Abstract
TAHRE, the least abundant of the three retrotransposons forming telomeres in Drosophila melanogaster, has high sequence similarity to the gag gene and untranslated regions of HeT-A, the most abundant telomere-specific retrotransposon. Despite TAHRE's apparent evolutionary relationship to HeT-A, we find TAHRE Gag cannot locate to telomere-associated “Het dots” unless collaborating with HeT-A Gag. TAHRE Gag is carried into nuclei by HeT-A or TART Gag, but both TART and TAHRE Gags need HeT-A Gag to localize to Het dots. When coexpressed with the appropriate fragment of HeT-A and/or TART Gags, TAHRE Gag multimerizes with either protein. HeT-A and TART Gags form homo- and heteromultimers using a region containing major homology region (MHR) and zinc knuckle (CCHC) motifs, separated by a pre_C2HC motif (motifs common to other retroelements). This region's sequence is strongly conserved among the three telomeric Gags, with precise spacing of conserved residues. Nontelomeric Gags neither interact with the telomeric Gags nor have this conserved spacing. TAHRE Gag is much less able to enter the nucleus by itself than HeT-A or TART Gags. The overall telomeric localization efficiency for each of the three telomeric Gag proteins correlates with the relative abundance of that element in telomere arrays, suggesting an explanation for the relative rarity of TAHRE elements in telomere arrays and supporting the hypothesis that Gag targeting to telomeres is important for the telomere-specific transposition of these elements.
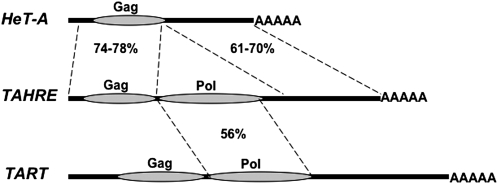
DROSOPHILA telomeres are maintained by a remarkable variant of the telomerase mechanism that maintains telomeres in almost all organisms (Pardue and DeBaryshe 2003; Melnikova and Georgiev 2005). As in other organisms, Drosophila telomeres are elongated by tandem repeats that are reverse transcribed onto the ends of the chromosomes. What makes Drosophila telomeres unusual is the RNA template that is reverse transcribed to produce these repeats: Drosophila telomere repeats are copied from full-length retrotransposons (HeT-A, TART, and TAHRE), rather than from a short segment of the RNA molecule that makes up part of the telomerase holoenzyme (Figure 1).
Figure 1.—
The telomere retrotransposons of D. melanogaster. These three non-LTR retrotransposons are drawn, approximately to size, as the RNA molecules that are reverse transcribed onto the telomere. Thick solid lines, untranslated regions; shaded ovals, Gag and Pol coding sequences; AAAAA, poly(A) 3′ tail. Dotted lines enclose regions of significant nucleotide identity between TAHRE and the other elements. The range of identity in pairwise comparisons of TAHRE with different copies of HeT-A or TART is given for each region. The TAHRE 5′-UTR is shorter and the 3′-UTR is longer than the UTRs of HeT-A. The extra sequences were not included in calculating identity.
Although clearly related to other retrotransposons in the Drosophila melanogaster genome, the three retrotransposons that make up telomeres have several characteristics that set them apart from the more typical retrotransposable elements. One of these characteristics is their localization to telomere arrays. The euchromatic regions of the D. melanogaster genome have been completely sequenced (Celniker et al. 2002). Analysis of these gene-rich regions reveals no sequence from any of the three telomeric elements (George et al. 2006), although these euchromatic regions are littered with other retrotransposons (Kaminker et al. 2002). Conversely, the long arrays of telomeric retrotransposons do not contain their nontelomeric relatives. Thus, the telomeric and nontelomeric elements have distinctly different genomic distributions, except for small “transition zones” at the proximal ends of telomere arrays where fragments of both kinds of elements are mingled (George et al. 2006).
The telomere-specific transposition of HeT-A and TART appears to depend on the intranuclear targeting of the Gag proteins encoded by each element. These Gags share amino acid sequence motifs with retroviral Gags, proteins known to be important in intracellular transport of viral RNA. The sequence similarities with retroviral Gags suggest that telomeric Gags are important in intracellular transport of the retrotransposon RNA, a suggestion supported by studies of the intracellular localization of HeT-A and TART Gag proteins. Transient expression of tagged Gag proteins in D. melanogaster cells showed that Gags of both HeT-A and TART localize to nuclei very efficiently. Gags of nontelomeric retrotransposons were also tested in these experiments and found predominantly, if not entirely, in the cytoplasm (Rashkova et al. 2002b). Preventing Gags of nontelomeric retrotransposons from entering the nucleus may be one of the mechanisms cells use to protect their genomes from parasitic invaders. In contrast, the telomeric retrotransposons have an essential role in the nucleus and the cell benefits from facilitating nuclear localization of these Gags.
After moving from the cytoplasm into the nucleus, HeT-A Gags form aggregates (Het dots) associated with telomeres in interphase nuclei. HeT-A and TART are intermingled in D. melanogaster telomere arrays so it was surprising that TART Gags formed loose intranuclear clusters with no obvious telomere associations. However, cotransfection experiments showed that when the two Gags are expressed in the same cells, HeT-A Gag dominates the localization and moves TART Gag into telomere-associated Het dots (Rashkova et al. 2002a). Presumably this localization is necessary for transposition to telomeres.
The collaborative localization of the two Gags suggests an explanation for two puzzling observations. The first observation is that all D. melanogaster stocks and cell lines have both HeT-A and TART in their telomeres, suggesting that both elements are needed by the cell. However, the two elements seem to be distributed randomly in telomere arrays, giving no indication that either one has a special role. The second observation is that HeT-A elements do not encode reverse transcriptase, while TART does. Most, if not all, other retrotransposons encode this enzyme. Having the enzyme sequence encoded by the element's RNA would be expected to allow more efficient transposition, as has been shown for human Lines-1 elements (Wei et al. 2001). Nevertheless HeT-A transposes efficiently and is significantly more abundant than TART in telomeres of all D. melanogaster stocks and cell lines studied (George et al. 2006). The finding that HeT-A Gag positions TART for transposition to telomeres suggested that TART might provide the reverse transcriptase for both elements, thereby explaining the need for both elements in the genome. We suggest that HeT-A is more abundant than TART because HeT-A has stronger telomere targeting.
After these localization studies were finished, a third D. melanogaster telomeric retrotransposon, TAHRE, was reported (Abad et al. 2004). TAHRE has both a HeT-A-related Gag protein and a reverse transcriptase closely related to that of TART and thus presumably with the same activity. TAHRE's sequence predicted that it should combine the localization activity of HeT-A with the enzyme activity of TART and transpose more efficiently than either of the other elements, yet TAHRE is actually much less abundant than either HeT-A or TART. Only one full-length copy of this element has been reported and only one full-length copy of its Gag gene is found in the D. melanogaster database. In this study we have examined the intracellular localization of TAHRE Gag to see whether the sequence similarity to HeT-A Gag yields a protein with the remarkable telomere targeting of HeT-A Gag and to shed light on TAHRE's relative rarity in telomeric arrays.
MATERIALS AND METHODS
Cultured Drosophila cells:
Drosophila Schneider line 3 cells (S3) were maintained at room temperature in Schneider's medium (GIBCO Invitrogen) with heat-inactivated 10% fetal bovine serum (Hyclone) and 10 units/ml penicillin, 10 μg/ml streptomycin solution (Mediatech).
Recombinant DNA and plasmid construction:
TAHRE gag coding sequence was amplified by PCR from BACR40C07, a kind gift from Alfredo Villasante, Centro de Biología Molecular Severo Ochoa, Madrid. PCR primers (forward 5′-GAAGAATTCCCACCATGTCCACGTCCGACCAC-3′ and reverse 5′-GATGGATCCTATCCGGAGGTCATGAGGTGTGCT-3′) amplified the entire coding region, preceded by a Drosophila Kozak sequence and minus the stop codon. This was flanked by 5′ EcoRI and 3′ KpnI sites to insert the gene in frame with the gene for fluorescent protein (green fluorescent protein, GFP; yellow fluorescent protein, YFP; or cyan fluorescent protein, CFP) in expression vectors. The PCR product was TA cloned into a Strataclone PCR cloning vector (Stratagene) and verified by sequencing. The gag sequence was cut out with EcoRI and KpnI and inserted into pPL17 and its variants pSR24 and pSR25. The resulting constructs expressed Gag driven by the armadillo promoter and tagged at the C terminus with enhanced (E)GFP, YFP, and CFP, respectively.
Transfection:
Two milliliters of S3 cells (5 × 106 cells/ml) were seeded into wells of six-well tissue culture dishes and transfected with 1 μg of DNA plus 25 μl of Effectene Transfection Reagent (QIAGEN), used as the manufacturer directs. For cotransfections, 0.5 μg of each DNA was used. After 72 hr at room temperature, transfected cells were resuspended in their medium and 40 μl was used for cytological preparations while the rest was washed in PBS, pelleted, and frozen for immunoblotting.
Cytology:
Twenty microliters of transfected cells was spotted onto slides that had been dipped in 0.005% poly-l-lysine and dried. Cells were allowed to settle for 20 min; were fixed with 3.7% formaldehyde/PBS for 30 min; and were washed in PBS for 5 min, 0.2% Triton/PBS for 10 min, and PBS for 2 min. Preparations were blocked with 1% BSA/PBS for 1 hr and incubated with anti-lamin (Dmo ADL67.10-c; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) 1:200 in 1% BSA/PBS at 4° overnight. Slides were washed with PBS 4 × 5 min and incubated with Cy3-labeled anti-mouse IgG (Jackson Labs) 1:200 in BSA/PBS at room temperature for 1 hr, washed 4 × 5 min with PBS, and mounted in 70% glycerol/PBS. Slides were examined with a Nikon ECLIPSE E 600 microscope, photographed with a Spot RT slider camera, and false colored with Photoshop.
Immunoblots:
Cells from 2-ml cultures were washed in PBS and resuspended in 100 μl of PBS. One hundred microliters of 2× Laemmli buffer was added and samples were boiled for 5 min. Two microliters of β-mercaptoethanol was added to each 20 μl of sample. Proteins were resolved by SDS-polyacrylamide gel electrophoresis in 8% gels and transferred to nitrocellulose membranes. Fusion proteins were visualized by immunoblotting with anti-GFP antiserum from guinea pig (Rashkova et al. 2002b) and secondary anti-guinea pig antibody conjugated to alkaline phosphatase (Jackson ImmunoLaboratories).
Sequences used for analysis:
The following sequences were used: TAHRE, AJ542581; TART A, AY561850; TART B, U14101; TART C, AY600955; canonical HeT-A, U06920 (nt 1015–7097); HeT-A elements from the 4R telomere, AC010841 (4-5 = nt 27,992–33,790, 4-4 = nt 33,948–39,772, 4-3 = nt 40,382–46,298, 4-2 = nt 53,189–59,033, 4-1 = nt 60,023–65,592); HeT-A element from the XL telomere, CP000372 (nt 12,874–18,879); jockey, M22874; Doc, X17551; and I Factor, M14954.
RESULTS
The TAHRE gag gene and untranslated regions have strong nucleotide sequence similarity to the corresponding regions of HeT-A:
The nucleotide sequences suggest that TAHRE and HeT-A are derived from the same ancestor (Abad et al. 2004). These elements differ in that TAHRE has a reverse transcriptase coding sequence (pol gene) plus 200 bp of novel 3′-UTR sequence (see Figure 1). The ancestral sequence is unknown: the pol gene may have been either deleted to produce HeT-A or inserted to produce TAHRE. The pol gene has more sequence similarity to TART pol than to pol genes of other nontelomeric retrotransposons (data not shown).
Even within the same telomere, HeT-A elements differ in sequence, as shown by the six full-length elements in the partially assembled telomeres on chromosomes X and 4 (George et al. 2006). Surprisingly, all these Gag proteins show less conservation of the amino acid sequence than of the nucleotide sequence. Pairwise comparisons of the Gag proteins from these HeT-A elements show a broad distribution of between 75.5 and 99.7% amino acid identity. Interestingly, pairwise comparisons between TAHRE and these HeT-A Gags show that TAHRE has similar amounts of amino acid identity (69.5–72.3%) with each of the divergent HeT-A proteins.
There are three subfamilies of TART elements in the D. melanogaster genome. In pairwise comparisons of amino acids, Gags of these subfamilies differ by 82.5–94.7%. These TART Gags have less amino acid identity with TAHRE Gag (44.8–45.9%) than do the HeT-A proteins. HeT-A and TART Gags have less amino acid identity with each other (21.1–23.9%) than either one has with TAHRE.
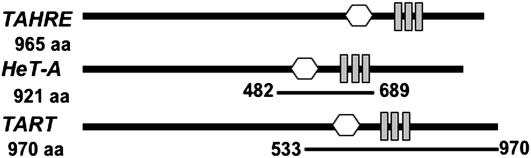
Amino acid identities between TAHRE and HeT-A Gags are most concentrated in the region containing the major homology region (MHR) and zinc knuckle motifs (see Figure 2 and supporting information, Figure S1, Figure S2, and Figure S3). This same region also has the highest concentration of identities of HeT-A and TAHRE Gags to TART Gags.
Figure 2.—
TAHRE Gag proteins. Comparison of TAHRE Gag with HeT-A and TART Gags showing locations of amino acid motifs shared with retroviral Gags. Open hexagon, major homology region (MHR); shaded bars, zinc knuckle motifs. The region between the hexagon and the shaded bars in insects has been named the pre_C2HC domain. Lines below HeT-A and TART diagrams indicate regions of those proteins expressed by deletion constructs designed to test specific parts of the Gags. First and last amino acids of each deletion construct are given. For both HeT-A and TART, Gag proteins can differ in length between different copies of the element. The amino acid numbers given here refer to the sequences used for the tagged proteins in this study.
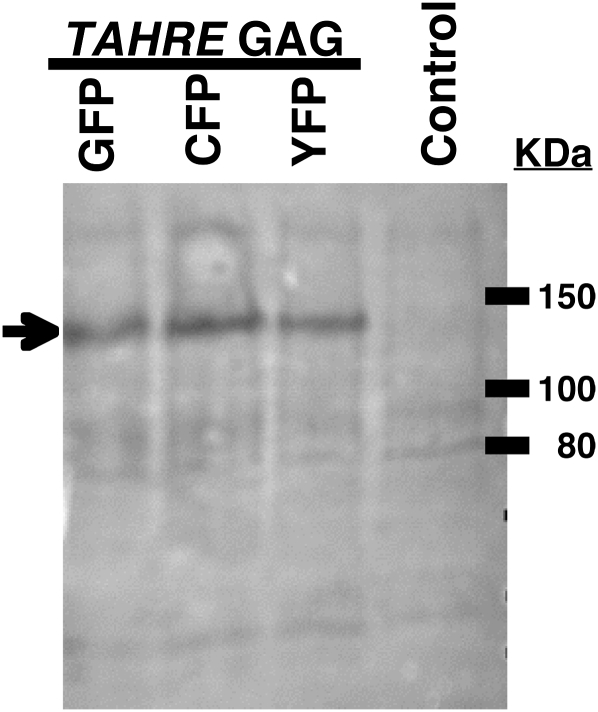
To investigate the significance of these sequence similarities, we subcloned the TAHRE Gag coding region (Figure 2), minus its stop codon, from BACR40C07. This coding region was fused in frame to sequence for a fluorescent protein tag in the expression vector, pPL17, that has been used to study the HeT-A and TART Gags (Rashkova et al. 2002a). Three constructs were made, adding GFP, CFP, or YFP to the C terminus of TAHRE Gag. When transfected into cultured Drosophila cells, these constructs expressed full-length fusion protein, as confirmed by immunoblot analysis (Figure 3).
Figure 3.—
Immunoblot of proteins expressed by cultured cells transfected with constructs carrying TAHRE sequences. Lanes are labeled with the fluorescent tag on the protein expressed by the transfected construct. Control cells were not transfected. The filter was probed with an antibody to GFP, which also recognizes CFP and YFP. All constructs express a protein of the size calculated from the sequence of the fusion protein, 133 kDa. The weaker bands of nonspecific antibody binding seen in all lanes serve as a loading control.
Transient transfection in cultured Drosophila cells:
S3 cells express HeT-A and TART RNA. We have not detected TAHRE transcripts, possibly a matter of detection sensitivity. There are no useful antibodies to any of these Gag proteins so we do not know whether any endogenous proteins are present. However, the tagged proteins studied here are sufficiently overexpressed that we do not expect endogenous proteins to affect the results. One of the remarkable findings obtained from overexpressing these telomere Gags is that the localization is very robust: it can accommodate a significant amount of protein without changing the localization of that protein. The system is also efficient; almost nothing is left behind, arguing that even a small amount of Gag protein would reach a telomere.
Unlike other telomeric Gags, TAHRE Gag is found mostly in the cytoplasm:
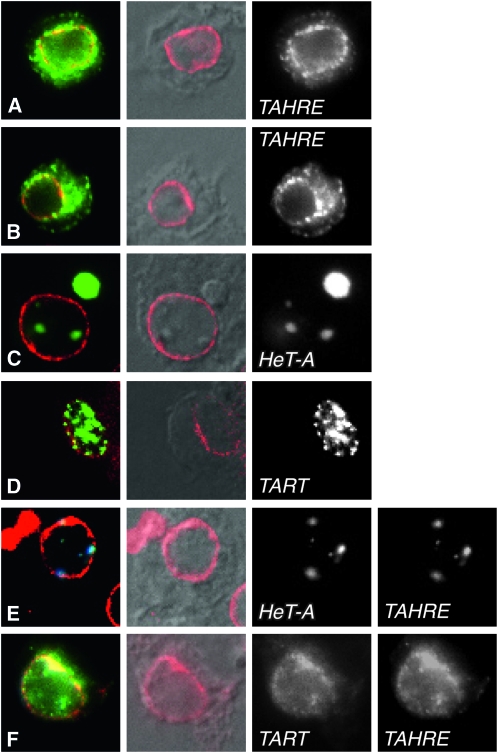
Because of the strong sequence similarity to HeT-A Gag, we assumed that TAHRE Gag, like HeT-A Gag, would enter the nucleus and localize to telomeres (Rashkova et al. 2002b). Surprisingly, TAHRE Gag formed clusters but almost all of these clusters were in the cytoplasm, although they tended to concentrate around the nucleus (Figure 4, A and B). Lamin staining to define the nuclear envelope showed many Gag clusters superimposed on the lamin stain, suggesting that they were stuck within the nuclear lamina. A few TAHRE Gag clusters were seen inside the lamin staining; however, these remained near the periphery of the nucleus, rather that moving to more interior regions. TAHRE Gag also differs from HeT-A Gag in not forming a Het body. These are characteristic cytoplasmic bodies, one per cell, sometimes seen in cells with nuclear Het dots. We have suggested that Het bodies may be the result of overload of the transport system in cells that are overexpressing HeT-A Gag (Rashkova et al. 2002b). Cytoplasmic TAHRE Gag does not form these distinctive bodies, a second indication that it is not transported as is HeT-A Gag.
Figure 4.—
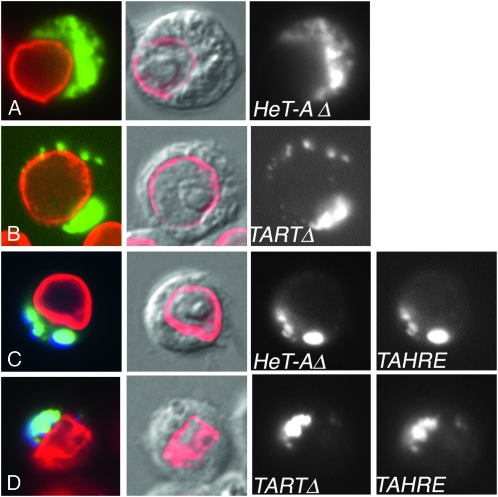
Intracellular localization of TAHRE, HeT-A, and TART Gag proteins in transiently transfected D. melanogaster cultured cells. (A and B) GFP-tagged TAHRE Gag in single transfections, showing most of the protein clustered in the perinuclear cytoplasm or superimposed on the lamin. (C) YFP-tagged HeT-A Gag, forming large round Het dots in the nucleus and a single round cytoplasmic Het body. (D) YFP-tagged TART Gag, forming small irregular dots completely contained within the nucleus or superimposed on the lamin. (E) CFP-tagged TAHRE Gag plus YFP-tagged HeT-A Gag, colocalizing to regular nuclear Het dots preferentially near the edge of the nucleus. (F) CFP-tagged TAHRE Gag plus YFP-tagged TART Gag, colocalizing in diffuse clusters mostly within the nucleus. Columns from right to left show (1) false-colored Gag fluorescence (green, yellow, or yellow and cyan) plus fluorescent antibody staining lamin (red), (2) anti-lamin staining (red) plus DIC of the cell, (3) black-and-white view of GFP channel in A and B and YFP channel in C–F, and (4) black-and-white view of CFP channel in E and F.
We note that both HeT-A and TART Gags move to the cytoplasm when chromosomes condense for mitosis but then move back into the nucleus when it reforms (Rashkova et al. 2002a). In contrast, TAHRE Gag is predominantly cytoplasmic throughout the cell cycle.
The cytoplasmic distribution of TAHRE Gag differs from that of the Gags of nontelomeric retrotransposons studied in our transient transfection assay. These nontelomeric Gags do not enter the nucleus but are distributed more evenly through the cytoplasm and do not concentrate near the nucleus like TAHRE Gag (Rashkova et al. 2002b). This difference suggests that TAHRE is actively localized toward the nucleus and has some ability to enter; however, either it is unable to move independently within the nucleus or it is impeded by interactions with the nuclear periphery.
HeT-A and TART Gags can move TAHRE Gag into the nucleus:
As noted above, amino acid identities between TAHRE Gag and HeT-A Gag are especially strong in the region containing the MHR–zinc knuckle motifs (Figure 2). This region of HeT-A and TART Gags is responsible for the protein–protein interactions that allow HeT-A Gag to move TART Gag to telomere-associated Het dots (Rashkova et al. 2003). The sequence similarity suggested that TAHRE Gag might interact with Gags of the other two telomeric retrotransposons to enable it to localize to the nucleus. We tested this possibility by coexpression experiments similar to those used to characterize the interaction of HeT-A and TART Gags.
HeT-A Gag has been shown to enter the nucleus and localize to telomere-associated Het dots. It also forms a single, distinctive cytoplasmic Het body in some cells that have several Het dots (Rashkova et al. 2002a). We saw similar localizations when we expressed HeT-A Gag (Figure 4C). When we coexpressed TAHRE Gag with HeT-A Gag, the two Gags colocalized to nuclear Het dots (Figure 4E) and, in some cells, to a cytoplasmic Het body, showing that HeT-A Gag could direct TAHRE Gag to telomeres. Interestingly, HeT-A Gag had also cleared TAHRE Gag from its diffuse distribution in the cytoplasm: any TAHRE Gag left in the cytoplasm was gathered into the single Het body. Although HeT-A Gag has a strong influence on TAHRE Gag, TAHRE Gag also affects the interaction. In the cotransfections, nuclear Het dots are less round and regular and most tend to be located near the nuclear membrane (Figure 4E), while cells expressing only HeT-A Gag have round Het dots both at the membrane and in interior positions (Figure 4C).
The similarity of the MHR–zinc knuckle regions (Figure S1, Figure S2, and Figure S3) suggested that TART Gag might also be able to localize TAHRE Gag to the nucleus. When expressed alone, TART Gag enters the nucleus and forms many small clusters. No Gag remains in the cytoplasm but the nuclear clusters do not show preferential association with telomeres (Rashkova et al. 2002a) (see also Figure 4D). Coexpression of TART and TAHRE Gags showed that TART Gag was able to move TAHRE Gag into the nucleus. In the nucleus the two Gags colocalized but their distribution is much more diffuse than the clusters that TART Gag makes when transfected alone (Figure 4F). TAHRE Gag changes the intranuclear organization of TART Gag more dramatically than it changes the intranuclear organization of HeT-A Gag. This suggests that HeT-A Gag, which dominates localization of the other two Gags, has stronger targeting within the nucleus than either of the other Gags.
TAHRE Gag association with HeT-A and TART Gags involves the region containing the MHR–zinc knuckle motifs:
Rashkova et al. (2003) have shown that the regions of TART and HeT-A Gags containing the MHR–zinc knuckle motifs facilitate both homologous and heterologous associations to form clusters and colocalize. Deletion derivatives of the two Gags that retain the MHR–zinc knuckle internal association region can be carried into the nucleus by full-length Gags of either element, even though the deletion derivatives lack the N-terminal residues necessary to independently enter the nucleus. The deletion derivatives used in those experiments were HeT-A Gag, amino acids 482–689, and TART Gag, amino acids 533–970 (see Figure 2). Neither deletion derivative was able to enter the nucleus when expressed individually (Rashkova et al. 2003). Instead they were distributed through the cytoplasm and formed aggregates of various sizes (Figure 5, A and B).
Figure 5.—
Intracellular localization of Gag protein deletion derivatives in transiently transfected D. melanogaster cultured cells. (A) YFP-tagged HeT-A Gag 482–689 in single transfection, forming irregular clusters completely excluded from the nucleus. (B) YFP-tagged TART Gag 533–970 in single transfection, forming varied size clusters in the cytoplasm. (C) Cotransfected YFP-tagged HeT-A Gag 482–689 plus CFP-tagged TAHRE Gag, colocalizing completely in cytoplasmic aggregates. (D) Cotransfected YFP-tagged TART Gag 533–970 plus CFP-tagged TAHRE Gag, colocalizing completely in cytoplasmic aggregates. Columns from right to left show (1) false-colored Gag fluorescence (yellow or superimposed cyan and yellow) plus fluorescent antibody staining lamin (red), (2) anti-lamin staining (red) plus DIC of the cell, (3) black-and-white view of YFP channel, and (4) black-and-white view of CFP channel in C and F.
To see whether TAHRE Gag's association with HeT-A and TART Gags involved the MHR–zinc knuckle regions, we coexpressed TAHRE Gag with the deletion derivatives of HeT-A Gag and TART Gag used by Rashkova et al. When coexpressed with either HeT-A Gag 482–689 or TART Gag 533–950, TAHRE Gag colocalized tightly with the deletion derivative (Figure 5, C and D). This colocalization shows that TART and HeT-A Gags interact with TAHRE Gag, using the same regions in which they interact with each other, the regions containing the MHR and zinc knuckles.
None of the proteins in these colocalization experiments were moved into the nucleus. However, each protein had some modulating effect on its partner's localization. When expressed with the HeT-A derivative, TAHRE Gag was pulled away from the nuclear membrane into HeT-A cytoplasmic aggregates, producing larger aggregates, some resembling Het bodies (Figure 5C). When TAHRE Gag was coexpressed with the TART Gag derivative, the proteins colocalized to form one or more large Het body-like aggregates in the cytoplasm (Figure 5D). These bodies sometimes became large enough to distort the shape of the nucleus but were not concentrated around the nuclear membrane or seen inside the nucleus. We observed a range of sizes of cytoplasmic clusters in both single transfections and cotransfections of TART Gag 533–970; however, the clusters of individually transfected TART Gag 533–970 were much more diffuse than the smooth, dense clusters seen in the cotransfectants. These results show that TAHRE Gag cannot localize other proteins to the nuclear membrane but can affect the conformation of the aggregates it forms with other proteins.
DISCUSSION
HeT-A, TART, and TAHRE are an unusual trio in terms of the intracellular localization of their Gag proteins:
Although the three retrotransposons appear to have similar roles in forming telomere arrays, each Gag protein has a different pattern of localization when expressed by itself. HeT-A Gag localizes to Het dots associated with telomeres in interphase nuclei. TART Gag moves into nuclei but does not show preferential association with telomeres. TAHRE Gag remains predominantly in the cytoplasm with a tendency to concentrate around the nucleus and to colocalize with nuclear lamin. Neither TAHRE nor TART Gags localize to telomeres independently. Both require interaction with HeT-A Gag to reach this localization.
Association between telomeric Gags is directed by a well-conserved segment of these highly variable proteins:
Studies of deletion derivatives of Gag proteins (Rashkova et al. 2003) show that association between HeT-A and TART Gags depends on a highly conserved region of each protein that contains the MHR and the zinc knuckle (CCHC box) motifs (Figure 2A). Our experiments show that this same region directs associations of these two telomeric Gags with TAHRE.
The MHR and zinc knuckle amino acid motifs are hallmarks of retroviral Gag proteins. The MHR (QGX2EX7R) is so named because it is the only region of significant homology among different groups of retroviruses (Craven et al. 1995). The zinc knuckle motif has the general formula CX2CX4HX7C, although the spacing of the conserved C and H residues may differ in different elements (Covey 1986; Summers et al. 1990). Retroviral Gags usually have one or two zinc knuckles; the D. melanogaster retrotransposons studied here each have three. In both retroviral and retrotransposon Gags, the MHR is slightly N terminal of the zinc knuckle region. These two regions and the sequence between them are strongly conserved, in contrast to the marked sequence variability seen in much of the amino acid sequence of Gag proteins. The MHR–zinc knuckle region appears to have several roles in the retroviral life cycle, including involvement in multimerization of Gags (Strambio-De-Castillia and Hunter 1992; Franke et al. 1994; Orlinsky et al. 1996; Singh et al. 2001). In the insect retrotransposons discussed here this region also contains a domain, pre_C2HC, of unknown function (Doerks et al. 2002). This domain occupies most of the sequence between the MHR and the zinc knuckles (see Figure S1).
HeT-A Gags in the same D. melanogaster genome can differ significantly in amino acid sequence, yet the 151 amino acids of their MHR–zinc knuckle regions align with no gaps in spacing and only 15 residues where one or more of the amino acids differ from the consensus (Figure S1). The only available TAHRE Gag sequence is very similar, having only 20 residues that are not identical to all of the HeT-A Gags in the alignment. Interestingly, 15 of these TAHRE residues are at sites where HeT-A Gags are not all identical and for most sites TAHRE has the amino acid found in the majority of the HeT-A Gags (Figure S1). Therefore most of the differences in the TAHRE sequence are ones that are tolerated in HeT-A Gag as well.
Sequence variation in TART elements is concentrated in the untranslated regions, which define three subfamilies, TART A, TART B, and TART C (Sheen and Levis 1994). The MHR–zinc knuckle regions in Gags of the TART subfamilies also have 151 amino acids, all identical except for two residues in TART C. The TART sequence in this region aligns with the sequences from HeT-A and TAHRE with no gaps and no misalignment of CCHC residues; however, there are more amino acid differences between TART and HeT-A than between TAHRE and HeT-A (Figure S2). TAHRE and the canonical HeT-A have 95% identity in this region but only 50 and 52% identity, respectively with TART. Because HeT-A Gag interacts efficiently with TART Gag, it appears that these amino acid differences are tolerated.
The D. melanogaster genome has many non-LTR retrotransposons that do not transpose into telomeric arrays (Kaminker et al. 2002). Gags of these nontelomeric elements also have a MHR–zinc knuckle region with three zinc knuckles. However, the MHR–zinc knuckle regions of Gag in the nontelomeric elements differ more from the regions in HeT-A, TART, and TAHRE than the regions in the telomeric Gags differ from each other. These differences are easily seen in the spacing of the CCHC residues and in their spacing relative to the MHR (Figure S3). All of the sequences from telomeric Gags have identical spacing while the other three sequences differ in spacing from the telomeric Gags and from each other. Jockey and Doc have only 27–31% amino acid identity with each other or any of the telomeric Gags, while I factor has ∼17% amino acid identity with any of the other sequences. HeT-A Gag does not form functional associations with Gag proteins from Doc, jockey, or I Factor (Pardue et al. 2005). This specificity is similar to that of the MHR–zinc knuckle region of retroviruses that forms heteromultimers only between genetically related retroviruses (Franke et al. 1994). The sequence differences between the nontelomeric Gags and telomeric Gags support the hypothesis that the MHR–zinc knuckle region is involved in the association between telomeric elements.
These sequence comparisons suggest that the MHR–zinc knuckle provides an amino acid code for formation of heteromultimers. They also raise questions about how degenerate the code is. Does the higher similarity of the HeT-A and TAHRE Gags indicate a stronger affinity than either one has for TART Gag or is the code degenerate enough to accommodate the differences seen in this region? The strong interactions between any two of these proteins seen in our experiments indicate that a rigorous answer to this question will require careful quantitative studies with purified proteins. However, as discussed below, the in vivo studies presented here suggest that TART Gag's interaction with HeT-A Gag may be favored by its presence in the nucleus in contrast to the more distant position of TAHRE Gag in the cytoplasm.
Conclusion:
These studies provide new evidence that Gag protein localization is important in the transposition of the three telomere-specific retrotransposons of D. melanogaster. Of the three telomere-specific retrotransposons only HeT-A encodes a Gag protein that specifically localizes to telomeres. Nevertheless both TAHRE and TART Gags can be directed to telomeres by association with HeT-A Gag. Interactions between any of the three Gag proteins depend on the segment containing the MHR, pre_ C2HC, and zinc knuckle motifs. The amino acid sequence in this region has a highly conserved pattern that is specific for the telomere retrotransposons. The conservation of this segment in these unusually variable proteins suggests the importance of Gag interactions between these retrotransposons.
Gags of the three telomere elements differ in their ability to localize to telomere Het dots. HeT-A Gag can localize to telomeres independently. TART Gag localizes to the nucleus independently but must have the help of HeT-A Gag to associate with telomeres (Rashkova et al. 2002a). Moving into the nucleus puts TART Gag into an optimal position to encounter HeT-A Gag for localization to Het dots. TAHRE Gag requires assistance to move from the cytoplasm so is less efficient than TART Gag in encountering HeT-A Gag for localization to Het dots. Thus TAHRE is less likely to have carried in its RNA for reverse transcription onto telomeres. This could explain the rarity of TAHRE in telomeres, one complete and three truncated copies in the D. melanogaster genome sequenced by the genome project (Abad et al. 2004). Similarly, the observation that HeT-A is consistently more abundant than TART in different stocks of D. melanogaster (George et al. 2006) may reflect the fact that TART Gag needs HeT-A Gag for telomere localization. This correlation between the abundance of each element and the efficiency of its Gag in localizing to Het dots provides additional support for the hypothesis that Gag localization is important for targeting telomere-specific transposition.
Acknowledgments
We are grateful to Alfredo Villasante for the BAC containing the complete TAHRE element. Members of the Pardue lab and Elena Casacuberta have provided helpful discussion and comments on this manuscript. We particularly thank Laura Croal for helpful advice. This work was initiated in a student laboratory supported by a grant to Massachusetts Institute of Technology from Howard Hughes Medical Institute and has been supported in part by National Institutes of Health grant GM50315 (to M.-L.P.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.109744/DC1.
References
- Abad, J. P., B. De Pablos, K. Osoegawa, P. J. De Jong, A. Martin-Gallardo et al., 2004. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol. Biol. Evol. 21 1620–1624. [DOI] [PubMed] [Google Scholar]
- Celniker, S. E., D. A. Wheeler, B. Kronmiller, J. W. Carlson, A. Halpern et al., 2002. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3 RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey, S. N., 1986. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 14 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr. and J. W. Wills, 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69 4213–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks, T., R. R. Copley, J. Schultz, C. P. Ponting and P. Bork, 2002. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 12 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, E. K., H. E. Yuan, K. L. Bossolt, S. P. Goff and J. Luban, 1994. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J. Virol. 68 5300–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, J. A., P. G. DeBaryshe, K. L. Traverse, S. E. Celniker and M.-L. Pardue, 2006. Genomic organization of the Drosophila telomere retrotransposable elements. Genome Res. 16 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker, J. S., C. M. Bergman, B. Kronmiller, J. Carlson, R. Svirskas et al., 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3 RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova, L., and P. Georgiev, 2005. Drosophila telomeres: the non-telomerase alternative. Chromosome Res. 13 431–441. [DOI] [PubMed] [Google Scholar]
- Orlinsky, K. J., J. Gu, M. Hoyt, S. Sandmeyer and T. M. Menees, 1996. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J. Virol. 70 3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue, M.-L., and P. G. DeBaryshe, 2003. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 37 485–511. [DOI] [PubMed] [Google Scholar]
- Pardue, M.-L., S. Rashkova, E. Casacuberta, P. G. DeBaryshe, J. A. George et al., 2005. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 13 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova, S., S. E. Karam, R. Kellum and M.-L. Pardue, 2002. a Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J. Cell Biol. 159 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova, S., S. E. Karam and M.-L. Pardue, 2002. b Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA 99 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashkova, S., A. Athanasiadis and M.-L. Pardue, 2003. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J. Virol. 77 6376–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, F. M., and R. W. Levis, 1994. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc. Natl. Acad. Sci. USA 91 12510–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. R., R. L. Hill and J. R. Lingappa, 2001. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology 279 257–270. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., and E. Hunter, 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J. Virol. 66 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers, M. F., T. L. South, B. Kim and D. R. Hare, 1990. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry 29 329–340. [DOI] [PubMed] [Google Scholar]
- Wei, W., N. Gilbert, S. L. Ooi, J. F. Lawler, E. M. Ostertag et al., 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]