Abstract
Bacterial actin homologue MreB is required for cell shape maintenance in most non-spherical bacteria, where it assembles into helical structures just underneath the cytoplasmic membrane. Proper assembly of the actin cytoskeleton requires RodZ, a conserved, bitopic membrane protein that colocalises to MreB and is essential for cell shape determination. Here, we present the first crystal structure of bacterial actin engaged with a natural partner and provide a clear functional significance of the interaction. We show that the cytoplasmic helix-turn-helix motif of Thermotoga maritima RodZ directly interacts with monomeric as well as filamentous MreB and present the crystal structure of the complex. In vitro and in vivo analyses of mutant T. maritima and Escherichia coli RodZ validate the structure and reveal the importance of the MreB–RodZ interaction in the ability of cells to propagate as rods. Furthermore, the results elucidate how the bacterial actin cytoskeleton might be anchored to the membrane to help constrain peptidoglycan synthesis in the periplasm.
Keywords: bacterial cytoskeleton, cell shape, helix-turn-helix motif, MreB, RodZ
Introduction
The bacterial actin homologue MreB is required for the maintenance of cell shape in the majority of species. Bacteria control their particular shape by the spatio-temporal regulation of cell wall synthesis. A crucial component of the cell wall is the murein sacculus, a network of glycan strands linked together by short peptides. Purified murein sacculi retain the shape of the cell from which they were derived and cells become misshapen and/or burst when integrity of the sacculus is compromised in vivo. Over the past 20 years, intensive research has given us a glance into how non-spherical bacteria manage to mould and maintain the shape of the murein sacculus (for reviews see Shih and Rothfield, 2006; Cabeen and Jacobs-Wagner, 2007; den Blaauwen et al, 2008). One of the first mutants identified in cells with aberrant cell morphology in Escherichia coli indicated the importance of the actin homologue MreB for cell morphogenesis (Wachi et al, 1987). Not much later, bitopic membrane protein MreC and integral membrane protein MreD (both encoded by the same operon as MreB) were also found to be required to control cell shape (Wachi et al, 1989), as was penicillin-binding protein 2 (PBP2) and integral membrane protein RodA (Tamaki et al, 1980; Wachi et al, 1989). PBP2 interacts with MreC (Divakaruni et al, 2005; Slovak et al, 2006; van den Ent et al, 2006) and is the only murein synthase that is specifically required for cylindrical growth of the sacculus during cell elongation (Spratt, 1975; de Pedro et al, 2001; Vollmer and Bertsche, 2008). Depletion of these cell shape proteins results in the formation of spherical cells that eventually lyse under standard conditions (Levin et al, 1992; Varley and Stewart, 1992; Lee et al, 2003; Leaver and Errington, 2005; Bendezú and de Boer, 2008). MreB forms cytoskeletal filaments that localise as patchy, helical structures just underneath the cell membrane (Jones et al, 2001; Kruse et al, 2003; Shih et al, 2003; Defeu Soufo and Graumann, 2004; Figge et al, 2004; Gitai et al, 2004). As the structure of both monomeric and polymeric MreB resembles that of actin (van den Ent et al, 2001; Amos et al, 2004; Löwe et al, 2004), it is now widely accepted that an actin-like cytoskeleton is involved in cell shape maintenance in prokaryotes.
As the insertion of newly synthesised peptidoglycan follows a helical path that is reminiscent of that of MreB and the other cell shape factors (Daniel and Errington, 2003; Figge et al, 2004; Divakaruni et al, 2005; Dye et al, 2005; Leaver and Errington, 2005; Slovak et al, 2006), it has been proposed that MreB filaments might form tracks for murein synthases and/or hydrolases (Daniel and Errington, 2003; Figge et al, 2004; Carballido-López et al, 2006; Yamamoto et al, 2008). But how does cytosolic MreB spatially constrain peptidoglycan synthesis in the periplasm? A possible solution comes from the recently discovered cell shape protein, RodZ (Shiomi et al, 2008; Alyahya et al, 2009; Bendezú et al, 2009). RodZ is a conserved, bitopic membrane protein that is required for cell shape maintenance. It is located in the inner membrane with its C-terminus positioned in the periplasm (Newitt et al, 1999; Shiomi et al, 2008). As for other cell shape factors in E. coli (Bendezú and de Boer, 2008), deletion of rodZ results in spherical cells that can be rescued by an overdose of FtsZ (hence its name). RodZ colocalises to MreB structures (Shiomi et al, 2008; Alyahya et al, 2009; Bendezú et al, 2009) and this colocalisation is independent of cell shape proteins MreC, MreD, PBP2 and RodA (Bendezú et al, 2009). Interestingly, Caulobacter crescentus RodZ is confined to MreB in space and time and marks future sites of peptidoglycan synthesis (Alyahya et al, 2009), indicating a possible link between cytoplasmic MreB filaments and periplasmic peptidoglycan synthases. Deletion analyses have shown that the conserved, cytoplasmic domain of RodZ is required for its helical organisation (Shiomi et al, 2008; Alyahya et al, 2009; Bendezú et al, 2009). Domains on either side of the membrane serve partially redundant roles in shape maintenance (Bendezú et al, 2009). Cytoplasmic and periplasmic domains of RodZ are therefore likely to engage cell shape proteins at both sides of the membrane independently. Partial redundancy in the function of RodZ domains might ensure a reliable mechanism for cell shape determination.
Here, we present structural and biochemical evidence for a direct interaction between RodZ and MreB, either in its monomeric or filamentous form. The cocrystal structure between the cytoplasmic domain of RodZ and MreB from Thermotoga maritima reveals that RodZ's helix-turn-helix domain forms a scaffold for the interacting helix binding to subdomain IIA of MreB. Mutations that affect this interaction impair RodZ's localisation in E. coli as well as its ability to impose a rod shape on cells.
Results
RodZ interacts with monomeric and filamentous MreB
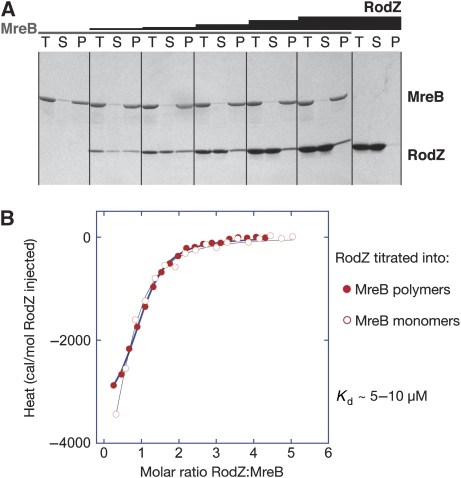
Proof of a direct interaction between purified MreB and the cytoplasmic domain of RodZ comes from pelleting assays that monitor MreB polymerisation in the presence of increasing amounts of RodZ(1−104). As shown in Figure 1A, RodZ(1−104) copellets with MreB filaments and does not pellet on its own (right panel, Figure 1A). While keeping the concentration of MreB constant and increasing the concentration of RodZ(1−104), more and more RodZ spins down in the pellet, reaching a saturation point of roughly one RodZ molecule per one MreB molecule.
Figure 1.
A direct interaction between purified cell shape proteins RodZ (1−104) and MreB from T. maritima. (A) MreB polymerisation assays. MreB (28 μM) was incubated at 37°C with nucleotide and MgCl2 in the absence (panel 1, lanes 1–3) and the presence of increasing amounts of RodZ (14 μM panel 2; 35 μM panel 3; 70 μM panel 4; 140 μM panel 5, 214 μM panel 6). In the absence of MreB, RodZ(1−104) remains in the supernatant (panel 7 with 280 μM RodZ). The reactions were centrifuged at 140 000 g and total (T), supernatant (S) and pellet (P) were analysed on a 10–20% SDS gel, stained with Coomassie. (B) Isothermal titration calorimetry shows RodZ(1−104) binds to monomeric and filamentous MreB with similar affinities. The cell contained 40 μM MreB and the syringe 1 mM RodZ(1−104), which was added over 18 injections of 2 uL. Before the experiments, proteins were dialysed to 20 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM sodium azide, 200 mM NaCl. The solid lines are the fit to the data and follow a simple binding model, showing a low micromolar affinity, exothermic binding enthalpy and 1:1 binding stoichiometry.
RodZ not only binds to polymers of MreB, but also to its monomeric form, as shown by isothermal titration calorimetry (ITC) (Figure 1B). RodZ(1−104) (1 mM) was slowly titrated into MreB (40 μM) that was either in its monomeric form (open circles) or polymeric form (closed circles). For the latter, MreB polymers were formed in the presence of ATPγS and MgCl2, separated from the supernatant by ultracentrifugation and resuspended in a buffer containing ATPγS/MgCl2. The binding curves suggest that RodZ's interaction with monomeric or filamentous MreB is exothermic at 10°C with a low micromolar affinity (Kd ∼5–10 μM) and a 1:1 stoichiometry. Therefore, RodZ(1−104) appears to interact with a single subunit of MreB without interfering with filament formation.
The molecular mass of RodZ and MreB before ITC was determined using size exclusion chromatography coupled to multi-angle light scattering measurements (SEC multi-angle light scattering (MALS), Wen et al, 1996). In the absence of nucleotide, both proteins elute as single peaks with the evaluated mass indicating that they are monomeric and highly monodisperse (see Supplementary Figure S1). After the ITC experiment, the peaks containing MreB and RodZ(1−104) elute at a slightly shorter retention time compared with the proteins alone, indicating that the two proteins interact (Supplementary Figure S1). Furthermore, there is no evidence for a higher order association of RodZ(1−104) when RodZ(1−104) by itself is examined over a wide concentration range (up to a maximum of 730 μM at injection, Supplementary Figure S1).
Crystal structure
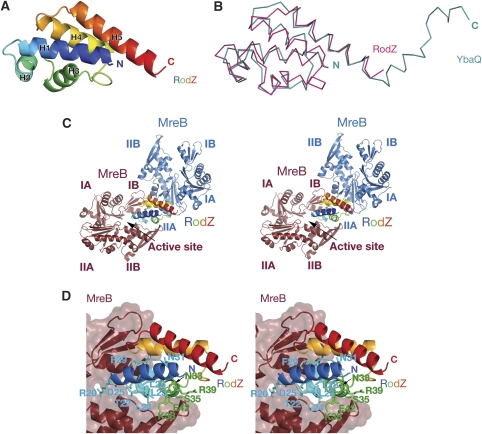
The structure of the heterodimer between RodZ and MreB reveals that RodZ's cytoplasmic domain (aa 1–88) has a classical helix-turn-helix motif (HTH) at its N-terminus followed by two additional α-helices (Figure 2A). The C-terminal 17 residues of RodZ(1−104) are not resolved in the crystals. The reason for the disorder can either be that the transmembrane region is missing, that a binding partner is needed or that it is intrinsically flexible. Earlier, it has been shown by deletion analyses that the juxtamembrane (JM) region (aa 85–111) in E. coli RodZ is required for cell shape maintenance (Bendezú et al, 2009). This region corresponds to the C-terminal 27 amino acids of T. maritima RodZ(1−104) (residues 78–104, Figure 5A) and includes the C-terminal half of helix H5. Deleting the JM region might impair the integrity of this helix. The N-terminal helix-turn-helix fold consists of three α-helices in a triangular arrangement, forming a loosely packed right-handed helical bundle. HTH motifs are mostly involved in transcriptional regulation, but have also been found to function in DNA replication as well as RNA metabolism and protein–protein interactions (for a review see Aravind et al, 2005). A similarity search among all structures in the Protein Databank (pdb) reveals that RodZ(1−88) superimposes on DNA-binding proteins with the putative transcription factor YbaQ having the best fit (pdb entry 2eby). It superimposes onto the five helices of RodZ with a root mean square deviation (rmsd) of 2.1 Å and a Z-score of 9.8 (Figure 2B). In fact, the DALI search (Holm and Sander, 1993) listed only putative DNA-interacting proteins, some of which were classified as HTH-type transcriptional regulators or belonged to the XRE family of transcription factors. Giving the high structural similarity to DNA-binding proteins, a pertinent question is whether the HTH motif of RodZ engages DNA or if it has a different function.
Figure 2.
(A) Ribbon representation of the crystal structure of the cytoplasmic part of RodZ(2−88) from T. maritima at 2.9 Å resolution. The secondary structure elements are labelled according to their appearance in the primary sequence (N-terminus in blue, C-terminus in red). (B) Superposition of the structures of RodZ(2−88) (in magenta) and helix-turn-helix type putative transcriptional regulator YbaQ (pdb entry 2eby, in turquoise). The two proteins overlap with a root mean square deviation (rmsd) of 2.1 Å over 77 residues, Z-score 9.8 (using structural similarity search DALI (Holm and Sander, 1993). (C) Stereograph of a ribbon representation of the cocrystal structure of MreB and RodZ(2−88) shows two possible interfaces. A larger interface where RodZ binds in the active site cleft of MreB between subdomains IB and IIB (MreB in brown) and a smaller interface, engaging subdomain IIA of MreB (MreB in blue). RodZ(2−88) is coloured in rainbow colours with the N-terminus in blue and C-terminus in red. Although the crystal packing shows the regular packing of heterodimers, only one molecule of RodZ(2−88) is shown for clarity reasons. (D) Binding mode of RodZ(2−88) within the active site cleft of MreB in stereo, as observed in the crystals. Substitution of the residues of RodZ depicted as sticks do not affect MreB binding in solution according to isothermal titration calorimetry or polymerisation assays.
Figure 5.
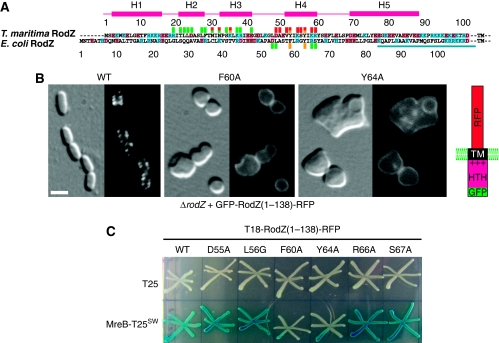
(A) Sequence alignment of RodZ cytoplasmic domain from T. maritima and E. coli. Numbers above the alignment refer to the T. maritima RodZ sequence, below the alignment to the E. coli sequence. TM refers to the transmembrane helix. Residues marked with a red square above the sequence contribute to the surface area buried by the interface between RodZ and MreB. Mutations in residues marked with a yellow square abolish MreB binding, whereas those marked with a green square do not interfere with MreB binding when mutated. The squares underneath the E. coli sequence indicate mutations that affect cell shape and MreB interaction in E. coli (yellow) or behave as the wild-type protein (green) (see Supplementary Table III). The bar (in turquoise) indicates the juxtamembrane region in E. coli RodZ. (B) The effect of interface mutations on RodZ1−138 localisation and its ability to impose rod shape on ΔrodZ cells in E. coli. Shown are ΔrodZ cells (FB60) expressing GFP-RodZ1−138-RFP (left panel), or a mutant version thereof (middle and right) from a construct integrated at the chromosomal attHK022 site. The middle and right panels show cells producing the F60A and Y64A version, respectively (corresponding to Y53 and Y57 in T. maritima RodZ, respectively). Note the inability of these cells to maintain rod-shape, as well as the even membrane distribution of fluorescence. Cells were grown in M9-mal with 250 μM IPTG to OD600=0.3–0.5, and were imaged live with DIC (left side of each panel) and GFP fluorescence (right side of each panel) optics. Bar equals 2 μ. The construct used, GFP-RodZ1−138-RFP, is shown in the cartoon on the right. (C) Effects of RodZ interface mutations on its in vivo interaction with MreB in E. coli. Strain BTH101 [cya-99] was cotransformed with plasmid pairs encoding the indicated T18- and T25-fusions, and individual colonies were patched on M9-agar containing 0.2% glucose, 40 mg/ml X-Gal and 250 μM IPTG. The plate was incubated at 30°C and imaged after 24 h. Plasmids used were the vector control pKNT25 [Plac∷lacZ′-t25] (upper row), pCH375 [Plac∷mreB′-t25-′mreB], encoding a sandwich fusion that carries the CyaA T25 domain inserted between helices 6 and 7 of MreB (Bendezú et al, 2009) (lower row), pCH395 [Plac∷t18-rodZ(1–138)-rfp], encoding the CyaA T18 domain appended to the N-terminus of a version of RodZ in which its periplasmic domain has been substituted with mCherry (Bendezú et al, 2009) (column 1), and mutant derivatives of pCH395 (columns 2–7).
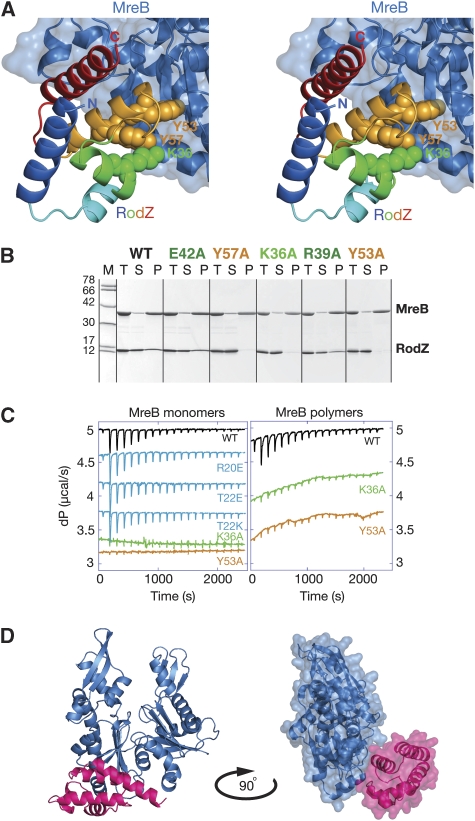
The cocrystal structure of MreB and cytoplasmic RodZ reveals an interesting crystal packing, where RodZ is sandwiched between two MreB molecules (Figure 2C). Hence, RodZ shares two distinct interfaces with MreB in the cocrystal structure: a substantial interface of 732 Å2 where RodZ is positioned between the two subdomains of MreB (in brown, Figure 2C) thereby closing off the active site cleft and a smaller interface of 552 Å2, where it binds to the side of subdomain IIA (in blue, Figure 2C). Mutational analyses of RodZ(1−104) show that single or multiple residues in the interface with the active site cleft (Figure 2D) can be changed without affecting MreB binding in vitro, as determined by ITC and polymerisation assays (Figure 3B and C and data not shown). This suggests that the binding site of RodZ(1−104) within the active site cleft of MreB is crystallisation enforced. In contrast, substitutions in the smaller interface (Figure 3A) abolish MreB binding; single substitutions including K36A, Y53A and Y57A (depicted as spheres in Figure 3A) severely impair RodZ's interaction with both filamentous and monomeric MreB (Figure 3B and C). This is consistent with the observation that wild-type RodZ(1−104) binds with similar affinities to either form of MreB (Figure 1B). Single or multiple mutations in RodZ(1−104) that do not affect copelleting with MreB are: R20A/E, T22A/E/K/R, L23D, L24A, D25A, L28A, F29A/K/R, N31A, N33A, S35W, K38A, R39A, E42A, [T22A, L23D, L24A, D25A, N31A, E99D], [T22A, L23D, L24A, D25A], [L28A, F29A], [N31A, N33A] (depicted as sticks in Figure 2D). Some of these single mutants were also tested by ITC showing the same binding kinetics as the wild-type protein (Figure 3C). Thus, mutational analyses reveal that the heterodimer of MreB and RodZ(1−104) as shown in Figure 3D is the functionally relevant complex.
Figure 3.
(A) Stereograph of RodZ's interaction with subdomain IIA of MreB (blue). Residues K36, Y53 and Y57, shown in spheres, are required for MreB interaction. (B) MreB polymerisation assays were used to test the interface mutants of RodZ for MreB binding. MreB (28 μM) was incubated with RodZ(1−104) (35 μM) in the presence of nucleotide and MgCl2 at 37°C and the pellet was separated from the supernatant by centrifugation at 140 000 g. Total (T), supernatant (S) and pellet (P) was analysed on a 10–20% gradient gel and stained with Coomassie. Mutations in RodZ(1−104) are shown above the panels in colours corresponding to the labelled residues in Figures 2D and 3A. Protein marker (M) depicts molecular weights of 78, 66, 42, 30, 17 and 12 kDa. (C) Isothermal titration calorimetry (ITC) confirmed the behaviour of the interface mutants. Wild-type (WT) or mutant RodZ (1−104) was titrated into a cell containing monomeric MreB (left panel) or filamentous MreB (right panel). Colour code as for Figures 2D and 3A. The cell contained 40 μM MreB and the syringe 1 mM RodZ, which was added over 18 injections of 2 μl. Before the experiments, proteins were dialysed to 20 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM NaN3, 200 mM NaCl. (D) Ribbon representation of the cocrystal structure of MreB (in blue) and RodZ(2−88) (in magenta) (left). A surface representation of the contacts between the two proteins is shown on the right, where the structure is rotated by 90°.
Light microscopy
Purified, alexa-labelled MreB and RodZ(1−104) can form large structures that are visible under the light microscope (Figure 4). The two proteins colocalise irrespective of preformation of MreB filaments; RodZ(1−104) is either incubated with MreB and ATPγS together (middle panel) or is added after MreB polymers are formed (in the presence of the crowding agent PEG 4000 (top panel)). Substitution of Y53A in RodZ(1−104) impairs colocalisation of the pure proteins (lower panel).
Figure 4.
Colocalisation of purified proteins MreB and RodZ(1−104) from T. maritima. Alexa 555-labelled MreB polymers (red) were formed in the presence of crowding agent PEG 4000, ATPγS and MgCl2. Subsequently, Alexa 488-labelled RodZ(1−104) was added (green) (top panel). In the absence of PEG 4000, Alexa 555-labelled MreB (red) and Alexa 488-labelled RodZ(1−104) (green) colocalise as large structures from which filaments grow when mixed together in the presence of nucleotide and MgCl2 (middle panel). RodZ's mutation Y53A impairs MreB interaction and does not colocalise with the large MreB structures (lower panel). Left images taken with mCherry filter showing MreB alone, middle with YFP filter showing RodZ(1−104) alone, right panel merged image. Images were taken using a 100 × microscope oil immersion objective (Nikon) and a Photometrics CoolSNAP HQ2 camera. Typical exposures were 50–100 msec.
The importance of the MreB interface for RodZ's function in vivo was investigated by introducing two of the critical mutations in E. coli RodZ—F60A and Y64A, equivalent to T. maritima Y53A and Y57A (Figure 5A). The mutants were expressed as a GFP fusion to the cytoplasmic and transmembrane region of RodZ (GFP-RodZ1−138-RFP) in a ΔrodZ strain (Figure 5B; Supplementary Table III). The non-mutated fusion restores cell shape in ΔrodZ cells and localises in a helical pattern (Figure 5B, left panel) (Bendezú et al, 2009). The mutants however fail to correct cell shape and localise evenly along the membrane (Figure 5B, middle and right panels). Other amino acid changes (D55A, L56G, R66A or S67A) have no noticeable effects on MreB interaction and cells producing any one of these versions of the GFP-RodZ1−138-RFP fusion resemble those of the wild-type sequence (see Supplementary Table III). In the context of the full-length protein, the two mutations that affect MreB interaction, F60A and Y64A, cause RodZ to localise evenly along the membrane with some associated spots of higher intensity (Supplementary Table III). These mutants partially correct cell shape, though most cells are wider than normal. The previously reported phenotype of a HTH deletion strain (Bendezú et al, 2009) is consistent with these results, suggesting that the most important function of the HTH domain is the interaction with MreB. Bacterial two hybrid experiments confirm that mutants containing the critical substitutions in helix H4, F60A and Y64A, no longer interact with MreB in vivo, whereas mutations in loops adjacent to helix H4 do not affect MreB binding (Figure 5C; Supplementary Table III).
How does the MreB–RodZ heterodimer compares to actin complexes?
As mentioned earlier, MreB is structurally and functionally related to actin and superimposes on actin with an rmds of 3.7 Å. To determine whether the homology extends towards their binding partners, the MreB–RodZ heterodimer structure is compared with actin complexes as known to date (see Supplementary Figure 2). The comparison shows that except for DNAse that disrupts actin polymerisation by blocking off the top of actin, all other binding partners engage the barbed end of the molecule, just like RodZ. However, none of them have the same binding mode as RodZ, nor share its fold, suggesting that they evolved separately to fulfil their specific functions.
Discussion
The similarity in binding thermodynamics of RodZ(1−104) to monomeric or filamentous MreB (Figure 1) suggests that RodZ's binding site within the heterodimer is unaffected by subunit contacts within the MreB protofilament. It also indicates that conformational changes induced by nucleotide binding by MreB and/or filament formation do not affect MreB–RodZ interaction. To determine whether the binding mode observed in the cocrystal structure is indeed compatible with filament formation, the structure of the RodZ–MreB heterodimer was superimposed on the protofilament structure present in the original MreB crystals (van den Ent et al, 2001). The superposition is compatible with the possibility that RodZ(1−104) interacts with MreB protofilaments in the same manner as it does with monomeric MreB (Figure 6A and B). The front-end view (Figure 6B) shows that the C-terminal helix of RodZ(1−104) is not perpendicular to the filament axis; the magenta arrow indicating the direction of the C-terminal helix sits at an angle of ∼20° of the normal to the filament axis. This raises the interesting possibility that the membrane anchoring of MreB filaments by RodZ promotes a helical arrangement of the filaments. A basic requirement is that both the single transmembrane helix and the linker extending from helix H5 become rigid with respect to the membrane, for instance by additional protein–protein interactions. Thus far, there is no evidence for such an interaction. Earlier work has shown that the JM region (aa 85–111 in E. coli RodZ) is required in providing rod shape and might provide additional protein–protein interactions necessary to obtain rigidity (Bendezú et al, 2009). Alternatively, the role of the JM region might lie in the presence of the positively charged residues adjacent to the transmembrane region. This could restrict angular movement of the transmembrane helix by interacting with the negatively charged head groups of the polar lipids.
Figure 6.
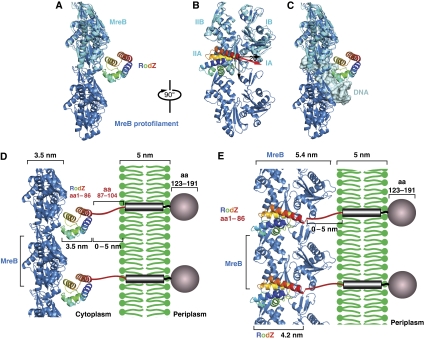
(A, B) Model of RodZ cytoplasmic domain in complex with an MreB protofilament. The crystal structure of the heterodimer of RodZ(1−88) (rainbow colours, N-terminus in blue, C-terminus in red) and MreB (cyan) was superimposed on the protofilament structure of MreB (blue) (van den Ent et al, 2001). The black, downwards-pointing arrow shows the MreB filament axis. The red arrow indicates the direction of the C-terminal helix of RodZ's cytoplasmic domain that is positioned at ∼20° of the normal to the filament axis. MreB subdomains IA, IB, IIA, IIB are labelled in cyan. (C) Model showing that RodZ's HTH domain is unable to bind MreB and the nucleoid simultaneously. The helix-turn-helix motif of RodZ is modelled onto a classical DNA-binding protein (phage 343 repressor, pdb entry 2OR1), revealing its hypothetical DNA-binding site. Superimposing this imaginary complex on to the heterodimer structure of MreB and RodZ(1−88) shows a steric clash between MreB and DNA. Colour code as in (B). Double-stranded DNA in grey (taken from pdb entry 2OR1). (D, E) Model showing how RodZ might anchor the bacterial actin cytoskeleton in the membrane. As the orientation of MreB relative to the membrane is not known, the MreB filaments could adopt any position between the two extremes depicted in (D, E), depending on the flexibility of the linker that connects the helix-turn-helix motif of RodZ to the transmembrane region. (D) The side-view shows RodZ(1−104) positioned between MreB and the membrane. In that case, the distance between MreB and the membrane could vary between ∼3.5 and 8.5 nm, depending on the orientation of the linker of RodZ. (E) Front-end view: the heterodimer between MreB and RodZ(1−104) is rotated by 90° relative to (D). In this model, the spacing between MreB and the membrane could be between 0 nm (if part of the linker folds back) and ∼4 nm (in case the linker is fully extended). As MreB would face the membrane with subdomain I, the lateral contacts between the protofilaments would be limited to subdomain II and/or the back of the molecule (E).
The present heterodimer structure shows MreB in a slightly more open conformation than in the original protofilament structure (compare the MreB molecule in cyan, coming from the heterodimer structure with the structure in sky blue that is in the straight protofilament configuration, Figure 6A and B, top). As RodZ(1−104) has an additional, crystallisation-enforced interface with the active site cleft of MreB, it is plausible that the slightly more open configuration in MreB is the result of crystal contacts with RodZ, which may not be functionally relevant.
Classical helix-turn-helix DNA-binding proteins bind their substrate with the third helix, occupying the major groove of DNA (Aravind et al, 2005). In RodZ(1−104), the third helix only provides a minor contact with its substrate MreB. The main contacts here are formed by the fourth α-helix that itself is buried in a conserved cleft formed by the HTH motif (see Supplementary movie). The highly conserved aromatic residue Tyr53 sits in a hydrophobic and reasonably conserved pocket of MreB, formed by L141, N149, R279, G280, F282 and I306. Replacing I306 with a Trp abolishes RodZ(1−104) binding in vitro (see Supplementary Figure S3). In the few organisms that lack RodZ, the pocket is shaped by similar, hydrophobic residues, suggesting that they are important for the fold of MreB and that RodZ has taken advantage of this pocket to insert the aromatic side chain of Y53 (T. maritima) or F60 (E. coli). Additional residues of T. maritima MreB that contribute to the binding surface of RodZ(1−104) are S139, N140, N142, I277, E278, D294, G302, S304, V305, E309.
It has been postulated that the HTH motif of RodZ might bind DNA and could possibly bring the MreB cytoskeleton in contact with the nucleoid (Gerdes, 2009). If RodZ were to bind DNA as a classical HTH motif, then it cannot bind MreB simultaneously, as shown in Figure 6C. The hypothetical DNA-binding mode of RodZ is derived from its superposition on the DNA complex of phage 343 repressor (PDB entry 2or1; Aggarwal et al, 1988). Combining this superposition with the heterodimer structure shows a steric clash between MreB (blue) and DNA (grey, Figure 6C), making it rather unlikely that RodZ would bring MreB in contact with the nucleoid. Rather, RodZ seems to use the HTH fold to position its fourth helix that then interacts with MreB.
Our biochemical and structural evidence shows that RodZ(1−104) binds to monomeric MreB. This binding event is compatible with MreB filament formation as shown by biochemical experiments and by light microscopy (Figures 1, 3 and 4). Through this interaction, the cytoplasmic HTH domain might enhance the local concentration of MreB raising its critical concentration that would enable filament formation in the vicinity of the membrane. As RodZ is a bitopic membrane protein, it will assist in anchoring the actin cytoskeleton to the membrane (Figure 6D and E). Assuming that the linker between the HTH domain and the transmembrane region is fairly rigid, MreB would face the membrane with subdomain I (Figure 6E). Lateral contacts between the protofilaments would then be limited to subdomain II and the back of the molecule. The orientation of MreB to the membrane depends on the flexibility of the linker in RodZ (Figure 6D and E). The spacing might vary from organism to organism, as the length of the linker attaching the C-terminal helix to the transmembrane region differs (Alyahya et al, 2009). In the past, it has been proposed that the morphogenetic membrane-linked proteins MreC and MreD would tie cytoplasmic MreB to the periplasmic peptidoglycan synthesis machinery. MreD is an integral membrane protein with only short stretches of amino acids exposed to the cytoplasm. Bitopic membrane protein MreC has a larger periplasmic domain and an N-terminal tail sticking into the cytoplasm that varies in length between 5 and 51 residues among different bacterial species. This makes it questionable whether MreC or MreD on their own could provide a large enough interface for a stable interaction with MreB, at least in some organisms. In E. coli, the cytoplasmic tail of MreC is predicted to be ∼9 residues long (http://www.enzim.hu/hmmtop/), which is most likely too short to ensure a stable interaction with MreB. Once brought into the vicinity of the membrane, MreB might well establish additional contacts with MreC and/or MreD. Taken together, a picture emerges that describes how the cytoplasmic MreB cytoskeleton dictates cell shape through its interaction with bitopic membrane proteins, such as RodZ and possibly MreC, both of which involve the peptidoglycan synthesis machinery in the periplasm to mould and maintain the shape of the cell.
Materials and methods
In vitro work
Cloning, expression and purification. The gene encoding RodZ (NCBI Accession number NP_229660) from T. maritima genomic DNA (ATCC 43589D_5) was used as template to amplify the cytoplasmic region (aa 1–104) by PCR. The PCR product generated with forward primer (5′-AGTCTACCATATGAGCGAAAAATGGAAGGAACTCGGTGAGAC) and reverse primer (5′-TGACTACGGATCCCAGGTCCCTTGTTTTCTCTTTCTCC) was cut with NdeI and BamHI and ligated into NdeI/BamHI digested pHis17 (B. Miroux personal communication). The newly obtained plasmid, pFE305 contains the rodZ gene downstream of the T7 promoter, encoding the cytoplasmic part of RodZ (aa 1–104) with a C-terminal His6-tag to facilitate purification.
Site-directed mutagenesis was carried out using QuickChange (Stratagene) on pFE305 to introduce interface mutations in RodZ(1−104). To label RodZ and MreB with alexa fluor maleimide, mutagenesis was carried out to introduce a KCK motif following the C-terminal His-tag, leaving the 3′ BamHI site intact, creating pFE344 encoding RodZ(1−104) (Y53A), pFE346 encoding RodZ(1−104) and pFE347 encoding MreB.
RodZ(1−104) and T. maritima MreB were expressed in C41(DE3) cells (Miroux and Walker, 1996) for 4 h at 37°C and MreB was purified as described earlier (van den Ent et al, 2001). Following affinity purification using a Histrap HP column (GE Healthcare) at pH 6.0, RodZ was further purified over a Sephacryl S100 column (GE Healthcare) in 20 mM Tris–HCl 7.0, 1 mM EDTA and 1 mM NaN3 and concentrated to 94 mg/ml before flash freezing in liquid nitrogen. Typical yields were ∼15 mg/l culture.
The cysteine-containing mutants were purified in the presence of 5 mM β-mercaptoethanol (Histrap HP column) and 2 mM TCEP (size-exclusion chromatography). MreB containing KCK was able to form filaments as the wild-type protein as seen under the electron microscope (data not shown). MreB was labelled with Alexa fluor 555 C2 maleimide (Invitrogen) and RodZ(1−104) (WT as well as Y53A mutant) were labelled with Alexa fluor 488 C5 maleimide (Invitrogen) according to the manufacturer's instructions. Proteins were mixed with a 5 × molar excess dye for 2–5 h at room temperature or left overnight at 4°C. Alexa-labelled MreB was separated from unbound dye on a Superdex 200 10/30 column (GE Healthcare) in 20 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM NaN3, 200 mM NaCl, 2 mM TCEP. Alexa-labelled RodZ was purified over a Superdex75 column (GE Healthcare) in 20 mM Tris–HCl, pH 7.0, 1 mM EDTA, 1 mM NaN3, 2 mM TCEP. Labelled proteins were checked on a gel, pooled and concentrated by ultrafiltration using Vivaspin concentrators (Vivascience).
Crystallisation and structure determination. Cocrystals of RodZ(1−104) and MreB were obtained in the initial screen of 1440 conditions (Stock et al, 2005). Final crystals were optimised by hanging-drop vapour diffusion in 12% PEG 1000 MME, 478 mM sodium thiocyanate, 147 mM CaCl2, 100 mM Tris–HCl, pH 7.7, 6.5% EtOH with 5.6 mg/ml (415 μM) RodZ(1−104) (MW=13.48 kDa) and 6.6 mg/ml (180 μM) MreB (MW=36.75 kDa) (2.3 molar excess of RodZ(1−104)). The crystals were cryoprotected with mother liquor containing 24.8% PEG400 and flash frozen in liquid nitrogen. Crystals diffracted to 2.9 Å and belong to spacegroup P22121 with two heterodimers in the asymmetric unit. Cell constants and crystallographic data are summarised in Table 1. Datasets were collected at ESRF beamline ID23–1. All data were indexed and integrated with Mosflm (Leslie, 1991). The structure was solved by molecular replacement using Phaser (McCoy et al, 2004) and MreB (PDB entry code 1JCE), with subdomains I and II split. The model was manually built with MAIN (Turk, 1992) and refined using Phenix (Adams et al, 2002). Details of the refined models are shown in Table 1.
Table 1. Crystallographic data and refinement statistics.
| P21212: a=81.81Å, b=109.84Å, c=112.27Å | |||||
|---|---|---|---|---|---|
| Crystal | l [Å] | Resol. [Å] | I/sIa | Rmb | Multipl.c |
| PEAK | 0.9762 | 2.9 | 8.5 (2.7) | 0.087 | 3.0 |
| MreB residues | A: 2–336; B: 4–336 | ||||
| RodZ residues | R: 2–88; S:3–84 | ||||
| Resolution | 2.9 Å | ||||
| R-factor, R-freed | 0.197, 0.288 | ||||
| B averagee | 46.6Å2 | ||||
| Geometry bonds/anglesf | 0.009Å, 1.367° | ||||
| Ramachandrang | 84.0%/0.0% | ||||
| PDB IDh | 2wus/r2wussf | ||||
| aSignal-to-noise ratio of intensities, highest resolution bin in brackets. | |||||
| bRm: ShSj∣I(h,i)−I(h)∣/ShSi I(h,i), where I(h,i) are symmetry-related intensities and I(h) is the mean intensity of the reflection with unique index h. | |||||
| cMultiplicity for unique reflections, for MAD datasets I(+) and I(−) are kept separate. | |||||
| d5% of reflections were randomly selected for determination of the free R-factor, before any refinement. | |||||
| eTemperature factors averaged for all atoms. | |||||
| fRMS deviations from ideal geometry for bond lengths and restraint angles (Engh and Huber, 1991). | |||||
| gPercentage of residues in the ‘most favoured region' of the Ramachandran plot and percentage of outliers (PROCHECK (Laskowski, 1993)). | |||||
| hProtein Data Bank identifiers for coordinates and structure factors, respectively. | |||||
Polymerisation assays. Purified, prespun T. maritima MreB (35.8 kDa) was mixed with or without prespun RodZ(1−104) (13.4 kDa) in the presence of 50 mM Tris–HCl, pH 7.0, 1 mM ATPγS, 2 mM MgCl2 and incubated for 20 min at 37°C. Final concentration of MreB was 28 and 35 μM for RodZ(1−104), unless stated otherwise. The reactions were centrifuged at 140 000 g in a TLA100 rotor (Beckman) for 20 min at 20°C. The supernatant was removed for analysis and the pellet was washed once with reaction buffer before it was dissolved in SDS loading dye. Equivalent amounts of the total reaction, the supernatant and pellet were analysed on a 10–20% SDS–polyacrylamide gel, which was stained with Coomassie.
In vitro colocalisation of RodZ(1−104) and MreB Purified, alexa-labelled MreB was mixed with non-labelled MreB (equimolar ratio) at a concentration of 2.2 mg/ml in 20 mM Tris–HCl, pH 9.0, 1 mM EDTA, 1 mM NaN3, 200 mM NaCl, 2 mM TCEP and prespun at 138 000 g for 10 min, 4°C. MreB was then polymerised in the presence of 2.5 molar excess of alexa-labelled RodZ(1−104) (either wild type or the Y53A mutant) in polymerisation buffer (50 mM Tris–HCl, pH 7.0, 1 mM ATPγS, 2 mM MgCl2) for 20 min at 37°C. Larger filaments are formed when the reaction is supplemented with 3% PEG4000.
Isothermal titration calorimetry. ITC measurements were made using manual and automated versions of a Microcal iTC200 calorimeter. In all measurements, RodZ(1−104) in the syringe was injected into MreB in the cell as it was not possible to concentrate solutions of MreB to the level required for the alternate configuration. Typical experiments used 40 μM MreB in the cell and 1 mM RodZ(1−104) in the syringe, which was added over 18 injections of 2 μl. Experiments were run at 10°C. Before each experiment, proteins were dialysed over night at 4°C to 20 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 mM NaN3, 200 mM NaCl (TEN200). Before ITC experiments with polymerized MreB, filaments were formed as described for the pelleting assay: prespun MreB was incubated for 20 min at 37°C in 1 × polymerization buffer (50 mM Tris–HCl, pH 7.0, 1 mM ATPγS, 2 mM MgCl2), spun at 140 000 g for 20 min. The pellet was washed with polymerization buffer and resuspended in TEN200 complemented with 1 mM ATPγS and 2 mM MgCl2. Excess heat for each injection was determined by integration following manual adjustment of baselines and normalisation relative to the RodZ(1−104) concentration. The control heat of dilution of RodZ(1−104) was determined from the small excess heat at the end points of the titrations (molar ratio RodZ(1−104): MreB >4) and these were subtracted from the data before final fitting. Data analysis and curve fitting using a model with a simple one set of binding sites were performed using the Microcal Origin software provided with the ITC.
Multi-angle light scattering. SEC MALS was performed using a Wyatt Dawn Heleos-II angle light scattering instrument coupled to a Wyatt Optilab rEX online refractive index detector. Samples for analysis were resolved on a Superdex S-200 analytical gel filtration column (GE Healthcare) running at 0.5 ml/min before passing through the light scattering and refractive index detectors in a standard SEC MALS format. Protein concentration was determined from the refractive index based on 0.19 RI for 1 mg/ml, and combined with the observed scattered intensity to calculate absolute molecular mass using Wyatt's ASTRA analysis software.
In vivo work
E. coli strains. For plasmid construction see Supplementary data. Strains MG1655 [ilvG rfb50 rph1], TB28 [MG1655, lacIZYA<>frt] and FB60 [TB28, rodZ<>aph] were described earlier (Bernhardt and de Boer, 2003; Bendezú et al, 2009). BTH101 [cya-99 araD139 galE15 galK16 rpsL1 hsdR2 mcrA1 mcrB1] was provided by Gouzel Karimova.
Strains used for studying the effects of mutations on the in vivo properties of RodZ were obtained in two steps. First, pFB273 [Plac∷gfp-t-rodZ], pLP39 [Plac∷gfp-t-rodZ(1–138)-rfp] or each of their mutant derivatives was integrated at the chromosomal HK022 att site of strain TB28 [wt] as described (Haldimann and Wanner, 2001), resulting in strains TB28(iFB273) and TB28(iLP39), and their mutant versions. In each case, the corresponding FB60 [ΔrodZ] strain was then created by P1-mediated introduction of the rodZ<>aph allele from FB60/pFB233 [ΔrodZ/ Plac∷rodZ(1–319)], with selection for Kanr transductants on M9-maltose agar supplemented with 250 μM IPTG.
Growth conditions. Cells were routinely grown at 30°C in LB (0.5% NaCl) or M9 minimal medium supplemented with 0.2% maltose, 0.2% casamino acids and 50 μM thiamine (M9-mal). When appropriate, medium was supplemented with 15 (for strains with bla integrated in the chromosome) or 50 μg/ml ampicillin (Amp), 25 μg/ml kanamycin (Kan) or 25 μg/ml chloramphenicol (Cam). Other details are specified in the text.
Bacterial two hybrid assay. For BACTH analyses (Karimova et al, 1998), plasmid pairs encoding the indicated T18- and T25-fusions were cotransformed into BTH101 [cya-99], and individual colonies were patched on M9-agar containing 0.2% glucose, 25 μg/ml Kan, 40 μg/ml X-Gal and 250 μM IPTG. The plate was incubated at 30°C and imaged after 24 h.
ΔrodZ complementation assays and microscopy. Cells were grown overnight in M9-mal supplemented with 15 μg/ml Amp and 250 μM IPTG, diluted to OD600=0.05 in M9-mal with 250 μM IPTG, and grown further to OD600=0.3–05. Aliquots were applied to microscope slides containing thin pads of 1.2% agarose in M9, and cells were imaged immediately with differential interference contrast (DIC) and fluorescence (GFP specific) optics, as described (Bendezú et al, 2009).
Supplementary Material
Acknowledgments
We thank beamline scientists at ESRF's beamline ID23-1 and thank Felipe Bendezú for help in identifying T. maritima rodZ and Cynthia Hale for help in plasmid construction. This work was partially supported by NIH GM57059 (to PdB).
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC (1988) Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242: 899–907 [DOI] [PubMed] [Google Scholar]
- Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C (2009) RodZ, a component of the bacterial core morphogenic apparatus. Proc Natl Acad Sci USA 106: 1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA, van den Ent F, Löwe J (2004) Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol 16: 24–31 [DOI] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM (2005) The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29: 231–262 [DOI] [PubMed] [Google Scholar]
- Bendezú FO, de Boer PA (2008) Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, de Boer PA (2009) RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2003) The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48: 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C (2007) Skin and bones: the bacterial cytoskeleton, cell wall, and cell morphogenesis. J Cell Biol 179: 381–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J (2006) Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell 11: 399–409 [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113: 767–776 [DOI] [PubMed] [Google Scholar]
- de Pedro MA, Donachie WD, Holtje JV, Schwarz H (2001) Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol 183: 4115–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL (2004) Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep 5: 789–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA (2008) Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32: 321–344 [DOI] [PubMed] [Google Scholar]
- Divakaruni AV, Loo RR, Xie Y, Loo JA, Gober JW (2005) The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. Proc Natl Acad Sci USA 102: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z (2005) Two independent spiral structures control cell shape in Caulobacter. Proc Natl Acad Sci USA 102: 18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh RA, Huber R (1991) Accurate bond and angle parameters for x-ray protein-structure refinement. Acta Crystallogr A 47: 392–400 [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW (2004) MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Gerdes K (2009) RodZ, a new player in bacterial cell morphogenesis. EMBO J 28: 171–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L (2004) An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA 101: 8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183: 6384–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-López R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95: 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K (2003) Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J 22: 5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Leaver M, Errington J (2005) Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol 57: 1196–1209 [DOI] [PubMed] [Google Scholar]
- Lee JC, Cha JH, Zerbv DB, Stewart GC (2003) Heterospecific expression of the Bacillus subtilis cell shape determination genes mreBCD in Escherichia coli. Curr Microbiol 47: 146–152 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1991) Recent changes to the MOSFLM package for processing film and image plate data. (ed) SERC Laboratory: Daresbury, Warrington WA44AD, UK
- Levin PA, Margolis PS, Setlow P, Losick R, Sun D (1992) Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol 174: 6717–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, van den Ent F, Amos LA (2004) Molecules of the bacterial cytoskeleton. Annu Rev Biophys Biomol Struct 33: 177–198 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Storoni LC, Read RJ (2004) Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr D Biol Crystallogr 60: 1220–1228 [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260: 289–298 [DOI] [PubMed] [Google Scholar]
- Newitt JA, Ulbrandt ND, Bernstein HD (1999) The structure of multiple polypeptide domains determines the signal recognition particle targeting requirement of Escherichia coli inner membrane proteins. J Bacteriol 181: 4561–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YL, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100: 7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YL, Rothfield L (2006) The bacterial cytoskeleton. Microbiol Mol Biol Rev 70: 729–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H (2008) Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slovak PM, Porter SL, Armitage JP (2006) Differential localization of Mre proteins with PBP2 in Rhodobacter sphaeroides. J Bacteriol 188: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG (1975) Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA 72: 2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock D, Perisic O, Löwe J (2005) Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol 88: 311–327 [DOI] [PubMed] [Google Scholar]
- Tamaki S, Matsuzawa H, Matsuhashi M (1980) Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol 141: 52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D (1992) Weiterentwicklung eines Programms für Molekülgrafik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD Thesis, Technische Universität München, Munich, Germany.
- van den Ent F, Amos LA, Löwe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Leaver M, Bendezu F, Errington J, de Boer P, Löwe J (2006) Dimeric structure of the cell shape protein MreC and its functional implications. Mol Microbiol 62: 1631–1642 [DOI] [PubMed] [Google Scholar]
- Varley AW, Stewart GC (1992) The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (minCD) and cell shape (mreBCD) determinants. J Bacteriol 174: 6729–6742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U (2008) Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta 1778: 1714–1734 [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Okada Y, Matsuhashi M (1989) New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol 171: 6511–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M (1987) Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol 169: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Arakawa T, Philo JS (1996) Size-exclusion chromatography with on-line light-scattering, absorbance, and refractive index detectors for studying proteins and their interactions. Anal Biochem 240: 155–166 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J (2008) The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol Microbiol 70: 297–310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.