 4: 394–398 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Malerod L, Pedersen NM, Stenmark H (2008) Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp Cell Res 314: 801–813 [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458: 445–452 [DOI] [PubMed] [Google Scholar]
- Ren X, Kloer DP, Kim YC, Ghirlando R, Saidi LF, Hummer G, Hurley JH (2009) Hybrid structural model of the complete human ESCRT-0 complex. Structure 17: 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, Piper RC (2004) GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol 6: 252–259 [DOI] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z (1999) Overcoming expression and purification problems of RhoGDI using a family of ″parallel″ expression vectors. Prot Expr Purific 15: 34–39 [DOI] [PubMed] [Google Scholar]
- Shiba T, Takatsu H, Nogi T, Matsugaki N, Kawasaki M, Igarashi N, Suzuki M, Kato R, Earnest T, Nakayama K, Wakatsuki S (2002) Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415: 937–941 [DOI] [PubMed] [Google Scholar]
- Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R (2009) ESCRT ubiquitin binding domains function cooperatively during MVB cargo sorting. J Cell Biol 185: 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L (2002) Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol 4: 389–393 [DOI] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of Rap80. Mol Cell 33: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Prot Expr Purific 41: 207–234 [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I (2003) Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J 22: 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Katoh Y, Shiba Y, Nakayama K (2001) Golgi-localizing, gamma-adaptin ear homology domain, ADP- ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J Biol Chem 276: 28541–28545 [DOI] [PubMed] [Google Scholar]
- Umebayashi K, Stenmark H, Yoshimori T (2008) Ubc4/5 and c-Cbl continue to ubiquitinate EGF receptor after internalization to facilitate polyubiquitination and degradation. Mol Biol Cell 19: 3454–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr SD (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128: 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Hurley JH (2010) Molecular mechanism of multivesicular body biogenesis by the ESCRT complexes. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YX, Doray B, Poussu A, Lehto VP, Kornfeld S (2001) Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292: 1716–1718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.