Abstract
The endoplasmic reticulum (ER)-associated protein degradation (ERAD) pathway eliminates aberrant proteins from the ER. The key role of Cdc48p–Ufd1p–Npl4p is indicated by impaired ERAD in Saccharomyces cerevisiae with mutations in any of this complex's genes. We identified SSZ1 in genetic screens for cdc48-10 suppressors and show that it upregulates Cdc48p via the pleiotropic drug resistance (PDR) network. A pSSZ1 plasmid restored impaired ERAD-M of 6myc-Hmg2 in cdc48-10, ufd1-2, and npl4-1, while SSZ1 deletion had no effect. Ssz1p activates Pdr1p, the PDR master regulator. Indeed, plasmids of PDR1 or its target gene RPN4 increased cdc48-10p levels and restored ERAD-M in cdc48-10. Rpn4p regulates transcription of proteasome subunits and CDC48, thus RPN4 deletion abolished ERAD. However, the diminished proteasome level in Δrpn4 was sufficient for degrading a cytosolic substrate, whereas the impaired ERAD-M was the result of diminished Cdc48p and was restored by expression of pCDC48. The corrected ERAD-M in the hypomorphic strains of the Cdc48 partners ufd1-2 and npl4-1 by the pCDC48 plasmid, and in cdc48-10 cells by the pcdc48-10 plasmid, combined with the finding that neither pSSZ1 nor pcdc48-10 restored ERAD-L of CPY*-HA, support our conclusion that Ssz1p suppressing effects is brought about by upregulating Cdc48p.
SECRETORY and membrane proteins are synthesized, folded, and assembled in the endoplasmic reticulum (ER) and are transported along the secretory pathway to their final destinations. Essential quality control mechanisms ensure that misfolded or damaged proteins are retained in the ER and eliminated by the ER-associated protein degradation (ERAD) pathway. Such proteins are dislocated back to the cytosol, where they are tagged and degraded by the ubiquitin–proteasome system (Bonifacino and Weissman 1998; Bar-Nun 2005).
The cytosolic Cdc48p–Ufd1p–Npl4p complex is one of the key ERAD players, and mutations in any of this complex's genes result in stabilization of ERAD-M and ERAD-L substrates, as shown for 6myc-Hmg2 and CPY*, respectively (Bays et al. 2001; Ye et al. 2001; Braun et al. 2002; Jarosch et al. 2002; Rabinovich et al. 2002). The yeast Cdc48p and its mammalian homolog p97, members of the AAA-ATPases family, participate in many cellular processes, including cell cycle regulation, homotypic membrane fusion, and proteasomal degradation (Ghislain et al. 1996; Patel and Latterich 1998; Vale 2000). Cdc48p/p97 provides the driving force for pulling ERAD substrates out of the ER (Ye et al. 2001; Elkabetz et al. 2004). This function concurs with the ability of AAA-ATPases to translate ATP hydrolysis into mechanical forces (Rouiller et al. 2002) and with the underlying activity of AAA-ATPases to unfold and disassemble protein complexes (Lupas and Martin 2002).
In our search for novel ERAD factors, we screened for suppressors that, when overexpressed, restored the impaired ERAD-M of 6myc-Hmg2 in the Saccharomyces cerevisiae cdc48-10 conditional mutant. In our screen, we identified Ssz1p, a cytosolic member of the Hsp70 family. Ssz1p is tightly associated with the J-protein zuotin (Zuo1p) and the stable Zuo1p:Ssz1p complex (also known as the ribosome-associated complex, RAC) binds to the ribosome via Zuo1p, and, together with Ssbs, facilitates folding of nascent polypeptides as they exit the ribosome (Gautschi et al. 2001; Hundley et al. 2002; Gautschi et al. 2002; Shaner and Morano 2007).
Here we show that the restored ERAD-M in cdc48-10 is brought about by the RAC-independent participation of Ssz1p in the pleiotropic drug resistance (PDR) network. PDR regulates the expression of many genes in response to various cytotoxic compounds, including cycloheximide, canavanine, and cadmium. Ssz1p is a post-translational activator of the transcription factor Pdr1p (Hallstrom et al. 1998; Eisenman and Craig 2004), a PDR master regulator (Balzi et al. 1987; DeRisi et al. 2000; Devaux et al. 2001; Mamnun et al. 2002; Dohmen et al. 2007; Gulshan and Moye-Rowley 2007). Among others, the upregulated genes are involved in efflux of cytotoxic compounds (ABC and MFS transporters), stress response and, lipid metabolism (DeRisi et al. 2000; Gulshan and Moye-Rowley 2007). In particular, Pdr1p controls expression of the proteasome by upregulating Rpn4p, a transcriptional activator that binds to the proteasome-associated control element (PACE). This nonamer sequence (GGTGGCAAA) is located in the promoters of many genes including all proteasome subunits (Mannhaupt et al. 1999; Jelinsky et al. 2000; Kapranov et al. 2001; Xie and Varshavsky 2001; Owsianik et al. 2002; Ju et al. 2004; Dohmen et al. 2007; Gulshan and Moye-Rowley 2007; Hanna and Finley 2007;). Interestingly, the CDC48 gene also contains the Rpn4p-binding PACE (Mannhaupt et al. 1999; Kapranov et al. 2001). Indeed, CDC48 mRNA levels decrease upon RPN4 deletion (Karpov et al. 2008). Here we provide evidence that the pSSZ1 plasmid restores the impaired ERAD-M in mutants of the Cdc48p–Ufd1p–Npl4p complex and attribute this effect to upregulation of Cdc48p via the Pdr1p-dependent activation of Rpn4p.
MATERIALS AND METHODS
Strains and plasmids:
Yeast strains used in this study are listed in Table 1. SBN100 and SBN194 were generated by replacing the TRP1 gene with LEU2 in KFY100 (CDC48) and KFY194 (cdc48-10), respectively. Strains Δssz1, Δpdr1, Δrpn4, and their parental BY4741 were obtained from the deletion library of all nonessential genes (Giaever et al. 2002). Yeast YEp24-based 2μ genomic libraries (Carlson/Botstein library) were used in the genetic screen for cdc48-10 suppressors. The protein 6myc-Hmg2 was expressed either from the genome as indicated (Table 1) or from plasmids pRH244 (Rabinovich et al. 2002) or pER244 (generated by replacing the URA3 gene with LEU2 in pRH244). CPY*-HA (prc1-1 allele) was expressed from plasmid pBG15 (Elkabetz et al. 2004) and ΔssCPY*-GFP was expressed from plasmid POW0668 (Lipson et al. 2008). Ssz1p was overexpressed either as an untagged protein (pHE31; generously provided by E. Craig) or as an N-terminally HA-tagged version (HA-Ssz1; generously provided by J. Frydman). The hyperactive Pdr1-3p mutant (F815S; Carvajal et al. 1997), was expressed from plasmid pSK (generously provided by W. S. Moye-Rowley). Low-copy plasmids were used to express Rpn4p either as an untagged protein (p314CUP1RPN4) or as a C-terminally FLAG-tagged Rpn4 (p314CUP1RPN4-FLAG) (generously provided by Y. Xie). Cdc48p was expressed from a high-copy plasmid pKF700 (generously provided K. U. Fröhlich) or from plasmid pOO700. The latter was generated by replacing the LEU2 gene with URA3: pKF700 and a URA3-containing pBluescriptIISK+ (Stratagene) digested with BstXI were filled in by T4 polymerase, and then digested with KpnI and the excised URA3 was inserted into pKF700. The myc-tagged cdc48-10p was expressed from plasmid pDS194 generated by amplifying cdc48-10 from KFY194 genomic DNA with primers 5′-CCC GGA TCC ATG GGT GAA GAA CAT AAA CC-3′ and 5′-CCC GGT ACC CG ACTATACAAATCATCATCTTCC-3′. The PCR product was digested with BamHI and KpnI and inserted into these sites in pAMT20. All construct were sequenced.
TABLE 1.
Yeast strain list
| Strain | Genotype | Source |
|---|---|---|
| CDC48 (KFY100) | MATahis4-619 leu2-3,112 ura3-52 | Rabinovich et al. (2002) |
| cdc48-10 (KFY194) | MATalys2-801 leu2-3,112 ura3-52 cdc48-10ts | Rabinovich et al. (2002) |
| SBN100 (CDC48) | MATaKFY100 trp1∷LEU2 | This study |
| SBN194 (cdc48-10) | MATaKFY194 trp1∷LEU2 | This study |
| DF5a | MATahis3-Δ200 leu2-3,2-112 lys2-801 trp1-1(am) ura3-52 | Finley et al. (1987) |
| ufd1-2 | MATahis3-Δ200 leu2-3,2-112 lys2-801 trp1-1(am) ura3-52ufd1-2 | Finley et al. (1987) |
| NPL4 (RHY400) | MATaade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 | Bays et al. (2001) |
| npl4-1 (RHY554) | MATaade2-101 lys2-801 his3Δ200 hmg1Δ∷LYS2 hmg2Δ∷HIS3 met2 ura3-52∷PTDH3-6MYC-HMG2 hrd4-1 | Bays et al. (2001) |
| Δhrd1 (RHY965) | MATahis3Δ200 lys2-801 ade2-101 met2hmg1∷LYS2 hmg2∷HIS3 trp1∷hisG leu2Δura3-52∷6MYC HMG2 hrd1Δ∷URA3 | Hampton et al. 1996) |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Giaever et al. (2002) |
| Δrpn4 | MATaBY4741 rpn4∷KanR | Giaever et al. (2002) |
| Δpdr1 | MATaBY4741 pdr1∷KanR | Giaever et al. (2002) |
| Δssz1 | MATaBY4741 ssz1∷KanR | Giaever et al. (2002) |
Genetic screens for cdc48-10 suppressors:
The cdc48-10 strain was transformed with Yep24-based genomic libraries and grown for 3 days at 30° on SD plates lacking uracil. The resulting colonies were replica plated and incubated for an additional 3 days at 37°. Survivors were collected and transformed with plasmid pER244 expressing 6myc-Hmg2, and turnover of 6myc-Hmg2 was measured at 37°. The DNA from cells that exhibited restored ERAD was extracted and reintroduced into naive cdc48-10 cells. Plasmid DNA from the secondary transformants that exhibited restored ERAD was recovered and sequenced using primers flanking the inserts.
Growth sensitivity to cadmium:
Yeast cells, grown at 30° to 1.0 A600 in the appropriate selective media, were spotted as 10-fold serial dilutions on plates supplemented with increasing concentrations of CdCl2 (Jungmann et al. 1993; Wang and Chang 2003).
Degradation rates of 6myc-Hmg2, CPY*-HA, and ΔssCPY*-GFP:
Degradation at either 30° or 37° was followed by immunoblotting of cells (3 A600) collected and lysed at indicated time points after addition of cycloheximide (150 μg/ml), as previously described for 6myc-Hmg2 (Rabinovich et al. 2002), CPY*-HA (Elkabetz et al. 2004), or ΔssCPY*-GFP (Lipson et al. 2008). Ten percent of total cellular proteins was resolved by SDS–PAGE and electroblotted onto nitrocellulose. Blots were probed with anti-myc (clone 9E10), anti-HA (clone 12CA5), or anti-CPY (clone 10A5-B5; New Biotechnology) mouse antibodies followed by horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Jackson). The HRP was visualized by enhanced chemiluminescence (ECL) and the images were quantified by densitometry (ImageMaster 1D).
Levels of Cdc48p and Rpt1p:
Equal amounts of cells (1.5 A600) were lysed, protein levels were estimated, and equal amounts of cellular proteins were resolved by SDS–PAGE and electroblotted onto nitrocellulose. Blots were probed by the following primary antibodies: mouse anti-myc (clone 9E10) to follow the levels of myc-tagged cdc48-10p; rabbit anti-Cdc48 (generously provided by K. U. Fröhlich); chicken anti-Rpt1 (generously provided by M. Glickman). A mouse anti-actin antibody (clone C4, MP Biomedicals) was used to monitor gel loading. The HRP conjugated to the secondary antibodies goat anti-mouse IgG (Jackson), goat-anti-rabbit IgG (Sigma), or rabbit anti-chicken (Chemicon) was visualized by ECL and quantified by densitometry.
RESULTS
A genetic screen for suppressors of cdc48-10 defects in ERAD:
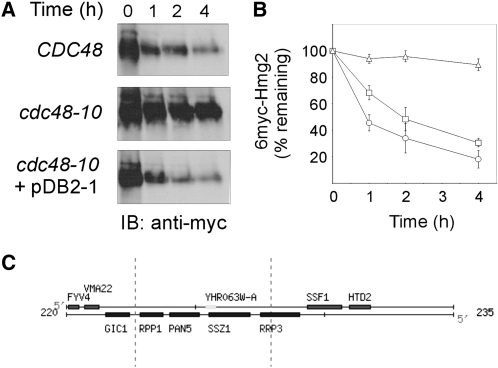
The CDC48 gene was originally identified as being involved in the cell-division cycle (Moir et al. 1982; Frohlich et al. 1991). Since mutations in CDC48 that lead to cell-division arrest were also found to hamper ERAD (Rabinovich et al. 2002), we sought to identify novel ERAD factors by screening for suppressors that, when overexpressed, allow growth of the temperature-sensitive cdc48-10 mutant under nonpermissive conditions. We transformed cdc48-10 cells with a YEp24-based 2μ yeast genomic library, and following 3 days incubation at the permissive temperature (30°), transformants were replica plated and further incubated for 3 days at the restrictive temperature (37°). Out of ∼27,000 initial transformants, 17 colonies survived the restrictive temperature and were tested for degradation of the ERAD-M substrate 6myc-Hmg2. As previously reported (Rabinovich et al. 2002), cdc48-10 cells failed to degrade 6myc-Hmg2 at 37° (Figure 1, A and B). Four out of the 17 surviving colonies exhibited restored ERAD-M at 37° (data not shown), suggesting that the temperature sensitivity of growth was not necessarily the outcome of impaired ERAD. The plasmids from these 4 colonies were isolated and individually reintroduced into naive cdc48-10 cells, to confirm their capacity to restore ERAD. One of these secondary transformants, which contained the pDB2-1 plasmid, degraded 6myc-Hmg2 at 37° at rates similar to those measured in CDC48 wild type (Figure 1, A and B).
Figure 1.—
Plasmid pDB2-1, identified in a genetic screen, restores ERAD-M in cdc48-10 mutant. Turnover of 6myc-Hmg2 was measured in wild-type CDC48 (KFY100; circles), cdc48-10 (KFY194; triangles), and cdc48-10 expressing plasmid pDB2-1 identified in the genetic screen (squares). The protein 6myc-Hmg2 was expressed from pER244. (A) Following a 90-min preincubation at 37°, cycloheximide (150 μg/ml) was added and cells further incubated at 37° were collected at the indicated time points. Cells (3 A600) were lysed, total cellular proteins were resolved by SDS-PAGE, electroblotted and probed (IB) with a mouse anti-myc antibody followed by HRP-conjugated anti-mouse IgG, and HRP was visualized by ECL. (B) Blots represented by A were quantified by densitometry and 6myc-Hmg2 decay was plotted. The remaining 6myc-Hmg2 was calculated as the percentage of its level at the time of cycloheximide addition (100%). Values shown are means ± SEM of at least five independent experiments. (C) A genetic map of the insert in plasmid pDB2-1 from the Saccharomyces Genome Database (http://www.yeastgenome.org).
The insert in pDB2-1 was identified by sequencing its boundaries and aligning the sequences against the yeast genomic database using the BLAST program (Altschul et al. 1997). pDB2-1 contained an insert of ∼5 kb of contiguous genomic DNA derived from chromosome VIII, spanning positions 222,686 to 227,948. Three full-length genes were found within this region (Figure 1C): RPP1, which encodes ribonuclease P protein 1 required for tRNA and 35S rRNA precursors processing (Stolc and Altman 1997); PAN5/YHR063C, a structural homolog of Escherichia coli panE 2-dehydropantoate 2-reductase that participates in the pantothenic acid pathway (White et al. 2001); and SSZ1, a cytosolic member of the Hsp70 family that associates with Zuo1p to form RAC, a stable complex that associates with ribosomes, and, together with Ssbs, is involved in facilitating folding of nascent polypeptides (Gautschi et al. 2001; Gautschi et al. 2002; Hundley et al. 2002; Shaner and Morano 2007). None of these genes has been previously reported to be associated with Cdc48p functions or with ERAD.
Ssz1 restores impaired ERAD in cdc48-10 but it is not required for ERAD:
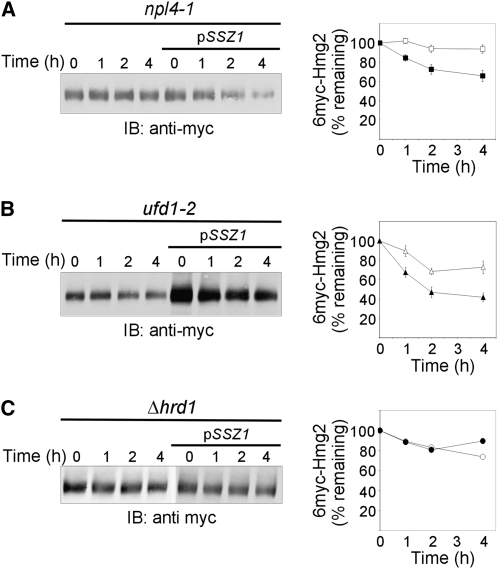
Of the proteins encoded by pDB2-1, we focused on Ssz1p, speculating that it was the most plausible candidate to be involved in ERAD because, similar to Cdc48p, Ssz1p is a cytosolic protein that contains an ATPase domain (Hallstrom et al. 1998). To directly test this hypothesis, a plasmid harboring only the SSZ1 gene was examined for its ability to restore growth and ERAD. The pSSZ1 allowed growth of cdc48-10 as well as cdc48-3, another temperature-sensitive cdc48 allele, at the restrictive 37° (data not shown). Importantly, pSSZ1 partially restored 6myc-Hmg2 degradation at 37° in cdc48-10 cells (Figure 2A). We considered the possibility that the Hsp70 Ssz1p is a novel ERAD component. To examine whether it was essential for ERAD, 6myc-Hmg2 turnover was assessed in Δssz1 cells. Clearly, deletion of SSZ1 had no effect on the degradation of 6myc-Hmg2, which still turned over rapidly in Δssz1 cells (Figure 2B).
Figure 2.—
pSSZ1 plasmid restores ERAD-M in cdc48-10 but Ssz1p is not required for ERAD-M. (A) Turnover of 6myc-Hmg2 was measured in cdc48-10 (KFY194; open triangles) and cdc48-10 expressing pSSZ1 (closed circles). The degradation of 6myc-Hmg2 was followed at 37° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments. (B) Turnover of 6myc-Hmg2 was measured in wild-type BY4741 (closed circles) and Δssz1 deletion strain derived from it (open circles). The degradation of 6myc-Hmg2 was followed at 30° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments.
Ssz1 restores impaired ERAD in ufd1-2 and npl4-1 but not in Δhrd1:
We next hypothesized that Ssz1p may act as a cytosolic chaperone, replacing the defective Cdc48p in its role in ERAD. This possibility was directly tested by introducing pSSZ1 to strains harboring mutations in Ufd1p and Npl4p, the ERAD-dedicated Cdc48p partners. Evidently, pSSZ1 partially restored the impaired 6myc-Hmg2 degradation in npl4-1 (Figure 3A) and ufd1-2 (Figure 3B) strains. This compensatory effect of pSSZ1 was restricted to members of the Cdc48p–Ufd1p–Npl4p complex, as it did not correct the impaired 6myc-Hmg2 degradation in Δhrd1 cells (Figure 3C). We could have concluded that Ssz1p participated directly in an alternative pathway bypassing Cdc48p–Npl4p–Ufd1p altogether. However, Ssz1p was not an essential ERAD component (Figure 2B), and if it acted directly, Ssz1p should interact with ERAD substrates. Nonetheless, interactions of Ssz1p with 6myc-Hmg2 was never detected (data not shown), contrary to the physical interaction of Cdc48p with 6myc-Hmg2 (Rabinovich et al. 2002). This may reflect a weak association of Ssz1p with ERAD substrates or suggests that Ssz1p plays another role in restoring ERAD when the Cdc48p–Ufd1p–Npl4p complex malfunctions.
Figure 3.—
pSSZ1 plasmid restores ERAD-M in npl4-1 and ufd1-2 mutant strains but not in Δhrd1. Turnover of 6myc-Hmg2 was measured in (A) npl4-1 (open squares) and npl4-1 expressing pSSZ1 (closed squares), (B) ufd1-2 (open triangles) and ufd1-2 expressing pSSZ1 (closed triangles), and (C) Δhrd1 (open circles) and Δhrd1 expressing pSSZ1 (closed circles). The degradation of 6myc-Hmg2 was followed at 30° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments.
The ability of Ssz1p to restore impaired ERAD could have been attributed to Cdc48p activation, facilitating proper folding of the temperature-sensitive cdc48-10p protein at the restrictive temperature. Such an activity should rely on interactions of Ssz1p with cdc4810p. Yet again, no such interaction was detected (data not shown). This, together with the unusual properties of Ssz1p as a chaperone, which functions within the Zuo1p:Ssz1p RAC complex as a J-protein that activates ATPases of other Hsp70s (Gautschi et al. 2001, 2002; Hundley et al. 2002; Shaner and Morano 2007), suggested that Ssz1p plays an indirect role in restoring ERAD.
Pdr1 and Rpn4 restore impaired ERAD in cdc48-10 but only Rpn4 is required for ERAD:
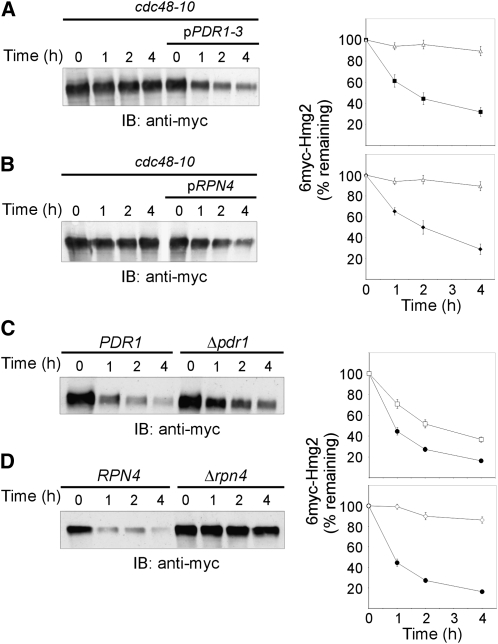
An interesting function of Ssz1p that could account for its involvement in ERAD is its participation in the PDR network. Ssz1p is a post-translational activator of the transcription factor Pdr1p (Hallstrom et al. 1998; Eisenman and Craig 2004), a master regulator of PDR (Balzi et al. 1987; DeRisi et al. 2000; Devaux et al. 2001; Mamnun et al. 2002; Dohmen et al. 2007; Gulshan and Moye-Rowley 2007). Importantly, in PDR activation Ssz1p neither functions as a chaperone nor as part of RAC, since Ssz1p and Zuo1p activate PDR only if their binding to the ribosome is abrogated (Hundley et al. 2002; Eisenman and Craig 2004). To determine whether pSSZ1 restored ERAD-M in cdc48-10 cells due to Ssz1p's role in PDR, we examined whether Pdr1p itself could restore ERAD. Indeed, when a plasmid expressing the Ssz1p-independent hyperactive PDR1-3 mutant (Carvajal et al. 1997) was introduced into cdc48-10 cells, 6myc-Hmg2 degradation at 37° was markedly accelerated (Figure 4A).
Figure 4.—
Plasmids pPDR1 and pRPN4 restore ERAD-M in cdc48-10 but only Rpn4p is required for ERAD-M. Turnover of 6myc-Hmg2 was measured in (A) cdc48-10 (KFY194; open triangles) and cdc48-10 expressing pPDR3-1 (closed squares) and (B) cdc48-10 (SBN194; open triangles) and cdc48-10 expressing pRPN4 (closed diamonds). The degradation of 6myc-Hmg2 was followed at 37° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments. Turnover of 6myc-Hmg2 was measured in wild-type BY4741 (closed circles) and the deletion strain derived from it: (C) Δpdr1 (open squares) and (D) Δrpn4 (open diamonds). The degradation of 6myc-Hmg2 was followed at 30° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments.
One of the target genes of Pdr1p is RPN4, encoding a transcriptional activator of the proteasome (Mannhaupt et al. 1999; Kapranov et al. 2001; Xie and Varshavsky 2001; Owsianik et al. 2002; Ju et al. 2004; Dohmen et al. 2007; Gulshan and Moye-Rowley 2007; Hanna and Finley 2007). Therefore, we surmised that the link of Ssz1p to ERAD was mediated by Rpn4p. This possibility was confirmed by introducing the pRPN4 plasmid into cdc48-10 cells and showing that the impaired ERAD-M of 6myc-Hmg2 at 37° was restored (Figure 4B). Taken together, these results suggest that the suppressing effect of Ssz1p in cdc48-10 can be mediated by Pdr1p and Rpn4p.
The effect of pPDR1-3 or pRPN4 plasmids in restoring 6myc-Hmg2 degradation in cdc48-10 cells could have resulted from their direct participation in ERAD. RPN4 was already demonstrated to be essential for the proteasomal elimination of two model substrates of the N-end rule and the UFD pathways (Johnson et al. 1995) and the ERAD-L substrate CPY* (Ng et al. 2000). However, Pdr1p, the upstream activator of Rpn4p, was never implicated in ERAD. Deletion of PDR1 caused some effect on ERAD-M, as indicated by the twofold prolonged half-life of 6myc-Hmg2 in Δpdr1 cells (Figure 4C). Conversely, ERAD-M was completely inhibited in Δrpn4 cells and 6myc-Hmg2 turned into a practically stable protein (Figure 4D). These results suggest that the ability of Ssz1p to restore ERAD-M when the Cdc48p is defective is mediated by Rpn4p, most likely via Pdr1p.
Cdc48p is a major limiting ERAD component in Δrpn4:
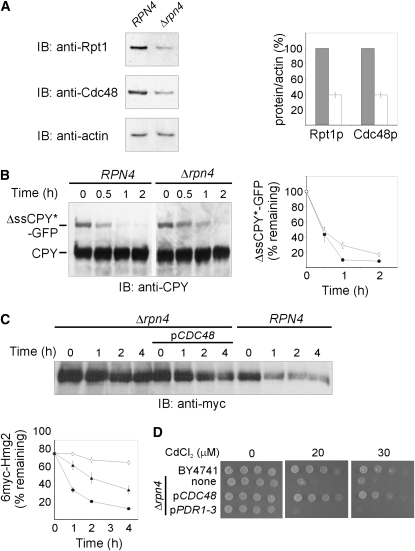
The critical function of Rpn4p in proteasomal degradation in general and ERAD in particular could be a direct consequence of its established role in regulating the expression of the proteasome subunits (Mannhaupt et al. 1999; Kapranov et al. 2001; Xie and Varshavsky 2001; Owsianik et al. 2002; Ju et al. 2004; Dohmen et al. 2007; Gulshan and Moye-Rowley 2007; Hanna and Finley 2007; Karpov et al. 2008). Hence, the impaired ERAD-M observed in Δrpn4 cells (Figure 4D) could have resulted from limiting amounts of the proteasome. Indeed, proteasomal Rpt1p was diminished to ∼40% of its wild-type levels in Δrpn4 cells (Figure 5A), consistent with decreased mRNA levels of two additional proteasome subunits, RPT4 and PRE1, in Δrpn4 cells (Karpov et al. 2008). However, CDC48 is among the many genes that contain the Rpn4p-binding PACE sequence (Mannhaupt et al. 1999; Kapranov et al. 2001) and Cdc48p was also diminished to ∼40% of its wild-type level in Δrpn4 cells (Figure 5A), consistent with a 2.5-fold decrease in its mRNA (Karpov et al. 2008). Hence, the impaired ERAD upon RPN4 deletion could be the outcome of lower levels of Cdc48p. To distinguish whether the proteasome or Cdc48p was the major ERAD limiting factor in Δrpn4 cells, proteasomal activity was examined by the turnover of the cytosolic substrate ΔssCPY*-GFP (Medicherla et al. 2004), whose degradation was independent of Cdc48p (Lipson et al. 2008). Evidently, the 2.5-fold lower levels of the proteasome in Δrpn4 cells (Figure 5A and Karpov et al. 2008) were not limiting for proteasomal proteolysis of ΔssCPY*-GFP, as its short half-life was hardly affected by RPN4 deletion (Figure 5B).
Figure 5.—
Cdc48p is a major limiting ERAD component in Δrpn4. (A) Equal amounts of wild-type RPN4 (BY4741) and Δrpn4 cells were lysed, equal amounts of total cellular proteins were resolved by SDS–PAGE, electroblotted and probed (IB) with the indicated primary antibodies, followed by HRP-conjugated secondary antibodies, and HRP was visualized by ECL. Anti-actin antibody was used to monitor cellular protein loads. Blots were quantified by densitometry, and the bars represent the ratio of Cdc48p or Rpt1p to actin in wild-type (100%; gray bars) or Δrpn4 cells (white bars). Values shown are means ± SEM of four independent experiments. (B) Proteasomal degradation of the cytosolic substrate ΔssCPY*-GFP was measured in wild-type RPN4 (BY4741; closed circles) and Δrpn4 (open diamonds) cells. Equal amounts of RPN4 or Δrpn4 cells expressing ΔssCPY*-GFP were lysed, total cellular proteins were resolved by SDS–PAGE, electroblotted and probed (IB) with a mouse anti-CPY antibody followed by HRP-conjugated anti-mouse IgG, and HRP was visualized by ECL. Endogenous CPY indicates protein loads. Blots were quantified by densitometry and the decay of ΔssCPY*-GFP was plotted. The remaining ΔssCPY*-GFP was calculated as the percentage of its level at the time of cycloheximide addition (100%). Values shown are means ± SEM of at least three independent experiments. (C) Turnover of 6myc-Hmg2 was measured in wild-type RPN4 (BY4741; closed circles), Δrpn4 (open diamonds) and Δrpn4 cells expressing pCDC48 (pKF700; closed triangles). The degradation of 6myc-Hmg2 was followed at 30° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least three independent experiments. (D) The growth of 10-fold serial dilutions of wild-type RPN4 (BY4741), Δrpn4 and Δrpn4 expressing either pCDC48 or pPDR1-3 plasmids was monitored on plates supplemented with the indicated concentrations of CdCl2.
The diminished level of Cdc48p in Δrpn4 cells (Figure 5A), along with the efficient proteasomal proteolysis of ΔssCPY*-GFP, which was independent of either Rpn4p (Figure 5B) or Cdc48p (Lipson et al. 2008), suggested that Cdc48p might be a major ERAD-M limiting component in Δrpn4 cells. Indeed, expression of the pCDC48 plasmid significantly accelerated ERAD-M in Δrpn4 cells (Figure 5C). However, the shortened half-life of 6myc-Hmg2 was still longer than that measured in the wild-type RPN4 strain, indicating that Cdc48p may not be the only limiting Rpn4p target involved in ERAD. Impaired degradation of abnormal proteins by the ubiquitin–proteasome system is also reflected by sensitivity to cadmium (Jungmann et al. 1993). Cadmium reacts with thiol groups and can displace zinc, iron, or copper from certain metalloproteins (Vido et al. 2001). The correlation between cadmium sensitivity and ER stress was inferred from activation of the unfolded protein response (Urano et al. 2002) and upregulation of ERAD-related proteins, such as Cdc48p, Kar2/BiP/GRP78, and 26S proteasome subunits, in response to cadmium (Vido et al. 2001). Increased cadmium sensitivity upon SSZ1 deletion was previously reported but Ssz1p-mediated copper tolerance indicated that neither PDR1 nor PDR5 was involved (Kim et al. 2001). We found that Δrpn4 cells were highly sensitive to cadmium (Figure 5D). In accordance with the remarkable sensitivity of Δrpn4 cells to many xenobiotics, which exceeds the sensitivity of PDR1 deletion mutants (Teixeira et al. 2008), we show that this increased sensitivity was not alleviated by the pPDR1-3 plasmid (Figure 5D), although Pdr1p upregulates ABC transporters and other genes that participate in efflux of cytotoxic compounds (DeRisi et al. 2000; Gulshan and Moye-Rowley 2007). Conversely, the cadmium hypersensitivity of Δrpn4 cells was alleviated by the pCDC48 plasmid (Figure 5D), correlating the cadmium sensitivity of Δrpn4 cells with impaired ERAD that results from limiting amounts of Cdc48p rather than insufficient proteasome. Interestingly, we found that the cdc48-10 and the Δrpn4 are synthetically lethal mutations (data not shown), indicating that cells can tolerate defective Cdc48p or diminished levels of this essential AAA-ATPase but cannot survive when the amounts of the compromised Cdc48p are limiting.
ERAD-M but not ERAD-L is restored by excess cdc48-10p, or Cdc48p in cdc48-10, or the hypomorphic ufd1-2 and npl4-1 strains:
Our results thus far demonstrate that Cdc48p level is regulated by Rpn4p and that Cdc48p is the major limiting ERAD-M factor in Δrpn4 cells. We also show that pRPN4, pPDR1, and pSSZ1 similarly restore ERAD-M in cdc48-10 cells. Combined, these findings could be explained by the ability of Ssz1p to increase the level of cdc48-10p via the post-translational activation of Pdr1p, which, in turn, activates RPN4 transcription, hence upregulating cdc48-10 expression. Indeed, we observed ∼70% increase in the levels of the cdc48-10p protein in cdc48-10 cells expressing pSSZ1, ∼40% in cells expressing pPDR1-3 and nearly 180% increase in cells expressing pRPN4 (Figure 6A). These findings suggested that ERAD-M could be restored by an excess of the mutated cdc48-10p protein that compensated for its poor activity and somewhat reduced level in cdc48-10 cells (Figure 6A). To directly test this possibility, we cloned the cdc48-10 gene and introduced the pcdc48-10 plasmid into cdc48-10 cells. Clearly, the approximately twofold excess cdc48-10p (Figure 6A) partially restored the impaired ERAD-M of 6myc-Hmg2 at 37° (Figure 6B). Sequencing of pcdc48-10 identified two missense mutations, P257L and T413R. These substitutions may hamper hexamerization, since P257 in the D1 Walker A motif is highly conserved and proximal to the ATP-binding K261. In mammalian p97, D1 and its ATP-binding lysine participate in hexamerization (Wang et al. 2003; DeLaBarre et al. 2006). Thus, hexamerization of cdc48-10p may nevertheless be driven by its excess upon pcdc48-10 expression.
Figure 6.—
Increased cdc48-10p protein levels restore ERAD-M in cdc48-10 cells. (A) Equal amounts of total cellular proteins from CDC48 wild-type and cdc48-10 cells or cdc48-10 cells expressing pPDR1-3, pSSZ1, pRPN4, or pcdc48-10 plasmids were analyzed as describe in Figure 5A. Total amounts of wild-type Cdc48p or cdc48-10p proteins were probed with anti-Cdc48. Anti-actin was used to monitor protein load. Blots were quantified by densitometry, and the bars represent the ratio of Cdc48p or cdc48-10p to actin, with the ratio in naive cdc48-10 set as 100%. Values shown are means of three independent experiments ± SEM (B) Turnover of 6myc-Hmg2 was measured in cdc48-10 (SBN194; open symbols) and cdc48-10 expressing excess cdc48-10 from pcdc48-10 (pDS194; closed symbols). The degradation of 6myc-Hmg2 was followed at 37° (triangles), as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments. The myc-tagged cdc48-10 expressed from the plasmid is marked by an asterisk.
The ERAD-dedicated partners of Cdc48p are Npl4p and Ufd1p. Therefore, the ability of pSSZ1 to restore ERAD-M in the npl4-1 or ufd1-2 strains (Figure 3) could have similarly resulted from upregulating npl4-1 and ufd1-2 expression. Although no PACE was found in the NPL4 or UFD1 promoters, UFD1 is considered among the Rpn4p-target genes (Beyer et al. 2006; Metzger and Michaelis 2009). Alternatively, the reduced activity of the entire Cdc48p–Npl4p–Ufd1p complex in the npl4-1 or ufd1-2 strains may have resulted from impaired interaction of the mutated hypomorphic component with Cdc48p. If this was the case, the activity of the Cdc48p–Npl4p–Ufd1p complex should be rescued by an excess of Cdc48p. Indeed, our results show that the impaired ERAD-M of 6myc-Hmg2 in npl4-1 (Figure 7A) and ufd1-2 (Figure 7B) was restored upon expression of the pCDC48 plasmid. This suggests that excess Cdc48p protein may compensate for its weakened interaction with the mutant npl4-1p and ufd1-2p proteins, thus providing a plausible explanation for the restoring effect of the pSSZ1 plasmid by upregulating Cdc48p.
Figure 7.—
CDC48 plasmid restores ERAD in npl4-1 and ufd1-2 hypomorphic mutant strains. Turnover of 6myc-Hmg2 was measured in (A) wild-type NPL4 (open circles), npl4-1 (open squares), and npl4-1 expressing pCDC48 (pOO700; closed squares). (B) Wild-type UFD1 (DF5a; open diamonds), ufd1-2 (open triangles), and ufd1-2 expressing pCDC48 (pOO700; closed triangles). The degradation of 6myc-Hmg2 was followed at 30° as described in Figure 1A and plotted as described in Figure 1B. Values shown are means ± SEM of at least four independent experiments.
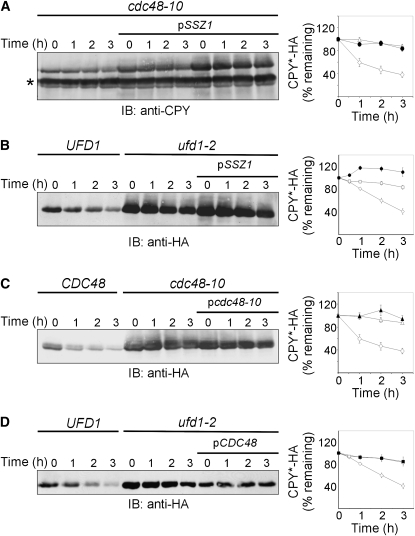
ERAD-L and ERAD-M are distinguished by their substrates' topology and although they share some components, other proteins are specific to ERAD-L and are not required for ERAD-M (Carvalho et al. 2006; Ismail and Ng 2006; Nakatsukasa et al. 2008; Nakatsukasa and Brodsky 2008). Our initial attempt to restore the impaired ERAD-L with pSSZ1 by following degradation of CPY*-HA has failed in either cdc48-10 (Figure 8A) or ufd1-2 (Figure 8B) strains. Thus, we speculated that we might have identified an ERAD-M-specific component. However, as the evidence accumulated that pSSZ1 restored ERAD-M by upregulating Cdc48p, the disparate effects of pSSZ1 on ERAD-M and ERAD-L were puzzling because both pathways converge at the Cdc48p–Ufd1p–Npl4p complex (Ismail and Ng 2006; Nakatsukasa and Brodsky 2008). This conundrum was resolved by our finding that neither pcdc48-10 (Figure 8C) nor pCDC48 (Figure 8D) restored the impaired degradation of CPY*-HA in cdc48-10 or ufd1-2 strains, respectively. Therefore, the correlation between excess Ssz1p and excess Cdc48p stands firm, suggesting that the ability of Ssz1p to restore ERAD is attributed mainly to its capacity to upregulate Cdc48p.
Figure 8.—
Not pSSZ1 or pcdc48-10 or pCDC48 plasmids restore ERAD-L in either cdc48-10 or ufd1-2 strains. Turnover of CPY*-HA was measured at 37° (A and C) in wild-type (C; CDC48; open diamonds), cdc48-10 (A and C; open triangles), cdc48-10 expressing pSSZ1 (A; closed circles) or cdc48-10 expressing pcdc48-10 (C; closed triangles). Turnover of CPY*-HA was measured at 30° (B and D) in wild-type (UFD1; open diamonds), ufd1-2 (open squares), ufd1-2 expressing pSSZ1 (B; closed circles), or ufd1-2 expressing pcdc48-10 (D; closed squares). The degradation of CPY*-HA was monitored by the cycloheximide chase, as described in Figure 1A. Cellular proteins resolved by SDS–PAGE were probed (IB) with mouse anti-CPY (A) or anti-HA (B–D) antibodies. Endogenous CPY, marked by an asterisk, indicates protein loads. The remaining CPY*-HA was calculated as described in Figure 1B. Values are means ± SEM of at least three independent experiments.
DISCUSSION
Our genetic screen for suppressors that restore defective ERAD in cdc48-10 cells identified Ssz1p. Being a member of the Hsp70 family, we considered the possibility that Ssz1p acted as a cytosolic chaperone that functions in ERAD by either activating or replacing the defective Cdc48p. The ability of Ssz1p to partially restore the impaired 6myc-Hmg2 degradation in ufd1-2 and npl4-1 mutants could have suggested that Ssz1p replaced the entire Cdc48p–Ufd1p–Npl4p complex in its role in ERAD. However, Ssz1p was not an essential ERAD factor and we could not detect any interaction of Ssz1p with the ERAD-M substrate 6myc-Hmg2. Moreover, Ssz1p is an unusual chaperone (Gautschi et al. 2001; Gautschi et al. 2002; Hundley et al. 2002; Shaner and Morano 2007), and therefore, we attributed its ability to correct ERAD to other functions of this Hsp70 member.
On the basis of the RAC-independent participation of Ssz1p in the PDR network, we examined whether other PDR members might be linked to ERAD. Indeed, Pdr1p and Rpn4p, which operate downstream of Ssz1p along the same PDR activation path, exerted similar suppression of the ERAD defects in the cdc48-10 cells. Thus, PDR upregulation could compensate for the defective Cdc48p in ERAD. A connection between ERAD and PDR was previously suggested by the observation that the cytosolic Hsp70 Ssa1p, which was implicated in ERAD (Huyer et al. 2004), interacted with Pdr3p, repressed its activity, and downregulated the expression of its target gene PDR5 (Shahi et al. 2007). Clearly, Pdr1p and Rpn4p, which are transcriptional activators that operate in the nucleus, are highly unlikely to play a direct role in ERAD. However, our finding that Rpn4p was essential for ERAD suggested that the suppressing effects of Ssz1p and Pdr1p were mediated through Rpn4p.
The involvement of Rpn4p in ERAD-M of 6myc-Hmg2 (Figure 4) and ERAD-L of CPY*-HA (Ng et al. 2000) is consistent with its well-established role as a transcriptional activator of proteasome subunit genes. Hence, stabilization of ERAD substrates in the Δrpn4 could have resulted from a globally reduced proteasomal degradation, as was shown for N-end rule and ubiquitin fusion degradation (UFD) model substrates (Johnson et al. 1995). However, in Δrpn4 cells, the Cdc48p-independent proteasomal degradation of the cytosolic ΔssCPY*-GFP proceeded unabated and expression of CDC48 plasmid partially restored the impaired ERAD-M and alleviated the hypersensitivity to cadmium (Figure 5), strongly suggesting that Cdc48p rather than the proteasome was a major limiting ERAD factor in Δrpn4 cells. Indeed, the level of the Cdc48 mRNA (Karpov et al. 2008) and protein (Figure 5) was similarly diminished in Δrpn4 cells. This agrees with the presence of Rpn4p-binding PACE sequence in the CDC48 promoter and its contribution to the CDC48 gene expression (Mannhaupt et al. 1999; Kapranov et al. 2001). Interestingly, the beneficial effects of Cdc48p in Δrpn4 cells were manifested mainly in ERAD, since CDC48 expression in these cells did not correct their cell cycle abnormalities (Xie and Varshavsky 2001).
Inasmuch as the critical outcome of SSZ1 expression is the Rpn4p-dependent upregulation of Cdc48p but not the proteasome, it is puzzling that Pdr1p and Ssz1p are not essential for ERAD, since Rpn4p itself is a short-lived substrate of the proteasome (Xie and Varshavsky 2001) that needs to be continuously replenished. This can be explained by the multitude of stress-related factors that regulate RPN4 gene transcription (Owsianik et al. 2002; Hahn et al. 2006; Dohmen et al. 2007; Gulshan and Moye-Rowley 2007; Hanna and Finley 2007). These include Pdr1p and its inducible homolog Pdr3p (Gulshan and Moye-Rowley 2007), Yap1p, a key transcriptional regulator of oxidative stress response (Rodrigues-Pousada et al. 2004) and cadmium tolerance (Wu et al. 1993) that contains PACE (Fleming et al. 2002) and requires Rpn4p for its full activation (Teixeira et al. 2008), and the heat shock transcription factor HSF, which regulates PDR3, RPN4, and Rpn4p target genes in response to heat shock (Hahn et al. 2006).
The notion that Ssz1 restores ERAD-M in cdc48-10, ufd1-2, and npl4-1 cells as a result of Rpn4p-dependent upregulation of Cdc48p was supported by our findings that the levels of the cdc48-10p protein indeed increased upon expression of the pSSZ1, pPDR1-3, and pRPN4 plasmids, and by the partially restored degradation of 6myc-Hmg2 in the cdc48-10 mutant upon a twofold increase in cdc48-10p level when expressed from pcdc48-10 plasmid (Figure 6). This indicates that an excess of this mutant protein can compensate for its sluggish activity and somewhat reduced levels in the cdc48-10 strain, possibly driving hexamerization of the cdc48-10 (P257L/T413R) mutant. Likewise, the pCDC48 plasmid partially restored the impaired ERAD-M in the ufd1-2 and npl4-1 mutant strains (Figure 7), indicating that excess wild-type Cdc48p protein can compensate for weak interactions with its hypomorphic partners in forming the Cdc48p–Ufd1p–Npl4p complex.
Finally, we show that pSSZ1 can restore ERAD-M but not ERAD-L, as 6myc-Hmg2 is classified as an ERAD-M substrate (Hampton et al. 1996; Sato and Hampton 2006). Although ERAD-L, ERAD-M, and ERAD-C converge at the p97/Cdc48p–Ufd1p–Npl4p complex (Carvalho et al. 2006; Ismail and Ng 2006; Christianson et al. 2008; Nakatsukasa and Brodsky 2008; Wang and Ng 2008), and Ssz1p restores ERAD-M by upregulating a key component of this common complex, its inability to restore ERAD-L may suggest a quantitative difference. Namely, larger amounts of Cdc48p are required for ERAD-L as compared to ERAD-M. This possibility is supported by the effect of p97 on ERAD-M of cystic fibrosis transmembrane conductance regulator (CFTR) in a reconstituted cell-free system, where p97 augmented degradation but was not obligatorily required (Carlson et al. 2006). We found that neither excess cdc48-10p in cdc48-10 strain nor excess Cdc48p in ufd1-2 strain could restore ERAD-L (Figure 8). Nonetheless, the correlation between pSSZ1 expression, excess Cdc48p protein, and restored ERAD-M remains. Interestingly, a unique role for Rpn4p in tolerating ERAD-M substrates was recently reported (Metzger and Michaelis 2009). Among the 67 genes induced by the misfolded membrane Ste6p* (Metzger and Michaelis 2009), 23 are regulated by Rpn4p (Mannhaupt et al. 1999; Fleming et al. 2002; Beyer et al. 2006), and many of them encode for proteasome subunits and additional putative members of the ubiquitin–proteasome related pathways (Metzger and Michaelis 2009). By showing that Δrpn4 cells are not sensitive to tunicamycin, these authors concluded that only misfolded membrane proteins require Rpn4p, further suggesting that the particular sensitivity of Δrpn4 cells to ER stress is due to the limiting proteasome (Metzger and Michaelis 2009). Our results propose that the inability of Δrpn4 cells to handle ERAD-M and to tolerate cadmium or defective Cdc48p is due to the role of Rpn4p in regulating Cdc48p. Thus, misfolded membrane proteins may provide a particular challenge to cells with impaired or limiting Cdc48p, which can be restored by Ssz1p, whereas ERAD-L requires even higher levels of Cdc48p that cannot be provided by Ssz1p.
Acknowledgments
We are indebted to W. S. Moye-Rowley, R. Hampton, E. Craig, J. Brodsky, J. Frydman, Y. Xie, M. Glickman, and K. U. Fröhlich for the gifts of plasmids, strains, and antibodies. We are grateful to E. Grinboim, Y. Manaster, F. Yerusalemski, M. Shaked, G. Alalouf, H. Nelson, Y. Mazor, and G. Bosis for their excellent technical help. We thank J. Roitelman, N. Nelson, E. Nachliel, and members of our laboratory for critical reading of the manuscript. Initial stages of this study were supported (in part) by grants from the Israel Science Foundation (to E.R.) and the United States–Israel Binational Science Foundation (to S.B.) and the Chief Scientist Office of the Ministry of Health, Israel (grant no. 3-3211 to S.B.).
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux and A. Goffeau, 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 262 16871–16879. [PubMed] [Google Scholar]
- Bar-Nun, S., 2005. The role of p97/Cdc48p in endoplasmic reticulum-associated degradation: from the immune system to yeast. Curr. Top. Microbiol. Immunol. 300 95–125. [DOI] [PubMed] [Google Scholar]
- Bays, N. W., S. K. Wilhovsky, A. Goradia, K. Hodgkiss-Harlow and R. Y. Hampton, 2001. HRD4/NPL4 Is Required for the Proteasomal Processing of Ubiquitinated ER Proteins. Mol. Biol. Cell 12 4114–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, A., C. Workman, J. Hollunder, D. Radke, U. Moller et al., 2006. Integrated assessment and prediction of transcription factor binding. PLoS Comput. Biol. 2 e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J. S., and A. M. Weissman, 1998. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu. Rev. Cell. Dev. Biol. 14 19–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, S., K. Matuschewski, M. Rape, S. Thoms and S. Jentsch, 2002. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, E. J., D. Pitonzo and W. R. Skach, 2006. p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J. 25 4557–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal, E., H. B. Van Den Hazel, A. Cybularz-Kolaczkowska, E. Balzi and A. Goffeau, 1997. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol. Gen. Genet. 256 406–415. [DOI] [PubMed] [Google Scholar]
- Carvalho, P., V. Goder and T. A. Rapoport, 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126 361–373. [DOI] [PubMed] [Google Scholar]
- Christianson, J. C., T. A. Shaler, R. E. Tyler and R. R. Kopito, 2008. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 10 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre, B., J. C. Christianson, R. R. Kopito and A. T. Brunger, 2006. Central Pore Residues Mediate the p97/VCP Activity Required for ERAD. Mol. Cell. 22 451–462. [DOI] [PubMed] [Google Scholar]
- DeRisi, J., H. B. Van Den, P. Marc, E. Balzi, P. Brown et al., 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470 156–160. [DOI] [PubMed] [Google Scholar]
- Devaux, F., P. Marc, C. Bouchoux, T. Delaveau, I. Hikkel et al., 2001. An artificial transcription activator mimics the genome-wide properties of the yeast Pdr1 transcription factor. EMBO Rep. 2 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R. J., I. Willers and A. J. Marques, 2007. Biting the hand that feeds: Rpn4-dependent feedback regulation of proteasome function. Biochim. Biophys. Acta 1773 1599–1604. [DOI] [PubMed] [Google Scholar]
- Eisenman, H. C., and E. A. Craig, 2004. Activation of pleiotropic drug resistance by the J-protein and Hsp70-related proteins, Zuo1 and Ssz1. Mol. Microbiol. 53 335–344. [DOI] [PubMed] [Google Scholar]
- Elkabetz, Y., I. Shapira, E. Rabinovich and S. Bar-Nun, 2004. Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplamic reticulum-bound p97/Cdc48p and proteasome. J. Biol. Chem. 279 3980–3989. [DOI] [PubMed] [Google Scholar]
- Finley, D., E. Ozkaynak and A. Varshavsky, 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48 1035–1046. [DOI] [PubMed] [Google Scholar]
- Fleming, J. A., E. S. Lightcap, S. Sadis, V. Thoroddsen, C. E. Bulawa et al., 2002 Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. USA 99 1461–1466. [DOI] [PMC free article] [PubMed]
- Frohlich, K. U., H. W. Fries, M. Rudiger, R. Erdmann, D. Botstein et al., 1991. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi, M., H. Lilie, U. Funfschilling, A. Mun, S. Ross et al., 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. USA 98 3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi, M., A. Mun, S. Ross and S. Rospert, 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. USA 99 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain, M., R. J. Dohmen, F. Levy and A. Varshavsky, 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin- mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15 4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Gulshan, K., and W. S. Moye-Rowley, 2007. Multidrug resistance in fungi. Eukaryot. Cell 6 1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, J. S., D. W. Neef and D. J. Thiele, 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 60 240–251. [DOI] [PubMed] [Google Scholar]
- Hallstrom, T. C., D. J. Katzmann, R. J. Torres, W. J. Sharp and W. S. Moye-Rowley, 1998. Regulation of transcription factor Pdr1p function by an Hsp70 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton, R. Y., R. G. Gardner and J. Rine, 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3- methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7 2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, J., and D. Finley, 2007. A proteasome for all occasions. FEBS Lett. 581 2854–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley, H., H. Eisenman, W. Walter, T. Evans, Y. Hotokezaka et al., 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. USA 99 4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer, G., W. F. Piluek, Z. Fansler, S. G. Kreft, M. Hochstrasser et al., 2004. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279 38369–38378. [DOI] [PubMed] [Google Scholar]
- Ismail, N., and D. T. Ng, 2006. Have you HRD? Understanding ERAD is DOAble! Cell 126 237–239. [DOI] [PubMed] [Google Scholar]
- Jarosch, E., C. Taxis, C. Volkwein, J. Bordallo, D. Finley et al., 2002. Protein dislocation from the ER requires polyubiquitination and the AAA- ATPase Cdc48. Nat. Cell Biol. 4 134–139. [DOI] [PubMed] [Google Scholar]
- Jelinsky, S. A., P. Estep, G. M. Church and L. D. Samson, 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., P. C. Ma, I. M. Ota and A. Varshavsky, 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270 17442–17456. [DOI] [PubMed] [Google Scholar]
- Ju, D., L. Wang, X. Mao and Y. Xie, 2004. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 321 51–57. [DOI] [PubMed] [Google Scholar]
- Jungmann, J., H. A. Reins, C. Schobert and S. Jentsch, 1993. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature 361 369–371. [DOI] [PubMed] [Google Scholar]
- Kapranov, A. B., M. V. Kuriatova, O. V. Preobrazhenskaia, V. V. Tiutiaeva, R. Shtuka et al., 2001. Isolation and identification of PACE-binding protein rpn4–a new transcription activator, participating in regulation of 26S proteosome and other genes. Mol. Biol. 35 420–431. [PubMed] [Google Scholar]
- Karpov, D. S., S. A. Osipov, O. V. Preobrazhenskaya and V. L. Karpov, 2008. Rpn4p is a positive and negative transcriptional regulator of the ubiquitin–proteasome system. Mol. Biol. (Mosk) 42 456–462. [PubMed] [Google Scholar]
- Kim, D. Y., W. Y. Song, Y. Y. Yang and Y. Lee, 2001. The role of PDR13 in tolerance to high copper stress in budding yeast. FEBS Lett. 508 99–102. [DOI] [PubMed] [Google Scholar]
- Lipson, C., G. Alalouf, M. Bajorek, E. Rabinovich, A. Atir-Lande et al., 2008. A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J. Biol. Chem. 283 7166–7175. [DOI] [PubMed] [Google Scholar]
- Lupas, A. N., and J. Martin, 2002. AAA proteins. Curr. Opin. Struct. Biol. 12 746–753. [DOI] [PubMed] [Google Scholar]
- Mamnun, Y. M., R. Pandjaitan, Y. Mahe, A. Delahodde and K. Kuchler, 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 46 1429–1440. [DOI] [PubMed] [Google Scholar]
- Mannhaupt, G., R. Schnall, V. Karpov, I. Vetter and H. Feldmann, 1999. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450 27–34. [DOI] [PubMed] [Google Scholar]
- Medicherla, B., Z. Kostova, A. Schaefer and D. H. Wolf, 2004. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 5 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger, M. B., and S. Michaelis, 2009. Analysis of quality control substrates in distinct cellular compartments reveals a unique role for Rpn4p in tolerating misfolded membrane proteins. Mol. Biol. Cell 20 1006–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir, D., S. E. Stewart, B. C. Osmond and D. Botstein, 1982. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics 100 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa, K., and J. L. Brodsky, 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa, K., G. Huyer, S. Michaelis and J. L. Brodsky, 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D. T., E. D. Spear and P. Walter, 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianik, G., I. L. Balzi and M. Ghislain, 2002. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol. Microbiol. 43 1295–1308. [DOI] [PubMed] [Google Scholar]
- Patel, S., and M. Latterich, 1998. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8 65–71. [PubMed] [Google Scholar]
- Rabinovich, E., A. Kerem, K. U. Frohlich, N. Diamant and S. Bar-Nun, 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada, C. A., T. Nevitt, R. Menezes, D. Azevedo, J. Pereira et al., 2004. Yeast activator proteins and stress response: an overview. FEBS Lett. 567 80–85. [DOI] [PubMed] [Google Scholar]
- Rouiller, I., B. DeLaBarre, A. P. May, W. I. Weis, A. T. Brunger et al., 2002. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 9 950–957. [DOI] [PubMed] [Google Scholar]
- Sato, B. K., and R. Y. Hampton, 2006. Yeast Derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast 23 1053–1064. [DOI] [PubMed] [Google Scholar]
- Shahi, P., K. Gulshan and W. S. Moye-Rowley, 2007. Negative transcriptional regulation of multidrug resistance gene expression by an Hsp70 protein. J. Biol. Chem. 282 26822–26831. [DOI] [PubMed] [Google Scholar]
- Shaner, L., and K. A. Morano, 2007. All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones 12 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V., and S. Altman, 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 11 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, M. C., P. J. Dias, T. Simoes and I. Sa-Correia, 2008. Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem. Biophys. Res. Commun. 367 249–255. [DOI] [PubMed] [Google Scholar]
- Urano, F., M. Calfon, T. Yoneda, C. Yun, M. Kiraly et al., 2002. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 158 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale, R. D, 2000. AAA Proteins. Lords of the ring. J. Cell Biol. 150 F13–F20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vido, K., D. Spector, G. Lagniel, S. Lopez, M. B. Toledano et al., 2001. A proteome analysis of the cadmium response in Saccharomyces cerevisiae. J. Biol. Chem. 276 8469–8474. [DOI] [PubMed] [Google Scholar]
- Wang, Q., and A. Chang, 2003. Substrate recognition in ER-associated degradation mediated by Eps1, a member of the protein disulfide isomerase family. EMBO J. 22 3792–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., C. Song, X. Yang and C. C. H. Li, 2003. D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J. Biol. Chem. 278 32784–32793. [DOI] [PubMed] [Google Scholar]
- Wang, S., and D. T. Ng, 2008. Lectins sweet-talk proteins into ERAD. Nat. Cell Biol. 10 251–253. [DOI] [PubMed] [Google Scholar]
- White, W. H., P. L. Gunyuzlu and J. H. Toyn, 2001. Saccharomyces cerevisiae is capable of de Novo pantothenic acid biosynthesis involving a novel pathway of beta-alanine production from spermine. J. Biol. Chem. 276 10794–10800. [DOI] [PubMed] [Google Scholar]
- Wu, A., J. A. Wemmie, N. P. Edgington, M. Goebl, J. L. Guevara et al., 1993. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J. Biol. Chem. 268 18850–18858. [PubMed] [Google Scholar]
- Xie, Y., and A. Varshavsky, 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl. Acad. Sci. USA 98 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., H. H. Meyer and T. A. Rapoport, 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414 652–656. [DOI] [PubMed] [Google Scholar]