Abstract
Objective
Plasma adiponectin is strongly associated with various components of metabolic syndrome, type 2 diabetes and cardiovascular outcomes. Concentrations are highly heritable and differ between men and women. We therefore aimed to investigate the genetics of plasma adiponectin in men and women.
Methods
We combined genome-wide association scans of three population-based studies including 4659 persons. For the replication stage in 13795 subjects, we selected the 20 top signals of the combined analysis, as well as the 10 top signals with p-values less than 1.0*10-4 for each the men- and the women-specific analyses. We further selected 73 SNPs that were consistently associated with metabolic syndrome parameters in previous genome-wide association studies to check for their association with plasma adiponectin.
Results
The ADIPOQ locus showed genome-wide significant p-values in the combined (p=4.3*10-24) as well as in both women- and men-specific analyses (p=8.7*10-17 and p=2.5*10-11, respectively). None of the other 39 top signal SNPs showed evidence for association in the replication analysis. None of 73 SNPs from metabolic syndrome loci exhibited association with plasma adiponectin (p>0.01).
Conclusions
We demonstrated the ADIPOQ gene as the only major gene for plasma adiponectin, which explains 6.7% of the phenotypic variance. We further found that neither this gene nor any of the metabolic syndrome loci explained the sex differences observed for plasma adiponectin. Larger studies are needed to identify more moderate genetic determinants of plasma adiponectin.
Keywords: adiponectin, genome-wide association study, polymorphism, cardiovascular disease, metabolic syndrome
Introduction
Plasma adiponectin is a quantitative parameter, which has a strong role in modulating insulin sensitivity and glucose homeostasis. It has been found to be decreased in humans with type 2 diabetes and cardiovascular disease (CVD) (1,2) and decreased plasma adiponectin was found to be associated with deteriorated levels of virtually all parameters of the metabolic syndrome (3-5). Experiments in mice transgenic or deficient for the adiponectin gene have underscored the functional role of adiponectin on various components of the metabolic syndrome and diabetes mellitus (3,6,7).
Concerning CVD outcomes the observations on adiponectin are heterogeneous as recently reviewed extensively (8): experimental data demonstrate that adiponectin stimulates the production of nitric oxide, positively affects inflammatory mechanisms, has anti-apoptotic properties and is involved in vascular remodeling. Clinical data are diverse depending mainly on the disease stage when investigated. Low levels seem to be associated with worse outcomes when measured in healthy conditions. However, there is accumulating data that in diseased states such as chronic heart failure or existing CVD high rather than low levels predict CVD and non-CVD mortality. Knowing the genes which affect plasma adiponectin might be helpful to disentangle adiponectin as cause or consequence of disease states using a Mendelian randomization approach (9).
Plasma adiponectin shows pronounced differences between men and women with about 1.5 times higher concentrations in women (10). An explanation for these differences is lacking as plasma adiponectin is only moderately influenced by nutritional behavior, physical activity or other environmental components (5,8,11). However, there is clear evidence for a high heritability of about 50% (4,12-14) which one study even suggested to be sex-dependent (14). In line with lower plasma adiponectin in men, higher prevalences of type 2 diabetes and impaired fasting glucose were also reported in men (15).
Recent genome-wide association (GWA) scans have highlighted the potential of genetic factors with differential sex effects on concentrations of uric acid (16-18) and lipids (19), waist circumference (20) or schizophrenia (21). Many of these phenotypes show pronounced sex-specific differences in plasma concentrations or prevalence. A sex-differential SNP association with a quantitative phenotype can even mask a real association if data are analyzed without stratification. One example is a SNP near the LYPLAL1 gene which recently showed a strong association with waist-hip-ratio in women but not in men and would have been missed in the sex-combined analysis (20). To our knowledge, sex-specific differences for genetic effects on plasma adiponectin have not been investigated so far.
In the study at hand, we aimed to identify not only novel genes modulating plasma adiponectin but also whether genetic effects are differential between men and women. We combined this meta-analysis with a candidate gene approach considering all genes which have recently been associated with singular components of the metabolic syndrome in GWA studies.
Methods
Study cohorts and Genotyping
Our gene discovery included 4659 subjects (women=2562, men=2097) derived from three population-based studies, the Erasmus Rucphen Family Study (ERF, n=1820) (22), the follow-up of the third survey from the “Kooperative Gesundheitsforschung in der Region Augsburg” Study (KORA-F3, n=1644) (23), and the MICROS Study (n=1195) (24). The replication contained 13795 subjects (women=7673, men=6122 from the study cohorts CoLaus (n=5381), Framingham (n=2228), GEMS (n=1780), ALSPAC (n=1415), TWINS UK (n=1399), InChianti (n=1027) and BLSA (n=565).
All studies had genotypes available from genome-wide SNPs imputed based on the HapMap Ceu r22 reference sample after quality control. Measurement of adiponectin was made by ELISAs (from Mercodia, BioVendor and R&D Systems) or RIA (Linco). Details on study cohorts including the phenotyping for adiponectin measurements, genotyping methods, statistical analysis, and descriptive statistics are provided in the Supplementary Material and Supplementary Table S1.
Study design and statistical analysis
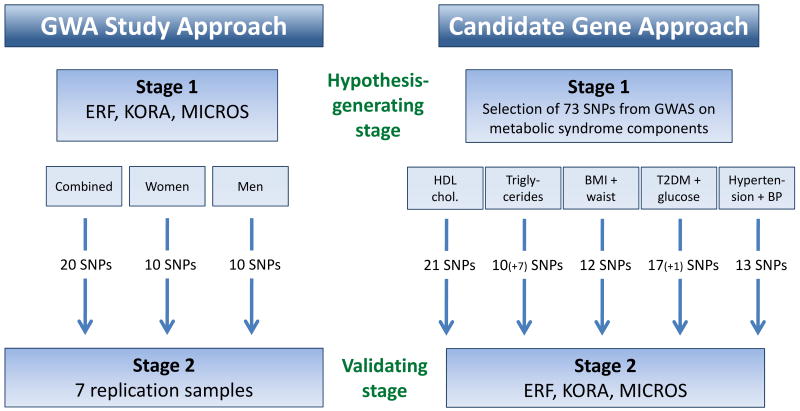
The study design is summarized in Figure 1.
Figure 1.
Study design illustrating the genome-wide association (GWA) study approach and the candidate gene approach.
GWA analyses (stage 1)
GWA analyses were conducted using a standardized protocol in each of the three stage 1 studies. For each of the 2,585,854 SNPs, linear regression using an additive genetic model was performed for log-transformed adiponectin values adjusting for age, sex, and BMI and accounting for the uncertainty in the inferred genotype from the imputation by utilizing the estimated genotype probabilities (implemented in MACH2QTL and GenABEL/ProABEL, respectively). All analyses were repeated for men and women separately. Relatedness between study participants was accounted for where appropriate (ERF, MICROS). Genomic control was applied when appropriate with study-specific lambda factors being 1.05, 1.05, and 0.99 for ERF, KORA, and MICROS, respectively. The beta-estimates of the three cohorts were combined using a fixed effect model. Also, a scaling-invariant p-value pooling meta-analysis using a weighted Z-score method was applied. For each SNP, we tested for significant differences between pooled men-specific beta-estimates across the three GWA studies as well as women-specific beta-estimates (see Supplementary Material for details).
GWA SNP selection
We selected three types of interesting regions to identify potentially novel signals for plasma adiponectin: (1) from the sex-combined sample (20 loci), (2) from the analysis in women (10 loci) and (3) in men (10 loci). Loci were considered as interesting and one SNP per locus was selected, if the combined p-values were less than 1*10-4 and if study-specific MAF was greater than 5% and imputation quality r2 greater than 0.2.
Replication analysis (stage 2)
For the selected 40 SNPs, we attempted replication based on 7 studies with the same study-specific SNP analysis as for stage 1 studies. A stage 2 only and a joint analysis of stage 1 and 2 (n=18454) was performed using the scaling-invariant weighted Z-score method.
Further statistical issues
In stage 1, we had 92% power to detect a variant that explains 1% of the variance of plasma adiponectin with genome-wide significance (alpha=5*10-8). In the stage 1 and stage 2 combined analysis, we had 99% power to yield genome-wide significant evidence for the 40 selected SNPs if they explained 1% of more of the variance in plasma adiponectin.
Candidate gene approach
From the literature, we identified loci associated with metabolic syndrome parameters in large GWA studies to obtain a list of candidate gene SNPs for adiponectin levels. We examined the association of these SNPs with plasma adiponectin from our stage 1 sex-combined and sex-stratified meta-analyses. For this candidate gene approach, we had 92% power to detect a SNP association that explains 0.5% of the variance accounting for the 73 SNPs tested (alpha=0.0007).
Percentage of variance explained
The general population design of KORA enabled computation of the proportion of the adiponectin variance explained by all analyzable ADIPOQ SNPs (i.e. SNPs available in all three GWA studies in the 50kb region of the ADIPOQ locus with MAF>5%), by an independent SNP set of these (i.e. selecting the SNP with the lowest p-value in the meta-analysis for each bin of SNPs with pairwise r2>0.2; r2 information was taken from HapMap), or by the top SNP alone. Computations were performed by linear regression on the standardized residuals (log of adiponectin concentrations adjusted for age, BMI and – if appropriate – for sex) and computing the r2 measure of the model adjusting for the SNP(s) using PROC REG by SAS.
Heritability
The family-based design of MICROS allowed us to compute heritability of plasma adiponectin using a polygenic model for standardized residuals of plasma adiponectin (adjusted for age and BMI - and sex if applicable). Heritability was also computed with additional adjustment of the top ADIPOQ SNP, with the independent SNP set as described above (see above). Computations were performed using the R library GenABEL(25).
Bioinformatic analysis
Bioinformatic analysis for potential functional SNPs was done in two stages, using bioinformatic tools outlined in the GenEpi Toolbox (26) (Supplementary Material).
Results
GWA analysis (stage 1)
Figure 2 shows the p-value, ADIPOQ-region and q-q-plots from the meta-analysis results of plasma adiponectin of the three GWA studies, ERF, KORA and MICROS cohorts. Results are presented for the sex-combined (n=4659) analysis as well as stratified for women (n=2562) and men (n=2097). The combined analysis yielded one genome-wide significant locus (Figure 2A), the ADIPOQ locus (p=4.3*10-24), which was consistent in women (p=8.7*10-17) and men (p=2.5*10-11) (Figure 2B). The q-q plot did not show evidence for bias due to population stratification in any of the analyses (Figure 2C). The top ADIPOQ SNP rs17366568 (Table 1) exhibited low imputation quality in ERF and MICROS that was genotyped using the Illumina platform in contrast to high imputation quality in KORA genotyped using the Affymetrix platform. However, other SNPs in this region such as rs3774261 reached genome-wide significance in the combined analysis (p=3.0*10-16) and had good imputation quality in all three stage 1 samples (0.82<r2<0.97).
Figure 2.

The analyses in panel A-C are provided for the combined sex analysis as well as the analysis stratified for women and men. A. Manhattan plots showing p-values of association of each SNPs in the meta-analysis with plasma adiponectin levels. SNPs are plotted on the X-axis to their position on each chromosome against association with plasma adiponectin on the Y-axis (shown as −log10 P-value). B. Regional Manhattan plots showing significance of association of all SNPs in the ADIPOQ region (3q27). SNPs are plotted on the X-axis to their position on chromosome 3 against association with plasma adiponectin on the Y-axis (shown as −log10 P-value). In each panel, the top-SNP rs17366568 is shown as red diamond. The SNPs surrounding this top-SNP are color-coded (see inset) to reflect their LD with the top-SNP using pair-wise r2 values from the KORA study. Estimated recombination rates from HapMap-CEU are plotted in blue to illustrate the local LD structure on a secondary Y-axis. Genes and their direction of transcription are provided below the plots using data from the UCSC genome browser. C. Quantile-quantile (QQ) plots of SNPs. Expected p-values are plotted on X-axis against the observed p-values plotted on the Y-axis.
Table 1.
Genome-wide significant association of the rs17366568 (G>A) SNP in the ADIPOQ locus
| Combined | Women | Men | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | EAF* | Rsqr† | n | Beta‡ | P | n | Beta‡ | P | n | Beta‡ | P | |||||||||||
| Stage 1 | ||||||||||||||||||||||
| ERF | 0.91 | 0.37 | 1817 | 0.103 | 2.7E-07 | 1052 | 0.115 | 1.0E-05 | 765 | 0.088 | 0.004 | |||||||||||
| KORA | 0.89 | 0.91 | 1643 | 0.173 | 1.7E-15 | 830 | 0.204 | 1.9E-11 | 813 | 0.142 | 5.8E-06 | |||||||||||
| MICROS | 0.90 | 0.27 | 1195 | 0.114 | 3.0E-06 | 678 | 0.102 | 4.1E-04 | 517 | 0.182 | 1.6E-05 | |||||||||||
| Combined** | 0.90 | - | 4655 | 4.3E-24 | 2560 | 8.7E-17 | 2095 | 2.5E-11 | ||||||||||||||
| Stage 2 | ||||||||||||||||||||||
| Colaus | 0.88 | 1.00 | 5261 | 0.132 | 3.0E-13 | 2759 | 0.119 | 1.1E-06 | 2502 | 0.146 | 5.1E-08 | |||||||||||
| Framingham | 0.88 | 1.00 | 2220 | 0.072 | 0.003 | 1213 | 0.050 | 0.108 | 1007 | 0.094 | 0.012 | |||||||||||
| GEMS | 0.87 | 1.00 | 1780 | 0.149 | 2.9E-06 | 732 | 0.084 | 0.095 | 1048 | 0.194 | 2.1E-06 | |||||||||||
| ALSPAC | 0.92 | 0.37 | 1415 | 0.395 | 2.9E-14 | 691 | 0.453 | 9.4E-09 | 724 | 0.351 | 3.5E-07 | |||||||||||
| TWINS UK | 0.998 | NA | 1399 | 0.154 | 0.078 | 1399 | 0.154 | 0.078 | - | - | - | |||||||||||
| InChianti | 0.94 | NA | 1027 | -0.056 | 0.481 | 562 | -0.130 | 0.268 | 465 | 0.007 | 0.95 | |||||||||||
| BLSA | 0.92 | 0.61 | 565 | 0.263 | 0.004 | 266 | -0.028 | 0.822 | 299 | 0.488 | 2.5E-04 | |||||||||||
| Combined** | 0.89 | - | 13667 | - | 5.2E-22 | 7622 | - | 2.7E-10 | 6045 | - | 8.1E-14 | |||||||||||
| Stage 1 + 2 | ||||||||||||||||||||||
| Combined** | 0.89 | - | 18322 | 1.1E-41 | 10182 | 2.8E-22 | 8140 | 7.8E-23 | ||||||||||||||
EAF = effect allele frequency (i.e. frequency of G) for sex-combined analysis
Rsqr = imputation certainty
Beta estimate from linear regression adjusted for age, BMI, and (if appropriate) for sex per unit change [log(μg/mL)] for the risk allele G
Results are provided for a beta-pooling meta-analysis using the fixed effect model weighting for the inverse variance. When a scaling-invariant p-value pooling meta-analysis using the sample size weighted z-score method was applied for sensitivity analysis, we found no major differences between both methods.
Replication analysis (stage 2)
Characteristics of the 40 SNPs taken forward for replication are provided in Supplementary Table S2. From the combined, women-, and men-specific GWA-analyses (n=13795, 7673, and 6122, respectively), only the ADIPOQ SNP remained significant in the combined analyses (Supplementary Table S3). P-values for rs17366568 were 1.09*10-41, 2.8*10-22 and 7.8*10-23 for the combined and the analysis stratified for women and men, respectively (Table 1).
Sex-specific analyses
In line with previous reports, plasma adiponectin in women was approximately 1.5 times higher than in men in each of the three stage 1 studies (Supplementary Table S1). Heritability computations in the family-based MICROS study showed slightly higher estimates of 65.1% for women and 54.0% for men (Table 2).
Table 2. Heritability and percentage of variance explained by the ADIPOQ locus SNPs.
Heritability of plasma adiponectin in the family-based study MICROS and percentage of plasma adiponectin variance (KORA) explained by the ADIPOQ locus SNPs in KORA (region on chr 3, position 188.030 – 188.080kb).
| Combined | Women | Men | |
|---|---|---|---|
| Heritability (%) in MICROS | |||
| no SNP adjustment | 59.6 | 65.1 | 54.0 |
| adjusted for top hit rs17366568 | 58.4 | 64.6 | 51.5 |
| adjusted for “independent ”SNPs (n=9) a | 52.9 | 55.1 | 48.1 |
| % of variance of plasma adiponectin in KORA explained by | |||
| top hit rs17366568 | 3.8 | 5.3 | 2.4 |
| for “independent” SNPs (n=9) a | 5.9 | 6.3 | 5.1 |
| all SNPs with MAF >5% (n=33) b | 6.7 | 6.4 | 5.5 |
Computations were based on standardized sex-combined or sex-specific residuals of plasma adiponectin adjusted for age (and sex if applicable) and BMI without and with additional SNP adjustment; includes only SNPs with MAF>5% available in all three studies.
Among the SNPS of the ADIPOQ region with MAF > 5% and available in all three GWA studies: selecting the SNP with the smallest p-value from each bin of SNPs with pairwise r2>0.2: rs1063539, rs16861194, rs17300539, rs17366568, rs17366743, rs3774261, rs6810075, rs7615090, rs822394
All SNPS of the ADIPOQ region with MAF > 5% and available in all three GWA studies: rs6810075, rs10937273, rs12637534, rs1648707, rs864265, rs822387, rs16861194, rs17300539, rs266729, rs182052, rs16861205, rs16861209, rs822391, rs16861210, rs822394, rs822396, rs12495941, rs7649121, rs17366568, rs2241767, rs3821799, rs3774261, rs3774262, rs17366743, rs6773957, rs1063537, rs2082940, rs1063539, rs7639352, rs6444175, rs7628649, rs17373414, rs9860747, rs1501296, rs7615090
For each SNP, we evaluated whether the sex-specific beta-estimates combined across the three stage 1 studies were significantly different between men and women pointing towards a gender-SNP interaction. The q-q plot for the p-values of sex differences indicated some observed sex difference of genetic effects beyond that expected by chance (Supplementary Figure S1A), but not due to differences in the ADIPOQ region. For none of the SNPs in the GWA studies, the sex-specific beta-estimates were significantly different between men and women on a genome-wide level (Supplementary Figure S1B). For the ADIPOQ top SNP rs17366568 the p-value for sex difference was 0.62.
Association of metabolic syndrome candidate gene SNPs with adiponectin
From the literature, we identified loci associated with metabolic syndrome parameters in large GWA studies to obtain a list of candidate gene SNPs for adiponectin levels (Figure 1). These were partially overlapping for the various metabolic syndrome components and included 21 SNPs for HDL cholesterol, 17 for triglycerides (7 of them were also found for HDL cholesterol and were therefore only counted once), 12 for BMI and/or waist circumference, 18 for type 2 diabetes and/or glucose levels (one of them was already mentioned for BMI and is therefore only counted once), and 13 for hypertension and blood pressure. Details on these SNPs are given in Supplementary Table S4.
Only 3 out of the 73 SNPs showed p-values between 0.01 and 0.05 for example for the gender-combined analysis (with 3.65 expected under the assumption of no association). No p-value was below the Bonferroni-adjusted significance level of 0.007. Thus, our data indicated no association of these metabolic syndrome parameter SNPs with plasma adiponectin.
Sensitivity analyses
Sensitivity analyses repeating all analyses without the adjustment for BMI showed the same results regarding the ADIPOQ genome-wide significant results, the lack of sex difference, the lack of other SNPs in the replication stage to show replication, and the lack of metabolic syndrome SNPs to show association with plasma adiponectin.
ADIPOQ region
A closer look at the ADIPOQ region revealed that the top SNP rs17366568 was completely independent of all other SNPs in that region. A linkage disequilibrium (LD) plot depicting D′ and r2 measures (Figure 3) revealed that for many SNPs in the ADIPOQ region the r2 was weak even if they were located in the same LD block (as defined by D′). At least nine SNP groups were significantly and independently associated with plasma adiponectin. The percentage of plasma adiponectin variance explained by the top hit was 3.8% and increased to 5.9% when including an independent SNP set (selecting the SNP with the smallest p-value in each bin of pairwise r2<0.2) and peaked at 6.7% when including all SNPs with MAF>5% in the 50 kb region covering the three LD blocks (Table 2).
Figure 3.

Linkage disequilibrium (LD) plot of SNPs in the ADIPOQ region spanning 50kb (positions 188030-188080kb). The grey shading of the diamonds represent the pair-wise D′ and the numbers in the diamonds represent the pair-wise r2 between the two SNPs defined by the top left and the top right sides of the diamond. The figure clearly shows that the top-hit rs17366568 is located within an own LD block and shows virtually no correlations with any other SNP in the entire 50kb region. The columns on the right side of the Figure show i) whether a particular SNP is genotyped by the Affymetrix 500K chip (A) or the Illumina HumanHap300 chip (I); all other SNPs are imputed; ii) the z-scores and iii) the p-values for each SNP-adiponection association for the combined analysis of the cohorts ERF, KORA and MICROS; iv) SNPs that are correlated with an r2>0.60 are grouped in groups 1-9.
Bioinformatic analysis revealed two main putative functional elements located in the second and the third LD block as depicted in Figure 3. Three SNPs located immediately up-and downstream of rs17366568 (for details see Supplementary Table S5) are predicted to affect 10, 6 or 4, respectively, transcription factor binding sites (using adipose tissue-specific analysis). No transcription factor binding sites or splicing regulation elements were detected for rs17366568 itself. Therefore, it is likely that rs17366568 is not the functional variant, but relates to a functional element located in the immediate vicinity (although regulatory potential was very low throughout the region).
Analysis of LD block 3 (encompassing exon 3 and a large intergenic region downstream of the ADIPOQ locus) revealed three putative regulatory promoter regions located approximately 5.1 kb, 6.3 kb and 15.8 kb downstream of the ADIPOQ locus. Interestingly, especially the proximal two regulatory regions are known to be affected by several copy number regions (see Figure 4 and Supplementary Table S6). However, no SNP in our data set was located directly in these CNVs, whose functional relevance may therefore require further investigation. More generally, the whole genomic region of ADIPOQ seems to be highly affected by copy number variations (Figure 4).
Figure 4.

Predicted regulatory regions in the ADIPOQ downstream region and their affection by copy number variations. Panel A: Genomatix Software Suite: Position of the regulatory promoter regions predicted by PromoterInspector (red boxes) and their position relative the ADIPOQ gene region (green box). Panel B: UCSC Browser: Position of the predicted regulatory regions (red boxes) relative to known copy number variations in the ADIPOQ gene region (represented by bold blue lines). The numbers on the left side correspond to the accession number of the respective copy number variation in the Database of Genomic Variants.
Discussion
In the meta-analysis of genome-wide SNP association with plasma adiponectin in three population-based studies including a total of 4655 subjects, we found genome-wide significant evidence for the association with the ADIPOQ locus, which is a known locus for plasma adiponectin (10,27). Furthermore, we did not identify any genome-wide significant evidence for association in any other locus when replicating the other 39 most strongly associated loci in 13795 independent samples. Despite the clear sex difference in plasma adiponectin, there was no sex difference observed for the ADIPOQ SNP associations. Finally, we found, despite the strong association between plasma adiponectin and the metabolic syndrome, no significant association with adiponectin for any of the chosen variants within reported loci for metabolic syndrome parameters.
Our GWA study identified only one major locus for plasma adiponectin, the ADIPOQ gene region. The only other GWA study on adiponectin was performed in 1845 individuals of the GEMS Study and identified also only the ADIPOQ locus with genome-wide significance (28). The other top seven hits from that study could not be replicated in our GWA study, neither in the combined (all p-values >0.28) nor in the sex-specific analysis (p>0.16). In our GWA discovery stage, the power was more than 90% to detect novel loci which explain 1% of the adiponectin variance, and, including the replication stage, over 99% to show genome-wide significant evidence of the 40 SNPs in the 18454 subjects. Therefore, our data suggests a lack of a major gene locus other than ADIPOQ.
ADIPOQ was studied earlier as a candidate gene and the relationship to plasma levels has long been recognized. The SNP rs17366568 showing the strongest association in our GWA study explained 3.8% of the variance and this number increased to 6.7% if all analyzable SNPs in the ADIPOQ region were included into the model. This pronounced difference of the explained variance between the two models can be explained by a large number of SNPs independently contributing to adiponectin levels. The SNPs contributing most to the explained variance are not only located in the three different LD blocks but also several genetic variants within each of at least two of the three blocks contribute to the explained variance. In total, the explained variance was very similar to the 8% reported earlier (10). Functional studies within the promoter of the ADIPOQ gene revealed a pronounced influence of three SNPs also investigated in our study and the corresponding haplotypes on the promoter activity which was accompanied by changes in the DNA binding activity interfering with transcription factor bindings sites (29). Other studies showed that histone acetylation might influence the transcriptional regulation of the ADIPOQ gene (30) and that pioglitazone increases plasma adiponectin by posttranscriptional regulation (31). Finally, an extensive bioinformatic analysis revealed that the ADIPOQ region might be a highly copy number variable region. It remains to be determined how strong the effect of these CNVs on plasma adiponectin is.
Since adiponectin has been viewed as a marker for the metabolic syndrome, we have also studied 73 SNPs that have been associated with any of the major determinants of metabolic syndrome in previous GWA studies. This candidate gene-based analysis did not yield any convincing associations with plasma adiponectin. This was surprising due to the strong link between plasma adiponectin and the metabolic syndrome or any of its components (3-5), but in-line with previous reports on a lack of association of the ADIPOQ SNPs with metabolic syndrome parameters (10). Whether plasma adiponectin affects metabolic syndrome parameters or metabolic syndrome parameters modulate adiponectin is highly debated as illustrated in Supplementary Figure S2. If the association of any of these 73 SNPs had been very strong with adiponectin - stronger than with the metabolic syndrome parameters - this would have pointed towards a gene locus primarily affecting plasma adiponectin and consecutively modulating the metabolic syndrome parameters. This is not suggested by our data (panel A of Supplementary Figure S2). Our data on these 73 metabolic syndrome SNPs lacks association with adiponectin beyond that expected by chance. This would rather support the idea that genetic pathways for plasma adiponectin are different from the pathways depicted by these 73 loci (panel B), or, alternatively, that pathways depicted by these 73 loci affect plasma adiponectin via the metabolic syndrome parameter and the lack of association was due to loss of power for a parameter further down the road (panel C). Both ideas (panel B and C) would point towards the hypothesis that genetically determined adiponectin does not modulate metabolic syndrome parameters directly.
The present data suggests that the sex differences in plasma adiponectin can not be explained by any major gene. The GWA approach yielded no genome-wide significant difference between men and women for any SNP, not even the ADIPOQ locus. In fact, none of the variants studied in the replication or in our candidate gene approach based on metabolic syndrome loci showed a significant sex difference. Therefore, the sex-difference in plasma adiponectin might rather be explained by sex hormones (5) or sex-specific epigenetic programming that could be transmitted to subsequent generations in a sex-specific manner leading to transgenerational effects as recently suggested (32).
The heritability estimates of plasma adiponectin are high with roughly 50-60% (4,12-14). The ADIPOQ locus accounts for 6.7% of the variance in our populations-based KORA Study, which is in-line with previous reports (10). This is also in-line with 6.6% of the heritability accounted for by this locus in our family-based MICROS Study. While the ADIPOQ locus association with plasma adiponectin is thus among the strongest associations for quantitative phenotypes in genetic epidemiology, it explains only a small proportion of the overall heritability, a puzzle observed for many other phenotypes (e.g. lipids or obesity measures) (19,20,33). Potential explanations of this gap between explained and estimated heritability are unknown rare variants with strong effects on adiponectin (34), unknown common loci influencing adiponectin with small effects, or deflation of association estimates due to heterogeneity between studies, uncertainties in the genotypes from imputation or uncertainties in the phenotype assessment. Our study suggests that these other genetic variants influencing plasma adiponectin are variants that explain less than 1% of the phenotypic variance. To localize these loci and to build up gene networks which identify even trans-acting quantitative trait loci, will require substantially larger data sets in combination with gene expression analysis.
Strengths and Limitations of the Study
A limitation of our study is the limited sample size for gene discovery for small genetic effects, in particular when conducting stratified analyses. Furthermore, our top hit in the ADIPOQ locus had limited imputation quality in two of the included GWA studies, which can be explained by the fact that KORA used a different SNP-panel (Affymetrix 500K chip) for GWAs genotyping than ERF and MICROS (Illumina HumanHap300). For most of the other SNPs followed in replication samples, the imputation quality was quite high. The relatively low imputation quality of our top-hit in two of the studies explains the lower (but still genome-wide significant) p-values in these two studies compared to KORA. This is entirely in-line with measurement error theory: a “measurement error” (like the uncertainty induced by the imputation) that does not depend on the phenotype (as the case here assuming that genotyping does not depend on adiponectin in the plasma) is expected to attenuate the precision of an underlying association yielding larger p-values. Therefore, the association in ERF and MICROS was rather underestimated than false positive. Finally, it can be considered a limitation of most GWAS studies that gonsomes are not analyzed due to technical issues not yet solved concerning the imputation of SNPs which, however, is a prerequisite to allow meta-analysis of data over various genotyping platforms used.
The strong point of our study is the population-based design, in which the participants have not been ascertained based on the presence of pathology. Hypothesizing a genetic basis of sex differences in plasma adiponectin, a further advantage is the sex-stratified analysis since a sex-combined analysis would otherwise mask an association. Further, the family-based MICROS study enables us to estimate heritability.
Conclusions
We present a genome-wide association study on adiponectin which the first time attempts to explain adiponectin sex difference by the underlying genetics. We conclude that there is no major gene involved in modulating plasma adiponectin other than the known ADIPOQ locus and that there is no major gene explaining the differences of plasma adiponectin between men and women.
Supplementary Material
Acknowledgments
We thank all staff members involved in the MONICA/KORA Augsburg Studies as well as the general practitioner and other clinicians for compiling the Genetic Research in Isolated Populations, Erasmus Rucphen Family (ERF) study. The technical assistance of Barbara Luhan for measurement of adiponectin in the KORA Study is highly appreciated. We also thank Julia Müller for help in table management. For the MICROS study, we thank the primary care practitioners Raffaela Stocker, Stefan Waldner, Toni Pizzecco, Josef Plangger, Ugo Marcadent and the personnel of the Hospital of Silandro (Department of Laboratory Medicine) for their participation and collaboration in the research project. Finally, we express our appreciation to all study participants.
The acknowledgements of financial and other support for each study is provided in the Supplementary Material.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li S, Shin HJ, Ding EL, van Dam RM. Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2009;302:179–188. doi: 10.1001/jama.2009.976. [DOI] [PubMed] [Google Scholar]
- 2.Tilg H, Moschen AR. Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol. 2006;6:772–783. doi: 10.1038/nri1937. [DOI] [PubMed] [Google Scholar]
- 3.Kadowaki T, Yamauchi T, Kubota N, et al. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–1792. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hivert MF, Manning AK, McAteer JB, et al. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes. 2008;57:3353–3359. doi: 10.2337/db08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henneman P, Janssens ACJW, Zillikens MC, et al. Menopause impacts the relation of plasma adiponectin levels with the metabolic syndrome. J Intern Med. 2009 doi: 10.1111/j.1365-2796.2009.02162.x. [DOI] [PubMed] [Google Scholar]
- 6.Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi T, Kamon J, Waki H, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 8.Antoniades C, Antonopoulos AS, Tousoulis D, Stefanadis C. Adiponectin: from obesity to cardiovascular disease. Obes Rev. 2009;10:269–279. doi: 10.1111/j.1467-789X.2009.00571.x. [DOI] [PubMed] [Google Scholar]
- 9.Davey SG, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 10.Heid IM, Wagner SA, Gohlke H, et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006;55:375–384. doi: 10.2337/diabetes.55.02.06.db05-0747. [DOI] [PubMed] [Google Scholar]
- 11.Mantzoros CS, Williams CJ, Manson JE, Meigs JB, Hu FB. Adherence to the Mediterranean dietary pattern is positively associated with plasma adiponectin concentrations in diabetic women. Am J Clin Nutr. 2006;84:328–335. doi: 10.1093/ajcn/84.1.328. [DOI] [PubMed] [Google Scholar]
- 12.Comuzzie AG, Funahashi T, Sonnenberg G, et al. The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metab. 2001;86:4321–4325. doi: 10.1210/jcem.86.9.7878. [DOI] [PubMed] [Google Scholar]
- 13.Patel SR, Larkin EK, Redline S. Shared genetic basis for obstructive sleep apnea and adiposity measures. Int J Obes (Lond) 2008;32:795–800. doi: 10.1038/sj.ijo.0803803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miljkovic-Gacic I, Wang X, Kammerer CM, et al. Genetic determination of adiponectin and its relationship with body fat topography in multigenerational families of African heritage. Metabolism. 2007;56:234–238. doi: 10.1016/j.metabol.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathmann W, Haastert B, Icks A, et al. High prevalence of undiagnosed diabetes mellitus in Southern Germany: target populations for efficient screening. The KORA survey 2000 Diabetologia. 2003;46:182–189. doi: 10.1007/s00125-002-1025-0. [DOI] [PubMed] [Google Scholar]
- 16.Döring A, Gieger C, Mehta D, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 17.Vitart V, Rudan I, Hayward C, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 18.Dehghan A, Kottgen A, Yang Q, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372(9654):1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aulchenko YS, Ripatti S, Lindquist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindgren CM, Heid IM, Randall JC, et al. Genome-Wide Association Scan Meta-Analysis Identifies Three Loci Influencing Adiposity and Fat Distribution. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000508. e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shifman S, Johannesson M, Bronstein M, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henneman P, Aulchenko YS, Frants RR, et al. Prevalence and heritability of the metabolic syndrome and its individual components in a Dutch isolate: the Erasmus Rucphen Family study. J Med Genet. 2008;45:572–577. doi: 10.1136/jmg.2008.058388. [DOI] [PubMed] [Google Scholar]
- 23.Wichmann HE, Gieger C, Illig T, MONIKA/KORA Study Group KORA-gen: Resource for Population Genetics, Controls and a Broad Spectrum of Disease Phenotypes. Gesundheitswesen. 2005;67:S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 24.Pattaro C, Aulchenko YS, Isaacs A, et al. Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int. 2009 doi: 10.1038/ki.2009.135. [DOI] [PubMed] [Google Scholar]
- 25.Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 26.Coassin S, Brandstätter A, Kronenberg F. Lost in the space of bioinformatic tools: a constantly updated survival guide for genetic epidemiology The GenEpi Toolbox. Atherosclerosis. doi: 10.1016/j.atherosclerosis.2009.10.026. in press. [DOI] [PubMed] [Google Scholar]
- 27.Vasseur F, Helbecque N, Dina C, et al. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11:2607–2614. doi: 10.1093/hmg/11.21.2607. [DOI] [PubMed] [Google Scholar]
- 28.Ling H, Waterworth DM, Stirnadel HA, et al. Genome-wide Linkage and Association Analyses to Identify Genes Influencing Adiponectin Levels: The GEMS Study. Obesity (Silver Spring) 2009;17(4):737–744. doi: 10.1038/oby.2008.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laumen H, Saningong AD, Heid IM, et al. Functional characterization of promoter variants of the adiponectin gene complemented by epidemiological data. Diabetes. 2009;58:984–991. doi: 10.2337/db07-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musri MM, Corominola H, Casamitjana R, Gomis R, Parrizas M. Histone H3 lysine 4 dimethylation signals the transcriptional competence of the adiponectin promoter in preadipocytes. J Biol Chem. 2006;281:17180–17188. doi: 10.1074/jbc.M601295200. [DOI] [PubMed] [Google Scholar]
- 31.Rasouli N, Yao-Borengasser A, Miles LM, Elbein SC, Kern PA. Increased plasma adiponectin in response to pioglitazone does not result from increased gene expression. Am J Physiol Endocrinol Metab. 2006;290:E42–E46. doi: 10.1152/ajpendo.00240.2005. [DOI] [PubMed] [Google Scholar]
- 32.Gabory A, Attig L, Junien C. Sexual dimorphism in environmental epigenetic programming. Mol Cell Endocrinol. 2009;304:8–18. doi: 10.1016/j.mce.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen JC, Kiss RS, Pertsemlidis A, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.