Abstract
The development of asexual spores, that is, the process of conidiation, in the fungus Neurospora crassa is increased by light. The fluffy (fl) gene, encoding a major regulator of conidiation, is activated by light. We describe here a detailed characterization of the regulation by blue light of fl in vegetative hyphae. This induction requires the white collar complex (WCC) while the FLD protein acts as a dark repressor of fl transcription. We show that the WCC directly regulates fl transcription in response to blue light after transiently binding the promoter. We propose that fl is repressed by FLD in vegetative mycelia and that the repression is lost after light exposure and WCC activation. The increase in fl mRNA in vegetative mycelia after light exposure, and the corresponding increase in the amount of the regulatory FL protein, should promote the activation of the conidiation pathway. The activation by light of fl provides a simple mechanism for the activation of conidiation by blue light in Neurospora that may be at work in other fungi.
ASEXUAL sporulation in the fungus Neurospora crassa leads to the development of asexual spores called macroconidia. Other types of conidia, microconidia and arthroconidia, are produced by Neurospora (Springer 1993), but macroconidia are most abundant, and we will focus on the regulation of macroconidiation, or conidiation, from here on. Conidiation is induced by several environmental signals, including desiccation, carbon and nitrogen starvation, and exposure to blue light (Springer 1993; Davis 2000). In addition, conidiation is controlled by an endogenous circadian clock (Dunlap and Loros 2004; Tan et al. 2004; Heintzen and Liu 2007; Brunner and Káldi 2008). During conidial development, vegetative hyphae grow away from the substrate to form a mass of aerial hyphae. About 4 hr after conidial induction, hyphal growth changes from apical elongation to apical budding, leading to the formation of chains of proconidia divided by minor constrictions. Budding continues in proconidial chains, and major constrictions appear ∼8 hr after the induction of conidiation. Interconidial junctions are cleaved several hours later, but conidia are held together by fragile connective threads until they are dispersed by wind currents (Springer 1993).
Several genes are required for conidiation. Strains with mutations in aconidiate-2 (acon-2) or fluffyoid (fld) are blocked in the transition from filamentous to budding growth. Mutations in aconidiate-3 (acon-3) or fluffy (fl) allow the production of minor, but not major, constriction chains. Mutations in two conidial separation genes (csp-1 and csp-2) prevent the separation of cross walls to release free conidia (Springer 1993). Two developmental genes, fl (NCU08726) and csp-1 (NCU02713), have been identified, and the corresponding proteins are putative zinc-finger transcription factors (Bailey and Ebbole 1998; Lambreghts et al. 2009).
The fl gene has been investigated in some detail. The FL protein is a 792-amino-acid polypeptide containing a Zn2Cys6 binuclear zinc cluster domain belonging to the Gal4p family (Bailey and Ebbole 1998). A mutation in fl blocks conidiation at the minor constriction stage, ∼4 hr after induction (Springer and Yanofsky 1989), and fl mRNA is observed 6 hr after the initiation of conidiation when major constrictions appear in proconidial chains (Bailey and Ebbole 1998). The presence of fl mRNA in aerial hyphae where conidiation-specific genes are expressed suggests a major role for FL in conidiation and in conidial-specific gene expression (Bailey-Shrode and Ebbole 2004). However, fl is transiently induced 30 min after the induction of conidiation, suggesting an additional role for FL in the formation of aerial hyphae (Correa and Bell-Pedersen 2002). The relevance of FL as a major regulator of conidiation in Neurospora is supported by the observation of conidial development when fl expression is forced in vegetative hyphae (Bailey-Shrode and Ebbole 2004), a condition that leads to the expression of eas (Bailey-Shrode and Ebbole 2004), the gene for the hydrophobin rodlet protein that is located on the surface of matured conidia (Bell-Pedersen et al. 1992; Lauter et al. 1992). This is consistent with the observed binding of FL to the eas promoter (Rerngsamran et al. 2005). Other genes are upregulated in the fl-overexpressing strain, including the conidiation-specific genes con-6 and con-10 (Rerngsamran et al. 2005), supporting the proposal of FL as a conidiation-specific transcription factor.
The aconidial phenotype of an fl strain is partially suppressed by a mutation in vib-1, a gene involved in heterokaryon incompatibility, suggesting that FL may regulate conidiation through the repression of VIB-1 (Xiang and Glass 2004). Light regulation of conidiation has been described via the activity of the proteins WC-1 and WC-2 (Lauter et al. 1997). WC-1 contains a zinc finger, a chromophore-binding domain, and PAS domains for protein–protein interactions (Ballario et al. 1996; Crosthwaite et al. 1997). The chromophore-binding domain binds the flavin FAD, allowing WC-1 to act as a blue-light photoreceptor (Froehlich et al. 2002; He et al. 2002). The protein WC-2 contains a zinc finger and a PAS domain and interacts with WC-1 (Linden and Macino 1997) to form a white collar complex (WCC). This complex, upon light exposure, binds transiently to the promoters of light-inducible genes, presumably to activate their transcription (Froehlich et al. 2002; He and Liu 2005; Belden et al. 2007b).
The increase in conidiation observed in Neurospora cultures exposed to light suggests that light may activate the transcription of key regulatory genes, such as fl. Indeed, mRNAs for both fl and csp-1 accumulate after light exposure, suggesting that the corresponding genes are activated by light (Belden et al. 2007a; Chen et al. 2009). We describe here a detailed characterization of the regulation by light of fl in vegetative hyphae. We show that the WCC directly regulates fl transcription in response to blue light after transiently binding the promoter and that developmental regulators are not required for light regulation of fl. We propose that light regulates conidiation in Neurospora through the activation of fl, a key developmental regulatory gene.
MATERIALS AND METHODS
Strains and culture conditions:
We used the standard N. crassa wild-type strain 74-OR23-1VA [Fungal Genetics Stock Center (FGSC) 2489 matA] and the mutant strains FGSC 4397 (wc-1ER53 matA), FGSC 4407 (wc-2ER33 matA), FGSC 3263 (acon-2 mata), FGSC 5074 (acon-3 mata), FGSC 7431 (flL mata), FGSC 7023 (fld mata), FGSC 2555 (csp-1 mata), FGSC 1690 (flP mata), FGSC 1616 (flP961 matA), FGSC 11044 (flKO mata), FGSC 9504 (flRIP matA), and FGSC 4241 (flY mata). N. crassa strains were obtained from the FGSC (http://www.fgsc.net). All strains were maintained by growth in Vogel's minimal media with 1.5% sucrose as the carbon source. Strain manipulation and growth media preparation followed standard procedures and protocols (Davis 2000). See also the Neurospora protocol guide (http://www.fgsc.net/Neurospora/NeurosporaProtocolGuide.htm).
Light induction experiments:
Cultures were grown and mycelia were illuminated for the times indicated to measure regulation of gene expression by light or to detect the binding of the WCC to the promoters of light-regulated genes. Cultures were prepared by inoculating ∼106 viable conidia into 25 ml of liquid Vogel's minimal medium containing 0.2% Tween 80 as the wetting agent. Cultures for developmental mutants that did not produce any or few conidia were started using 0.5 ml of hyphal homogenates obtained from mycelia that had grown on 40 ml of Vogel's liquid medium containing 0.2% Tween 80. Mycelial pads were then homogenized by two 0.5-min pulses in a mini-beadbeater (Biospec) with 1.5 g of zirconium beads (0.5-mm diameter) in 1.9-ml screw-cap tubes prior to inoculation. The plates were incubated in the dark for 48 hr (22°) inside a dark box and were then exposed to white light provided by a set of fluorescent bulbs (the active blue-light component was 1 W/m2). Light exposures with different intensities were obtained using an illumination chamber that allowed the simultaneous irradiation of three plates in a temperature-controlled room set at 22°. The illumination chamber was a black wooden box with plate holders placed at the bottom. White light from a quartz halogen lamp installed in a slide projector passed through an upper window carrying a filter holder with two heat filters and neutral-density filters, as required to obtain the desired light intensity. Additional filters were used for blue-light exposure (Corning broadband blue filter with maximum transmission at 440 nm) or for red-light exposure (two Plexiglas red filters). After light exposure, mycelia were collected with the help of tweezers, dried on filter paper, wrapped in aluminum foil, frozen in liquid nitrogen, and stored at −80°, unless otherwise indicated. Control cultures were kept in the dark prior to collection. All the manipulations in the dark were performed under red light. Light intensities were measured with a calibrated photodiode.
RNA isolation and quantitative RT–PCR:
Neurospora mycelia were disrupted by two 0.5-min pulses in a mini-beadbeater (Biospec) in an RNA extraction buffer (47% guanidinium thiocyanate) with 1.5 g of zirconium beads (0.5-mm diameter) in 1.9-ml screw-cap tubes. The samples were cooled on ice for 4 min after the first pulse of the mini-beadbeater. The extracts in screw-cap tubes were clarified by centrifugation in a microcentrifuge (13,000 rpm) for 5 min prior to RNA purification. Total RNA from mycelia was obtained using the Perfect RNA Eukaryotic mini kit (Eppendorf). Quantitative PCR experiments were performed to determine relative mRNA abundance using one-step RT–PCR with 25 μl 1× Power SYBR Green PCR Master Mix (Applied Biosystems), 6.25 units MultiScribe Reverse Transcriptase (Applied Biosystems), 1.25 units RNase Inhibitor (Applied Biosystems), 0.2 μm of each primer (con-10 5′-CAGCCACAGCGGAGGC-3′ and 5′-TTGGAAGCAATTTCGCGC-3′, fl 5′-GGCGATTCCCGCTATGTT-3′ and 5′-TTGCAGGCCTTTCCCAAA-3′, and tub-2 5′-CCCGCGGTCTCAAGATGT-3′ and 5′-CGCTTGAAGAGCTCCTGGAT-3′), and 100 ng of RNA. Quantitative PCR analyses were performed using a 7500 real time PCR system (Applied Biosystems). The reaction included retrotranscription (30 min at 48°), denaturation (10 min at 95°), and 40 PCR cycles (15 sec at 95° and 1 min at 60°). After each PCR we performed a melting curve analysis to show the specific amplification of a single DNA segment and the absence of nonspecific amplified DNA. The results for each gene were normalized to the corresponding results obtained with tub-2 to correct for sampling errors. Then the results obtained with each sample were normalized to the RNA sample obtained from wild-type mycelia kept in the dark. The gene identification numbers in the Neurospora genome database (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html) are fl (NCU08726), con-10 (NCU07325), and tub-2 (NCU04054).
Chromatin immunoprecipitation:
Mycelia were transferred to 25 ml of liquid Vogel's minimal medium with 0.2% Tween 80 as the wetting agent and 1% formaldehyde and kept under moderate agitation (room temperature, 15 min) to allow protein–DNA crosslinking. The crosslinking reactions were stopped by adding glycine (125 mm final concentration) and moderate shaking (at room temperature for 5 min). For chromatin immunoprecipiation (ChIP), mycelia were ground in liquid nitrogen using a mortar and pestle, and ∼100 mg from each mycelial sample was transferred to 0.5 ml of lysis buffer (50 mm HEPES, pH 7.5, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) and mixed prior to sonication (30 sec on/30 sec off for 15 min in an ice-filled water bath and a Bioruptor sonicator at maximum power). The average DNA size obtained after sonication was 500–600 bp. Then each sample was clarified by centrifugation (15 min at 13,000 rpm), and a fraction (1/10) of each supernatant was transferred to a new tube and marked as each “input” sample. Each remaining supernatant was precleared by the addition of 60 μl of salmon sperm DNA–protein A agarose mix (Millipore) and gentle rotation (4°, 60 min). After centrifugation (1 min at 13,000 rpm), each supernatant was collected and divided in two; one-half was treated with the antibody, the immunoprecipitation (IP) sample, while the other half remained as the “no-IP” control. Then 50 μl of anti-WC-2 antibody (2 μg) (Neiss et al. 2008) was added to ∼200 μl from each IP supernatant and incubated overnight at 4° with gentle rotation. Then, 60 μl of salmon sperm DNA–protein A agarose mix (Millipore) was added to each sample (IP and no-IP) and incubated with gentle rotation (for 2 hr at 4°). The agarose beads were collected by centrifugation and cleaned twice with 100 μl of lysis buffer, once with high-salt buffer (lysis buffer with 0.5 m NaCl), once with wash buffer (0.1 m Tris–HCl, pH 7.5, 1 mm EDTA, 0.25 m LiCl, 0.5% IGEPAL-CA630, 0.5% sodium deoxycholate), and once with TE buffer (10 mm Tris–HCl, pH 7.5, 1 mm EDTA). DNA was eluted from the agarose beads by treating each sample with 100 μl of elution buffer (50 mm Tris–HCl, pH 7.5, 1 mm EDTA, 1% SDS) (at 65° for 15 min) and collected after centrifugation. Additional DNA was collected from each pellet after further treatment with 150 μl of TE/0.67% SDS (at 65° for 10 min) and centrifugation. The “input” samples were then included in the remaining protocol after increasing their volume by adding 200 μl TE/1% SDS. Each sample received NaCl (0.2 m final concentration) prior to treatment with RNAse A (1.2 μg) and protein–DNA de-crosslinking (at 65° for 5 hr). Then each sample was treated with proteinase K (10 μg) after the addition of EDTA (10 mm final concentration) and Tris–HCl, pH 8 (40 mm final concentration) (at 50° for 1 hr). Finally, DNA from each sample was purified using GFX columns (GE Healthcare). Protease inhibitors (1 mm PMSF, pepstatin 1 μg/ml, aprotinin 1 μg/ml) were included in all the buffers up to the DNA elution step.
DNA quantification by PCR:
The amount of DNA in each ChIP sample was estimated after quantitative PCR using primers specific for the putative WCC binding sites in each promoter and to a segment of the fl ORF as a control (fl 5′-CGGCCTTGGCTTCGA-3′ and 5′-GCCATTGGGCTTTGGT-3′, al-3 5′-CCCGCACGCTATGACGATA-3′ and 5′-ATAGCAAAGTGAGGTCGATTGCT-3′, frq-p 5′-CATCACTGCCCAGGTTCCA-3′ and 5′-GACGACGGCTGGCCAAT-3′, frq-d 5′-GTATCTTGAGCCTCCAGATCTCAAT-3′ and 5′-CCCGAGGCGTCCTGATG-3′, and fl-ORF 5′-GGCTTCATCGTCTTTTCCTTCA-3′ and 5′-CTTCCGAGCACCCAAGCTT-3′). Quantitative PCRs were performed with SYBR Premix Ex Taq (Perfect Real Time) (TaKaRa) in a reaction volume of 10 μl using a LightCycler 480 II (Roche). The reaction included denaturation (10 sec at 95°) and 40 cycles (5 sec at 95° and 20 sec at 60°). After each PCR, we performed a melting curve analysis to show the specific amplification of a single DNA segment and the absence of nonspecific amplified DNA. For each set of primers, we prepared a standard curve using serial dilutions of one input sample to allow the absolute quantification of the DNA amplified in each sample. The amount of DNA obtained from each sample was normalized to the amount obtained with each input sample to obtain the percentage of DNA purification by immunoprecipitation. The gene identification numbers in the Neurospora genome database (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html) are fl (NCU08726), al-3 (NCU01427), and frq (NCU02265).
RESULTS AND DISCUSSION
The fl gene is activated by short light exposures:
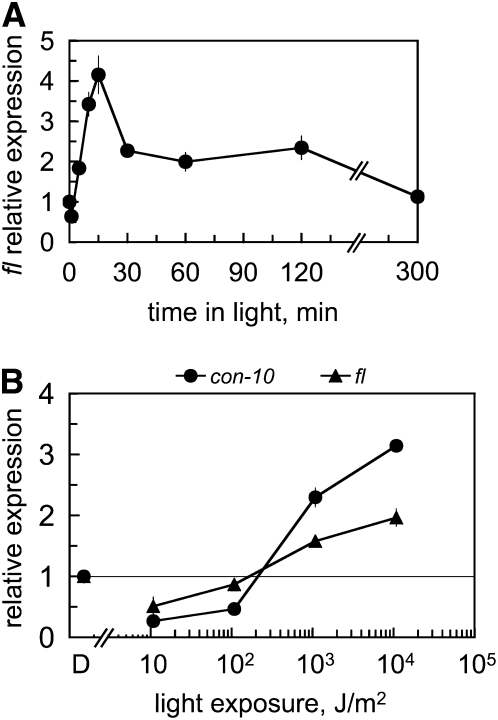
To characterize the regulation by light of fl, we exposed dark-grown vegetative mycelia of wild-type N. crassa to broad-spectrum white light. We used liquid cultures to grow Neurospora and ensure that hyphae were kept in the vegetative stage for the duration of each experiment. The exposure time ranged from 2 min to 5 hr, and light-exposed mycelia were then collected and used for RNA isolation. The presence of fl mRNA was assayed by quantitative RT–PCR relative to the amount observed in vegetative mycelia kept in the dark. We observed up to four times more fl mRNA in mycelia that had been exposed to light than in mycelia kept in the dark, confirming that fl was regulated by light in vegetative hyphae (Figure 1A). The activation of fl by light was very rapid since we detected light-dependent fl mRNA accumulation after 5 min of light. fl mRNA accumulation reached a maximum after 15 min of light and decreased with longer light exposures (Figure 1A). Light-dependent fl mRNA accumulation was not observed after 5 hr of light, presumably due to photoadaptation, a phenomenon described for other light-regulated genes like con-10 or con-6 (Lauter and Yanofsky 1993).
Figure 1.—
The fl gene is activated by light. (A) Total RNA was isolated from vegetative mycelia of the wild-type strain that had been exposed to white light (the active blue light component was 1 W/m2 blue light) for various periods or kept in the dark (0) prior to RNA isolation. (B) Threshold of gene photoactivation. Wild-type vegetative mycelia were exposed to white light of various intensities for 10 min or kept in the dark (D) prior to RNA isolation. The light exposures used were 1.08 × 104, 1.08 × 103, and 1.08 × 102 and 10.8 J/m2. A horizontal line is drawn at the position that marks the absence of light-dependent mRNA accumulation (light/dark value equal to 1) to help identify the threshold. The amount of fl, con-10, and tub-2 RNAs were determined by quantitative RT–PCR. Each fluorescent signal was first normalized to the corresponding tub-2 signal to correct for loading errors and then was normalized to the RNA sample from mycelia kept in the dark. The plot shows the average and standard error of the mean of the relative mRNA accumulation in three to nine experiments (A) or in two experiments (B). Each RNA sample was quantified in one PCR experiment.
Threshold of fl gene activation by light:
The kinetics of fl activation, in particular the detection of fl mRNA after only 5 min of light, suggested a direct activation by the WCC acting on the fl promoter. To investigate further the activation of fl by light, we measured light-dependent fl mRNA accumulation after exposing mycelia to light of different intensities and compared the threshold with that for the light-regulated gene con-10 (Lauter and Yanofsky 1993; Corrochano et al. 1995; Lee and Ebbole 1998). The activation by light of both genes had a similar threshold, ∼102 J/m2 (Figure 1B), suggesting that the same photoreceptor, WC-1 in the WCC, was involved in the regulation by light of both genes.
Blue light activates fl, and light regulation requires a white collar complex:
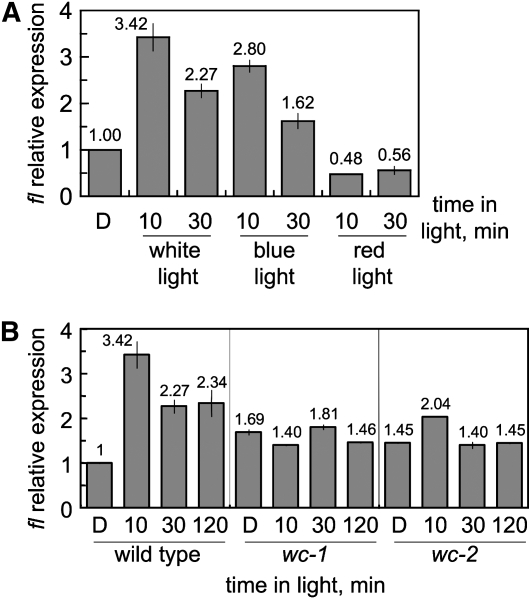
As further confirmation of the role of the blue-light photoreceptor WC-1 in fl activation, we detected light regulation of fl after exposure to blue light, but not to red light, a feature exhibited by most light responses in Neurospora (Linden et al. 1997) (Figure 2A). In addition, mutations in wc-1 or wc-2 prevented the light-dependent accumulation of fl mRNA (Figure 2B) and confirmed that the WCC is required for the activation by light of fl. We used a wc-1 allele (wc-1ER53) with a nonsense mutation that resulted in early termination and a wc-2 allele (wc-2ER33) with a single mutation that replaced a conserved glycine with glutamic acid in the zinc finger (Lee et al. 2003; Linden and Macino 1997). Both alleles should yield nonfunctional WC-1 or WC-2 proteins, and strains with these alleles have been shown to be defective in the activation by light of gene transcription (Lauter and Russo 1991; Arpaia et al. 1993; Ballario et al. 1996). The amount of fl mRNA in the wc mutants was slightly increased as compared to the amount of fl mRNA accumulated in the wild-type strain in the dark (∼1.6-fold on average), suggesting a mild repressive role for the WCC in the fl promoter in the dark (Figure 2B).
Figure 2.—
Activation of fl by blue light and the white collar complex. (A) The fl gene is activated by blue light. Total RNA was isolated from vegetative mycelia of the wild type exposed to white light (16.5 W/m2), blue light (2.6 W/m2), or red light (1.8 W/m2) for 10 or 30 min or kept in the dark (D) prior to RNA isolation. (B) The activation of fl requires the WCC. Total RNA was isolated from vegetative mycelia of the wild type and the wc mutants that had been incubated in the dark for 48 hr (22°C) and exposed to white light during 10, 30, or 120 min or kept in the dark (D) prior to RNA isolation. The amount of fl and tub-2 RNAs were determined by quantitative RT–PCR. Each fluorescent signal was first normalized to the corresponding tub-2 signal to correct for loading errors and then was normalized to the RNA sample from wild-type mycelia kept in the dark. The plot shows the average and standard error of the mean of the relative photoactivation in two to nine experiments (A) or in two experiments (B) except one experiment for the wc strains after 10 or 120 min of light. Each RNA sample was quantified in one PCR experiment.
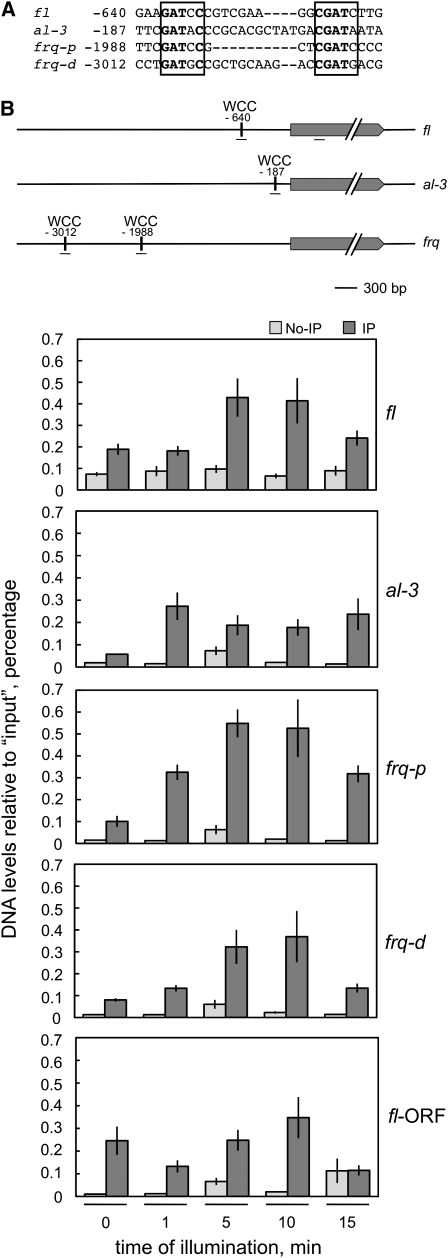
The white collar complex binds transiently to the promoter of fl:
The consensus DNA sequence for WCC binding to the promoters of the light-regulated genes frq, al-3, and vvd is GATNC–CGATN, where “N” is the same nucleotide in both repeats (He and Liu 2005), and a similar putative regulatory sequence has been identified in the upstream DNA of a set of fast light-regulated genes (Chen et al. 2009). We identified a putative WCC binding site at position −640 (from the initiation ATG) in the fl promoter (Figure 3A), which further suggested that the WCC might activate the transcription of fl after blue-light exposure. To detect light-dependent binding of the WCC to the fl promoter, we exposed wild-type mycelia to light and then performed ChIP experiments using an antibody against WC-2. Antibodies against WC-1 or WC-2 have been used routinely to detect the WCC (Froehlich et al. 2002; He et al. 2002; He and Liu 2005; Belden et al. 2007b). The amount of DNA around the putative WCC binding site of fl obtained after each ChIP experiment was assayed by quantitative PCR (Figure 3B). As controls we assayed the amount of DNA around the WCC binding sites in the promoters of al-3, the distal (frq-d) and proximal (frq-p) sites in the promoter of frq (Froehlich et al. 2002; He and Liu 2005; Belden et al. 2007b), and within the fl ORF (fl-ORF) in the same ChIP experiments (Figure 3B). WCC binding to the fl promoter was observed in mycelia that had been exposed to 5–10 min of light, but WCC binding was transient since binding was reduced to dark levels after 15 min of light (Figure 3B). As a control we observed some WCC binding within the fl ORF, but this WCC binding was not regulated by light and was not explored further (Figure 3B). The absence of WCC binding after 15 min of light was consistent with the reduced fl mRNA accumulation after 15 min of light that we have observed previously (Figure 1A) and suggests that transient binding of the WCC is responsible for the transient accumulation of fl mRNA after light exposure, as proposed for other light-regulated genes (He and Liu 2005). Transient WCC binding was observed in the distal and proximal binding sites of frq, but binding to the al-3 promoter remained high during the 15-min duration of the illumination experiments (Figure 3B). Transient WCC binding had been reported for the promoters of al-3 and frq (He and Liu 2005) and seems to occur as a consequence of the rapid phosphorylation of the WCC, which inhibits DNA binding and results in photoadaptation (He and Liu 2005). Our observation of similar transient binding of the WCC to the promoter of fl suggests that a similar molecular mechanism may regulate the activation by light of fl by the WCC.
Figure 3.—
The WCC binds transiently to the promoter of fl. (A) A putative WCC binding site in the fl promoter. The WCC binding sites in the promoters of the light-regulated genes frq (proximal site, frq-p; distal site, frq-d) and al-3 (in the complementary strand) (He and Liu 2005) are compared to a putative WCC binding site in the fl promoter. Conserved nucleotides are shown in boldface type, and the putative WCC binding sites are boxed. The nucleotide position is shown relative to the initiator ATG. (B) Chromatin immunoprecipation assays. Wild-type mycelia were exposed to white light (the active blue-light component was 1 W/m2 blue light) for the indicated times and chromatin immunoprecipitated with an antibody against WC-2 (IP) or treated without any antibody (no-IP) as a control. After immunoprecipiation, the amount of DNA around the WCC binding site of fl, al-3, frq-p, and frq-d was measured by quantitative PCR and plotted relative to the amount obtained in each corresponding “input” sample. As a control, we assayed the amount of DNA of a segment located within the fl ORF. A scheme showing the relative position of each putative WCC binding site and the corresponding ORF is included. The short horizontal lines under each gene indicate the position of the DNA segments amplified by PCR. The plot shows the average and standard error of the mean in three experiments. Each DNA sample was quantified in three PCR experiments and averaged.
The activation of fl by light does not require an active developmental pathway:
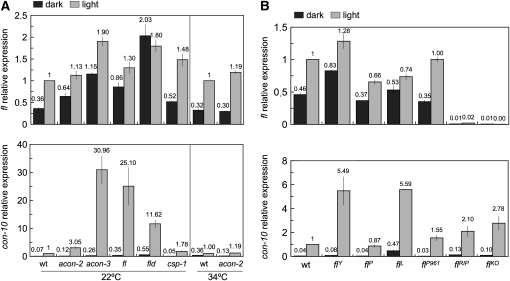
Since conidiation is induced by light, the proteins that regulate the development of conidia might participate in the regulation by light of fl. We thus examined the light-dependent accumulation of fl mRNA in the acon-2 mutant at both permissive (22°) and nonpermissive (34°) temperatures (the acon-2 mutation is a temperature-sensitive allele) and in other developmental mutants including fld, acon-3, and csp-1. In addition, we assayed light-dependent fl mRNA accumulation in different fl mutants. As a control, we assayed the light-dependent accumulation of con-10 mRNA since light regulation for this gene has been observed in several developmental mutants (Lauter and Yanofsky 1993). We observed that fl was activated by light in all the developmental mutants that we tested, with the exception of fld (Figure 4A). Interestingly, the absence of fl photoactivation in the fld strain was due to an increased accumulation of fl mRNA in the dark since the light-dependent mRNA accumulation of fl was slightly larger than in the wild-type strain (Figure 4A). This observation suggested that the fld gene product may operate as a specific repressor of fl transcription in the dark and reveals a key role for transcriptional repression in WCC-mediated photobiology in Neurospora.
Figure 4.—
Photoactivation of the fl gene in the wild-type and developmental mutants. Mycelia of the wild-type and developmental mutants (A) or different fl strains (B) were exposed to white light (the active blue-light component was 1 W/m2 blue light) for 10 min or kept in the dark prior to RNA isolation. The amount of fl, con-10, and tub-2 RNAs was determined by quantitative RT–PCR. Each fluorescent signal was first normalized to the corresponding tub-2 signal to correct for loading errors and then was normalized to the signal obtained with the wild-type strain after exposure to light. The plot shows the average and standard error of the mean of the relative photoactivation in six experiments (A) or in two experiments (B). Each RNA sample was quantified in one PCR experiment. The fl strain in A was FGSC 4241.
Developmental regulation of fl requires the acon-2 gene (Bailey-Shrode and Ebbole 2004), as expected since acon-2 is epistatic over fl (Springer and Yanofsky 1989). In addition to developmental regulation, fl expression is under the control of the circadian clock, and this clock regulation requires the product of the acon-2 gene (Correa and Bell-Pedersen 2002). However, acon-2 was not required for the activation of fl by light as we observed light-dependent fl mRNA accumulation in the acon-2 strain at both permissive and nonpermissive temperatures (Figure 4A). Our results suggest that ACON-2 specifically participates in the regulation of fl expression by development and the circadian clock, but not by light.
FL acts as a repressor of con-10 activation by light:
The amount of fl mRNA in mycelia exposed to light did not change significantly in any of the mutants blocked in conidiation (Figure 4). However, the amount of con-10 mRNA that accumulated in light-exposed mycelia was increased in strains with mutations in acon-3, fl, and fld, which suggested that the corresponding gene products should repress con-10 photoactivation (Figure 4A). The increase in the accumulation of con-10 mRNA after light exposure in these developmental mutants had been reported after RNA hybridization experiments (Lauter and Yanofsky 1993), further supporting our results.
Several alleles of fl are available, and they may help to characterize further the effect of FL on con-10 regulation by light. A strain with a complete deletion of fl, flKO, is available from the collection of N. crassa gene knockouts (Colot et al. 2006), and an additional null allele, flRIP, was obtained by repeat-induced point mutation (RIP) (Bailey and Ebbole 1998). A small deletion in flL removes the first 20 amino acids, including the translational start codon. This is probably a null allele unless translation reinitiates downstream. Two alleles produce truncated proteins that retain the zinc finger and may have some activity. They are allele flP961 with a 1-bp insertion after codon D96 that causes a frameshift and premature termination and flP with a duplication that results in a frameshift after codon T230 and a premature stop codon. Allele flY results from two point mutations that change two amino acids that must be relevant for FL activity (Bailey and Ebbole 1998). The amount of fl mRNA in fl mycelia incubated in the dark or exposed to light varied within the expected experimental variation, further confirming the absence of any role of FL in the regulation of its own gene by light (Figure 4B). As expected, we did not detect fl mRNA in the deletion strain or in the strain with the flRIP allele. However, we observed a two- to fivefold increase in the amount of light-dependent con-10 mRNA in all the fl strains except in the strain with the flP allele (Figure 4B). The presence of the first 230 amino acids, including the zinc finger, in the truncated FL protein synthesized by the flP allele may still provide sufficient activity for con-10 regulation, although this allele resulted in the absence of conidiation that is typical of the fluffy phenotype (Springer and Yanofsky 1989). On the contrary, the strain with the flY allele had a mild conidiation phenotype (Bailey and Ebbole 1998), but a high accumulation con-10 mRNA after light exposure (Figure 4B). These results suggest that different forms of FL may play different roles in the biology of Neurospora. Most of the protein is necessary for completion of development, but the amino end of the protein may still act as a repressor of gene regulation as we have observed for con-10.
Conclusion:
Our results suggest that blue light, through the WCC, induces the transcription of fl. Furthermore, we show a role for FLD as a repressor of fl transcription in the dark. We thus propose that fl is repressed by FLD in vegetative mycelia growing in the dark and that this repression is lost after light exposure and WCC activation. An additional repressing role for FL in the regulation by light of the conidiation gene con-10 is supported by the increased light-dependent mRNA accumulation in different fl mutants. The increase in fl mRNA after light exposure in vegetative mycelia should produce an increase in the amount of FL protein that should promote the activation of the conidiation pathway, as already observed in forced-expression experiments in vegetative hyphae (Bailey-Shrode and Ebbole 2004). The activation by light of fl may provide a simple mechanism for the activation by blue light of conidiation in Neurospora. Light activates sporulation in many types of fungi (Corrochano and Galland 2006), and we expect that the induction by light of genes for developmental regulators may be part of the mechanism for the regulation by light of sporulation in other fungi.
Acknowledgments
We thank W. Belden (Dartmouth Medical School), M. Freitag (Oregon State University), F. Gómez-Herreros and A. Rodríguez-Gil (University of Seville), J. Rodríguez-Romero (University of Karlsruhe), and G. Sancar (University of Heidelberg) for ChIP protocols and advice. We thank J. Loros (Dartmouth Medical School) for wc strains and M. Brunner (University of Heidelberg) for the WC-2 antibody. We thank M. Merrow (University of Groningen) and J. Avalos, D. Cánovas, and V. G. Tagua (University of Seville) for critically reading the manuscript. This work was supported by European funds (European Regional Development Fund), the Spanish Ministerio de Educación y Ciencia (BIO2006-14897), and the Regional Government (Junta de Andalucía, CVI 0119).
References
- Arpaia, G., J. J. Loros, J. C. Dunlap, G. Morelli and G. Macino, 1993. The interplay of light and the circadian clock: independent dual regulation of clock-controlled gene ccg-2(eas). Plant Physiol. 102 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, L. A., and D. J. Ebbole, 1998. The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 148 1813–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Shrode, L., and D. J. Ebbole, 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 166 1741–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballario, P., P. Vittorioso, A. Magrelli, C. Talora, A. Cabibbo et al., 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15 1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Belden, W. J., L. F. Larrondo, A. C. Froehlich, M. Shi, C. H. Chen et al., 2007. a The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 21 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden, W. J., J. J. Loros and J. C. Dunlap, 2007. b Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25 587–600. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen, D., J. C. Dunlap and J. J. Loros, 1992. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 6 2382–2394. [DOI] [PubMed] [Google Scholar]
- Brunner, M., and K. Káldi, 2008. Interlocked feedback loops of the circadian clock of Neurospora crassa. Mol. Microbiol. 68 255–262. [DOI] [PubMed] [Google Scholar]
- Chen, C. H., C. S. Ringelberg, R. H. Gross, J. C. Dunlap and J. J. Loros, 2009. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 28 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew et al., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, A., and D. Bell-Pedersen, 2002. Distinct signaling pathways from the circadian clock participate in regulation of rhythmic conidiospore development in Neurospora crassa. Eukaryot. Cell 1 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrochano, L. M., and P. Galland, 2006. Photomorphogenesis and gravitropism in fungi, pp. 233–259 in The Mycota. I. Growth, Differentiation and Sexuality, edited by U. Kües and R. Fischer. Springer-Verlag, Berlin.
- Corrochano, L. M., F. R. Lauter, D. J. Ebbole and C. Yanofsky, 1995. Light and developmental regulation of the gene con-10 of Neurospora crassa. Dev. Biol. 167 190–200. [DOI] [PubMed] [Google Scholar]
- Crosthwaite, S. K., J. C. Dunlap and J. J. Loros, 1997. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276 763–769. [DOI] [PubMed] [Google Scholar]
- Davis, R. H., 2000. Neurospora. Contributions of a Model Organism. Oxford University Press, Oxford.
- Dunlap, J. C., and J. J. Loros, 2004. The Neurospora circadian system. J. Biol. Rhythms 19 414–424. [DOI] [PubMed] [Google Scholar]
- Froehlich, A. C., Y. Liu, J. J. Loros and J. C. Dunlap, 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297 815–819. [DOI] [PubMed] [Google Scholar]
- He, Q., and Y. Liu, 2005. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 19 2888–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q., P. Cheng, Y. Yang, L. Wang, K. H. Gardner et al., 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297 840–843. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., and Y. Liu, 2007. The Neurospora crassa circadian clock. Adv. Genet. 58 25–66. [DOI] [PubMed] [Google Scholar]
- Lambreghts, R., M. Shi, W. J. Belden, D. Decaprio, D. Park et al., 2009. A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181 767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter, F. R., and V. E. A. Russo, 1991. Blue light induction of conidiation-specific genes in Neurospora crassa. Nucleic Acids Res. 19 6883–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter, F. R., and C. Yanofsky, 1993. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 90 8249–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter, F. R., V. E. Russo and C. Yanofsky, 1992. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 6 2373–2381. [DOI] [PubMed] [Google Scholar]
- Lauter, F. R., C. T. Yamashiro and C. Yanofsky, 1997. Light stimulation of conidiation in Neurospora crassa: studies with the wild-type strain and mutants wc-1, wc-2 and acon-2. J. Photochem. Photobiol. B 37 203–211. [Google Scholar]
- Lee, K., and D. J. Ebbole, 1998. Analysis of two transcription activation elements in the promoter of the developmentally regulated con-10 gene of Neurospora crassa. Fungal Genet. Biol. 23 259–268. [DOI] [PubMed] [Google Scholar]
- Lee, K., J. C. Dunlap and J. J. Loros, 2003. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, H., and G. Macino, 1997. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 16 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, H., P. Ballario and G. Macino, 1997. Blue light regulation in Neurospora crassa. Fungal. Genet. Biol. 22 141–150. [DOI] [PubMed] [Google Scholar]
- Neiss, A., T. Schafmeier and M. Brunner, 2008. Transcriptional regulation and function of the Neurospora clock gene white collar 2 and its isoforms. EMBO Rep. 9 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerngsamran, P., M. B. Murphy, S. A. Doyle and D. J. Ebbole, 2005. Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene. Mol. Microbiol. 56 282–297. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. BioEssays 15 365–374. [DOI] [PubMed] [Google Scholar]
- Springer, M. L., and C. Yanofsky, 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3 559–571. [DOI] [PubMed] [Google Scholar]
- Tan, Y., M. Merrow and T. Roenneberg, 2004. Photoperiodism in Neurospora crassa. J. Biol. Rhythms 19 135–143. [DOI] [PubMed] [Google Scholar]
- Xiang, Q., and N. L. Glass, 2004. The control of mating type heterokaryon incompatibility by vib-1, a locus involved in het-c heterokaryon incompatibility in Neurospora crassa. Fungal Genet. Biol. 41 1063–1076. [DOI] [PubMed] [Google Scholar]