Abstract
Drosophila Raf (DRaf) contains an extended N terminus, in addition to three conserved regions (CR1–CR3); however, the function(s) of this N-terminal segment remains elusive. In this article, a novel region within Draf's N terminus that is conserved in BRaf proteins of vertebrates was identified and termed conserved region N-terminal (CRN). We show that the N-terminal segment can play a positive role(s) in the Torso receptor tyrosine kinase pathway in vivo, and its contribution to signaling appears to be dependent on the activity of Torso receptor, suggesting this N-terminal segment can function in signal transmission. Circular dichroism analysis indicates that DRaf's N terminus (amino acids 1–117) including CRN (amino acids 19–77) is folded in vitro and has a high content of helical secondary structure as predicted by proteomics tools. In yeast two-hybrid assays, stronger interactions between DRaf's Ras binding domain (RBD) and the small GTPase Ras1, as well as Rap1, were observed when CRN and RBD sequences were linked. Together, our studies suggest that DRaf's extended N terminus may assist in its association with the upstream activators (Ras1 and Rap1) through a CRN-mediated mechanism(s) in vivo.
EVOLUTIONARILY conserved receptor tyrosine kinase (RTK) signaling pathways function in fundamental cellular processes including differentiation, proliferation, and cell survival in eukaryotes (Schlessinger 2000). The Raf serine/threonine kinase, as a key component of RTK signaling modules, plays a central role in transmitting upstream stimuli to the nucleus (Daum et al. 1994). Cyclic control of Raf depends on activities of GTPases, kinases, phosphatases, and scaffold proteins (Kolch 2000; Chong et al. 2001; Morrison 2001; Dhillon et al. 2002; Raabe and Rapp 2002). Clues to these regulatory events were derived from the identification of conserved regions/motifs/sites. However, the mechanisms that modulate Raf serine/threonine kinases are complicated and remain elusive. Mammals have three Raf isoforms, ARaf, Braf, and CRaf. They share a similar primary structure consisting of three conserved regions (CR1, CR2, and CR3). Conserved region 1 (CR1), where a Ras binding domain (RBD) and a cysteine-rich domain (CRD) reside, is required for Ras–Raf interaction. CR2, a serine/threonine-rich region, contains a 14-3-3 binding site. CR1 and CR2 are embedded in the regulatory N-terminal half of Raf proteins, while CR3, including the catalytic kinase region and an additional 14-3-3 binding site, resides in the C terminus (reviewed by Wellbrock et al. 2004). In addition to these three conserved regions, BRaf has an extended amino-terminal segment followed by CR1 (Terai and Matsuda 2006; Fischer et al. 2007). However, studies of BRaf regulation have mainly focused on CR1, CR2, and CR3 with little attention, thus far, given to the role of this N-terminal region. Translocation of Raf proteins to the membrane, a critical step in their activation, can be mediated through different mechanisms. It is reported that direct interaction between a basic motif in CRaf's kinase region and phosphatidic acid (PA) can recruit Raf to the membrane (Rizzo et al. 2000; Kraft et al. 2008). This PA-binding site is conserved in ARaf and BRaf proteins. Also, association with Ras, a major regulator of Raf kinases, plays a crucial role(s) in translocation and activation of Raf. However, the molecular mechanisms of Ras–Raf coupling are not completely understood. Raf's RBD can directly interact with the switch 1 region of GTP–Ras and is thought to be the core element for Ras binding (Nassar et al. 1995). CRD is involved in Ras–Raf coupling, as well, through interaction between its hydrophobic patch and the lipid moiety of Ras (Williams et al. 2000; Thapar et al. 2004). Thus, both RBD and CRD contribute to Ras–Raf interaction and the effects are likely additive. Disabling either RBD or CRD is thought to reduce but not completely eliminate Raf activity (Hu et al. 1995). Recently, Fischer et al. (2007) found BRaf's interaction with HRas was also facilitated by the extended N terminus, in vitro. At the present time, however, the identity of residues/sites that participate in this process are unknown and the biological implications of this N-terminal region in vivo have not been defined. Drosophila has one Raf gene first described genetically as l(1) pole hole, and later referred to as DRaf or Raf. As a member of the MAP kinase signaling module, DRaf plays an essential role in numerous RTK pathways in Drosophila development (Perrimon 1994; Van Buskirk and Schüpbach 1999; Duffy and Raabe 2000; Brennan and Moses 2000). On the basis of its primary structure, the DRaf protein is more similar to BRaf than either ARaf or CRaf (Morrison and Cutler 1997; Dhillon and Kolch 2002; Chong et al. 2003). DRaf and BRaf have two acidic residues (E420–E421 in DRaf; D447–D448 in BRaf) preceding the kinase region that correspond to residues Y301–Y302 in ARaf and Y340–Y341 in CRaf, respectively. These negative charged acidic residues mimic constitutive phosphorylation and are thought to be related to the higher basal activity of BRaf (Mason et al. 1999; Mishra et al. 2005). Both DRaf and BRaf have an extended amino terminus, when compared to ARaf and CRaf, in addition to CR1, CR2, and CR3. DRaf and BRaf also share parallels in their modes of regulation. Rap1 can activate both BRaf and DRaf, but not ARaf or CRaf (Ohtsuka et al. 1996; Mishra et al. 2005). Like the Raf proteins in mammals, the activity of DRaf is regulated through phosphorylation/dephosphorylation (Baek et al. 1996; Rommel et al. 1997; Radke et al. 2001; Laberge et al. 2005), interaction with scaffold proteins or other binding partners (Roy et al. 2002; Roy and Therrien 2002; Douziech et al. 2003, 2006; Roignant et al. 2006; Rajakulendran et al. 2008). These regulatory events occur within the three conserved regions (CR1–CR3) of Draf; however, the role of DRaf's N-terminal region has not been elucidated.
Development of both embryonic termini in Drosophila is dependent on DRaf-mediated Torso RTK signaling. Binding of Trunk or Torso-like with the Torso receptor initiates Ras1–DRaf–MEK signaling at the poles of early staged embryos, and in turn, triggers expression of at least two gap genes, tailless and huckebein, which specify terminal structures and help to establish segmental identities in the embryo (reviewed by Furriols and Casanova 2003). The domain of tailless (tll) expression in the embryonic posterior region has been used as a quantitative marker to measure the strength of the Torso RTK signal in early embryos. At the cellular blastoderm stage, embryos from wild-type (WT) mothers show posterior tll expression from approximately 0–15% embryo length (EL). At a later stage embryos exhibit normal internal head structures, three thoracic segments (T1–T3), eight abdominal denticle belts (A1–A8), as well as the Filzkörper (Fk) tail structure. Decreased or loss of Torso RTK pathway activity results in a reduced posterior expression domain of tll and consequently absence of embryonic tail structures. In contrast, gain-of-pathway activity can lead to expanded tll expression domains at both poles, and subsequently enlarged head and tail structures, accompanied by deletion of central abdominal segments (Ghiglione et al. 1999; Jiménez et al. 2000).
In this study, using the Drosophila embryonic termini as both a qualitative and quantitative in vivo assay system, we examined the role played by DRaf's N terminus in Torso signaling in different genetic backgrounds. We observed a subtle, but consistent, higher signaling potential for full-length DRaf proteins when compared with those lacking amino-terminal residues 1–114 (DRafΔN114). Furthermore, a novel region within DRaf's N terminus that is conserved in RAF genes of most invertebrates and BRaf genes of vertebrates was identified and termed conserved region N-terminal (CRN). Our studies suggest that DRaf's extended N terminus may assist in its association with the upstream activators Ras1 and Rap1 in vivo and thus, potentially play a regulatory role(s) in DRaf's activation through a CRN-mediated mechanism(s). Minor adjustment by CRN on Ras1 and Rap1 binding may help to fine tune DRaf's activity and consistently provide optimal signal output.
MATERIALS AND METHODS
Drosophila strains and genetics:
In this study, y w, Draf11-29 (Draf −; DRaf protein null, Melnick et al. 1993), trunk1 (trk−; loss-of-function allele, lacks C-terminal 16 amino acids, Schüpbach and Wieschaus 1989; Casanova et al. 1995), torsoXR1 (tor−; Torso protein null allele, tor gene deletion, Sprenger et al. 1989), and torsoRL3 (torRL3; gain-of-function allele, H242L amino acid replacement in the extracellular domain, Sprenger et al. 1993) strains were used. The flippase dominant female sterile (FLP-DFS) technique was utilized to generate Draf11-29 germline clones (Chou and Perrimon 1996). Drosophila stocks were raised at 25° on standard cornmeal medium. To study the gain-of-function effects of the temperature-sensitive torRL3 allele (Figure 3), virgin females were collected and mated with wild-type males at 25° for 3–4 days and then moved into a 29° incubator. Eggs were collected at 29° during the first 1–2 days for Western analysis and phenotypic characterization.
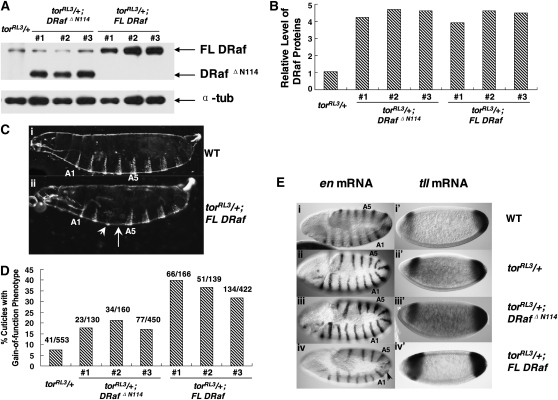
Figure 3.—
Gain-of-function effects of torRL3 are differentially enhanced by expression of FL DRaf and DRaf ΔN114 transgenes. (A) Western analysis of embryonic DRaf proteins from eggs (0–3 hr) produced by torRL3/+, torRL3/+; DRaf ΔN114 (three independent lines, #1, #2, and #3), or torRL3/+;FL DRaf (three independent lines, #1, #2, and #3) females at 29°. Full-length DRaf (∼90 kDa) and DRafΔN114 (∼77 kDa) proteins are denoted by arrows. α-Tubulin was used as the loading control. (B) Normalized DRaf protein level from A is shown as a bar graph. (C) Cuticles of mature embryos are shown. (i) A wild-type (WT) embryo exhibits normal cuticle pattern with 8 abdominal denticle belts. (ii) An embryonic cuticle derived from torRL3/+; FL DRaf mother has one broken abdominal denticle band (arrow head) and is missing one central abdominal denticle belt (arrow). (D) Percentage of embryonic cuticles with gain-of-function phenotypes is shown. Gain-of-function effects of torRL3 were differentially enhanced by FL DRaf and DRafΔN114 proteins (χ2 = 51.063837, P < 0.001). (E) Expression of engrailed (en) at approximately stage 11 (left) and accumulation of tailless (tll) mRNA at cellular blastoderm stage (right) in embryos from WT, torRL3/+, torRL3/+;DRaf ΔN114, or torRL3/+;FL DRaf mothers: Examples of embryos derived from (i) WT, (ii) torRL3/+, and (iii) torRL3/+;DRaf ΔN114 mothers exhibit normal en mRNA pattern with three thoracic (T1–T3) and nine abdominal (A1–A9) expression stripes. (iv) An embryo from a torRL3/+; FL DRaf mother is shown with partial deletion of en stripes (arrow) in a region that gives rise to central abdominal segmental pattern. Examples of embryos derived from (i′) WT and (ii) torRL3/+ mothers exhibiting a normal tll mRNA pattern. (iii′) An embryo from a torRL3/+; DRaf ΔN114 mother shows slightly expanded posterior expression domain of tll. (iv′) An embryo derived from torRL3/+;FL DRaf females exhibits expanded domain of tll expression for both anterior and posterior regions.
Transgene design:
Full-length and truncated DNAs were amplified using wild-type DRaf cDNA (GenBank no.AY089490, obtained from Drosophila Genomics Research Center) as template, and inserted into the polylinker site of the P-element transformation vector pCaSpeR-HS83. The full-length cDNA sequence (FL DRaf) encodes a DRaf protein with 739 residues, while the truncated cDNA sequence (DRafΔN114) corresponds to amino acids 115–739 of the FL DRaf protein. The constitutively active heat-shock 83 gene (HS83) promoter was used to drive the expression of DRaf transgenes to simplify the generation of transgenic lines with various genetic backgrounds. Transgenic lines were generated by Genetic Services (Sudbury, MA).
Multiple lines derived for each transgene were used in this study. DRafΔN114 (L1, #a and #b), FL DRaf (#a and #b) were used to generate germline clones bearing females (Figure 1). Lines #1, #2, #3 of DRafΔN114 and #1, #2, #3 of FL DRaf were used in torRL3, trk1, and torXR1 backgrounds. The DRafΔN114 line #1 is homozygous lethal, thus we generated trk1/trk1; DRafΔN114#1/DRafΔN114#3, and torXR1/torXR1; DRafΔN114#1/DRafΔN114#3 lines that produce DRaf protein levels equivalent with other lines (Figure 4, Table 1).
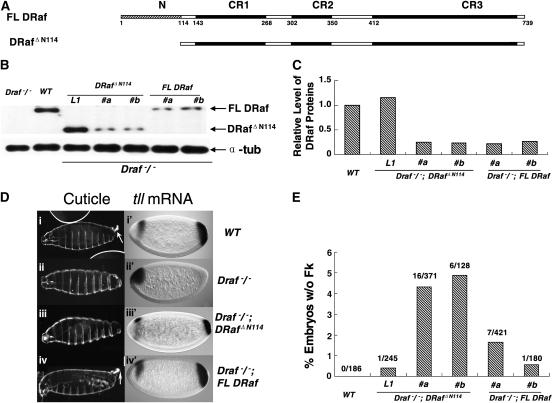
Figure 1.—
Rescue of posterior structures in embryos derived from Draf−/− female germ cells by expression of full-length DRaf or truncated DRaf ΔN114 transgenes. (A) Schematic representations of full-length DRaf (FL DRaf) with 739 amino acids and truncated DRafΔN114 proteins. In addition to the three conserved regions (CR1, CR2, and CR3), FL DRaf has an extended N terminus. (B) Western analysis of embryonic DRaf proteins from eggs produced by Draf 11-29/Draf 11-29 (Draf−/−), wild type (WT), Draf−/−; DRaf ΔN114 (three independent lines, L1 with ∼1× endogenous DRaf level, #a and #b with ∼1/4 endogenous DRaf level) and Draf−/−; FL DRaf (two independent lines, #a and #b with ∼1/4 endogenous DRaf level) germline clone bearing females. Lysate was prepared from eggs at 0–3 hr after egg deposition. Full-length DRaf (∼90 kDa) and DRafΔN114 (∼77 kDa) proteins are denoted by arrows. Lysate of eggs from Draf−/− germ cells was used as a negative control. α-Tubulin (α-tub) was used as the loading control. (C) A bar graph representing relative levels of DRaf proteins normalized with α-tubulin. (D) Cuticles of mature embryos derived from wild type (WT), Draf−/−, Draf−/−; DRaf ΔN114, and Draf−/−; FL DRaf female germ cells (left). Accumulation of tll mRNA in cellular blastoderm embryos was detected by in situ hybridization (right). Posterior tll expression is solely dependent on the Torso pathway and used as a marker for Torso RTK signaling. Anterior expression of tll is regulated by another pathway(s) in addition to Torso signaling, is more complex, and is used as an internal control for staining here. Wild-type embryos show (i) normal Filzkörper (Fk) structure (arrow), (i′) tll mRNA accumulation at the posterior (∼0–15% embryo length, EL), and an anterior head “stripe” (∼75–85% EL). Embryos derived from Draf−/− germ cells lack (ii) posterior structures (A8 denticle belt, Filzkörper) and (ii′) posterior tll mRNA expression. (iii) An embryonic cuticle derived from Draf−/−; DRaf ΔN114 germ cell lacks the Filzkörper structure. (iii′) A reduced posterior tll expression domain is at ∼0–8% EL in an embryo from Draf−/−; DRaf ΔN114 germline clone bearing mother. (iv) Filzkörper structure (arrow) and (iv′) normal expression pattern of tll mRNAs are rescued by FL DRaf expression for embryos derived from Draf−/−; FL DRaf maternal germ cells. (E) A bar graph showing the percentage of embryos without Fk structures. When expressed at low maternal level (∼1/4 endogenous level), embryos without Fk were found more often for those that inherited truncated DrafΔN114 rather than full-length DRaf proteins (χ2 = 9.91976318, P < 0.01).
Figure 4.—
Effects of FL DRaf and DRafΔN114 expression on posterior development in embryos derived from trk 1/trk 1 mothers. (A) Western analysis of embryonic DRaf proteins from eggs (0–3 hr) produced by Draf 11-29/Draf 11-29 (Draf−/−), wild type (WT), trk 1/trk 1 (trk−/−), trk−/−; DRaf ΔN114/DRaf ΔN114 (three lines, #1/#3, #2/#2, and #3/#3), and trk−/−; FL DRaf/FL DRaf (three lines, #1/#1, #2/#2, and #3/#3) females. Full-length DRaf (∼90 kDa) and DRafΔN114 (∼77 kDa) proteins are denoted by arrows. Embryonic lysate from Draf−/− germline clone females was used as a negative control. α-Tubulin was used as the loading control. (B) Normalized DRaf protein level from A is shown in the bar graph. (C) Representative cuticles of mature embryos derived from wild type (WT), trk−/−, trk−/−; DRaf ΔN114/DRaf ΔN114, or trk−/−; FL DRaf/FL DRaf females are shown (left). Accumulation of tll (right) mRNAs was detected by in situ hybridization. (i) A wild-type (WT) embryo has normal cuticle pattern with eight abdominal denticle belts and Filzkörper structure. (i′) A WT embryo at cellular blastoderm stage exhibits a normal posterior expression domain of tll. (ii) Cuticle of a mature embryo from a trk−/− mother is missing posterior structures (A8 segment, Filzkörper). (ii′) A cellular blastoderm embryo from a trk−/− mother lacks posterior tll expression. (iii) An embryonic cuticle from a trk−/−; DRaf ΔN114/DRaf ΔN114 mother lacks posterior structures (A8 denticle belt, Filzkörper). (iii′) A cellular blastoderm embryo from a trk−/−; DRaf ΔN114/DRaf ΔN114 mother lacks posterior expression of tll mRNA. Expression of FL DRaf (iv) restores the A8 denticle belt (arrow) and (iv′) posterior tll expression (arrow) in embryos lacking maternal Trk activity. (D) Effect of FL DRaf or DRaf ΔN114 transgene expression on A8 denticle development in embryos derived from trk−/− mothers (percentage of embryonic cuticles with A8 denticle belt). Shown are results using transgenic DRaf ΔN114 or FL DRaf lines that express DRaf protein at similar levels. Expression of exogenous FL DRaf, but not DRafΔN114, results in partial rescue of A8 denticle belt in some embryos derived from trk−/− mothers (χ2 = 82.8574882, P < 0.001).
TABLE 1.
Expression of FL DRaf or DRafΔN114 did not result in rescue of the A8 denticle belt in embryos produced by torXR1/torXR1 females
| No. (%) embryos whose most posterior structure belongs to: |
|||
|---|---|---|---|
| Maternal genotype | A6/A7 segment | A8 denticle belt | Total no. |
| torXR1/torXR1 | 117 (100) | 0 (0) | 117 |
| torXR1/torXR1; DRafΔN114#1/DRafΔN114#3 | 72 (100) | 0 (0) | 72 |
| torXR1/torXR1; DRafΔN114#2/DRafΔN114#2 | 77 (100) | 0 (0) | 77 |
| torXR1/torXR1; DRafΔN114#3/DRafΔN114#3 | 98 (100) | 0 (0) | 98 |
| torXR1/torXR1; FL Draf#1/FL DRaf#1 | 61 (100) | 0 (0) | 61 |
| torXR1/torXR1; FL Draf#2/FL DRaf#2 | 70 (100) | 0 (0) | 70 |
| torXR1/torXR1; FL Draf#3/FL DRaf#3 | 81 (100) | 0 (0) | 81 |
Western analysis:
To produce protein extracts, 100 eggs were collected and homogenized in 36-μl lysis buffer containing 20 mm Tris-Cl (pH 8.0), 150 mm sodium chloride, 0.2% Triton-X 100, 0.2% Nonidet P-40, 10 mm EDTA, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 0.15 units/ml aprotinin and 20 mm leupeptin. Insoluble material was removed by centrifugation (13,000 rpm, 10 min) at 4°. Protein extracts were separated by 8% SDS–PAGE, and electrophoretically transferred to nitrocellulose membrane. DRaf proteins were probed with rabbit anti-Raf antibody (70.1, Sprenger et al. 1993) and horseradish peroxidase-coupled goat anti-rabbit secondary antibody (Thermo Scientific). α-Tubulin proteins probed with mouse antibody (Sigma) and horseradish peroxidase-coupled goat anti-mouse secondary antibody (Thermo Scientific) were used as an internal control. The membranes were developed using SuperSignal West Pico kit (Thermo Scientific). Protein level was quantified with Image J.
In situ hybridization:
tailless and engrailed probes were generated from wild-type cDNA clones (tailless: GenBank no. BT022195; engrailed: GenBank no. AY069448, obtained from the Drosophila Genomics Research Center) using the PCR DIG probe synthesis kit (Roche Applied Science). Whole-mount mRNA in situ hybridizations were performed in embryos according to the protocol of Tautz and Pfeifle (1989) with minor modifications.
Circular dichroism spectral measurement:
DNA corresponding to amino acids 1–117 of DRaf (DRafN117) was recombined into the pGEX vector. The GST–DRafN117 fusion protein was produced by expression in Escherichia coli BL21 and purified by standard affinity chromatography. Purified GST–DRafN117 protein was digested with thrombin. The DRafN117 protein (∼13 kDa) was purified by a size-exclusive column (Amersham Biosciences) and verified by mass spectra and N-terminal sequencing. Protein sample (0.05 mg/ml in 10 mm sodium phosphate buffer) was loaded to 0.1-cm quartz circular dichroism (CD) cuvette. CD spectra was measured by Jasco J-710 spectropolarimeter (Protein facility at Iowa State University) at room temperature. Data were collected with 0.2-nm resolution and at a scan rate of 1.5 nm min−1. The ellipcity value of the blank buffer at each wavelength was substracted from each point.
Yeast two-hybrid analysis:
The R174 to L mutation in DRaf (DRafR174L) was generated by PCR-based site-directed mutagenesis and confirmed by sequencing. DNA sequences corresponding to amino acids 1–117 (N), 1–212 (NRBD), 18–212 (Δ17NRBD), 78–212 (Δ77NRBD), and 115–212 (RBD) were obtained by PCR using wild-type DRaf as the template, while DNAs encoding NRBDR174L and RBDR174L were amplified from DRafR174L DNA. Amplified DNAs were cloned into pGADT7 vector (Clontech). DNA sequences encoding amino acids 1–183 of Ras1 (Ras1ΔCAAX) and 1–180 of Rap1 (Rap1ΔCAAX) were amplified from cDNAs of wild-type Ras1 and Rap1 (Ras1: GenBank no. AF186648; Rap1: NCBI Reference no. NM_057509, obtained from the Drosophila Genomics Research Center), respectively, and inserted into the pGBKT7 vector (Clontech).
Constructed pGADT7 and pGBKT7 plasmids were transformed into yeast Y187 strain. Protein–protein interactions were tested by β-galactosidase assays using X-gal (5-bromo-4-chloro-3-indolyl-b-d-galactopyranoside, Sigma; solid-support assay) or ONPG (ortho-nitrophenyl-b-d-galactopyranoside, Sigma; liquid quantitative assay) as substrates. β-Galactosidase units in quantitative assays were calculated according to the Yeast Protocol Handbook (Clontech). All yeast two-hybrid experiments are confirmed by reciprocal bait–prey assays and repeated at least four times.
RESULTS
To study the potential function of DRaf's N-terminal residues (amino acids 1–114), we generated transgenic flies expressing full-length DRaf (FL DRaf) or DRaf proteins lacking amino-terminal residues 1–114 (DRafΔN114; Figure 1A). The constitutive heat-shock 83 (HS83) promoter was selected to drive transgene expression to simplify the generation of complex genetic backgrounds required to test the functionality of N-terminal residues. We used the Torso pathway to test the signaling potential of these maternally expressed DRaf proteins. Since the Torso signaling system is solely dependent on activity of maternal DRaf proteins, we could readily determine and verify the quantity of DRaf proteins available for Torso signal transduction in early staged embryos by Western blot analysis. Thus, at equivalent protein concentrations, we compared the signal potential of FL DRaf and DRafΔN114 proteins to characterize the role of the N terminus in a well-defined RTK pathway in vivo.
DRaf's N terminus can contribute to RTK signaling in Drosophila embryos:
Embryos that were deficient for maternal DRaf protein (derived from Draf11-29/Draf11-29 female germ cells, see materials and methods) lack posterior tll expression at ∼2.5 hr after egg deposition and subsequently exhibit abnormal cuticle pattern with deletion of posterior structures due to loss of Torso RTK signaling (Figure 1D, ii and ii′). We generated females with germ cells homozygous mutant for the Draf11-29 allele (Draf−/−) but expressing FL DRaf or DRafΔN114 proteins using the “FLP-DFS” technique (Chou and Perrimon 1996). Cuticles of embryos produced by Draf−/−; DRafΔN114 female germline clones expressing maternal truncated proteins equal to or greater than endogenous wild-type DRaf levels were essentially equivalent to those of wild-type embryos with only one (1/245) lacking posterior Filzkörper (line L1, Figure 1, B and E). However, when DRafΔN114 was expressed at low maternal levels (∼1/4 of endogenous DRaf level; two independent transgenic lines #a and #b; Figure 1B), ∼4.5% of the embryos assayed lacked posterior Filzkörper (Figure 1, Diii and E). At such a reduced expression level (two independent lines #a and #b, Figure 1B), FL DRaf showed rescue of posterior pattern with Filzkörper development observed for a higher percentage of embryos (∼98.5%, χ2 = 9.91976318, P < 0.01; Figure 1, D and E). In agreement with the cuticle phenotype, an abnormal posterior tll expression pattern (<13% EL) was observed more often for embryos that inherited truncated DRafΔN114 (21.2%, n = 52) rather than full-length DRaf proteins (9.0%, n = 78; χ2 = 3.9386844, P < 0.05, Figure 1D), suggesting that DRafΔN114 was less active than FL DRaf in Torso RTK signaling.
To test our protein quantification assay, a more rigorous examination was conducted using Western blot analysis. Three samples representing lysates of 6, 12, and 18 eggs from each line (Draf−/−; DRaf ΔN114#a and Draf−/−; FL DRaf #a) were loaded on to a SDS–PAGE gel. As shown in Figure 2A, the intensity of DRaf and corresponding tubulin bands exhibits a roughly linear correlation with the number of eggs lysed (Figure 2B). In addition, the normalized DRaf protein level was consistent among the three samples loaded for the same transgenic line (Figure 2C), suggesting our Western blots analysis was reliable. Importantly, we also addressed the question of maternal DRaf protein stability and whether deletion of the N-terminal region altered DRaf accumulation levels during Torso signal transduction. Embryonic lysates from eggs collected at 0–1, 1–2, and 2–3 hr after deposition were prepared (Draf−/−; DRaf ΔN114#a and Draf−/−; FL DRaf #a). As shown in Figure 2, D and E, DRaf protein levels remained roughly constant, indicating both FL DRaf and DRafΔN114 proteins are stable throughout the 0- to 3-hr period when the Torso pathway is active.
Figure 2.—
Verification of DRaf protein quantitation assays and stability of DRaf proteins in early embryos. (A) Three samples representing lystes of 6, 12, and 18 eggs for each line (Draf−/−; DRaf ΔN114#a and Draf−/−; FL DRaf #a) were loaded for Western blot analysis. Full-length DRaf (∼90 kDa) and DRafΔN114 (∼77 kDa) proteins are denoted by arrows. Lysate of eggs from Draf 11-29/Draf 11-29 (Draf−/−) germ cells was used as a negative control. α-Tubulin levels were probed as a loading control. (B) Bar graph showing relative intensity of DRaf (solid bar) and α-tubulin (shaded bar) bands. (C) A bar graph depicting normalized DRaf protein level from A. (D) Western analysis of embryonic DRaf proteins from eggs collected at 0–1, 1–2, and 2–3 hr after deposition and produced by Draf−/−, Draf−/−; DRafΔN114 (line #a) and Draf−/−; FL DRaf (line #a) germline-bearing females. Full-length DRaf (∼90 kDa) and DRafΔN114 (∼77 kDa) proteins are denoted by arrows. α-Tubulin was used as the loading control. (E) Normalized DRaf protein level from D is shown in this bar graph depicting the stable accumulation of these DRaf proteins.
Next, we genetically altered the Torso pathway to create a sensitized signaling environment and compared the potential of DRafΔN114 and FL Raf proteins in this background. torRL3 is a temperature-sensitive, recessive, gain-of-function allele of the Torso receptor. At the nonpermissive temperature 25–29°, torRL3/torRL3 mothers produce embryos that show broad tll expression at both anterior and posterior ends. These embryos develop and show deletion of central abdominal segments and do not hatch (Strecker et al. 1989). Eggs derived from females heterozygous for torRL3 (torRL3/+) can hatch as larvae; however, some of these larvae show a gain-of-function phenotype with deletion of an abdominal segment(s). At 29°, we found 7.4% (n = 553, Figure 3D) of the larvae from torRL3/+ mothers showed deletion, fusion, or broken abdominal denticle bands. In this genetic background, when expressed at comparable protein levels (Figure 3, A and B), FL DRaf enhanced the torRL3 phenotype much more significantly than DRafΔN114, resulting in a greater number of embryos with central abdominal defects. We found 31.8% (n = 422) embryos with FL DRaf proteins showed the gain-of-function phenotype (Figure 3, C and D), while only 17.1% (n = 450) of the DRafΔN114 embryos showed such defects (Figure 3, C and D). We repeated these experiments using two additional, independently derived transgenic FL DRaf and DRafΔN114 lines and observed similar results (χ2 = 51.063876, P < 0.001; Figure 3D).
To test whether the cuticle phenotypes observed were due to alterations in the embryonic fate map, we determined the mRNA accumulation pattern for the engrailed (en) segmentation gene in approximately stage 11 embryos. The en mRNA wild-type pattern is dependent on normal signaling in the Torso pathway. We found 36.1% (n = 169) of the embryos from torRL3/+; FL DRaf mothers had at least one deleted, fused, or broken en central abdominal stripe(s) (Figure 3E, iv). The segmentation defects observed were most likely due to the expansion of head and/or tail domains and indicative of the gain-of-function phenotype. In contrast, only 33 of 141 (23.4%) embryos from torRL3/+; DRafΔN114 mothers had such defects (χ2 = 6.38030206, P < 0.02). Consistent with the cuticular phenotypes and en expression patterns, expansion in the domain of tll expression was observed more often for embryos from torRL3/+; FL DRaf mothers (32.0%, n = 50) compared with those from torRL3/+; Draf ΔN114 females (14.5%, n = 76; χ2 = 5.50220096, P < 0.02, Figure 3E, iii′ and iv′). Together, these in vivo studies consistently indicated that deletion of the N terminus reduces the ability of DRaf to enhance the ectopic gain-of-function effects of torRL3 and that these N-terminal residues could participate in Torso RTK signaling.
The contribution of DRaf's N terminus to signaling appears to be dependent on the activity of the Torso receptor:
Embryos lacking normal maternal Trunk (Trk) activity show little or no posterior tll expression (occasionally, trace-level tll expression was detected in 3–4 cells at the posterior embryo tip) and exhibit terminal defects with deletion of all posterior structures (A8 denticle belt and Filzkörper; Figure 4Cii). Interestingly, overexpression of FL DRaf partially restores the A8 denticle belt structure in embryos from trunk1/trunk1 (trk−/−; lacks the last 16 amino acids) mothers (Figure 4Civ). This result is consistent with our unpublished findings (L. Ambrosio and K. H. Baek) using the trk3 allele (encodes the first 89 amino acids) and a different method. Rescue of posterior structures for some Trk-deficient embryos was found after injection of wild-type DRaf mRNA, also suggesting that accumulation of exogenous DRaf proteins promotes signaling in this trk− background. However, expression of the DRaf ΔN114 transgene at a similar level failed to rescue the A8 denticle band defect in embryos from trk−/− mothers (Figure 4, A and Ciii). We repeated these experiments using two additional FL DRaf and DRaf ΔN114 transgenic lines and observed similar results (χ2 = 82.8574882, P < 0.001; Figure 4D). This indicated that addition of FL DRaf but not DRafΔN114 proteins partially restored posterior Torso RTK signaling in the trk− background and FL DRaf appears to possess greater activity compared with DRafΔN114. Consistent with this hypothesis, as shown in Figure 4C, iii′ and iv′, partial rescue of posterior tll mRNA expression was detected in some cellular blastoderm embryos derived from trk−/−; FL DRaf/FL DRaf females (8.1%, n = 37) but not for those derived from trk−/−; DRaf ΔN114/DRaf ΔN114 mothers (n = 52, χ2 = 4.36329353, P < 0.05). Together, these data consistently suggest that the absence of the N-terminal segment reduces the signaling potential of DRaf and the N terminus can contribute to Torso RTK terminal signaling in a positive manner.
Sprenger et al. (1993) previously observed a low level of Torso receptor phosphorylation in eggs derived from trk loss-of-function, but not tor loss-of-function females. Therefore, a small amount of Torso signal activity may exist in our trk− background. This may be due to (1) the presence of active Torso-like (Tsl) ligand; (2) potential residual Trunk activity, considering the molecular lesion of the trk1 allele we used (lacks only the last 16 amino acids); or (3) the intrinsic activity of the Torso receptor. This activity could allow rescue of posterior structures by FL DRaf expression. If so, the contribution of the N terminus to terminal signaling is likely sensitive to such upstream events. Thus, we examined the consequences of DRaf expression in embryos from torXR1/torXR1 (tor−/−; protein null) mothers that lacked the Torso receptor. We found that expression of FL DRaf or DRafΔN114 failed to restore the posterior structure (A8 denticle belt) for these embryos (Table 1). This indicates the contribution of DRaf's N-terminal residues to rescue the A8 denticle belt is dependent on activity of the receptor.
The N terminus of DRaf contains a novel conserved region and has a high content of helical secondary structure:
We analyzed the amino acid sequence of DRaf's N terminus using several bioinformatics tools to obtain hints regarding its structure, and perhaps mechanism(s) of its functional role(s). A PROSITE motif search showed a putative protein kinase C (PKC) phosphorylation site within the “T-S-K” motif of the N terminus (positions 60–62; Sigrist et al. 2002). Phosphorylation site prediction by NetPhos 2.0 suggested that the Thr in this T-S-K motif had a high phosphorylation potential (Figure 5A; Blom et al. 1999). Predictions of secondary structure for the N-terminal region using GORV, PHD, and Predator indicated a high α-helical propensity (Figure 5A) (Rost et al. 1994; Frishman and Argos 1996; Garnier et al. 1996; Combet et al. 2000). A blastp search of other organisms with DRaf's N-terminal sequence identified honeybee Raf, chick C-Rmil, and BRaf proteins of sea urchin, zebrafish, frog, and human. The region containing amino acids 19–77 of DRaf showed homology between candidates. These sequences were aligned using ClustalW, and are shown in Figure 5A (Combet et al. 2000). Overall, the amino acids showed 18.6% identity and 47.5% similarity, and we term this region CRN. Interesting features of CRN include the putative phosphorylation site and a propensity to form two α-helical structures. This suggests that the N-terminal region of DRaf may have function(s) shared by other BRaf proteins.
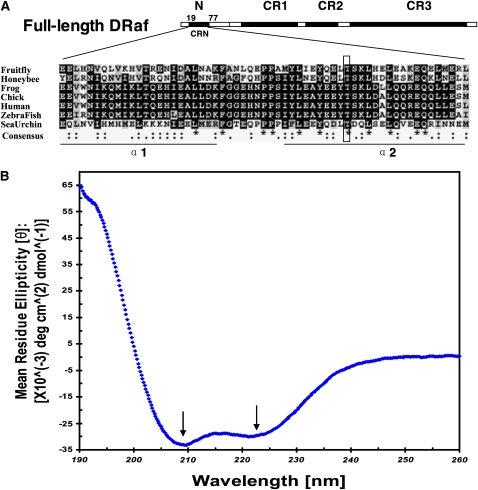
Figure 5.—
The N terminus of DRaf contains a novel conserved region and has a high content of helical secondary structure. (A) Drosophila Raf (NP_525047; 739 amino acids) has in addition to its three conserved regions (CR1–CR3), an extensive N terminus. A novel region (amino acids 19–77) within the N terminus is conserved in honeybee Raf (Apis mellifera XP_396892), frog BRaf (Xenopus laevis AAU29410), chicken C-Rmil (Gallus gallus CAA47436), human BRaf (Homo sapiens NP_004324), zebrafish BRaf (Danio rerio BAD16728), and sea urchin BRaf (Strongylocentrotus purpuratus XP_781094) and termed conserved region N-terminal (CRN). Sequences of CRN were aligned using ClustalW (identities were denoted as *; strong and weak similarities were denoted as : and .," respectively, in consensus line, http://www.ebi.ac.uk/tools/clustalw/), and the conserved residues were shaded using BOXSHADE (identities, solid; similarities, shaded, http://www.ch.embnet.org/software/BOX_form.html). Secondary structure prediction with GORV indicates CRN has the propensity to form two α-helices (α1 and α2). The putative PKC phosphorylation site DRaf's Thr60 is framed. (B) Circular dichroism (CD) spectral measurement of DRaf's N terminus (amino acids 1–117) in vitro: The bilobed spectrum (arrows, local minima at ∼209.4 nm and at ∼221.4 nm) indicative of helical secondary structure is shown.
The conserved structural features, including α-helical propensity, may be related to the functional role(s)/regulatory mechanism(s) of DRaf's N terminus. To confirm the prediction attained by bioinformactics tools, CD spectral measurement of the N-terminal part of DRaf (amino acids 1–117, DRafN117) was performed after its expression and purification in vitro (see materials and methods). As shown in Figure 5B, a bilobed spectrum with local minima at ∼209.4 nm and at ∼221.4 nm was observed, indicating the relatively high content of helical secondary structure for DRaf's N terminus. The estimated helix content of DRaf N117 is ∼77% on the basis of the CD spectra data analysis using DICHROWEB (Whitmore and Wallace 2008, http://dichroweb.cryst.bbk.ac.uk/html/home.shtml). This result bolsters the predictions by GORV, PHD, and Predator.
The N terminus assists in association of DRaf's RBD with small GTPases Ras1 and Rap1 in vitro:
Fischer et al. (2007) found that association of BRaf with HRas was facilitated by N-terminal sequences, in vitro. To examine whether the presence of DRaf's N terminus can affect Ras1 binding, we tested interaction between Ras1ΔCAAX and DRaf's RBD (Ras binding domain) using the yeast two-hybrid assay. A stronger interaction with Ras1ΔCAAX was detected when N-terminal residues were linked to RBD in both solid-support (data not shown) and liquid quantitative β-galactosidase assays (P < 0.05, t-test; Figure 6B), suggesting that the N terminus may assist in association of DRaf with Ras1. This is consistent with results obtained for BRaf (Fischer et al. 2007).
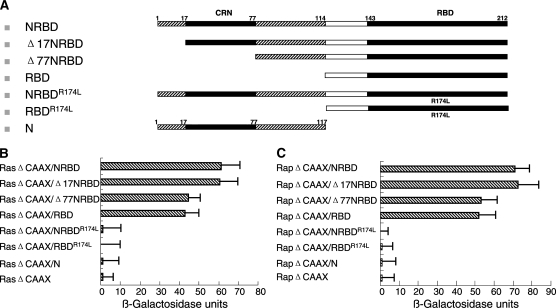
Figure 6.—
Effects of the extended N terminus of DRaf on Ras1 and Rap1 binding. (A) Schematic representations of different DRaf constructs used for yeast two-hybrid analysis. (B) Interactions between DRaf's RBDs and Ras1ΔCAAX: Removal of CRN or the entire N-terminal region reduces Ras1ΔCAAX binding (P < 0.05, t-test). (C) Interactions between DRaf's RBDs and Rap1ΔCAAX: Removal of CRN or the entire N-terminal region reduces Rap1ΔCAAX binding (P < 0.05, t-test).
No direct interaction was detected between Ras1ΔCAAX and isolated N-terminal residues of DRaf (Figure 6B, and solid-support data not shown). Thus, the N terminus appears to contribute to Ras1 binding, but as an isolated protein fragment, cannot directly interact with Ras1. Arg174 located in DRaf's RBD region is essential for its association with Ras1 and substitution of Arg174 to Leu in RBD (RBDR174L) abolishes Ras1 binding (Fabian et al. 1994; Li et al. 1998). We found that N-terminal residues cannot restore Ras1 interaction when linked with RBDR174L (Figure 6B and solid-support data not shown). This indicated the effects of the N terminus were dependent on interaction between RBD and Ras1.
Moreover, we tested the idea that the conserved CRN region (19–77) might be essential for the contribution of DRaf's N terminus to Ras1ΔCAAX binding. Deletion of the first 17 N-terminal amino acids (Δ17NRBD) did not change Ras1ΔCAAX binding. However, if N-terminal amino acids including CRN were removed (Δ77NRBD), interaction with Ras1ΔCAAX was reduced to a level similar to that observed by deletion of the entire N terminus (amino acids 1–114; Figure 6, A and B). Together, these findings suggested the hypothesis that N-terminal residues of DRaf can assist in Ras1 interaction through a CRN-mediated mechanism(s). The small GTPase Rap1, a close relative of Ras1, is known to interact with DRaf and play a role in Torso RTK signaling in vivo (Mishra et al. 2005). To examine if DRaf's N terminus affects its association with Rap1, we tested interaction between Rap1ΔCAAX and DRaf's RBD (Figure 6C). A stronger interaction with Rap1ΔCAAX was detected when the N terminus was linked to RBD, similar to our findings with Ras1 (P < 0.05, t-test). Furthermore, the conserved CRN region (19–77) seems essential for the contribution of the N terminus to this interaction with Rap1, suggesting a CRN-mediated mechanism(s) may be a general feature for its binding to both Ras1 or Rap1.
DISCUSSION
In our study, a novel region (amino acids 19–77) within Draf's N terminus, conserved for Raf genes of most invertebrates and BRaf genes of vertebrates, was identified and termed CRN. This conserved region has not been described by others, but potential roles for the extended N terminus have been proposed in two reports. Terai and Matsuda (2006) found that in HeLa cells, the N terminus of BRaf may mediate Raf dimerization to generate BRaf–BRaf or BRaf–CRaf complexes, and play an important regulatory role in calcium-induced BRaf activation. However, Fischer et al. (2007) reported that deletion of BRaf's N terminus did not affect BRaf–CRaf dimer formation. Instead, they found that N-terminal residues appeared to facilitate interaction with HRas in vitro. In accordance with their data, stronger interactions between DRaf's RBD (Ras binding domain) and the small GTPase Ras1ΔCAAX were observed when N-terminal and RBD sequences were linked in our yeast two-hybrid analysis. This suggested that the N terminus might assist in Ras1 binding. Furthermore, the identity of specific residues in the N terminus that might participate in Ras1 binding were mapped to the CRN region (amino acids 19–77). Two known Raf motifs, RBD and CRD, are involved in Raf's interaction with Ras. Our studies, and results obtained by Fischer et al. (2007) using BRaf, suggest that the N-terminal residues of DRaf and BRaf proteins, particularly the CRN region, might be another element that plays a role(s) in Ras–Raf coupling.
The small GTPase Rap shares with Ras nearly identical Raf binding regions that comprise switch 1 and the lipid moiety (Hariharan 2005). Rap functions as an antagonist of Ras in regulating CRaf activity (Cook et al. 1993), but can activate BRaf in a parallel way with Ras (Ohtsuka et al. 1996). Isoform-specific features of different Raf family members may explain their distinct responses to Rap. In flies, both Ras1 and Rap1 can interact with and activate DRaf (Mishra et al. 2005). Thus, it was reasonable to test whether DRaf's N terminus including CRN might also assist in Rap1 binding. In agreement with this idea, stronger interaction between RBD and Rap1ΔCAAX was observed when DRaf's CRN and RBD sequences were linked in vitro, further suggesting that the N terminus may contribute to both Ras1 and Rap1 binding potentially through a CRN-mediated mechanism(s) in vivo.
What is/are the molecular mechanism(s)?:
No direct interaction between Ras1 or Rap1 and the isolated DRaf N-terminal segment (amino acids 1–117) was detected, or when the N terminus was linked with the Ras1/Rap1 binding-deficient RBDR174L. Thus, the contribution of DRaf's N-terminal residues to Ras1 and Rap1 binding requires the presence of RBD. It is possible that the CRN-containing N terminus may assist in Raf–Ras interaction by making RBD more accessible to Ras1 and/or in a sequential manner, subsequent to RBD–Ras1 interaction, by stabilizing the RBD–Ras1 complex. Deletion of CRN may result in conformational or structural changes that reduce Ras1 binding affinity. Structural analysis of these complexes may provide important clues and help to understand the molecular mechanism(s) by which CRN assists in Ras–Raf interaction. Our computational analysis suggested conserved CRN has the propensity to form two α-helical structures (α1 and α2; Figure 5A) and contains a putative phosphorylation motif T-S-K located in α2. In agreement, DRaf's N terminus (amino acids 1–117) was folded in vitro and had a high content of helical secondary structure (Figure 5B). These findings may help to establish a basis for future determination of molecular structure.
Although no verified binding partner(s) for DRaf or BRaf's N terminus has been identified, it is still possible that CRN may interact with other regulatory factors in vivo, that may affect Ras or Rap binding and/or function in activation of DRaf and BRaf. If so, the conserved structural features of CRN most likely relate to these regulatory events in vivo. Site-directed mutagenesis of conserved sites/motifs could provide useful information regarding the molecular mechanism(s) of CRN's role in the activation of DRaf and BRaf.
Torso RTK signal is differentially elevated by overexpression of FL DRaf and DRafΔN114 in vivo:
We initiated our in vitro studies of DRaf's N terminus on the basis of our in vivo findings using both loss- and gain-of-function genetic assays that deletion of N-terminal residues consistently reduces DRaf's signal potential in the Torso pathway. When expressed at high levels, FL DRaf enhanced the gain-of-function effects of the torRL3 allele much more significantly than DRafΔN114. In embryos from trk−/− mothers, addition of FL DRaf, but not DRAFΔN114, partially restored the A8 denticle belt structure (Figure 4). These findings indicate that the N terminus can play a positive role(s) in Torso RTK signaling. Interestingly, the contribution of DRaf's N terminus in the Torso pathway appeared to be dependent on upstream receptor activity, suggesting its role in transmission of the signal. Together with our yeast two-hybrid data, as well as the results obtained by Fischer et al. (2007) for BRaf, we propose that the presence of N-terminal residues may facilitate the association of DRaf with the upstream regulators Ras1 and Rap1, thereby assisting in transmission of the RTK signal in vivo.
For instance, in the trk− background, a small amount of active GTP–Ras1 and GTP–Rap1 are likely present, mostly due to activation by residual upstream Trunk activity, the presence of Torso-like ligand, and/or the intrinsic activity of the Torso receptor. The trk1 mutation used in this analysis results in protein truncation at the last 16 amino acids. It is possible that overexpression of FL DRaf proteins in this background increases the likelihood of interaction between abundant DRaf proteins and membrane bound GTP–Ras1 or GTP–Rap1. This in turn, could elevate the RTK signal and partially restore development of the A8 denticle belt structure in some embryos. On the other hand, deletion of the N terminus could destabilize Ras1–DRaf (or Rap1–DRaf) coupling or decrease the duration of interaction, resulting in reduced DRaf signal transmission. This may explain why expression of DRafΔN114 failed to rescue the A8 denticle belt in embryos from trk−/− mothers.
Why are only minor differences detected in vivo between FL DRaf and DRafΔN114:
Previously, an auto-inhibitory role had been assigned to residues compromising the first half of the DRaf protein, in addition to their functions in promoting its activity. Deletion of the N-terminal amino acids 1–272 (including the N terminus and CR1) or 1–402 (including the N terminus, CR1, and CR2) of DRaf at least partially relieved these negative effects (Baek et al. 1996). Here, although removal of the N-terminal 1–114 residues did not result in constitutive DRafΔN114 activity in embryos lacking the maternal Torso receptor (Table 1), it is still possible that the N terminus may contribute to auto-inhibitory effects. Together with CR1 and CR2, these N-terminal residues (1–114) may help maintain DRaf's inactive conformation. If so, the N terminus might play dual roles, both positively and negatively regulating DRaf. Therefore, its contribution to signaling may be neutralized by this auto-inhibition and consequently result in a subtle in vivo effect. If so, selective mutagenesis of the “inhibitory” motifs/sites in the N-terminal region or removal of other cofactors involved in its negative regulation may amplify signaling differences between FL DRaf and DRafΔN114. Ras binding has been thought crucial to recruit Raf to the membrane and promote its RTK signaling activity. However, the Drosophila Torso pathway appears tolerant of alterations in Ras1–DRaf coupling (Hou et al. 1995). Draf C110 has a R174L point mutation in the RBD domain and likely comprised for Ras1 binding (Li et al. 1998). The RBDR174L is Ras binding deficient in our yeast two-hybrid assay (Figure 6B). However, tll expression patterns and cuticles of the embryos derived from mothers with Draf C110/Draf C110 germ cells were indistinguishable from those of wild-type embryos (Melnick et al. 1993), suggesting a mechanism(s) independent of RBD–Ras1 interaction might function in recruiting DRaf to the membrane. In agreement with this model, Rizzo et al. (2000) found membrane translocation of CRaf could be mediated by its interaction with PA and independent of Ras binding. This PA binding site is also conserved in ARaf, BRaf, and DRaf. Thus, DrafC110 could be recruited to the cell membranes by associating with PA. Moreover, it is known that Raf's CRD participates in Ras binding through its interaction with the lipid moiety of Ras (Williams et al. 2000; Thapar et al. 2004). Once at the membrane, it is also possible that the interaction between DrafC110's CRD and Ras1 could further promote its membrane attachment and result in relatively normal Torso signal production. In this study, the presence of RBD, CRD, and the potential PA binding site may be sufficient to promote DRaf's activation in Torso signaling. This may explain why at approximately endogenous wild-type protein level maternally expressed DRafΔN114 is able to rescue the embryonic terminal defects of Draf11-29 mutants (Figure 1, B, C, and E). Together, considering the Torso pathway's tolerance of alterations in Ras1–DRaf coupling and the minor role DRaf's N terminus plays in Ras1 binding, it is reasonable that the phenotypic consequences of removing these N-terminal residues (DRafΔN114) are not great in Torso signaling. The subtle phenotypic effects of DRaf's N terminus could also be due to compensation provided by potential autoregulatory feedback or alternative redundant processes in the in vivo system. In our study, the expression of DRaf proteins at a low level (∼1/4 endogenous wild-type level) appeared to sensitize the assay system. We found deletion of the N terminus seemed to increase the threshold of DRaf protein levels required for normal signaling. Furthermore, by adding one copy of the ectopic torRL3 allele or removing wild-type maternal Trunk activity we apparently increased the sensitivity of the Torso pathway. These allowed the embryonic terminal system to display enhanced differences between FL DRaf and DRafΔN114 proteins.
The biological implications of the N-terminal region:
Why is this N terminus with its “subtle” functional effects conserved during evolution, and what is its biological relevance? There are numerous RTK pathways functioning in Drosophila cellular and developmental processes. In spite of the identical Ras–Raf–MEK signal cassette they share, these RTK pathways can lead to different biological responses. Previous studies indicated that such specificity might be due to the difference in the intensity and/or duration of the signal (Woods et al. 1997, 2001; Kerkhoff and Rapp 1998; Ghiglione et al. 1999). This suggested that the magnitude of Raf signal could function as a critical determinant of biological responses. Participation of multiple DRaf elements in Ras1 or Rap1 binding could be a good strategy to modulate its activity. Normally, tight association with Ras1 or Rap1 through RBD and CRD regions is required and sufficient to initiate the activation of DRaf, while minor adjustments/regulation of interaction by the CRN region could optimize signaling potential and reduce variability. Thus, the extended N terminus including CRN may play a role(s) as one element in a multidomain effort to promote DRaf's interaction with Ras1 and Rap1, participating and assisting in regulation to reliably attain maximal signal output.
Acknowledgments
We gratefully acknowledge the gift of the Raf antibody from Deborah K. Morrison, fly strains from Drosophila Stock Center (Bloomington), and cDNA clones from Drosophila Genomics Research Center. We thank Yosef Scolnik for assistance in analysis of CD spectra and Clark Coffman and Lei Li for helpful discussions.
References
- Baek, K.-H., J. R. Fabian, F. Sprenger, D. K. Morrison and L. Ambrosio, 1996. The activity of D-raf in torso signal transduction is altered by serine substitution, N terminal deletion and membrane targeting. Dev. Biol. 175 191–204. [DOI] [PubMed] [Google Scholar]
- Blom, N., S. Gammeltoft and S. Brunak, 1999. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294 1351–1362. [DOI] [PubMed] [Google Scholar]
- Brennan, C. A., and K. Moses, 2000. Determination of Drosophila photoreceptors: timing is everything. Cell. Mol. Life Sci. 57 195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova, J., M. Furriols, C. A. Mccormick and G. Struhl, 1995. Similarities between trunk and spätzle, putative extracellular ligands specifying body pattern in Drosophila. Genes Dev. 9 2539–2544. [DOI] [PubMed] [Google Scholar]
- Chong, H., L. Jeeyong and K. L. Guan, 2001. Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20 3716–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, H., H. G. Vikis and K. L. Guan, 2003. Mechanisms of regulating the Raf kinase family. Cell. Signal. 15 463–469. [DOI] [PubMed] [Google Scholar]
- Chou, T. B., and N. Perrimon, 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet, C., C. Blanchet, C. Geourjon and G. Deléage, 2000. NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci. 25 147–150. [DOI] [PubMed] [Google Scholar]
- Cook, S. J., B. Rubinfeld, I. Albert and F. McCormick, 1993. RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J. 12 3475–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, G., I. Eisenmann-Tappe, H. W. Fries, J. Troppmair and U. R. Rapp, 1994. The ins and outs of Raf kinases. Trends Biochem. Sci. 19 474–480. [DOI] [PubMed] [Google Scholar]
- Dhillon, A. S., and W. Kolch, 2002. Untying the regulation of the Raf-1 kinase. Arch. Biochem. Biophys. 404 3–9. [DOI] [PubMed] [Google Scholar]
- Dhillon, A. S., S. Meikle, Z. Yazici, M. Eulitz and W. Kolch, 2002. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech, M., F. Roy, G. Laberge, M. Lefrancois, A. V. Armengod et al., 2003. Bimodal regulation of RAF by CNK in Drosophila. EMBO J. 22 5068–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech, M., M. Sahmi, G. Laberge and M. Therrien, 2006. A KSR/CNK complex mediated by HYP, a novel SAM domain-containing protein, regulates RAS-dependent RAF activation in Drosophila. Genes Dev. 20 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, J. B., and N. Perrimon, 1994. The torso pathway in Drosophila: lessons on receptor tyrosine kinase signaling and pattern formation. Dev. Biol. 166 380–395. [DOI] [PubMed] [Google Scholar]
- Fabian, J. R., A. B. Vojtek, J. A. Cooper and D. K. Morrison, 1994. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc. Natl. Acad. Sci. USA 91 5982–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, A., M. Hekman, J. Kuhlmann, I. Rubio, S. Wiese et al., 2007. B- and C-RAF display essential differences in their binding to Ras: the isotype-specific N terminus of BRAF facilitates Ras binding. J. Biol. Chem. 282 26503–26516. [DOI] [PubMed] [Google Scholar]
- Frishman, D., and P. Argos, 1996. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 9 133–142. [DOI] [PubMed] [Google Scholar]
- Furriols, M., and J. Casanova, 2003. In and out of Torso RTK signaling. EMBO J. 22 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier, J., J. F. Gibrat and B. Robson, 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266 540–553. [DOI] [PubMed] [Google Scholar]
- Ghiglione, C., N. Perrimon and L. A. Perkins, 1999. Quantitative variations in the level of MAPK activity control patterning of the embryonic termini in Drosophila. Dev. Biol. 205 181–193. [DOI] [PubMed] [Google Scholar]
- Hariharan, I. K., 2005. Ras and Rap: Are former enemies now friends? Dev. Cell 8 303–304. [DOI] [PubMed] [Google Scholar]
- Hou, X. S., T. B. Chou, M. B. Melnick and N. Perrimon, 1995. The torso receptor tyrosine kinase can activate Raf in a Ras-independent pathway. Cell 81 63–71. [DOI] [PubMed] [Google Scholar]
- Hu, C. D., K. Kariva, M. Tamada, K. Akasaka, M. Shitouzu et al., 1995. Cysteine-rich region of Raf-1 interacts with activator domain of post-translationally modified Ha-Ras. J. Biol. Chem. 270 30274–30277. [DOI] [PubMed] [Google Scholar]
- Jiménez, G., A. Guichet, A. Ephrussi and J. Casanova, 2000. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 14 224–231. [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff, E., and U. R. Rapp, 1998. High-intensity Raf signals convert mitotic cell cycling into cellular growth. Cancer Res. 58 1636–1640. [PubMed] [Google Scholar]
- Kolch, W., 2000. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351 289–305. [PMC free article] [PubMed] [Google Scholar]
- Kraft, C.A., J. L. Garrido, E. Fluharty, L. Leiva-Vega and G. Romero, 2008. Role of phosphatidic acid in the coupling of the ERK cascade. J. Biol. Chem. 283 36636–36645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge, G., M. Douziech and M. Therrien, 2005. Src42 binding activity regulates Drosophila RAF by a novel CNK-dependent derepression mechanism. EMBO J. 24 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. X., E. M. Skoulakis, R. L. Davis and N. Perrimon, 1997. The Drosophila 14–3-3 protein Leonardo enhances Torso signaling through D-Raf in a Ras1-dependent manner. Development. 124 4163–4171. [DOI] [PubMed] [Google Scholar]
- Li, W., M. Melnick and N. Perrimon, 1998. Dual function of Ras in Raf activation. Development 125 4999–5008. [DOI] [PubMed] [Google Scholar]
- Mason C. S., C. J. Springer, R. G. Cooper, G. Superti-Furga, C. J. Marshall et al., 1999. Serine and tyrosine phosphorylations cooperate in Raf-1, but not in B-Raf activation. EMBO J. 18 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick, M. B., L. A. Perkins, M. Lee, L. Ambrosio and N. Perrimon, 1993. Developmental and molecular characterization of mutations in the Drosophila-raf serine/threonine protein kinase. Development 118 127–138. [DOI] [PubMed] [Google Scholar]
- Mishra, S., S. M. Smolik, M. A. Forte and P. J. Stork, 2005. Ras-independent activation of ERK signaling via the torso receptor tyrosine kinase is mediated by Rap1. Curr. Biol. 15 366–370. [DOI] [PubMed] [Google Scholar]
- Morrison, D. K., 2001. KSR: A MAPK scaffold of the Ras pathway? J. Cell. Sci. 114 1609–1612. [DOI] [PubMed] [Google Scholar]
- Morrison, D. K., and R. E. Cutler, 1997. The complexity of Raf-1 regulation. Curr. Opin. Cell. Biol. 9 174–179. [DOI] [PubMed] [Google Scholar]
- Nassar, N., G. Horn, C. Herrmann, A. Scherer, F. McCormick et al., 1995. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase C-Raf1 in complex with Rap1A and a GTP analogue. Nature 375 554–560. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, T., K. Shimizu, B. Yamamori, S. Kuroda and Y. Takai, 1996. Activation of brain BRaf protein kinase by Rap1B small GTP-binding protein. Am. Soc. Biochem. Mol. Biol. 271 1258–1261. [DOI] [PubMed] [Google Scholar]
- Raabe, T., 2000. The sevenless signaling pathway: variations of a common theme. Biochim. Biophys. Acta 1496 151–163. [DOI] [PubMed] [Google Scholar]
- Raabe, T., and U. R. Rapp, 2002. KSR–a regulator and scaffold protein of the MAPK pathway. Sci. Sig. Trans. Know. Environ. 136 PE28. [DOI] [PubMed] [Google Scholar]
- Radke, K., K. Johnson, R. Guo, A. Davidson and L. Ambrosio, 2001. Drosophila-Raf acts to elaborate dorsoventral pattern in the ectoderm of developing embryos. Genetics 159 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakulendran, T., M. Sahmi, I. Kurinov, M. Tyers, M. Therrien et al., 2008. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc. Natl. Acad. Sci. USA 105 2836–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, M. A., K. Shome, S. C. Watkins and G. Romero, 2000. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J. Biol. Chem. 275 23911–23918. [DOI] [PubMed] [Google Scholar]
- Roignant, J.-Y., S. Hamel, F. Janody and J. E. Treisman, 2006. The novel SAM domain protein Aveugle is required for Raf activation in the Drosophila EGF receptor signaling pathway. Genes Dev. 20 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel, C., G. Radziwill, K. Moelling and E. Hafen, 1997. Negative regulation of Raf activity by binding of 14–3-3 to the amino terminus of Raf in vivo. Mech. Dev. 64 95–104. [DOI] [PubMed] [Google Scholar]
- Rost, B., C. Sander and R. Schneider, 1994. PHD—an automatic mail server for protein secondary structure prediction. Comput. Appl. Biosci. 10 53–60. [DOI] [PubMed] [Google Scholar]
- Roy, F., and M. Therrien, 2002. MAP kinase module: the Ksr connection. Curr. Biol. 12 R325–R327. [DOI] [PubMed] [Google Scholar]
- Roy, F., G. Laberge, M. Douziech, D. Ferland-Mccollough and M. Therrien, 2002. KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev. 16 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger, J., 2000. Cell signaling by receptor tyrosine kinases. Cell 103 211–225. [DOI] [PubMed] [Google Scholar]
- Schüpbach, T., and E. Wieschaus, 1989. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics. 121 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, C. J. A., L. Cerutti, N. Hulo, A. Gattiker, L. Falquet et al., 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 3 265–274. [DOI] [PubMed] [Google Scholar]
- Sprenger, F., L. M. Stevens and C. Nusslein-Volhard, 1989. The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature 338 478–483. [DOI] [PubMed] [Google Scholar]
- Sprenger, F., M. M. Trosclair and D. K. Morrison, 1993. Biochemical analysis of torso and D-raf during Drosophila embryogenesis: implications for terminal signal transduction. Mol. Cell. Biol. 13 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker, T. R., S. R. Halsell, W. W. Fisher and H. D. Lipshitz, 1989. Reciprocal effects of hyper- and hypoactivity mutations in the Drosophila pattern gene torso. Science 243 1062–1066. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98 81–85. [DOI] [PubMed] [Google Scholar]
- Terai, K., and M. Matsuda, 2006. The amino-terminal BRaf-specific region mediates calcium-dependent homo- and hetero-dimerization of Raf. EMBO J. 25 3556–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar, R., J. G. Williams and S. L. Campbell, 2004. NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation. J. Mol. Biol. 343 1391–1408. [DOI] [PubMed] [Google Scholar]
- Van Buskirk, C., and T. Schüpbach, 1999. Versatility in signalling: multiple responses to EGF receptor activation during Drosophila oogenesis. Trends Cell. Biol. 9 1–4. [DOI] [PubMed] [Google Scholar]
- Wellbrock, C., M. Karasarides and R. Marais, 2004. The RAF proteins take centre stage. Nat. Rev. Mol. Cell. Biol. 5 875–885. [DOI] [PubMed] [Google Scholar]
- Whitmore, L., and B. A. Wallace, 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89 392–400. [DOI] [PubMed] [Google Scholar]
- Williams, J. G., J. K. Drugan, G. S. Yi, G. J. Clark, C. J. Der et al., 2000. Elucidation of binding determinants and functional consequences of Ras/Raf-cysteine-rich domain interactions. J. Biol. Chem. 275 22172–22179. [DOI] [PubMed] [Google Scholar]
- Woods, D., D. Parry, H. Cherwinski, E. Bosch, E. Lees et al., 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 9 5598–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, D., H. Cherwinski, E. Venetsanakos, A. Bhat, S. Gysin et al., 2001. Induction of beta3-integrin gene expression by sustained activation of the Ras-regulated Raf-MEK-extracellular signal-regulated kinase signaling pathway. Mol. Cell. Biol. 21 3192–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]