Abstract
Postzygotic reproductive isolation evolves when hybrid incompatibilities accumulate between diverging populations. Here, I examine the genetic basis of hybrid male sterility between two species of Drosophila, Drosophila virilis and D. americana. From these analyses, I reach several conclusions. First, neither species carries any autosomal dominant hybrid male sterility alleles: reciprocal F1 hybrid males are perfectly fertile. Second, later generation (backcross and F2) hybrid male sterility between D. virilis and D. americana is not polygenic. In fact, I identified only three genetically independent incompatibilities that cause hybrid male sterility. Remarkably, each of these incompatibilities involves the Y chromosome. In one direction of the cross, the D. americana Y is incompatible with recessive D. virilis alleles at loci on chromosomes 2 and 5. In the other direction, the D. virilis Y chromosome causes hybrid male sterility in combination with recessive D. americana alleles at a single QTL on chromosome 5. Finally, in contrast with findings from other Drosophila species pairs, the X chromosome has only a modest effect on hybrid male sterility between D. virilis and D. americana.
SPECIATION occurs when populations evolve one or more barriers to interbreeding (Dobzhansky 1937; Mayr 1963). One such barrier is intrinsic postzygotic isolation, which typically evolves when diverging populations accumulate different alleles at two or more loci that are incompatible when brought together in hybrid genomes; negative epistasis between these alleles renders hybrids inviable or sterile (Bateson 1909; Dobzhansky 1937; Muller 1942). Classical and recent studies in diverse animal taxa have provided support for two evolutionary patterns that often characterize the genetics of postzygotic isolation (Coyne and Orr 1989a). The first, Haldane's rule, observes that when there is F1 hybrid inviability or sterility that affects only one sex, it is almost always the heterogametic sex (Haldane 1922). Over the years, many researchers have tried to account for this pattern, but only two ideas are now thought to provide a general explanation: the “dominance theory,” which posits that incompatibility alleles are generally recessive in hybrids, and the “faster-male theory,” which posits that genes causing hybrid male sterility diverge more rapidly than those causing hybrid female sterility (Muller 1942; Wu and Davis 1993; Turelli and Orr 1995; reviewed in Coyne and Orr 2004). In some cases, however, additional factors might contribute to Haldane's rule, including meiotic drive, a faster-evolving X chromosome, dosage compensation, and Y chromosome incompatibilities (reviewed in Laurie 1997; Turelli and Orr 2000; Coyne and Orr 2004).
The second broad pattern affecting the evolution of postzygotic isolation is the disproportionately large effect of the X chromosome on heterogametic F1 hybrid sterility (Coyne 1992). This “large X effect” has been documented in genetic analyses of backcross hybrid sterility (e.g., Dobzhansky 1936; Grula and Taylor 1980; Orr 1987; Masly and Presgraves 2007) and inferred from patterns of introgression across natural hybrid zones (e.g., Machado et al. 2002; Saetre et al. 2003; Payseur et al. 2004). However, in only one case has the cause of the large X effect been unambiguously determined: incompatibilities causing hybrid male sterility between Drosophila mauritiana and D. sechellia occur at a higher density on the X than on the autosomes (Masly and Presgraves 2007). Testing the generality of this pattern will require additional high-resolution genetic analyses in diverse taxa (Presgraves 2008). But whatever its causes, there is now general consensus that the X chromosome often plays a special role in the evolution of postzygotic isolation (Coyne and Orr 2004).
The contribution of the Y chromosome to animal speciation is less clear. Y chromosomes have far fewer genes than the X or autosomes, and most of these genes are male specific (Lahn and Page 1997; Carvalho et al. 2009). In Drosophila species, the Y chromosome is typically required for male fertility, but not for viability (Voelker and Kojima 1971). How often, then, does the Y chromosome play a role in reproductive isolation? In crosses between Drosophila species, hybrid male sterility is frequently caused by incompatibilities between the X and Y chromosomes (Schafer 1978; Heikkinen and Lumme 1998; Mishra and Singh 2007) or between the Y and heterospecific autosomal alleles (Patterson and Stone 1952; Vigneault and Zouros 1986; Lamnissou et al. 1996). In crosses between D. yakuba and D. santomea, the Y chromosome causes F1 hybrid male sterility, and accordingly, shows no evidence for recent introgression across a species hybrid zone (Coyne et al. 2004; Llopart et al. 2005). In mammals, reduced introgression of Y-linked loci (relative to autosomal loci) has been shown across natural hybrid zones of mice (Tucker et al. 1992) and rabbits (Geraldes et al. 2008), suggesting that the Y chromosome contributes to reproductive barriers.
Here I examine the genetic basis of hybrid male sterility between two species of Drosophila, D. virilis and D. americana. These species show considerable genetic divergence (Ks ∼0.11, Morales-Hojas et al. 2008) and are currently allopatric: D. virilis is a human commensal worldwide with natural populations in Asia, and D. americana is found in riparian habitats throughout much of North America (Throckmorton 1982; McAllister 2002). Nearly 70 years ago, Patterson et al. (1942) showed that incompatibilities between the D. americana Y chromosome and the second and fifth chromosomes from D. virilis cause hybrid male sterility, a result that was confirmed in a more recent study (Lamnissou et al. 1996). Another study suggested that the X chromosome might play the predominant role in causing hybrid male sterility between D. virilis and D. americana (Orr and Coyne 1989). But because previous genetic analyses had to rely on only a few visible markers to map hybrid male sterility, they lacked the resolution to examine the genomic distribution of incompatibility loci.
Using the D. virilis genome sequence, I have developed a dense set of molecular markers to investigate the genetic architecture of hybrid male sterility between D. virilis and D. americana. In this study, I perform a comprehensive set of crosses to address several key questions: What is the effect of the X chromosome on hybrid male sterility between D. virilis and D. americana? What is the effect of the Y chromosome? Approximately how many loci contribute to hybrid male sterility between these Drosophila species? Perhaps surprisingly, the answers to these questions differ dramatically from what has been found for other Drosophila species, including the well-studied D. melanogaster group.
MATERIALS AND METHODS
Fly lines and genetic crosses:
I performed crosses between two closely related species of Drosophila, D. virilis and D. americana. There is substantial premating isolation in one direction of the cross—D. americana females with D. virilis males (Stalker 1942). There is also strong postmating, prezygotic isolation that reduces hybrid offspring production between D. virilis females and D. americana males to 1% that of conspecific crosses (Sweigart 2010). Nevertheless, because these barriers are incomplete, male and female hybrids can be generated for genetic analysis.
The D. virilis parental line used here is the genome sequence strain, 15010-1051.87, an inbred line with a visible marker on each of the major autosomes (b; tb, gp-L2; cd; pe). Five D. americana lines were used in this study. Three of these originated as isofemale lines collected by B. McAllister from natural populations: SB 02.06 was collected in 2002 near the Cedar River in Muscatine County, Iowa; CB 05.14 was collected in 2005 near the Corney Bayou in the Kisatchie National Forest, Louisiana; and CD 04.02 was collected near the Columbia Lock and Dam on the Ouachita River in Caldwell Parish, Louisiana (see McAllister 2003; McAllister and Evans 2006; McAllister et al. 2008 for further details). Two D. americana lines were obtained from the Drosophila species stock center: 0951.09 was collected in 1961 in South Carolina and 0951.16 was collected in 2004 in Iowa. In this study, the SB 02.06 strain of D. americana was used for almost all genetic analyses, although the CB 05.14 was used in one mapping experiment. All remaining D. americana lines were used in a single experiment to characterize natural genetic variation (see results).
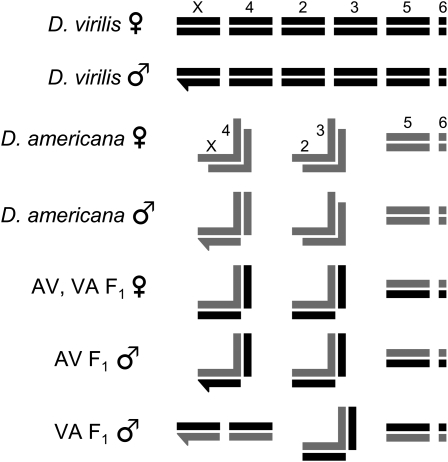
Both D. virilis and D. americana have six chromosome arms (including a dot chromosome; Figure 1). In D. americana, chromosomes 2 and 3 are fused and therefore do not segregate independently in crosses. In addition, D. americana is characterized by a polymorphic centromeric fusion between the X and fourth chromosomes that is positively correlated with latitude (McAllister 2002). The SB 02.06 strain carries the X–4 fusion (McAllister and Evans 2006), which affects segregation in certain crosses (see Figure 1). The X and fourth chromosomes are unfused in the CB 05.14 strain (McAllister et al. 2008). Several chromosomal regions are inverted between D. virilis and D. americana: three inversions are fixed between the species (two on the X chromosome and one on chromosome 2), and several inversions are polymorphic in D. americana, including 5a and 5b on chromosome 5 (Hughes 1939; Hsu 1952). The SB 02.06 strain of D. americana carries the 5b arrangement, whereas CB 05.14 carries 5a.
Figure 1.—
Schematic of D. virilis, D. americana (carrying the X–4 fusion), and F1 hybrid chromosomes. For D. virilis and D. americana females, X chromosomes and autosomes are labeled (order is the same for males and hybrids shown below). The Y and dot chromosomes are represented by hooked bars and small squares, respectively. The X–4 and 2–3 chromosomal fusions of D. americana are represented by connected bars that form backward “L” shapes. AV refers to an F1 hybrid with D. americana as the maternal parent, whereas VA refers to an F1 hybrid with D. virilis as the maternal parent. Note that D. americana males carry one unfused chromosome 4. Only in one direction of the interspecific cross (VA) does the F1 male inherit two unfused copies of chromosome 4, which allows the independent assortment of this chromosome in backcrosses.
To study the genetics of hybrid male sterility between D. virilis and D. americana, I generated reciprocal backcross (BC), F1 and F2 hybrid males. Species names are abbreviated as follows: “V” refers to D. virilis and “A” refers to D. americana. In crosses, the species abbreviation for females is listed first [e.g., VA = D. virilis females × D. americana males; V(VA) = D. virilis females × F1 males (D. virilis females × D. americana males)]. For all crosses, males and females were collected as virgins and maintained separately for 7–10 days to allow them to reach sexual maturity. Following this period, crosses were performed on fresh vials containing standard cornmeal medium at 20° ± 1°.
Assessment of male fertility:
To assay male fertility, I measured sperm motility. Testes were dissected in 1× PBS and examined under a compound microscope with dark-field optics. Following Coyne (1984), a male was scored as fertile if at least one motile sperm was observed and sterile if no motile sperm were observed.
Molecular analyses:
All but one of the molecular markers used in this study were microsatellites. Using the program Tandem Repeats Finder (Benson 1999), I identified candidate microsatellite markers from the D. virilis genome sequence. I then designed primers for these candidate regions using the program Primer3 (Rozen and Skaletsky 2000). One gene-based marker on chromosome 6 (the dot) was designed to amplify an intronic length polymorphism in the gene Caps (exon sequence obtained from Betancourt et al. 2009). I extracted genomic DNA from whole flies using the protocol of Gloor and Engels (1992) and amplified markers using standard touchdown PCR conditions (annealing temperatures incremented from 62° to 52° for the first 10 cycles and then an additional 30 cycles at 52°). Marker genotyping was performed by sizing PCR-amplified DNA fragments with an incorporated 5′ fluorescent-labeled primer on an ABI 3700 automated capillary sequencer (Applied Biosystems, Foster City, CA). Marker genotypes were assigned automatically using the program GeneMapper (Applied Biosystems) and then verified by eye.
Genetic mapping and QTL analyses:
In three separate mapping experiments, linkage groups that correspond to D. virilis chromosomes were constructed using JoinMap 4.0 (Van Ooijen 2006) by assessing the genotypes of F2 hybrid males. The Group function of JoinMap was used with a LOD score threshold of 10.0 to assign markers to linkage groups. The genetic map created for each linkage group used the Kosambi mapping function, a LOD threshold of 1.0, a recombination threshold of 0.400, a jump threshold of 5.00, and a “ripple” after the addition of each locus. In each of the three mapping experiments, marker order corresponded almost perfectly to known physical locations (based on the D. virilis genome sequence); the only two exceptions occurred in regions characterized by strong segregation distortion or low marker density. The order of a few markers on the affected linkage groups thus differed from physical locations. In these two cases, fixed marker orders were specified (on the basis of physical locations) before assembling linkage groups.
I mapped quantitative trait loci (QTL) for F2 hybrid male sterility between D. virilis and D. americana using composite interval mapping (CIM) (Zeng 1993, 1994) in Windows QTL Cartographer V. 2.5 (Wang et al. 2007). Co-factors included in each CIM model were determined with forward–backward stepwise regression, with the critical P-values set at 0.05. Tests were performed at 2-cM intervals with a flanking window size of 10 cM. Significance thresholds were set by permutation (experimentwise type I error rate of α = 0.05, n = 1000).
The X–4 chromosomal fusion carried by the SB 02.06 strain of D. americana affects patterns of segregation in F2 males, and thus, presents a challenge for linkage group assembly and QTL mapping. In F2 males that derive from a cross between VA F1 females and males, segregation of fourth chromosome markers is affected by the D. americana X–4 fusion in F1 mothers and independent assortment of the “free” fourth chromosome in F1 fathers (see Figure 1). As a result, in the (VA)(VA) F2 mapping population, markers on the X and fourth chromosomes show significant linkage (reflecting the D. americana X–4 fusion in F1 mothers), but map distances are high at markers that flank the fusion (reflecting independent assortment in F1 fathers). The situation is different in F2 males that derive from a cross between AV F1 females and males. In this case, all F2 males inherit a D. virilis fourth chromosome from their father (i.e., the X and fourth chromosomes do not assort independently in AV F1 males). Therefore, variation between markers flanking the X–4 fusion must be due to crossovers in F1 mothers. Note that heterozygous markers on the fourth chromosome indicate inheritance of a D. americana allele from the F1 mother. To reflect this fact, in the (AV)(AV) F2 mapping population, I recoded heterozygous genotypes on the fourth chromosome as D. americana homozygotes for linkage analysis and QTL mapping.
RESULTS
Hybrid male fertility:
To examine the genetic basis of hybrid male sterility, I crossed D. virilis and D. americana (strain SB 02.06) and compared the proportion of fertile males among F1, F2, BC, and parental classes. Males from both parental lines were highly fertile, as were VA and AV F1 hybrid males (Table 1). The lack of F1 hybrid male sterility implies a negligible role for incompatibilities between the X and Y chromosomes or between either sex chromosome and dominant heterospecific autosomal alleles.
TABLE 1.
Fertility of parental, F1, backcross and F2 hybrid males
| Males | Proportion fertile (N) |
|---|---|
| D. virilis (1051.87) | 0.94 (122) |
| D. americana (SB 02.06) | 1.00 (107) |
| VA | 0.94 (31) |
| AV | 0.97 (68) |
| V(VA) | 0.30 (67) |
| V(AV) | 0.94 (101) |
| A(VA) | 0.98 (57) |
| A(AV) | 0.61 (75) |
| (VA)(VA) | 0.49 (572) |
| (AV)(AV) | 0.68 (367) |
In this table and Tables 2 and 3, species names are abbreviated as follows: “V” refers to D. virilis and “A” refers to D. americana. In crosses, the species abbreviation for females is listed first [e.g., VA = D. virilis females × D. americana males; V(VA) = D. virilis females × F1 males (D. virilis females × D. americana males)]. Criteria for fertility are given in the text.
Backcross hybrid male sterility was common, but only in particular crosses. When crossed to D. virilis females, reciprocal F1 males produced very different patterns of male sterility among BC progeny: only 30% of V(VA) males were fertile, compared to 94% of V(AV) males. Similarly, when crossed to D. americana females, reciprocal F1 males gave different results: only 61% of A(AV) males were fertile, compared to 98% of A(VA) males. Because BC males are perfectly fertile when they carry the Y chromosome of the recurrent parent, D. virilis–D. americana hybrid male sterility clearly involves the Y chromosome. Indeed, the pattern of BC male sterility suggests that both species's Y chromosomes are incompatible with heterospecific recessive autosomal alleles.
Because all backcrosses were made using F1 hybrids as the paternal parents, I could determine whether autosomes carry incompatibility loci. Note that the lack of crossing over in Drosophila males means that a single marker identifies the species origin of an entire chromosome. All V(VA) males carry a D. americana Y chromosome; however, males were sterile only when they also carried D. virilis chromosomes 2 and 3 (which do not segregate independently because they are fused in D. americana) or chromosome 5 (C2–3: X2 = 48.2, P < 0.0001, N = 67; C5: X2 = 20.4, P < 0.0001, N = 66). Chromosome 4 had no significant effect on V(VA) male fertility (X2 = 0.5, P = 0.47, N = 67). The effect of chromosome 6 (the dot) was not measured for this cross, but was assessed in a later experiment (see D. virilis–D. americana F2 results below). In the A(AV) BC population, all males carried a D. virilis Y chromosome, but they were significantly more likely to be sterile if they also carried homozygous D. americana chromosomes 2, 3, and 5 (C2–3: X2 = 19.5, P < 0.0001, N = 74; C5: X2 = 10.6, P = 0.0012, N = 67; C6: X2 = 0.5, P = 0.55, N = 59; chromosome 4 does not segregate in this cross). Note that all A(AV) BC males are homozygous for the D. virilis fourth chromosome (see Figure 1), which, in addition to the Y, might be incompatible with D. americana alleles; however, the lack of hybrid sterility among A(VA) males (see above) rules out a role for such autosomal–autosomal incompatibilities. Taken together, these results suggest that BC male sterility is caused by several independent incompatibilities; both the D. americana and D. virilis Y chromosomes are incompatible with homozygous, heterospecific alleles on chromosomes 2–3 and 5.
I also examined the pattern of hybrid male sterility among F2 hybrids. Roughly half of all (VA)(VA) F2 hybrid males were sterile, which is close to the proportion expected if D. virilis alleles at only two autosomal loci are each incompatible with the D. americana Y chromosome [0.25 + (0.25(1–0.25)) = 0.44]. For (AV)(AV) F2 hybrids, 32% of males were sterile, which is close to the 25% expected if D. americana alleles at a single locus are incompatible with the D. virilis Y chromosome. This pattern of F2 hybrid male sterility is at least consistent with the idea that Y-autosomal incompatibilities are the primary cause of hybrid fertility problems between D. virilis and D. americana. Indeed, this idea is confirmed below by more direct experiments.
Genetic mapping of D. virilis–D. americana F2 hybrid male sterility:
To genetically dissect hybrid male sterility between D. virilis and D. americana, I generated a large F2 mapping population by crossing VA F1 females and males [(VA)(VA) F2, N = 572]. Because these (VA)(VA) F2 males carry a D. americana Y chromosome against a segregating genetic background, this population allows mapping of the incompatibility loci on chromosomes 2–3 and 5.
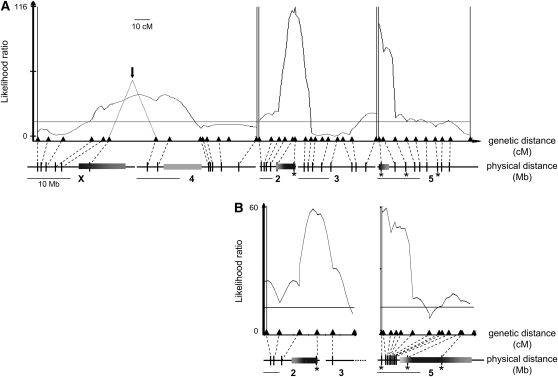
My analyses identified two highly significant QTL for hybrid male fertility on chromosomes 2 and 5 (Figure 2A). As predicted for loci that interact with the D. americana Y chromosome, when either QTL was homozygous for D. virilis alleles, F2 hybrid males were more likely to be sterile. On chromosome 2, the QTL mapped to an inversion that is fixed between D. virilis and D. americana; this genomic region is roughly half the length of the chromosome (Hughes 1939). On chromosome 5, the QTL mapped to the 5b inversion, which is polymorphic in D. americana (Hsu 1952). Using a different, noninverted strain of D. americana, I later dissect this chromosome 5 QTL (see below).
Figure 2.—
Genetic mapping of D. virilis–D. americana F2 hybrid male sterility. Likelihood ratio (LR) test statistic profiles from composite interval mapping (CIM) of male fertility in (VA)(VA) F2 mapping populations. Horizontal lines mark LR significance thresholds and vertical double lines delineate unlinked regions. The genetic positions of molecular markers are indicated by triangles, and the corresponding physical locations along chromosomes X, 4, 2, 3, and 5 (based on the D. virilis genome assembly) are indicated below by vertical bars. Physical distance is shown scaled to genetic distance, and therefore, differs among chromosomes; to the left of the label for each chromosome is a 10-Mb scale bar. The shaded horizontal bars on chromosomes denote inverted regions, with lighter shading representing uncertainty in precise physical locations. To facilitate comparisons of QTL peak positions between mapping experiments, a subset of molecular markers genotyped in both F2 mapping populations are indicated by asterisks. (A) Genomewide mapping of hybrid male sterility in (VA)(VA) F2 males generated using the SB 02.06 D. americana strain, which carries the 5b inversion on chromosome 5 (N = 572). Note that markers flanking the X–4 chromosomal fusion should be more tightly linked than they appear here because genotypic variation via recombination events are confounded by independent assortment of the paternal fourth chromosome. As a result, the QTL associated with these markers (indicated by dotted lines and a black arrow) appears somewhat wider than it should. LR significance threshold (indicated by a horizontal line) is equal to 12.9. See Figure S1 for a slightly different analysis that excludes paternal fourth chromosome haplotypes, but still yields similar QTL mapping results. (B) Targeted mapping of hybrid male sterility on chromosomes 2 and 5 in (VA)(VA) F2 males generated using the CB 05.14 D. americana strain, which carries the 5a inversion (but not 5b) on chromosome 5 (N = 459). LR significance threshold is equal to 11.3.
Three additional, marginally significant QTL for F2 hybrid male fertility mapped to chromosomes 3 and 5, and near the junction of the X–4 chromosomal fusion (Figure 2). Note, however, that the genetic distance covered by the X–4 QTL is artificially inflated as recombination events in females are confounded by independent assortment of the “free” fourth chromosome in males (see materials and methods and Figure 1). A slightly different analysis that excludes paternal fourth chromosome haplotypes (and, therefore, the effect of independent assortment in F1 fathers) yields similar QTL mapping results (Figure S1). Although the precise location of the X–4 QTL is uncertain, the marker with the largest individual effect on male fertility (SSR9: X2 = 41.6, P < 0.0001) is the one closest to the fusion on chromosome 4. In any case, D. virilis alleles at any of these three QTL resulted in only a modest reduction in hybrid male fertility. Also note that a marker on chromosome 6 (the dot) showed no association with hybrid male fertility when examined in a subset of the F2 mapping population (X2 = 1.8, P = 0.42, N = 168).
Taken together, these mapping results suggest that D. virilis–D. americana hybrid male sterility is caused primarily by two, potentially simple Y-autosomal incompatibilities.
Fine mapping of D. virilis–D. americana F2 hybrid male sterility on chromosome 5:
To further dissect the major male fertility QTL on chromosome 5, I generated a new F2 mapping population using a strain of D. americana that shows a similar pattern of hybrid male sterility (Table 2), but that does not carry the 5b inversion (CB 05.14 N = 459). As with the results from my previous mapping experiment, one highly significant QTL mapped to the fixed inversion on chromosome 2 and another mapped to the distal end of chromosome 5 (Figure 2B). In this new cross, however, the latter genomic region showed high rates of recombination. This analysis allowed me to map the QTL on chromosome 5 to a narrow interval of 15 cM corresponding to only 3 Mb. The D. americana Y chromosome thus interacts with a single small chromosomal region, and possibly a single locus, on chromosome 5 to cause hybrid male sterility.
TABLE 2.
Fertility of D. americana, F1, backcross and F2 hybrid males
| Males | Proportion fertile (N) |
|---|---|
| D. americana (CB 05.14) | 1.00 (42) |
| VA | 1.00 (20) |
| V(VA) | 0.20 (66) |
| (VA)(VA) | 0.53 (518) |
Genetic mapping of D. americana–D. virilis F2 hybrid male sterility:
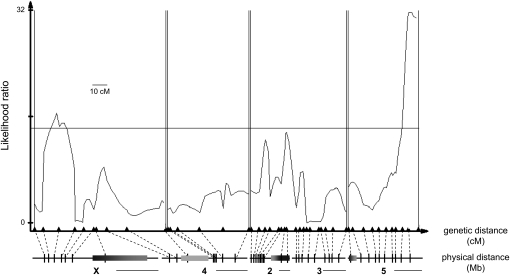
To produce the reciprocal F2 mapping population, I crossed AV F1 females to AV F1 males [(AV)(AV) F2, N = 347)]. The resulting (AV)(AV) F2 males carry a D. virilis Y chromosome against a segregating genetic background. In contrast to the A(AV) backcross analysis, which showed a phenotypic effect of chromosomes 2–3 and 5, I detected only a single autosomal QTL for F2 hybrid male fertility (Figure 3). This QTL mapped to a 12-cM interval (8 Mb) at the proximal end of chromosome 5. In addition, a marginally significant QTL also mapped to the X chromosome. When either QTL is homozygous or hemizygous for D. americana alleles, F2 hybrid males are more likely to be sterile. Aside from the modest effect of the X, these mapping results suggest that D. americana–D. virilis hybrid male sterility is due primarily to a simple incompatibility between the D. virilis Y chromosome and a single QTL on chromosome 5.
Figure 3.—
Genetic mapping of D. americana–D. virilis F2 hybrid male sterility. Likelihood ratio (LR) test statistic profile from composite interval mapping (CIM) of male fertility in the (AV)(AV) F2 mapping population (generated using the SB 02.06 D. americana strain, N = 347). A horizontal line marks the LR significance threshold of 14.3 and vertical double lines delineate unlinked regions. The genetic positions of molecular markers are indicated by triangles, and the corresponding physical locations along chromosomes X, 4, 2, 3, and 5 (based on the D. virilis genome assembly) are indicated below by vertical bars. Physical distance is shown scaled to genetic distance, and therefore, differs among chromosomes; to the right of the label for each chromosome is a 10-Mb scale bar. The shaded horizontal bars on chromosomes denote inverted regions, with lighter shading representing uncertainty in precise physical locations. Note that, in contrast to Figure 2A, markers flanking the X–4 chromosomal fusion are tightly linked. Because (AV)(AV) F2 males must inherit the D. virilis Y and fourth chromosomes together, crossover events at the junction of the X–4 fusion can be easily detected. Also note that markers on the fourth chromosome are split into two linkage groups; additional markers will be required to resolve the genetic map in this region.
Geographic distribution of Y-linked hybrid male sterility loci in D. americana:
To characterize the natural distribution of Y-linked incompatibility loci in D. americana, I generated three additional V(VA) BC male populations using parental strains of D. americana derived from different geographic regions (see materials and methods). As with the SB 02.06 and CB 05.14 strains, these three D. americana strains showed a pattern of hybrid male sterility consistent with the involvement of Y-autosomal incompatibilities (Table 3). In every case, V(VA) males were sterile only when they also carried D. virilis chromosomes 2–3 and/or chromosome 5. These results suggest that Y-linked hybrid male sterility loci are geographically widespread in D. americana.
TABLE 3.
Fertility of D. americana and backcross hybrid males
| Males | Proportion fertile (N) |
|---|---|
| D. americana (CD 04.02) | 0.96 (27) |
| V(VA) | 0.29 (38) |
| D. americana (0951.09) | 1.00 (77) |
| V(VA) | 0.21 (71) |
| D. americana (0951.16) | 1.00 (20) |
| V(VA) | 0.16 (37) |
DISCUSSION
Here I have characterized the genetic basis of hybrid male sterility between D. virilis and D. americana. Several findings emerge from this analysis. First, I observe no evidence for F1 hybrid male sterility in either direction of the cross: both VA and AV hybrid males are highly fertile. Second, in later-generation hybrids (BC and F2) male sterility is primarily caused by three independent, potentially simple genetic incompatibilities—two of these incompatibilities occur in hybrid males from the cross between D. virilis females and D. americana males, and one occurs in hybrid males from the reciprocal cross. Third, each of these incompatibilities involves the Y chromosome. Fourth, I detect only a modest effect of the X chromosome on hybrid male sterility between D. virilis and D. americana.
The results from my genetic analysis show that hybrid male sterility between D. virilis and D. americana can be explained by Y-autosomal incompatibilities. The D. americana Y is incompatible with loci on the second and fifth chromosomes; males that are homozygous for D. virilis alleles at either autosomal locus are sterile. These Y-linked incompatibility alleles appear to be geographically widespread in D. americana: in all of the five strains tested, I observed significant effects of chromosomes 2 and 5 on fertility in V(VA) BC males. Moreover, Y chromosomes sampled from two D. americana populations that are geographically separated by 1100 kb (SB 02.06 and CB 05.14) interact with D. virilis alleles at loci that map to the same genomic regions (Figure 2).
This strong effect of the Y chromosome on postzygotic isolation is not limited to D. americana; in the other direction of the cross, hybrid male sterility is caused by an incompatibility between the D. virilis Y and D. americana alleles at a locus on chromosome 5. This symmetry of Y-linked effects suggests there has been independent evolution of the Y chromosome affecting reproductive isolation within each lineage. Interestingly, the Y chromosome of D. novamexicana, another closely related North American species, causes hybrid male sterility when paired with the X chromosome from D. virilis (Heikkinen and Lumme 1998), revealing yet another Y-linked incompatibility in this species group. Taken together, these results suggest that the Y chromosome is particularly important for postzygotic isolation among species of the D. virilis group.
What mechanisms might account for the prevalence of Y-linked incompatibilities between D. virilis and D. americana? As with the X chromosome, Y-linked hybrid male sterility can be explained by the dominance and faster male theories. In addition, at least two other explanations are possible. First, a history of Y-linked meiotic drive could cause hybrid male sterility (Frank 1991; Hurst and Pomiankowski 1991); an analogous phenomenon has been seen in some instances of X-linked drive (Tao and Hartl 2003; Phadnis and Orr 2008). However, because of their relative strength and tendency to cause male-biased sex ratios, meiotic drivers on the Y might be less common than those on the X chromosome (Hamilton 1967). Second, a propensity for gene transposition on the Y relative to other chromosomes might explain the pattern of Y-linked hybrid male sterility. A recent survey of Y-linked genes among the 12 sequenced Drosophila species found low conservation of gene content (Koerich et al. 2008). If gene movement on and off the Y chromosome has occurred repeatedly in the D. virilis group, crosses between species might yield hybrids that lack a full complement of essential male fertility factors (see Masly et al. 2006; Lynch and Force 2000). To investigate this possibility, I designed primers to PCR amplify an exon from each of the six genes known to be Y-linked in D. virilis (kl-2, kl-3, kl-5, ORY, PPr-Y, and PRY: see Koerich et al. 2008). All amplicons were male specific in both D. virilis and D. americana (data not shown), suggesting that these six genes reside on the Y chromosome in both species. Until additional D. virilis Y-linked genes are identified, the question of whether Y-autosomal transpositions underlie postzygotic isolation must be addressed by fine mapping and screening for gene content variation between species.
Crosses between many other Drosophila species show Y-linked incompatibilities (Turelli and Orr 2000). Typically, however, these studies also find very large effects of the X chromosome on hybrid male sterility (e.g., Dobzhansky 1936; Sturtevant and Novitski 1941; Coyne and Kreitman 1984; Naveira and Fontdevila 1986; Orr 1987; Zouros et al. 1988; Masly and Presgraves 2007). In my analysis, however, I find no evidence for a “large X effect” in F2 hybrid males between D. virilis and D. americana. Accordingly, I find no evidence for a higher density of X-linked hybrid sterility factors, as has been found in crosses between D. mauritiana and closely related species (True et al. 1996; Tao et al. 2003; Masly and Presgraves 2007).
It also appears that hybrid male sterility between D. virilis and D. americana may be relatively genetically simple. In the well-studied D. melanogaster group, even closely related species carry many incompatibility loci: 17 genomic regions cause hybrid male sterility in advanced introgression lines between D. sechellia and D. mauritiana (Masly and Presgraves 2007; D. sechellia–D. mauritiana Ks ∼0.051, Kern et al. 2004), and all F1 hybrid males are sterile (Lachaise et al. 1986). The situation is even more dramatic in older species pairs. For example, in crosses between D. melanogaster and D. simulans (Ks ∼0.11, Shapiro et al. 2007), all F1 hybrids are dead or sterile (Sturtevant 1920). Advanced crosses using sophisticated genetic trickery (i.e., X-ray irradiation or hybrid rescue mutations) to overcome F1 isolation have shown that hybrid male sterility is highly polygenic and complex (Pontecorvo 1943; Sawumura et al. 2000). Indeed, even the tiny dot chromosome causes hybrid male sterility in these species (Muller and Pontecorvo 1940; Masly et al. 2006). In marked contrast, only a few QTL contribute to hybrid male sterility between D. virilis and D. americana, two species distinguished by roughly the same degree of synonymous genetic divergence as D. melanogaster and D. simulans (D. virilis–D. americana Ks ∼0.11, Morales-Hojas et al. 2008). In light of this difference, it is interesting to note that both species groups might be exceptional to some degree (and in the expected directions): comparing the rates at which postzygotic isolation evolves between Drosophila species pairs, D. melanogaster species are among the fastest evolving and D. virilis species the slowest (see figure 2 in Coyne and Orr 1989b).
All three autosomal hybrid male sterility QTL identified here map to regions associated with inversions in D. americana—one on chromosome 2 that is fixed, and two on chromosome 5 that are polymorphic. Recently, I showed the same inversion on chromosome 2 also carries incompatibility factors that disrupt the fertilization of D. virilis eggs by D. americana sperm (Sweigart 2010). Several empirical and theoretical studies have suggested that chromosomal rearrangements might facilitate the evolution of reproductive isolation (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003). This scenario may be unlikely for incompatibility loci on chromosome 5, as hybrid male sterility is not restricted to D. americana strains that carry the 5b inversion. However, it is possible that the fixed inversion on chromosome 2 affected male–female coevolution in fertilization molecules and/or the accumulation of hybrid sterility loci in D. americana. Unfortunately, the inability to perform recombinational mapping in this genomic region will make it difficult to disentangle this evolutionary history. Instead, future studies will focus on the two hybrid male sterility loci that map to chromosome 5, which carries the polymorphic 5a and 5b inversions. Using the appropriate, collinear strains of D. americana, it should be possible to fine map and identify isolation loci in these regions, a key step for understanding the genetic and evolutionary mechanisms of D. virilis–D. americana hybrid male sterility.
Acknowledgments
I am grateful to Bryant McAllister for generously providing the wild-collected strains of D. americana and to E. Landeen for technical help. I thank S. Kingan, E. Landeen, D. McNabney, C. Meiklejohn, D. Presgraves, and especially H. A. Orr for helpful discussions about this project. I also thank D. Begun, C. Meiklejohn, H. A. Orr, D. Presgraves, and two anonymous reviewers for helpful comments on a draft of this article. This research was supported by funds from a National Institutes of Health (NIH) Ruth L. Kirschstein National Research Service Award postdoctoral fellowship (GM-078974) to the author and by funds from the NIH (GM-51932) to H. A. Orr.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.112896/DC1.
References
- Bateson, W., 1909. Heredity and variation in modern lights, pp. 85–101 in Darwin and Modern Science, edited by A. C. Seward. Cambridge University Press, Cambridge, UK.
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt, A. J., J. J. Welch and B. Charlesworth, 2009. Reduced effectiveness of selection caused by a lack of recombination. Curr. Biol. 19 655–660. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. B., L. B. Koerich and A. G. Clark, 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 25 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., 1984. Genetic basis of male sterility in hybrids between two closely related species of Drosophila. Proc. Natl. Acad. Sci. USA 81 4444–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., 1992. Genetics and speciation. Nature 355 511–515. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and M. Kreitman, 1984. Evolutionary genetics of two sibling species, Drosophila simulans and D. sechellia. Evolution 40 673–691. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. a Two rules of speciation, pp. 180–207 in Speciation And Its Consequences, edited by D. Otte and J. Endler. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., and H. A. Orr, 1989. b Patterns of speciation in Drosophila. Evolution 43 362–381. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., S. Elwyn, S. Y. Kim and A. Llopart, 2004. Genetic studies of two sister species in the Drosophila melanogaster subgroup, D. yakuba and D. santomea. Genet. Res. 84 11–26. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T. H., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T. H., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Frank, S. H., 1991. Divergence of meiotic drive-suppressors as an explanation for sex-biased hybrid sterility and inviability. Evolution 45 262–267. [DOI] [PubMed] [Google Scholar]
- Geraldes, A., M. Carneiro, M. Delibes-Mateos, R. Villafuerte, M. W. Nachman et al., 2008. Reduced introgression of the Y chromosome between subspecies of the European rabbit (Oryctolagus cuniculus) in the Iberian Peninsula. Mol. Ecol. 17 4489–4499. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., and W. R. Engels, 1992. Single-fly DNA preps for PCR. Dros. Inf. Serv. 71 148–149. [Google Scholar]
- Grula, J. W., and O. R. Taylor, 1980. Some characteristics of hybrids derived from the sulfur butterflies Colias eurytheme and C. philodice: phenotypic effects of the X-chromosome. Evolution 34 673–687. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1922. Sex ratio and unisexual sterility in animal hybrids. J. Genet. 12 101–109. [Google Scholar]
- Hamilton, W. D., 1967. Extraordinary sex ratios. Science 156 477–488. [DOI] [PubMed] [Google Scholar]
- Heikkinen, E., and J. Lumme, 1998. The Y chromosomes of Drosophila lummei and D. novamexicana differ in fertility factors. Heredity 81 505–513. [DOI] [PubMed] [Google Scholar]
- Hsu, T. C., 1952. Chromosomal variation and evolution in the virilis group of Drosophila. Univ. Texas Publ. 5204 35–72. [Google Scholar]
- Hughes, R. D., 1939. An analysis of the chromosomes of the two sub-species Drosophila virilis virilis and Drosophila virilis americana. Genetics 24 811–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L. D., and A. Pomiankowski, 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics 128 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A. D., C. D. Jones and D. J. Begun, 2004. Molecular population genetics of male accessory gland proteins in the Drosophila simulans complex. Genetics 167 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerich, L. B., X. Wang, A. G. Clark and A. B. Carvalho, 2008. Low conservation of gene content in the Drosophila Y chromosome. Nature 456 949–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., J. R. David, F. Lemeunier, L. Tsacas and M. Ashburner, 1986. The reproductive relationships of Drosophila sechellia with D. mauritiana, D. simulans, and D. melanogaster from the Afrotropical region. Evolution 40 262–271. [DOI] [PubMed] [Google Scholar]
- Lahn, B. T., and D. C. Page, 1997. Functional coherence of the human Y chromosome. Science 278 675–680. [DOI] [PubMed] [Google Scholar]
- Lamnissou, K., M. Loukas and E. Zouros, 1996. Incompatibilities between Y chromosome and autosomes are responsible for male hybrid sterility in crosses between Drosophila virilis and Drosophila texana. Heredity 76 603–609. [DOI] [PubMed] [Google Scholar]
- Laurie, C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane's Rule. Genetics 147 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart, A., D. Lachaise and J. A. Coyne, 2005. Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics 171 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and A. G. Force, 2000. The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156 590–605. [DOI] [PubMed] [Google Scholar]
- Machado, C. A., R. M. Kliman, J. A. Markert and J. Hey, 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19 472–488. [DOI] [PubMed] [Google Scholar]
- Masly, J. P., and D. C. Presgraves, 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5 1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly, J. P., C. D. Jones, M. A. F. Noor, J. Locke and H. A. Orr, 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313 1448–1450. [DOI] [PubMed] [Google Scholar]
- Mayr, E., 1963. Animal Species and Evolution. Belknap Press, Cambridge, MA.
- McAllister, B. F., 2002. Chromosomal and allelic variation in Drosophila americana: selective maintenance of a chromosomal cline. Genome 45 13–21. [DOI] [PubMed] [Google Scholar]
- McAllister, B. F., 2003. Sequence differentiation associated with an inversion on the neo-X chromosome of Drosophila americana. Genetics 165 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, B. F., and A. L. Evans, 2006. Increased nucleotide diversity with transient Y linkage in Drosophila americana. PLoS ONE 1 e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister, B. F., S. L. Sheeley, P. A. Mena, A. L. Evans and C. Schlotterer, 2008. Clinal distribution of a chromosomal rearrangement: A precursor to chromosomal speciation? Evolution 62 1852–1865. [DOI] [PubMed] [Google Scholar]
- Mishra, P. N., and B. N. Singh, 2007. Assessing the putative roles of X-autosome and X-Y interactions in hybrid male sterility of the Drosophila bipectinata species complex. Genome 50 653–659. [DOI] [PubMed] [Google Scholar]
- Morales-Hojas, R., C. P. Vieira and J. Vieira, 2008. Inferring the evolutionary history of Drosophila americana and Drosophila novamexicana using a multilocus approach and the influence of chromosomal rearrangements in single gene analyses. Mol. Ecol. 17 2910–2926. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Muller, H. J., and G. Pontecorvo, 1940. Recombinants between Drosophila species, the F1 hybrids of which are sterile. Nature 146 199. [Google Scholar]
- Navarro, A., and N. H. Barton, 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57 447–459. [DOI] [PubMed] [Google Scholar]
- Naveira, H., and A. Fontdevila, 1986. The evolutionary history of Drosophila buzzatii. XII. The genetic basis of sterility in hybrids between D. buzzatii and its sibling D. serido from Argentina. Genetics 114 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor, M. A. F., K. L. Grams, L. A. Bertucci and J. Reiland, 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1987. Genetics of male and female sterility in hybrids of Drosophila pseudoobscura and D. persimilis. Genetics 116 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and J. A. Coyne, 1989. The genetics of postzygotic isolation in the Drosophila virilis group. Genetics 121 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J. T., and W. S. Stone, 1952. Evolution in the Genus Drosophila. Macmillan, New York.
- Patterson, J. T., W. S. Stone and R. K. Griffin, 1942. Genetic and cytological analysis of the virilis species group. Univ. Texas Publ. 4228 162–200. [Google Scholar]
- Payseur, B. A., J. G. Krenz and M. W. Nachman, 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mouse. Evolution 58 2064–2078. [DOI] [PubMed] [Google Scholar]
- Phadnis, N., and H. A. Orr, 2008. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo, G., 1943. Hybrid sterility in artificially produced recombinants between Drosophila melanogaster and D. simulans. Proc. R. Soc. Edinb. Sect. B (Biol.) 61 385–397. [Google Scholar]
- Presgraves, D. C., 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16 351–358. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Saetre, G.-P., T. Borge, K. Lindroos, J. Haavbie, B. C. Sheldon et al., 2003. Sex chromosome evolution and speciation in flycatchers. Proc. R. Soc. Lond. Ser. B 270 58–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawumura, K., A. W. Davis and C.-I. Wu, 2000. Genetic analysis of speciation by means of introgression into Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97 2652–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, U., 1978. Sterility in Drosophila hydei × D. neohydei hybrids. Genetica 49 205–214. [Google Scholar]
- Shapiro, J. A., W. Huang, C. Zhang, M. J. Hubisz, J. Lu et al., 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker, H. D., 1942. Sexual isolation studies in the species complex Drosophila virilis. Genetics 27 238–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., 1920. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster. Genetics 5 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A. H., and E. Novitski, 1941. Sterility in crosses of geographical races of Drosophila micromelanica. Proc. Natl. Acad. Sci. USA 27 392–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart, A. L., 2010. The genetics of postmating, prezygotic reproductive isolation between Drosophila virilis and D. americana. Genetics 184 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., and D. L. Hartl, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane's rule. Evolution 57 2580–2598. [DOI] [PubMed] [Google Scholar]
- Tao, Y., S. Chen, D. L. Hartl and C. C. Laurie, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164 1383–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throckmorton, L. H., 1982. The virilis species group, pp. 227–296 in The Genetics and Biology of Drosophila, Vol. 3b, edited by M. Ashburner, H. L. Carson and J. N. Thompson, Jr. Academic Press, London.
- True, J. R., B. S. Weir and C. C. Laurie, 1996. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, P. K., R. D. Sage, J. Warner, A. C. Wilson and E. M. Eicher, 1992. Abrupt cline for sex-chromosomes in a hybrid zone between 2 species of mice. Evolution 46 1146–1163. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 1995. The dominance theory of Haldane's rule. Genetics 140 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen, J. W., 2006. JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma B. V., Wageningen, The Netherlands.
- Vigneault, G., and E. Zouros, 1986. The genetics of asymmetrical male sterility in Drosophila mohavensis and Drosophila arizonensis hybrids: interactions between the Y-chromosome and autosomes. Evolution 40 1160–1170. [DOI] [PubMed] [Google Scholar]
- Voelker, R. A., and K.-I. Kojima, 1971. Fertility and fitness of XO males in Drosophila I. Qualitative study. Evolution 25 119–128. [DOI] [PubMed] [Google Scholar]
- Wang, S., C. J. Basten and Z.-B. Zeng, 2007. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC.
- Wu, C.-I., and A. W. Davis, 1993. Evolution of postmating reproductive isolation—the composite nature of Haldane's rule and its genetic bases. Am. Nat. 142 187–212. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. B., 1993. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros, E., K. Lofdahl and P. A. Martin, 1988. Male hybrid sterility in Drosophila: interactions between autosomes and sex chromosomes in crosses of D. mohavensis and D. arizonensis. Evolution 42 1321–1331. [DOI] [PubMed] [Google Scholar]