Abstract
The angiotensin-vasopressin receptor (AVR) responds with equivalent affinities to angiotensin II (ANG II) and vasopressin and is coupled to adenylate cyclase and hence a V2-type vasopressin receptor. AVR maps to the Nalp6 locus and overlaps with the larger Nalp6/PYPAF5 reported to be a T cell/granulocyte-specific, cytoplasmic-specific proapoptotic protein, thus questioning the existence of AVR. Here we confirm, through different experimental modalities, that AVR is distinct from Nalp6/PYPAF5 based on different mRNA and protein sizes, subcellular localization, and tissue-specific expression patterns. Binding studies of PYPAF5-specific Cos1 transfectants detect high-affinity binding to vasopressin but not ANG II, thus assigning PYPAF5 as a non-AVR (NAVR). Signaling array analysis reveals that AVP stimulation of AVR- and NAVR-specific Cos1 transfectants results in diametrical activation as well as coactivation of signaling pathways known to mediate renal sodium and water balance. Likewise, ANG II stimulation of Cos1-AVR transfectants reveals a signaling profile distinct from that of AVP-stimulated Cos1-AVR transfectants. Analysis of genomic organization of the AVR/NAVR locus shows an overlapping gene arrangement with alternative promoter usage resulting in different NH2 termini for NAVR and AVR. In addition to core promoter elements, androgen and estrogen response elements are detected. Promoter analysis of NAVR/AVR 5′-regulatory region detects transcriptional upregulation by testosterone and synergistic upregulation by testosterone and estrogen, thus suggesting that AVR and/or NAVR contribute to sex-specific V2-type vasopressin-mediated effects. Altogether, confirmation of AVR and identification of NAVR as vasopressin receptors are concordant with emerging vasopressin functions not attributable to V1a, V1b, or V2 receptors and add molecular bases for the multifunctional complexity of vasopressin-mediated functions and regulation.
Keywords: vasopressin sexual diergism, testosterone, estrogen, signaling pathway
The angiotensin II-vasopressin receptor (AVR) was originally isolated from a rat kidney cDNA library and shown through functional studies to respond with equivalent affinities to angiotensin II (ANG II) and vasopressin and coupled to adenylate cyclase (47). AVR has been implicated in salt-sensitive hypertension susceptibility through linkage analysis of an F2 rat intercross study and subsequent elucidation of an N119S/C163R variant in Dahl salt-sensitive rats that exhibits sodium-induced dysfunction (48). RNA blot analysis with the AVR probe, now known to be common to AVR and Nalp6 (NACHT, leucine-rich repeat and PYD-containing 6)/PYPAF5 (PYRIN-containing apoptotic protease-activating factor-5-like protein), detects different-sized transcripts, a 2.5-kb mRNA detected abundantly in kidney and larger transcripts detected in several tissues including liver, lung, adrenal gland, aorta, heart, brain, and kidney (47). Oligonucleotide in situ hybridization detects AVR transcripts in supraoptic neurons (25), suggesting a role in modulation of vasopressin-mediated neuronal functions. As a dual ANG II/AVP receptor, AVR provides a mechanism for cross talk between the ANG II and vasopressin receptor systems.
Recently Albrecht et al. (4) reported that AVR is contained in a larger protein, PYPAF5, which is identical to Nalp6 and belongs to a family of proteins comprised of PYRIN and caspase activating recruitment (CARD), nucleotide-binding oligomerization, and leucine-rich repeat domains. Human PYPAF5 has been reported to be recruited by PYRIN-CARD protein ASC (apoptosis-associated speck-like protein containing a CARD) to distinct cytoplasmic loci and to synergistically activate NF-κB and procaspase-1 when coexpressed with ASC (17). On the basis of computational analysis and the fact that PYPAF5 and other members of this family of proteins are described only as cytosolic proteins (17), the existence of AVR as a transmembrane receptor was questioned (4), although analysis of membrane protein isolates was not done.
However, in the context of the emerging complex paradigm of overlapping genes with different genomic organizations such as sense/antisense overlap or unistrand overlap, involving both exons and regulatory regions and involving not just two genes but three genes (53), the deduction by Albrecht et al. (4) that the larger PYPAF5 spanning AVR negates the existence of AVR, the shorter protein, is questioned. The concept of overlapping genes is further supported by the mechanism of alternative promoter usage in mammalian genes in which different transcripts and hence proteins are produced from the same genomic region sharing exons but not all, resulting in proteins with different NH2 termini as seen in the case of peroxisome proliferator-activated receptor (PPAR)-γ and p73 genes (35), and which would resemble the case for AVR and PYPAF5/Nalp6. The mechanism of alternative promoters also results in different proteins through different exon usage and translation frameshifts as seen in the case for INK4a/ARF and p21/p21B (35), thus supporting the argument that detection of overlapping transcripts/proteins is not valid grounds to negate the existence of the smaller transcript/protein.
Moreover, from a functional perspective, elucidation of vasopressin receptor functions that are not attributed to the currently identified vasopressin receptors V1a, V1b, or V2 provides physiological corroboration that indeed there must be other vasopressin receptors besides the prototype V1a, V1b, and V2 receptors (49).
We therefore investigated the hypothesis that AVR and PYPAF5 are overlapping genes in the Nalp6 locus with distinct physiological functions and that AVR is a membrane-bound vasopressin receptor. Stepwise analysis of DNA, RNA, protein, and cell-specific localization confirms AVR as a smaller AVP receptor and confirms that PYPAF5 is also membrane bound and binds vasopressin with high affinity but not ANG II, and hence is a non-AVR (NAVR). Moreover, signal transduction pathway analysis demonstrates differential regulation of signaling proteins by AVR and NAVR consistent with unique roles in modulating renal salt-water balance and other physiological functions. Finally, promoter function reporter analysis detects testosterone-mediated induction, as well as synergistic testosterone-estrogen induction, concordant with known vasopressin sexual diergic effects. Altogether, data confirm AVR and identify NAVR as V2-type vasopressin receptors—thus expanding the molecular basis of increasingly complex vasopressin-mediated functions not attributable to prototype V1a, V1b, or V2 receptors and mandating further investigation as receptors.
MATERIALS AND METHODS
Characterization of rat NAVR cDNA
NAVR cDNA was amplified by RT-PCR from Dahl R/JRHsd rat kidney poly(A)+ RNA (forward primer: 5′-TGC-ATC-TGA-GAG-CTA-GAT-CC-3′; reverse primer: 5′-AGA-CTA-ACC-TCC-GAG-TGT-CC-3′) and subsequently subcloned into the PT-vector system (Clontech, Palo Alto, CA). Primer sequences were obtained from the rat genomic sequence spanning NAVR and AVR transcription units (NCBI ID NC_005100.2). The 2,906-bp-long cDNA encompassing the entire NAVR amino acid coding region was then sequenced on both strands.
NAVR expression and membrane preparation
The NAVR cDNA was subcloned directionally (5′ to 3′) into the pcDNA(+) expression vector (Invitrogen, Carlsbad, CA) and transiently expressed in Cos1 cells (ATCC). Cos1 cells were transfected with the expression vector via lipofectin-mediated gene transfer, and cell membranes were isolated 72 h after transfection for hormone binding.
Rat kidney membranes were prepared essentially as described previously (26). Cos1 cell membranes were isolated as described previously (18). Briefly, cells were washed twice in phosphate-buffered saline (PBS) and homogenized in 10-fold ice-cold buffer (0.25 M sucrose, 1 mM EDTA, 50 µg/ml aprotinin, 10 µg/ml leupeptin, 100 µM phenylmethylsulfonyl fluoride, 25 mM imidazole HCl, pH 7.4). The homogenate was centrifuged at 5,000 g for 15 min, and the pellet was discarded. The supernatant was then centrifuged at 27,500 g for 30 min, and the resulting pellet was washed twice in ice-cold suspension buffer (mM: 5 MgCl2, 0.2 EDTA, 50 HEPES, pH 7.4). The final pellet was resuspended in the appropriate assay buffer and quickly frozen in liquid nitrogen. The membrane preparations were stored at −80°C until use. Protein concentrations of the membranes were determined by bicinchoninic acid (BCA) protein assay kit (Pierce).
Radioligand binding assays
Binding of 125I-Tyr4-ANG II to NAVR-expressing Cos1 membranes was performed by a rapid filtration method as described previously (28). Briefly, 125I-Tyr4-ANG II (1–10 nM) was incubated with membranes (100 µg) for 20 min at 37°C in 100 µl of buffer A (5 mM MgCl2, 0.2 mM EDTA, 10 mg/ml BSA, 10 mM HEPES, pH 7.4). Binding reactions were terminated by the addition of 1 ml of ice-cold buffer A, immediately filtered through a Whatman GF/C filter (presoaked overnight at 4°C in 10 mg/ml BSA), and subsequently washed with 15 ml ice-cold buffer A. Specific binding was determined as the difference between the total radioactivity bound to membranes and the radioactivity bound to blanks containing 10 µM ANG II. Binding of [3H]AVP to NAVR-expressing Cos1 membranes was performed by a rapid filtration method. Briefly, [3H]AVP (1–10 nM) was incubated with membranes (100 µg) for 60 min at 24°C in 100 µl of buffer B (1 mM MgCl2, 1 mg/ml BSA, 10 mM Tris · HCl, pH 7.5). Binding reactions were terminated by the addition of 1 ml of ice-cold buffer B, immediately filtered through a Whatman GF/C filter (presoaked overnight at 4°C in binding buffer B), and subsequently washed with 15 ml of ice-cold buffer B. Specific binding was determined as the difference between the total radioactivity bound to membranes and the radioactivity bound to blanks containing 10 µM AVP. The dissociation constant (Kd) and maximum ligand-binding site density (Bmax) were determined by Hill plot analysis (46). Hill coefficient (nH) values were calculated from the relationship ln[B/(Bmax − B)] = nHln[free radioligand] − lnKd. An F-test (P < 0.05) was used to determine whether the saturation binding curves best fitted one or two independent binding sites. The data were best fit by one affinity state determined by Scatchard plot analysis. Results are expressed as the means ± SE from five independent experiments.
Western blot analysis and immunohistochemistry
A polyclonal rabbit anti-peptide antibody raised against the AVR synthetic peptide K592DELKDEE599 (NAVR amino acid numbering; see Fig. 3) was utilized for Western blot analysis (47). Kidney membranes (40 µg protein/lane) were subjected to 10% SDS-PAGE, and the separated proteins were electrotransferred onto polyvinylidene difluoride (PVDF) membranes, which were incubated with blocking buffer (0.3% Tween 20, 5% nonfat milk, 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, and 1.5 mM KH2PO4, pH 7.4) for 2 h at room temperature and then incubated with primary antibody (1:200) for 16 h at 4°C. The PVDF membranes were then sequentially incubated with biotinylated goat anti-rabbit IgG, followed by immunostaining with horseradish peroxidase-linked streptavidin.
Fig. 3.
Deduced amino acid sequence of NAVR and AVR. NAVR spans 880 aa; AVR spans 457 aa. NAVR domains are depicted: NACHT domain, leucine-rich regions (LRR1-4). Predicted functional domains for AVR: AVP, vasopressin binding domain; ANG II, angiotensin binding domain; Gs, G protein interacting site by consensus sequence; ab, peptide sequence identified to be most antigenic and used for generating the antibody. Initiating methionines are marked (1NAVR and 1AVR). The 5′-RACE-determined transcription start site for AVR is noted (→+1AVR).
Immunohistochemistry was done as described previously (20) with the following specifics: anti-NAVR/AVR antibody was used at 1:20 dilution, primary antibody was incubated overnight at 4°C, diaminobenzidine (DAB) chromogen was used to detect immunostaining, Mayer’s hematoxylin was used as counterstain, preimmune serum (1:20) and antigenic peptide (K592DELKDEE599) at 100× molar excess concentration were used as negative controls, and rat-specific tissue array was used with duplicate tissue-specific cores. The exposure of multiple tissue cores in duplicates validates comparative analysis.
Northern blot analysis
Northern blot analysis was performed essentially as described previously (47). Poly(A)+ RNA was prepared from different tissues from Sprague-Dawley rats. Poly(A)+ RNA (4 µg/lane, except 1 µg for kidney and lung samples in blot hybridized with the common cDNA probe) was size separated on a 1% agarose formaldehyde gel, transferred to nitrocellulose, and hybridized to single-stranded 32P-labeled probe made by PCR in the presence of [α-32P]dCTP. For the common cDNA probe (nucleotides 1-1798 of the NAVR sequence; GenBank accession no. DQ631800) we used a reverse primer corresponding to nucleotides 5′-TCTGCTCCTACTGTCGCCAC-3′ (1798-1779 of the NAVR sequence; GenBank accession no. DQ631800), and for the NAVR-specific cDNA probe (nucleotides 1-316 of the NAVR sequence; GenBank accession no. DQ631800) we used the reverse primer 5′-TCCTTGAGTCGCAACGACAC-3′ (316-297 of the NAVR sequence; GenBank accession No. DQ631800). Blots were hybridized for 36 h and exposed for 2 and 7 days at −80°C with intensifying screens for the common and NAVR-specific blots, respectively.
Establishment of Cos1-AVR and Cos1-NAVR stable transfectants
For AVR and NAVR expression, the full-length rat AVR and rat NAVR cDNAs were subcloned directionally (5′ to 3′) into the pcDNA(+) expression vector with neomycin resistance as the basis for transfection-positive selection (Invitrogen). Cos1 cells were transfected with the expression vectors via lipofectin-mediated gene transfer and isolation of neomycin-resistant stable transfectants for AVR and Cos1-AVR, and for NAVR Cos1-NAVR was done in the presence of Geneticin (400 µg/ml, selection; 200 µg/ml, maintenance). Since Cos1 cells do not have endogenous AVR or NAVR, stable transfectants are specific for the receptor expressed. This allows the dissection of AVR-specific and NAVR-specific subcellular localization and ligand-activated signaling pathways in Cos1-AVR and Cos1-NAVR cell transfectants, respectively.
Confocal microscopy analysis
Stable Cos1-NAVR and Cos1-AVR transfectants were grown onto separate cell culture coverslips until the cultures reached 60% confluence. Cells were subsequently fixed with 4% paraformaldehyde and then briefly permeabilized for 10 min with 0.1% Triton X-100. Cells were then blocked with 3% normal goat serum (Invitrogen) for 30 min at room temperature, followed by incubation with polyclonal anti-NAVR/AVR antibody raised against the NAVR/AVR synthetic peptide K592DELKDEE599 (NAVR amino acid numbering; see Fig. 3) (47) at a concentration of 3.65 µg/ml for 30 min at room temperature. Excess antibody was washed with PBS, followed by secondary antibody incubation using Alexa Fluor 594 (Invitrogen) and finally nuclear counterstaining using Hoescht 33342 (Invitrogen). Slides were mounted with Prolong Gold Anti Fade mounting medium.
Immunofluorescent labeling of cells was viewed at ×60 oil immersion with the Nikon Eclipse TE-2000E high-precision multimode inverted epifluorescent microscope of the Boston University Medical Center Multi-Photon Imaging Core. To validate localization of the signals, automated Z stacks were recorded on the region of interest containing the viewed cell(s) at 0.30-µm intervals of x-y planes from the top to the bottom of the cell. This was followed with deconvolution of every frame with Nikon NIS Elements software using 20 iterations in a field with low noise. Resulting images were then documented.
Characterization of NAVR/AVR 5′-regulatory region and transcription start sites
Using a cDNA probe encompassing the 5′ region of the NAVR cDNA, we isolated a 28-kb λ genomic clone containing the rat NAVR/AVR gene from a Dahl R λ EMBL3 rat genomic library. A 3-kb XhoI-HindIII restriction fragment was subcloned into psp73 plasmid vector and sequenced in its entirety. Analysis for core promoter elements was then done with the following consensus sequences: TATA box, TATA A/T A A/T; TIIB-response element (BRE), G/C G/C PuCGCC; initiator (INR) at +1, PyPyA+1N T/A PyPy; downstream promoter element (DPE), PuG A/T C/T GTG; functional range set for DPE (DPE*), A/G/T C/G A/T C/T A/C/G C/T (12, 34). Analysis of Sp1 site, CCACCC or GGCGGG, is important since the presence of Sp1 stimulates TFIID interaction with the INR in the absence of a TATA box (29). To investigate putative sex-specific transcriptional regulation, the androgen response element (ARE) 5′-GGTAC A/G CGGTGTTCT-3′ (14) and the estrogen response element (ERE) 5′-GGTCANNNTGACC-3′ (32) were also analyzed. For all, N is any nucleotide, Pu is purine, and Py is pyrimidine.
Determination of the +1 transcription start site was done for AVR with Marathon-Ready rat kidney cDNA, which is ligated to the Marathon adaptor and ready for use as a template in 5′ Marathon rapid amplification of cDNA ends (RACE) reactions. The upstream adaptor primer (provided by Clontech: 5′-CCATCCTAATACGACTCACTATAGGGC-3′) and the downstream AVR reverse primer (5′-GAACTGTGCTTGATGCTTCAGGATGC-3′) were utilized for cDNA amplification per manufacturer’s specifications (Clontech). 5′-RACE cDNA fragments were then cloned and sequenced to identify the +1 start site. The +1 start site for NAVR was deduced from the identification of initiator site consensus sequence.
Multiplex analysis of signaling pathways by Ab-microarray
Analysis of ligand-dependent modulation of different signaling pathways by AVR and NAVR was custom performed by Kinexus, utilizing the Kinex antibody microarray system. We analyzed the effects of ANG II-AVR activation, AVP-AVR activation, and AVP-NAVR activation on multiplex signaling pathways after 30 min of ligand treatment, compared with the respective nonactivated AVR and NAVR in nontreated controls, using Cos1-AVR and Cos1-NAVR permanent cell transfectants, respectively. The Kinex antibody microarray system utilizes phospho-site-specific (tracking specific phosphorylation sites) and pan-specific (tracking protein expression) antibodies querying different signal transduction pathways implicated in multiple biological processes. For the present study, we used a single time point of 30 min and a single optimal dose for AVR and NAVR ligands based on binding affinity results (ANG II, 10 nM; AVP, 10 nM). Phospho-site-specific and pan-specific antibodies were arrayed in duplicates, with some in quadruplicates or six replicates. All fluorescent signals were normalized to background; only values exhibiting signal-to-noise ratios of 1.5-fold or greater and with percent error between duplicates <20% were accepted. Data are presented as percent change from control (% CFC) or change detected after 30 min of Ang II or AVP treatment compared with nontreated transfectant-matched controls, respectively (Table 1).
Table 1.
Signaling proteins phosphorylated/dephosphorylated by AVR and NAVR on ANG II and AVP stimulation
| Protein Name | Protein Symbol | Phospho Site | Role in Renal Physiology and Pathophysiology | AVR+ANG II % CFC |
AVR+AVP % CFC |
NAVR+AVP % CFC |
|---|---|---|---|---|---|---|
| Ataxia telangiectasia mutated | ATM | S1981 | Cellular response to increased osmolality/double strand break pathway (55) |
−21 | 8 | 25 |
| Protein-serine kinase Cβ2 | PKCb2 | T641 | reabsorption (44) | 106 | 28 | 36 |
| Protein-serine kinase Cγ | PKCg | T674 | reabsorption (44) | 23 | 21 | 50 |
| cAMP-dependent protein-serine kinase catalytic subunit β |
PKA Cb | S338 | Water balance/vasopressin-regulated water channels (39) | 108 | 7 | 10 |
| cAMP-dependent protein-serine kinase regulatory type 2 subunit α |
PKA R2a | S98 | Water balance/vasopressin-regulated water channels (39) | −20 | −6 | 68 |
| cAMP-dependent protein-serine kinase regulatory type 2 subunit β |
PKA R2b | S114 | Water balance/vasopressin-regulated water channels (39) | 39 | 43 | 6 |
| Janus protein-tyrosine kinase 2 | JAK2 | Y1007+Y1008 | ANG II-dependent (AT1-type receptor) modulation of water/salt balance (1, 38) |
175 | −8 | −26 |
| Jun protooncogene-encoded AP1 transcription factor | Jun* | S73 | ANG II-dependent (AT1-type receptor) modulation of water/salt balance (1, 38) |
91 | −24 | −20 |
| Jun protooncogene-encoded AP1 transcription factor | Jun | S63 | ANG II-dependent (AT1-type receptor) modulation of water/salt balance (1, 38) |
199 | 36 | −25 |
| Protein-serine kinase Cβ2 | PKCb2 | T641 | ANG II-dependent (AT1-type receptor) modulation of water/salt balance (1, 38) |
106 | 28 | 36 |
| Src proto-oncogene-encoded protein-tyrosine kinase | Src | Y418 | ANG II-dependent (AT1-type receptor) modulation of water/salt balance (1, 38) |
86 | 44 | −9 |
| Signal transducer and activator of transcription 1 | STAT1 | S727 | Diabetic nephropathy (50) | −33 | 173 | −14 |
| Signal transducer and activator of transcription 3 (acute phase response factor) |
STAT3 | S727 | Mesangial and proximal tubular epithelial cell proliferation; renal fibrosis; glomerular diseases; diabetic nephropathy (33, 36, 50) |
−31 | 37 | 51 |
| Jun NH2 terminus protein-serine kinases [stress-activated protein kinase (SAPK)] 1/2/3 |
JNK* | T183+Y185 | Renal ischemia-reperfusion injury (54) | 125 | −28 | −23 |
| Cyclin-dependent protein-serine kinase 1/2 | CDK1/2 | T161+ T160 | Parenchymal renal diseases (41) | −29 | 85 | −73 |
| Retinoblastoma-associated protein1 | Rb | S780 | Renal tubular cell senescence in renal ischemia- reperfusion injury (27) |
12 | 97 | −41 |
| BclII-antagonist of cell death protein | Bad | S75 | Renal tubular cell death (30) | 16 | −14 | −39 |
| BclII-antagonist of cell death protein | Bad | S91 | Renal tubular cell death (30) | 72 | −33 | −12 |
| Acetyl coenzyme A carboxylase | AcCoA | S80 | Fatty acid synthesis (3); chronic renal failure-induced hyperlipidemia (51) |
−65 | −33 | −49 |
| Insulin receptor | IR | Y999 | Glucose metabolism (2) | 29 | −36 | −17 |
| Insulin receptor substrate 1 | IRS1 | Y612 | Glucose metabolism (2, 11) | 126 | −25 | 31 |
| Insulin receptor substrate 1 | IRS1 | Y1179 | Glucose metabolism (2, 11) | 25 | −36 | −28 |
| Phosphoprotein-enriched in diabetes 15 | PED15 | S116 | Regulates glucose transporters (8) | −75 | 73 | 3 |
AVR, angiotensin-vasopressin receptor; NAVR, non-AVR; ANG II, angiotensin II; AVP, arginine-vasopressin; % CFC, percent change from control; phospho site, phosphorylation site (tracked with specified phospho-site-specific antibodies);
Performed with 6 replicas. % CFC ≥20 (bold) and ≤ −20 (italics) are highlighted; repeated-measures ANOVA P < 0.04.
Analysis of functionality of putative androgen and estrogen response elements
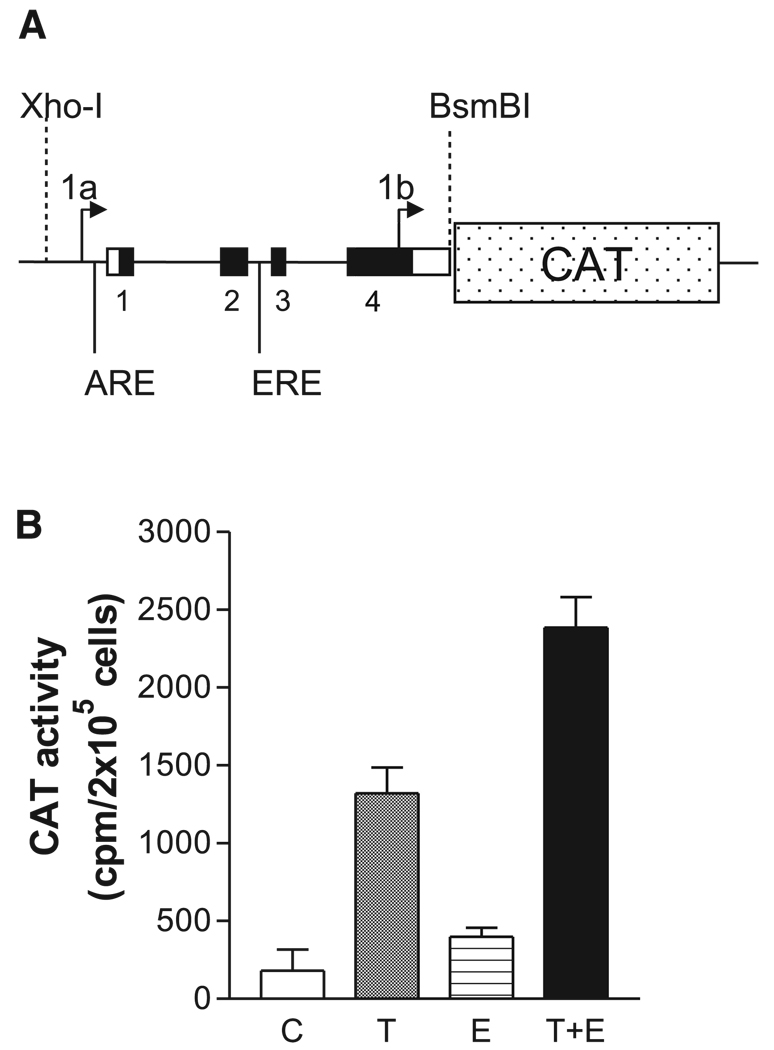
To evaluate transcriptional activity of putative ARE and ERE in the NAVR/AVR promoter region with sufficient 5′ flanking sequences in order to avoid a false-negative result, we investigated testosterone- and estrogen-induced transcriptional induction of a promoter-reporter construct spanning a 2,795-bp XhoI-BsmBI genomic restriction fragment encompassing core promoter elements of NAVR (promoter 1a) and of AVR (promoter 1b) (see Fig. 6) plus 126 bp of AVR 5′-untranslated region (5′-ut). The genomic fragment was subcloned into the classical promoter-reporter pSV0CAT vector in the appropriate orientation to produce the NAVR/AVR 5′-regulatory region-CAT construct that was utilized in the transient transfection experiments. Cells were maintained in DMEM supplemented with 5% fetal bovine serum (FBS). Cells were switched to DMEM containing 5% charcoal/dextran-stripped FBS for 48 h before transfection. Cells at 70% confluence (300,000 cells in P-35 dishes) were cotransfected with 3.0 µg of NAVR/AVR 5′-regulatory region-CAT construct plus 1.5 µg of pSV2-β-galactosidase (for internal control), using the DOTAP liposomal transfection reagent (Roche Molecular Biochemicals). After 6 h, cells were fed with fresh medium containing 5% charcoal/dextran-stripped FBS with testosterone at 1 µg/ml and/or estradiol at 1 µg/ml, or without hormones (control). After 48 h, cells were harvested and assayed for CAT activity with the CAT enzyme assay system with reporter lysis buffer (Promega). A portion of the protein extract was utilized for determination ofβ-galactosidase activity to normalize CAT activity.
Fig. 6.
Reporter function testing of androgen (ARE) and estrogen (ERE) response elements. A: diagram of promoter-reporter DNA construct testing functionality of sex steroid response elements ARE and ERE. A 2.795-kb XhoI to BsmBI restriction DNA fragment spanning both NAVR and AVR promoter regions, ARE and ERE (Fig. 1), was used for transcriptional regulatory element reporter function analysis. B: transcriptional activity of the NAVR/AVR promoters in Neuro2A cells. The experiment included 4 conditions: control conditions (no hormone added; C); testosterone added (final concentration = 1 µg/ml; T); estradiol added (final concentration = 1 µg/ml; E); 1 µg/ml testosterone + 1 µg/ml estradiol (T + E). Background CAT activity (determined from mock-transfected cells) was subtracted from each experimental data point. Each experimental condition was tested in quadruplicate. Data are shown as means ± SE.
RESULTS
Confirmation of AVR transcript start site and mRNA size
A recent report showing overlap of AVR and PYPAF5 (4) or Nalp6 (GenBank), and concluding that AVR does not exist as a membrane-bound receptor but rather is part of a larger cytosol-specific PYPAF5 protein expressed only in leukocytes (17), prompted us to investigate AVR and PYPAF (NAVR) RNA and protein size, cell-specific expression, subcellular localization, ligand affinities, and signal transduction pathways activated by AVR and NAVR on ligand-specific stimulation, respectively.
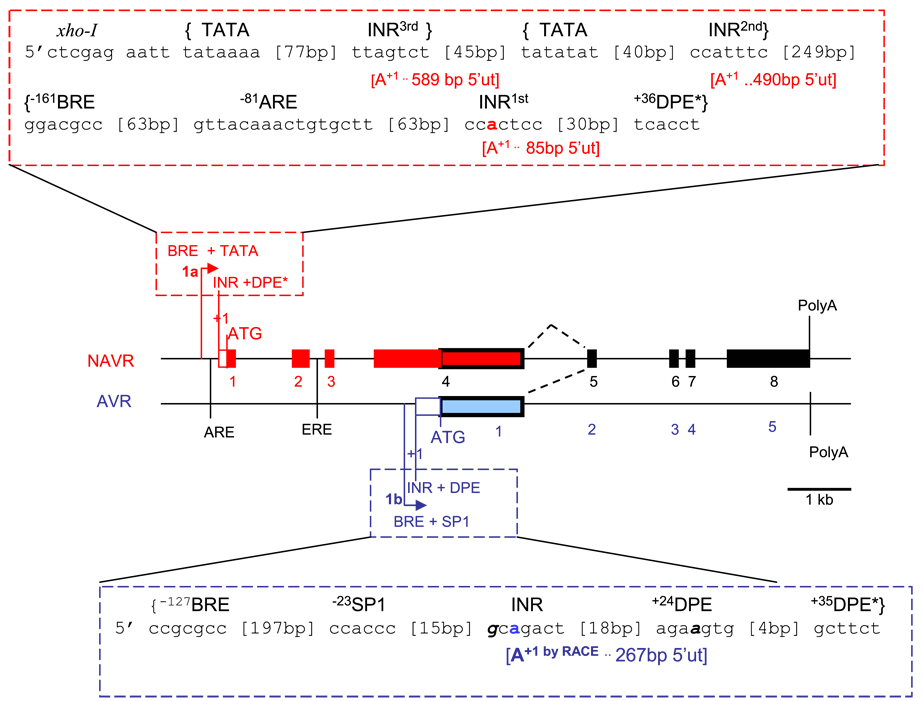
To determine that AVR is indeed a smaller and distinct transcript from the larger NAVR transcript, we identified the AVR transcription start (+1) site from the nucleotide sequence of 5′-RACE-PCR products obtained from rat kidney poly(A)+ RNA. This identified a 267-bp 5′-ut for AVR downstream from the initiating methionine of NAVR (Fig. 1) and determined a 2.354-kb transcript for AVR upstream from its cognate poly(A)+ tract.
Fig. 1.
Diagram of non-angiotensin-vasopressin receptor (NAVR, red) and angiotensin-vasopressin receptor (AVR, blue) overlapping transcripts and alternative promoter usage. Genomic organization depicting core promoter region (dashed box) and intron-exon organization of NAVR (red box) and AVR (blue box), with common exons (black box); exons are numbered 1–8 for NAVR, 1–5 for AVR. Alternative promoters are identified based on core promoter elements localization: 1a, NAVR promoter; 1b, AVR promoter. Core promoter elements were identified based on 100% homology or only 1 mismatch (bold italics) with corresponding consensus sequence: BRE, transcription factor II-B response element [5′ G/G G/G PuCGCC]; DPE, downstream promoter element [5′ PuG A/T C/T GTG 3′]; DPE*, functional range set [5′ Pu/T C/G A/T C/T Pu/C Py 3′]; INR at +1, initiator at transcription start site [5′ PyPyA+1N T/A PyPy]; TATA, TATA box [5′-TATA A/T A A/T-3′]. Based on 100% consensus sequence homology, there are several putative initiators for NAVR: INR1st to 3rd, with corresponding 5′ untranslated region lengths (5′-ut), and transcription start sites (+1 or A+1). AVR transcription start site was determined by rapid amplification of 5′ cDNA ends (5′-RACE) (A+1 by RACE). Other interacting response elements: ARE, androgen response element [5′-GGTACPuCGGTGTTCT-3′]; ERE, estrogen response element [5′-GGTCAnnnTGACC-3′]; Sp1 [5′-CCACCC or GGCGGG-3′]. bp, Base pairs; −n, upstream from transcription start site; +n, downstream from transcription start site. As reference, restriction digest enzyme XhoI site is 700 bp from NAVR initiating methionine (ATG); BsmBI is 135 bp from AVR initiating methionine (ATG). Pu, purine; Py, pyrimidine; n, any nucleotide.
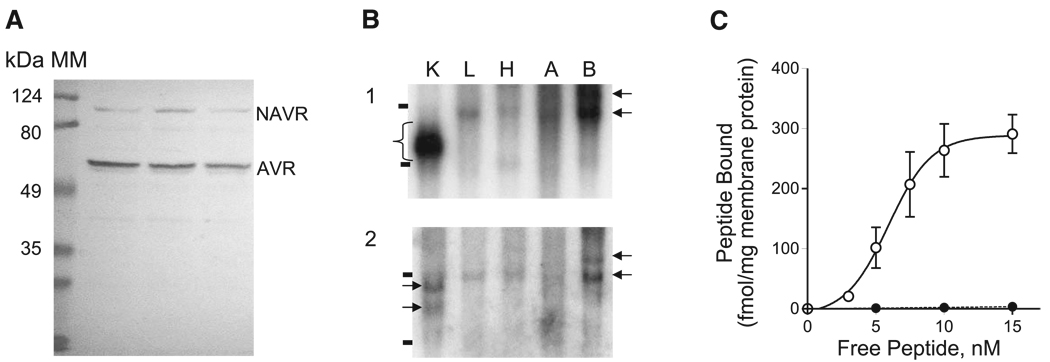
We next determined whether AVR exists as a transcript distinct from NAVR based on differences in size and tissue-specific expression. We performed RNA blot analysis of different rat tissue poly(A)+ RNA samples, using two single-strand antisense DNA probes. First, we probed with a probe common to both NAVR and AVR, encompassing nucleotides 1-1798 of NAVR, corresponding to amino acids (aa)1-589 of NAVR and aa1-aa165 of AVR (see Fig. 3). If indeed AVR does exist as a smaller transcript, we expect a smaller-sized mRNA for AVR and a larger mRNA for NAVR to be detected with this common probe. Second, we probed with a single-stranded cDNA probe spanning nucleotides 1-316 of NAVR (aa1-95 of NAVR), which is unique to NAVR and hence expected to hybridize only to NAVR mRNA sequences. As shown in Fig. 2B the common cDNA probe detects not just two but several mRNA isoforms ranging from 2.5 to 6 kb in all tissues tested. The NAVR-specific probe hybridizes to the larger mRNA species detected with the common cDNA probe, except for the abundant 2.5-kb transcript in kidney (Fig. 2B). This suggests that the abundantly expressed 2.5-kb transcript, present only in kidney, corresponds to AVR and not to NAVR, thus confirming the former as a distinct transcript. These results are consistent with the notion that NAVR is expressed in all tissues tested and is encoded by multiple larger mRNA species reflecting multiple transcription RNA initiation sites. This contrasts with the smaller mRNA size corresponding to AVR that appears to be expressed primarily in kidney. These observations differ from the report by Grenier et al. (17) stating that NAVR has expression restricted to immune cells: T cells and granulocytes. We note that the study by Grenier et al. (17) was limited to tissue culture cell lines. The detection of NAVR in the brain total RNA samples on RNA blot analysis is consistent with the report by Hurbin et al. (25) detecting neuronal expression with an oligonucleotide probe common to NAVR and AVR.
Fig. 2.
NAVR and AVR expression studies. A: Western blot analysis of purified rat kidney membrane proteins using polyclonal antibody raised against a peptide common to both NAVR and AVR: amino acids K592DELKDEE599 of NAVR or amino acids 168–175 of AVR detects two membrane-bound polypeptides (~100 and 50 kDa) in rat kidney membranes. Molecular mass (MM) markers (124, 80, 49, and 35 kDa) are shown on left. B: Northern blot analysis of poly(A)+ RNAs from different tissues. 1: RNA blot hybridized with a NAVR/AVR common probe (amino acids 1-589 of NAVR). 2: RNA blot hybridized with a NAVR-specific probe (amino acids 1–95 of NAVR). The 28S and 18S ribosomal RNAs are marked (−). Arrows indicate different-sized mRNAs detected by the common probe (1) and NAVR-specific probe (2), from the top: estimated 6 kb, 5 kb, 4 kb, 3 kb. Bracket in 1 indicates multiple mRNA sizes in kidney corresponding to 4- to 2.5-kb mRNA species. K, kidney; L, liver, H, heart, A, aorta, B, brain. C: saturation binding curves of ligand-binding studies of membrane proteins isolated from Cos1 cell transfectants expressing NAVR. ●, 125I-ANG II; ○, [3H]AVP. Values are presented as means ± SE from 5 independent experiments.
Western blot analysis detects two polypeptides accounting or a smaller AVR and a larger NAVR
To confirm the existence of different-sized proteins for NAVR and AVR, we performed Western blot analysis of rat kidney membranes, using a rabbit polyclonal antipeptide antibody raised against K592DELKDEE599 peptide (Fig. 3), which spans a predicted extracellular domain of NAVR/AVR (47). This antibody should be able to detect two distinct polypeptides if they share this particular epitope. As shown in Fig. 2A, two distinct proteins were detected, one at ~100 kDa and a second at ~50 kDa consistent with the predicted molecular masses for NAVR (98.2 kDa) and AVR (50.6 kDa), respectively. We note that this experiment further corroborates that AVR is indeed a membrane-bound polypeptide as described previously (47) and furthermore that NAVR is also a transmembrane protein, regardless of the number of transmembrane domains being predicted (4).
NAVR binds vasopressin but not ANG II
Having detected NAVR in membrane proteins by Western blot analysis, we next determined whether NAVR also exhibits ligand binding properties similar to those of AVR. We isolated NAVR cDNA, confirmed its sequence, and then expressed it in Cos1 tissue culture cells. To characterize the binding properties of NAVR for AVP and ANG II we performed ligand binding studies on Cos1 cell membranes expressing the NAVR polypeptide. As shown in Fig. 2C NAVR binds AVP with high affinity (Kd = 5.92 ± 0.23 nM, Bmax = 288.7 ± 8.26 fmol/mg membrane protein), but, unexpectedly, specific binding to ANG II was not detected. This result suggests that the additional NH2-terminal domain present in NAVR (aa1-423; Fig. 3) compared with AVR renders the ANG II binding domain (aa791-799; Fig. 3) inaccessible to ANG II, presumably because of an NH2-terminal domain-induced conformational change masking the ANG II binding site—a hypothesis that needs further study.
Confocal microscopy analysis of Cos1-AVR and Cos1-NAVR cell transfectants
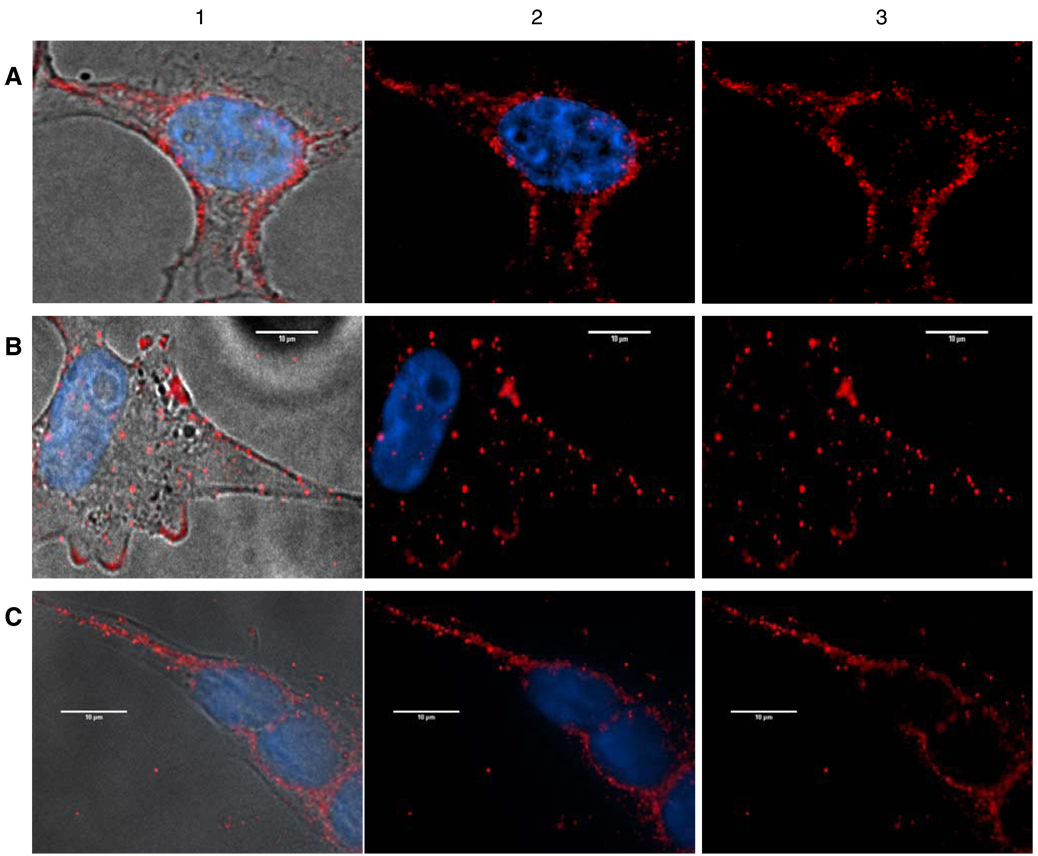
To further study the distinct identities of AVR and NAVR, as well as corroborate the observation that both are membrane-integrated proteins, we investigated the subcellular localization of AVR and NAVR. To distinguish AVR from NAVR, we developed permanent Cos1-transfectants expressing AVR alone (Cos1-AVR) and expressing NAVR alone (Cos1-NAVR). Confocal microscopy analysis using the anti-NAVR/AVR antibody detects AVR mostly in the cell membrane and to a lesser extent in the cytoplasm (Fig. 4A) in Cos1-AVR cells. In contrast, confocal microscopy analysis of Cos1-NAVR cells detects NAVR in the cytoplasm and cell membrane (Fig. 4B) and abundantly in the nuclear membrane as well (Fig. 4C). These data corroborate that both AVR and NAVR are integrated membrane proteins, while also being present in the cytoplasm. Additionally, AVR is distinct from NAVR, since NAVR is detected in the nuclear membrane, whereas AVR is not.
Fig. 4.
Confocal microscopy analysis of Cos1-AVR and Cos1-NAVR permanent cell transfectants. A: Cos1-AVR cells imaged in phase contrast (1); merged confocal image of Cos1-AVR cells with immunofluorescence of AVR in cytoplasm and cell membrane (red), in contrast to blue Hoechst stain for nuclei (2); red AVR+ fluorescence alone (3). B: Cos1-NAVR cells imaged in phase contrast (1); merged confocal image with NAVR immunofluorescence (red) detected in cytoplasm and cell membrane in contrast to blue nuclear stain (2); red NAVR+ fluorescence alone (3). C: Cos1-NAVR cells imaged in phase contrast (1); merged confocal image with NAVR immunofluorescence (red) detected in cytoplasm and nuclear membrane surrounding blue-stained nuclei (2); red NAVR+ fluorescence alone (3).
Tissue-specific expression of AVR and/or NAVR
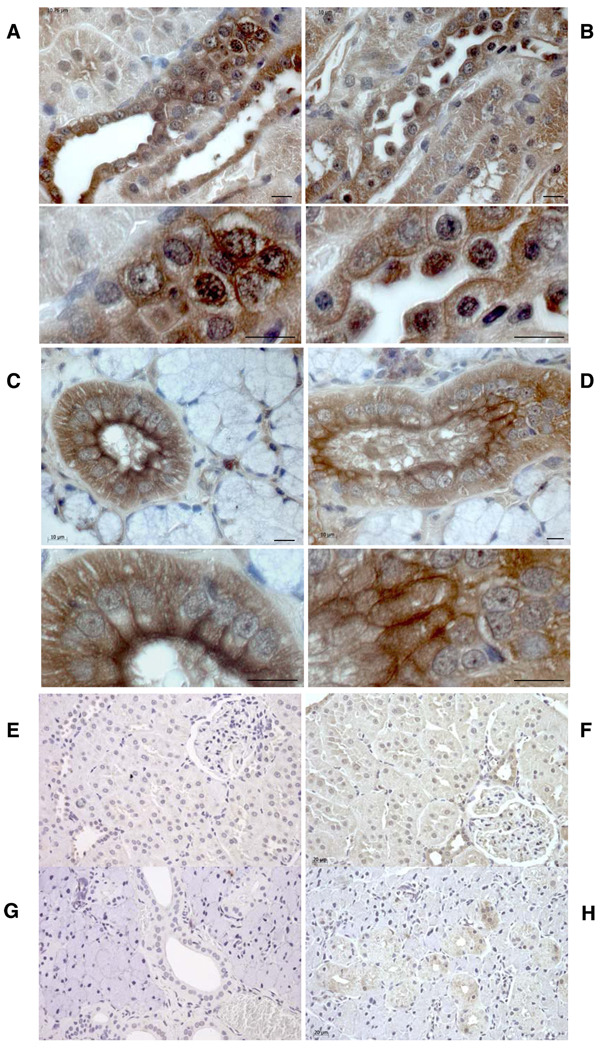
Since NAVR also exhibits vasopressin binding with high affinity similar to AVR, we next performed immunohistochemical analysis using a common AVR/NAVR immunogenic peptide-specific antibody. Analysis of multiple tissues detected membrane-specific and nuclear immunostaining of renal epithelial cells in medullary thick ascending limb of Henle (Fig. 5), confirming previous studies (15). Interestingly, we also detected NAVR/AVR immunostaining in salivary gland apical epithelium (Fig. 5), distinct from negative controls (Fig. 5, E–H), as well as in other tissues, confirming RNA-blot analysis (data not shown).
Fig. 5.
Immunohistochemistry analysis of NAVR/AVR protein. With an antibody raised against the NAVR/AVR common peptide sequence K592DELKDEE599, NAVR/AVR-positive immunostaining of renal epithelial plasma and nuclear membrane is detected in renal cortical collecting duct epithelial cells (A) and medullary thick ascending limb and collecting ducts (B). Corresponding high magnification reveals plasma and nuclear membrane staining. C and D: NAVR/AVR immunostaining is also detected in plasma membranes of salivary gland epithelial cells, with less nuclear membrane staining compared with renal epithelial cells. E–H: Negative controls for renal (E, F) and salivary gland (G, H) immunostaining done with preimmune serum and no anti-AVR/NAVR antibody (E, G) and done with 100× molar excess of AVR/NAVR common peptide sequence as competitor (F, H). NAVR/AVR-positive immunostaining, brown due to DAB chromogen; blue nuclei, Mayer’s hematoxylin counterstain. Bars: 10 µm (A–D), 40 µm (E–H).
Ligand- and receptor-specific signaling pathways
To demonstrate functionality of receptor binding, we investigated whether signal transduction pathways are activated/deactivated on ligand-specific stimulation of AVR and/or NAVR in respective permanent cell transfectants, Cos1-AVR and Cos1-NAVR cells. Multiplex analysis of signal transduction pathways using an Ab-microarray containing phosphorylation site-specific and pan-specific antibodies was done in order to detect and comparatively analyze phosphorylation or dephosphorylation of signaling proteins involved in multiple signal transduction pathways (40). Signaling arrays validate comparative analysis because of the simultaneous detection under identical experimental conditions (6, 40).
We analyzed Cos1-AVR and Cos1-NAVR signaling profiles, referenced to control nontreated Cos1-AVR and Cos1-NAVR cells, respectively, after 30-min treatments with ANG II (ANG II-Cos1AVR) and with AVP (AVP-Cos1AVR and AVP-Cos1NAVR). After normalizing to background, and taking only 1.5-fold changes with <20% error between duplicates or among four to six replicates, multiplex signaling analysis by Ab-microarray detected distinct phosphorylated/dephosphorylated/signaling profiles (repeated-measures ANOVA P < 0.04; Table 1). Interestingly, some signaling molecules are diametrically phosphorylated/dephosphorylated in Cos1-AVR cells by ANG II compared with AVP, while some are coactivated or co-deactivated as well (Table 1). These data demonstrate that AVR activates different signal transduction pathways in a ligand-specific manner, which could underlie ANG II- and AVP-specific renal functions. Additionally, AVP treatment of stable Cos1-NAVR cells activated its own unique set of signaling pathways compared with AVP-AVR activation (Table 1), although a significant number of signaling proteins are activated in a similar manner (Table 1). Together, the results show that AVR and NAVR are distinct receptors with differential ligand-specific signal transduction profiles.
Genomic organization of NAVR/AVR gene
Based on the rat genomic (NCBI ID NC_005100.2) and corresponding NAVR and AVR cDNA sequences, we delineated the intron/exon organization of rat NAVR and AVR transcription units (Table 2; Fig. 1). NAVR is encoded by 8 exons with a predicted 880-amino acid-long polypeptide. We note that AVR encompasses a portion of exon 4 plus exons 5–8 of NAVR containing 457 amino acids (Fig. 3). Comparative analysis suggests the AVR initiation methionine at aa424 of NAVR (Fig. 3) rather than aa400 as previously reported (47), since aa400 in the mouse sequence is not a methionine (unpublished data). We note that the AVR sequence (47) is smaller and contained within NAVR’s amino acid residues 424–880, depicting NAVR and AVR as another example of the growing list of overlapping genes (37). Based on the overlapping genes concept, the detection of AVR within NAVR in no way precludes AVR’s existence as a structurally and functionally distinct protein.
Table 2.
Genomic organization of exons encoding rat NAVR/AVR
| Exon Number | |||||||
|---|---|---|---|---|---|---|---|
| NAVR | AVR | Intron 3′ Seq | Exon Termini | Intron 5′ Seq | Exon Size, bp | Intron Size, bp | Functional Domain in Exon |
| 1 | From 1st (*), 2nd (**), 3rd (***) initiator seq (Fig. 1) |
85*/490**/589*** | NAVR +1 | ||||
| From +1 | |||||||
| 1 | gtgagt | 29b | 535 | Initiating M | |||
| From aa1 | ~GCTCCAG | ||||||
| 2 | cccag | CTCGGAG ~CTGCAGA |
gtaagc | 272 | 238 | ||
| 3 | tccag | GGCTCGG ~GTGTCGG |
gtaagt | 36 | 210 | ||
| 4 | tacag | AGTACAA ~TGAAGGG |
gtgagt | 1,753 | 746 | AVP bd/Gs | |
| 1 | AGACTTa ~TGAAGGG |
gtgagt | 1,088 | 746 | AVR +1 initiating M AVP bd/Gs |
||
| 5 | 2 | tccag | TTCTCAA ~TTCTGAC |
gtgagt | 93 | 1,027 | |
| 6 | 3 | cacag | CTTGTCA ~CACTCAG |
gtaagg | 171 | 75 | |
| 7 | 4 | cacag | GATACAG ~CCCTCAG |
gtaaag | 168 | 422 | ANG II bd |
| 8 | 5 | tgcag | TCTGACT ~GGTGGGC |
834 | Poly(A) | ||
| mRNA size, bp | |||||||
| 3,412* | 2,354 | ||||||
| 3,902** | |||||||
| 4,001*** | |||||||
Seq, nucleotide sequence;
AVR transcription RNA initiation site determined by rapid amplification of 5′ cDNA ends (5′-RACE);
NAVR exon 1 size determined from initiation-methionine consensus sequence; M, methionine; bd, binding domain; Gs, Gs activator domain.
To identify the mechanism for distinct transcripts with shared portions, we investigated the promoter regions for NAVR and AVR. Analysis of genomic sequences identified two promoter regions defined by core promoter elements within the NAVR/AVR gene: 1a and 1b (Fig. 1). Identification of two core promoter regions could account for the production of NAVR and AVR as distinct transcripts based on the mechanism of alternative promoter usage (35). Analysis shows that both promoter regions (1a and 1b, Fig. 1) contain core promoter elements, albeit with different compositions and arrangements. AVR promoter does not have a TATA box but contains an initiator consensus sequence in the +1 start site identified by RACE, thus corroborating this start site, a DPE, and an Sp1 site (Fig. 1). Both the DPE and Sp1 sites are known to enhance efficiency of transcription at initiator sites in TATA-less promoters (7, 10, 29, 52). Interestingly, the core promoter of AVR (promoter 1b, Fig. 1) localizes within NAVR’s exon 4.
Sex steroid hormones modulate NAVR/AVR transcription
Moreover, we detect consensus sequences for androgen (14) and estrogen (32) transcription regulatory elements (Fig. 1) within the NAVR/AVR gene, thus suggesting that sex steroid hormones might regulate NAVR/AVR transcription. Since vasopressin elicits sex-specific morphological and functional differences (45), we investigated the functionality of the putative ARE and ERE identified in the NAVR/AVR promoter region. While the ARE is upstream to both core promoter regions, the ERE is upstream to AVR’s promoter but intronic between exon 2 and 3 in NAVR’s. Nevertheless, because functional transcriptional regulatory elements are also documented to be intronic, we investigated functionality of both ARE and ERE, especially since sexual diergic effects of vasopressin have been described (45) but have not been elucidated at the transcriptional level.
Since NAVR/AVR transcripts were detected in neuronal systems in rat brain (25), and since sex-specific vasopressin effects on neurological functions have been reported (45), we assayed the NAVR/AVR 5′-regulatory region in brain-derived tissue culture cells, Neuro2A cells. Cells were grown in medium devoid of steroid hormones by using charcoal/dextran-stripped FBS. Either testosterone or estradiol was then added exogenously, alone or in combination, to test their ability to modulate NAVR/AVR promoter-CAT-driven activity. A priori, we would expect that cell lines expressing endogenous androgen receptors and/or estrogen receptors would be able to modulate NAVR/AVR promoter-CAT activity. We tested promoter activity in Neuro2A (mouse neuroblastoma cell line) cells essentially as described previously (19). For this purpose, the 2,795-bp XhoI-BsmBI restriction fragment (Fig. 6A) encompassing promoters 1a and 1b plus 126 bp of AVR 5′-ut, was subcloned into the pSV0CAT vector in the appropriate orientation to produce the NAVR/AVR 5′-regulatory region-CAT construct that was utilized in the transient transfection experiments.
As shown in Fig. 6B, the NAVR/AVR promoter-CAT construct was active in neuronal cells, showing a robust activity in the presence of testosterone with a mild activation in the presence of estradiol when compared with basal conditions (no hormone added). Interestingly, when Neuro2A cells were grown in the presence of both hormones, the hormone-stimulated CAT activity was significantly larger than the simple addition of the corresponding independent specific hormone-stimulated CAT activities (Fig. 6B). These results demonstrate that testosterone and estrogen can synergistically stimulate the NAVR/AVR promoters.
DISCUSSION
AVR and NAVR are distinct transcripts from type b-iv alternative promoter usage
Cumulative reports of multiple genes within a locus, be it on opposite DNA strands or overlapping on the same DNA strand, have solidified the concept of overlapping genes such that the definition of a one-gene/one-protein has been changed (37, 43, 53). In parallel, the use of alternative promoters by genes provides one mechanism for the generation of overlapping genes/transcripts from a single locus that could have different 5′-ut but identical amino acid sequence like CP19 and p18, or have different NH2 termini like PPAR-γ and p73, or altogether different proteins as seen for p21 and p21b (35). These precedents support our observations that AVR is distinct based on the identification of the +1 RNA start site for AVR by 5′-RACE, by the detection of differences in mRNA and protein sizes as well as in tissue-specific expression patterns that distinguish AVR from NAVR. Additionally, others have detected AVR/NAVR common sequence in neurons (25) and kidney (13), thus refuting the report that expression of NAVR is highly restricted to T cells and granulocytes (4, 17).
The pattern of alternative promoter usage seen in AVR and NAVR genomic organization is type b-iv, in which the use of multiple promoters produces mRNAs that encode protein isoforms differing in their NH2 termini (35). Interestingly, the ANG II-binding (AVR) and non-ANG II binding (NAVR) functional difference between AVR and NAVR recapitulates polar functional differences observed in other type b-iv alternative promoter usage genes, such as p73/deltaNp73 and p63/deltaN-p63 (35). Additionally, the distinct tissue-specific expression pattern between AVR and NAVR recapitulates the differential tissue specific expression of other type b-iv alternative promoter usage genes such as neuronal nitric oxide synthase (Nos1) and DNA (cytosine-5)-methyltransferase 3-α genes (35). Altogether, the existence of AVR as a distinct transcript and protein is confirmed.
AVR and NAVR as part of diverse vasopressin receptor family
Altogether, the detection of high-affinity binding to vasopressin for both AVR (47) and NAVR as well as cell membrane localization for both by confocal microscopy and Western blot analysis of membrane-bound protein isolates suggest that NAVR is also a V2-type vasopressin receptor like AVR. This is concordant with the fact that AVR’s deduced functional domains, vasopressin binding site, Gs interaction site, and phosphorylation sites are contained within the NAVR polypeptide. Additionally, multiplex signaling Ab-microarray analysis detects vasopressin-induced phosphorylation of PKA-R2a (cAMP-dependent protein-serine kinase regulatory type 2 subunit α) by NAVR and PKA-R2b (cAMP-dependent protein-serine kinase regulatory type 2 subunit β) by AVR (Table 1), both of which are components of cAMP-dependent protein kinases involved in modulation of vasopressin-regulated water channels (39), thus supporting their respective V2-type receptor status.
Importantly, the confirmation of AVR and identification of NAVR as V2-type receptors are concordant with emerging observations that indicate that there are V2-type AVP-mediated functions that are not accounted for by the prototype V2 vasopressin receptor. More specifically, Gouzenes et al. (16) demonstrated that V1a- and V2-type receptors mediate vasopressin responses in isolated vasopressinergic rat supraoptic neurons; however, the V2 receptor was not detected in supraoptic neurons (24), thus suggesting that other V2-type receptors must underlie V2-type mediated functions. Concordantly, Hurbin et al. (25) detected AVR/NAVR transcripts by oligonucleotide in situ hybridization in supraoptic magnocellular neurons along with another vasopressin isoreceptor, the vasopressin-activated calcium mobilizing-1 receptor.
Nevertheless, the emerging picture of multiple vasopressin functions not attributable to prototype V1a, V1b, and V2 receptors supports the emerging picture of other vasopressin receptors. Furthermore, the lack of homology and diverse molecular structures of all vasopressin receptors to date, V1-type receptors [V1a, V1b, vasopressin-activated calcium-mobilizing-1 or VACM-1 (5), single-transmembrane domain-AVP receptor (21)] as well as V2-type receptors [V2, AVR, and NAVR], recapitulates the multireceptor/coreceptor superfamily that binds and interacts with VEGF-A such as VEGFR1, VEGFR2, heterodimers of VEGFR1/R2, and neuropilin-1 (42). As with the VEGF multireceptor family, all vasopressin receptors should be studied comprehensively, individually, and in the context of each other. As with VEGF receptors, the existence of one does not negate the existence of the others.
Insights from signal transduction profiles
While more in-depth studies are needed, the specific activation of multiplex signaling reveals receptor functionality for both AVR and NAVR. Additionally, the distinct signaling pathway profiles activated by AVR and NAVR in Cos1 cells give insight into differential putative roles for these receptors in the modulation of physiological functions known to be mediated by said signaling pathways, such as renal sodium-water balance, glucose and lipid metabolism, apoptosis, and cell cycle regulation (Table 1).
Notably, different subunits of cAMP-dependent protein kinase (PKA) are differentially phosphorylated by ANG II-AVR, AVP-AVR, and AVP-NAVR activation (Table 1). These observations are consistent with V2-type receptors being coupled to cAMP (39) and with the report of Dulin et al. (9) that ANG II can activate PKA, albeit in a cAMP-independent manner. Moreover, the phosphorylation of JAK2 (Janus protein-tyrosine kinase 2), Jun (Jun proto-oncogene-encoded AP1 transcription factor), PKCb2 (Protein-serine kinase C2) and Src (Src protooncogene-encoded protein-tyrosine kinase) signaling proteins, known to be involved in ANG II-dependent regulation of water/salt balance (1, 38), on ANG II-AVR activation suggests that AVR, an AT1 receptor subtype (15), might contribute to as well as underlie these ANG II physiological effects (1, 38). Additionally, the phosphorylation of IR (insulin receptor) and IRS1 (insulin receptor substrate 1) on Ang II-AVR activation is concordant with reported “cross talk” between ANG II and insulin-signaling pathways (2, 11), thus suggesting the hypothesis that AVR could underlie the reported “cross talk” between ANG II and insulin-signaling pathways that occurs at the level of IRS proteins (11). These concordances, along with the data demonstrating that AVR is distinct from NAVR and that both are vasopressin receptors, validate the hypothesis that AVR and NAVR play important roles and could underlie complexities in ANG II- and AVP-mediated physiological effects not explained by prototype AT1a,b and AT2 ANG II receptors or V1a,b and prototype V2 AVP receptors.
Sex steroid hormone regulation of AVR/NAVR
The transcriptional modulation of AVR/NAVR 5′ flanking regulatory region through reporter function assays indicates sex-specific upregulation of AVR and/or NAVR. The robust induction by testosterone in contrast to estrogen is concordant with observations by Gerhold et al. (13) detecting upregulation of AVR in Dahl salt-sensitive male rats in response to salt but not in age-matched females. We note that in the microarray analysis, the V1a, V1b, and V2 receptors were not reported to be upregulated. Together, these observations suggest the hypothesis that renal AVR and/or NAVR play a key role in sodium response in a sex-specific manner. Furthermore, the synergistic increased induction by testosterone and estrogen of AVR/NAVR transcription suggests the hypothesis that the ratio of testosterone and estrogen plays a role in AVR/NAVR transcriptional regulation, since estrogen by itself did not induce AVR/NAVR transcription. Since AVR has been associated with salt-sensitive hypertension in F2 intercross male rats (22), the data suggest that the sex-specific upregulation of AVR/NAVR could play a role in salt-sensitive hypertension in males. Furthermore, when the ratio of testosterone to estrogen increases in postmenopausal women, the upregulation of AVR/NAVR could contribute to the increase in hypertension susceptibility. Further study of these putative mechanisms of sex-specific differences in hypertension progression becomes imperative.
AVR/NAVR in context of apoptosis of magnocellular neurons
Intuitively, AVR and/or NAVR, as the vasopressin-receptor(s) detected in magnocellular neurons of the supraoptic and paraventricular nuclei of the hypothalamus (25), could possibly underlie the apoptosis of said vasopressinergic neurons resulting from prolonged overstimulation in chronic diabetes mellitus observed by Klein et al. (31) through a putative autocrine/paracrine feedback loop. As putative proapoptotic vasopressin receptors, AVR/NAVR could present a counterbalance to the putative apoptosis inhibitory role of V1a receptor in vasopressinergic neurons, deduced from V1a’s apoptosis-inhibitory role in rat glomerular mesangial cells (23). These observations support the hypothesis that diabetes insipidus results from the loss of vasopressinergic neurons via the loss of homeostatic balance between antiapoptotic V1a receptors and proapoptotic AVR/NAVR receptors, both of which are expressed in vasopressinergic neurons in supraoptic and paraventricular nuclei (24, 25).
In summary, the confirmation of AVR and identification of NAVR as vasopressin receptors provide additional receptor diversity concordant with the emerging complexity of vasopressin functions not attributed to prototype V1a, V1b, and V2 receptors. Multiplex signaling pathway analyses suggest the hypothesis that AVR and NAVR contribute to ANG II- and AVP-mediated regulation of renal salt-water balance, glucose and lipid metabolism, apoptosis, and/or cell cycle. Moreover, the data provide new insight into vasopressin receptor regulation and sexual diergic effects through sex steroid-specific transcriptional regulation of AVR/NAVR promoter region. The detection of plasma and nuclear membrane expression suggests a more complex role involving nuclear translocation for NAVR. Clearly, parallel to the VEGF multireceptor system, vasopressin has multiple, diverse receptors concordant with its increasing complexity of functions. Much study needs to be done after this critical step forward confirming AVR and identifying NAVR as vasopressin receptors.
Acknowledgments
GRANTS
This work was supported by National Heart, Lung, and Blood Institute Grant HL-069937 awarded to N. Ruiz-Opazo.
REFERENCES
- 1.Abdulla HI, Pedraza PL, McGiff JC, Ferreri NR. CaR activation increases TNF production by mTAL cells via a Gi-dependent mechanism. Am J Physiol Renal Physiol. 2008;294:F345–F354. doi: 10.1152/ajprenal.00509.2006. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Basha EA, Yibchok-Anun S, Hsu WH. Glucose dependency of arginine vasopressin-induced insulin and glucagon release from the perfused rat pancreas. Metabolism. 2002;51:1184–1190. doi: 10.1053/meta.2002.34052. [DOI] [PubMed] [Google Scholar]
- 3.Ahima RS, Lazar MA. Adipokines and the peripheral and neural control of energy balance. Mol Endocrinol. 2008 January 17; doi: 10.1210/me.2007-0529. doi:10.1210/me.2007-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albrecht M, Domingues FS, Schreiber S, Lengauer T. Identification of mammalian orthologs associates PYPAF5 with distinct functional roles. FEBS Lett. 2003;538:173–177. doi: 10.1016/s0014-5793(03)00161-3. [DOI] [PubMed] [Google Scholar]
- 5.Burnatowska-Hledin MA, Spielman WS, Smith WL, Shi P, Meyer JM, Dewitt DL. Expression cloning of an AVP-activated, calcium-mobilizing receptor from rabbit kidney medulla. Am J Physiol Renal Fluid Electrolyte Physiol. 1995;268:F1198–F1210. doi: 10.1152/ajprenal.1995.268.6.F1198. [DOI] [PubMed] [Google Scholar]
- 6.Chan SM, Ermann J, Su L, Fathman CG, Utz PJ. Protein microarrays for multiplex analysis of signal transduction pathways. Nat Med. 2004;10:1390–1396. doi: 10.1038/nm1139. [DOI] [PubMed] [Google Scholar]
- 7.Chen BS, Hampsey M. Transcription activation: unveiling the essential nature of TFIID. Curr Biol. 2002;12:R620–R622. doi: 10.1016/s0960-9822(02)01134-x. [DOI] [PubMed] [Google Scholar]
- 8.Condorelli G, Vigliotta G, Iavarone C, Caruso M, Tocchetti CG, Andreozzi F, Cafieri A, Tecce MF, Formisano P, Beguinot L, Beghinot F. PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus. EMBO J. 1998;17:3858–3866. doi: 10.1093/emboj/17.14.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulin NO, Niu J, Browning DD, Ye RD, Voyno-Yasenetskaya T. Cyclic AMP-independent activation of protein kinase A by vasoactive peptides. J Biol Chem. 2001;276:20827–20830. doi: 10.1074/jbc.C100195200. [DOI] [PubMed] [Google Scholar]
- 10.Evans R, Fairley JA, Roberts SGE. Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes Dev. 2001;15:2945–2949. doi: 10.1101/gad.206901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folli F, Saad MJA, Velloso L, Hansen H, Carandente O, Feener EP, Khan CR. Crosstalk between insulin and angiotensin II signaling systems. Exp Clin Endocrinol Diabetes. 1999;107:133–139. doi: 10.1055/s-0029-1212088. [DOI] [PubMed] [Google Scholar]
- 12.Fukue Y, Sumida N, Nishikawa J, Ohyama T. Core promoter elements of eukaryotic genes have a highly distinctive mechanical property. Nucleic Acids Res. 2004;32:5834–5840. doi: 10.1093/nar/gkh905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerhold D, Bagchi A, Lu M, Figueroa D, Keenan K, Holder D, Wang Y, Jin H, Connolly B, Austin C, Alonso-Galicia M. Androgens drive divergent responses to salt stress in male versus female rat kidneys. Genomics. 2007;89:731–744. doi: 10.1016/j.ygeno.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Gobinet J, Poujol N, Sultan C. Molecular action of androgens. Mol Cell Endocrinol. 2002;198:15–24. doi: 10.1016/s0303-7207(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez CB, Herrera VLM, Ruiz-Opazo N. Renal immunocytochemical distribution and pharmacological properties of the dual angiotensin II/AVP receptor. Hypertension. 1997;29:957–961. doi: 10.1161/01.hyp.29.4.957. [DOI] [PubMed] [Google Scholar]
- 16.Gouzenes L, Sabatier N, Richard P, Moos FC, Dayanithi G. V1a- and V2-type vasopressin receptors mediate vasopressin-induced Ca2+ responses in isolated rat supraoptic neurons. J Physiol. 1999;517:771–779. doi: 10.1111/j.1469-7793.1999.0771s.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, Carlson A, Merriam S, Lora JM, Briskin M, DiStefano PS, Bertin J. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-κB and caspase-1. FEBS Lett. 2002;530:73–78. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 18.Hausdorff WP, Hnatowich M, O’Dowd BF, Caron MG, Lefkowitz RJ. A mutation of the β2-adrenergic receptor impairs agonist activation of adenylyl cyclase without affecting high affinity agonist binding. Distinct molecular determinants of the receptor are involved in physical coupling to and functional activation of GS. J Biol Chem. 1990;265:1388–1393. [PubMed] [Google Scholar]
- 19.Herrera VLM, Ruiz-Opazo N. Regulation of α-tropomyosin and N5 genes by a shared enhancer: modular structure and hierarchical organization. J Biol Chem. 1990;265:9555–9562. [PubMed] [Google Scholar]
- 20.Herrera VLM, Makrides SC, Xie HX, Adari H, Kraus RM, Ryan US, Ruiz-Opazo N. Spontaneous combined hyperlipidemia, coronary heart disease and decreased survival in Dahl salt-sensitive hypertensive rats transgenic for human cholesteryl ester transfer protein. Nat Med. 1999;5:1383–1389. doi: 10.1038/70956. [DOI] [PubMed] [Google Scholar]
- 21.Herrera VL, Ruiz-Opazo N. Identification of a novel V1-type AVP receptor based on the molecular recognition theory. Mol Med. 2001;7:499–506. [PMC free article] [PubMed] [Google Scholar]
- 22.Herrera VLM, Tsikoudakis A, Ponce LRB, Matsubara Y, Ruiz-Opazo N. Sex-specific QTLs and interacting loci underlie salt-sensitive hypertension and target organ complications in Dahl S/jrHS hypertensive rats. Physiol Genomics. 2006;26:172–179. doi: 10.1152/physiolgenomics.00285.2005. [DOI] [PubMed] [Google Scholar]
- 23.Higashiyama M, Ishikawa S, Saito T, Nakamura T, Kusaka I, Nagasaka S, Honda K, Saito T. Arginine vasopressin inhibits apoptosis of rat glomerular mesangial cells via V1a receptors. Life Sci. 2001;68:1485–1493. doi: 10.1016/s0024-3205(01)00942-0. [DOI] [PubMed] [Google Scholar]
- 24.Hurbin A, Boissin-Agasse L, Orcel H, Rabie A, Joux N, Desarmenien MG, Richard P, Moos FC. The V1a and V1b, but not V2, vasopressin receptor genes are expressed in the supraoptic nucleus of the rat hypothalamus, and the transcripts are essentially colocalized in the vasopressinergic magnocellular neurons. Endocrinology. 1998;139:4701–4707. doi: 10.1210/endo.139.11.6320. [DOI] [PubMed] [Google Scholar]
- 25.Hurbin A, Orcel H, Ferraz C, Moos FC, Rabie A. Expression of the genes encoding the vasopressin-activated calcium-mobilizing receptor and the dual angiotensin II/vasopressin receptor in the rat central nervous system. J Neuroendocrinol. 2000;12:677–684. doi: 10.1046/j.1365-2826.2000.00499.x. [DOI] [PubMed] [Google Scholar]
- 26.Jørgensen PL. Purification (Na+ plus K+)-ATPase: active site determinations and criteria of purity. Ann NY Acad Sci. 1974;242:36–52. doi: 10.1111/j.1749-6632.1974.tb19077.x. [DOI] [PubMed] [Google Scholar]
- 27.Kailong L, Du X, Yani H, Lin Z, Jvrong Y, Ruihua S, Lin C. P53-Rb signaling pathway is involved in tubular cell senescence in renal ischemia/reperfusion injury. Biocell. 2007;31:213–223. [PubMed] [Google Scholar]
- 28.Kaneko Y, Herrera VLM, Didishvili T, Ruiz-Opazo N. Sex-specific effects of dual ET-1/ANG II receptor (Dear) variants in Dahl salt-sensitive/resistant hypertension rat model. Physiol Genomics. 2005;20:157–164. doi: 10.1152/physiolgenomics.00108.2004. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann J, Smale ST. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 30.Kiley SC, Thornhill BA, Tang SS, Ingelfinger JR, Chevalier RL. Growth factor-mediated phosphorylation of proapoptotic BAD reduces tubule cell death in vitro and in vivo. Kidney Int. 2003;63:33–42. doi: 10.1046/j.1523-1755.2003.00706.x. [DOI] [PubMed] [Google Scholar]
- 31.Klein JP, Hains BC, Craner MJ, Black JA, Waxman SG. Apoptosis of vasopressinergic hypothalamic neurons in chronic diabetes mellitus. Neurobiol Dis. 2004;15:221–228. doi: 10.1016/j.nbd.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuratsune M, Masaki T, Hirai T, Kiribayashi K, Yokoyama Y, Arakawa T, Yorioka N, Kohno N. Signal transducer and activator of transcription 3 involvement in the development of renal interstitial fibrosis after unilateral ureteral obstruction. Nephrology (Carlton) 2007;12:565–571. doi: 10.1111/j.1440-1797.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 34.Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landry JR, Mager DL, Wilhelm BT. Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet. 2003;19:640–648. doi: 10.1016/j.tig.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Yang N, Zhang L, Huang Y, Zhang R, Wang F, Luo M, Liang Y, Yu X. Inhibition of Jak/STAT signaling ameliorates mice experimental nephrotic syndrome. Am J Nephrol. 2007;27:580–589. doi: 10.1159/000108102. [DOI] [PubMed] [Google Scholar]
- 37.Makalowska I, Lin CF, Makaloski W. Overlapping genes in vertebrate genomes. Comput Biol Chem. 2005;29:1–12. doi: 10.1016/j.compbiolchem.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Marrero MB, Banes-Berceli AK, Stern DM, Eaton DC. Role of the JAK/STAT signaling pathway in diabetic nephropathy. Am J Physiol Renal Physiol. 2006;290:F762–F768. doi: 10.1152/ajprenal.00181.2005. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen S, Frokler J, Marples D, Kwon TH, Agre P, Knepper MA. Aquaporins in the kidney: from molecules to medicine. Physiol Rev. 2002;82:205–244. doi: 10.1152/physrev.00024.2001. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc Natl Acad Sci USA. 2003;100:9330–9335. doi: 10.1073/pnas.1633513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obligado SH, Ibraghimov-Beskrovnaya O, Zuk A, Meijer L, Nelson PJ. CDK/GSK-3 inhibitors as therapeutic agents for parenchymal renal diseases. Kidney Int. 2008;73:684–690. doi: 10.1038/sj.ki.5002731. [DOI] [PubMed] [Google Scholar]
- 42.Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signaling—in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 43.Pearson H. What is a gene? Nature. 2006;441:399–401. doi: 10.1038/441398a. [DOI] [PubMed] [Google Scholar]
- 44.Perry C, Blaine J, Le H, Grichtchenko II. PMA- and AngII-induced PKC regulation of the renal Na+- cotransporter (hkNBCe1) Am J Physiol Renal Physiol. 2006;290:F417–F427. doi: 10.1152/ajprenal.00395.2004. [DOI] [PubMed] [Google Scholar]
- 45.Rhodes ME, Rubin RT. Functional sex differences (“sexual diergism”) of central nervous system cholinergic systems, vasopressin, and hypothalamic-pituitary-adrenal axis activity in mammals: a selective review. Brain Res Rev. 1999;30:135–152. doi: 10.1016/s0165-0173(99)00011-9. [DOI] [PubMed] [Google Scholar]
- 46.Rodbard D. Mathematics of hormone-receptor interaction. I. Basic principles. Adv Exp Med Biol. 1973;36:289–326. doi: 10.1007/978-1-4684-3237-4_14. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Opazo N, Akimoto K, Herrera VLM. Identification of a novel dual angiotensin II/vasopressin receptor on the basis of molecular recognition theory. Nat Med. 1995;1:1074–1081. doi: 10.1038/nm1095-1074. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Opazo N, Lopez LV, Herrera VLM. The dual Ang II/AVP receptor gene N119S/C163R variant exhibits sodium-induced dysfunction and cosegregates with salt-sensitive hypertension in the Dahl salt-sensitive hypertensive rat model. Mol Med. 2002;8:24–32. [PMC free article] [PubMed] [Google Scholar]
- 49.Sabatier N, Shibuya I, Dayanithi G. Intracellular calcium increase and somatodendritic vasopressin release by vasopressin receptor agonists in the rat supraoptic nucleus: involvement of multiple transduction signals. J Neuroendocrinol. 2004;16:221–236. doi: 10.1111/j.0953-8194.2004.01155.x. [DOI] [PubMed] [Google Scholar]
- 50.Shi YH, Zhao S, Wang C, Li Y, Duan HJ. Fluvastatin inhibits activation of JAK and STAT proteins in diabetic rat glomeruli and mesangial cells under high glucose conditions. Acta Pharmacol Sin. 2007;28:1938–1946. doi: 10.1111/j.1745-7254.2007.00653.x. [DOI] [PubMed] [Google Scholar]
- 51.Szolkiewicz M, Sucajtys E, Wolyniec W, Rutkowski P, Stelmanska E, Korczynska J, Swierczynski J, Rutkowski B. Mechanisms of enhanced carbohydrate and lipid metabolism in adipose tissue in uremia. J Ren Nutr. 2005;15:166–172. doi: 10.1053/j.jrn.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 52.Szutorisz H, Dillon N, Tora L. The role of enhancers as centers for general transcription factor recruitment. Trends Biochem Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Veeramachaneni V, Makalowski W, Galdzicki M, Sood R, Makalowska I. Mammalian overlapping genes: the comparative perspective. Genome Res. 2006;14:280–286. doi: 10.1101/gr.1590904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Ji HX, Xing SH, Pei DS, Guan QH. SP600125, a selective JNK inhibitor, protects ischemic renal injury via suppressing the extrinsic pathways of apoptosis. Life Sci. 2007;80:2067–2075. doi: 10.1016/j.lfs.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Woo SK, Lee SD, Kwon HM. TonEBP transcriptional activator in the cellular response to increased osmolality. Pflügers Arch. 2002;444:579–585. doi: 10.1007/s00424-002-0849-2. [DOI] [PubMed] [Google Scholar]