Abstract
Variation in maize for response to photoperiod is related to geographical adaptation in the species. Maize possesses homologs of many genes identified as regulators of flowering time in other species, but their relation to the natural variation for photoperiod response in maize is unknown. Candidate gene sequences were mapped in four populations created by crossing two temperate inbred lines to two photoperiod-sensitive tropical inbreds. Whole-genome scans were conducted by high-density genotyping of the populations, which were phenotyped over 3 years in both short- and long-day environments. Joint multiple population analysis identified genomic regions controlling photoperiod responses in flowering time, plant height, and total leaf number. Four key genome regions controlling photoperiod response across populations were identified, referred to as ZmPR1–4. Functional allelic differences within these regions among phenotypically similar founders suggest distinct evolutionary trajectories for photoperiod adaptation in maize. These regions encompass candidate genes CCA/LHY, CONZ1, CRY2, ELF4, GHD7, VGT1, HY1/SE5, TOC1/PRR7/PPD-1, PIF3, ZCN8, and ZCN19.
MAIZE (Zea mays L. subsp. mays) was domesticated in southern Mexico and its center of diversity is in tropical Latin America (Goodman 1999; Matsuoka et al. 2002), where precipitation rates and day lengths cycle annually. The presumed ancestor of maize, teosinte (Zea mays L. subsp. parviglumis), likely evolved photoperiod sensitivity to synchronize its reproductive phases to the wetter, short-day growing season (Ribaut et al. 1996; Campos et al. 2006). A critical event in the postdomestication evolution of maize was its spread from tropical to temperate regions of the Americas (Goodman 1988), requiring adaptation to longer day lengths. The result of this adaptation process is manifested today as a major genetic differentiation between temperate and tropical maize (Liu et al. 2003) and substantially reduced photoperiod sensitivity of temperate maize (Gouesnard et al. 2002). Tropical maize exhibits delayed flowering time, increased plant height, and a greater total leaf number when grown in temperate latitudes with daily dark periods <11 hr (Allison and Daynard 1979; Warrington and Kanemasu 1983a,b). Identifying the genes underlying maize photoperiod sensitivity will provide insight into the postdomestication evolution of maize and may reduce barriers to the use of diverse tropical germplasm resources for improving temperate maize production (Holland and Goodman 1995; Liu et al. 2003; Ducrocq et al. 2009).
Natural variation at key genes in flowering time pathways is related to adaptation and evolution of diverse plant species (Caicedo et al. 2004; Shindo et al. 2005; Turner et al. 2005; Cockram et al. 2007; Izawa 2007; Slotte et al. 2007). Identification of some of the genes controlling adaptation in numerous plant species relied on regulatory pathways elucidated in Arabidopsis (Simpson and Dean 2002). Many key genes in the Arabidopsis flowering time regulatory pathways are conserved across diverse plant species (Kojima et al. 2002; Hecht et al. 2007; Kwak et al. 2008), but their functions have diverged, resulting in unique regulatory pathways in some phylogenetic groups (Colasanti and Coneva 2009). For example, FRI and FLC control most natural variation for vernalization response in Arabidopsis (Caicedo et al. 2004; Shindo et al. 2005), but wheat and barley appear to lack homologs of these genes and regulate vernalization response with different genes (Yan et al. 2004).
Maize exhibits tremendous natural variation for flowering time (Gouesnard et al. 2002; Camus-Kulandaivelu et al. 2006), for which numerous QTL have been identified (Chardon et al. 2004). In contrast, only a few flowering time mutants are known and only a handful of flowering time genes, including DWARF8 (D8), DELAYED FLOWERING1 (DLF1), VEGETATIVE TO GENERATIVE TRANSITION1 (VGT1), and INDETERMINATE GROWTH1 (ID1), have been cloned in maize (Thornsberry et al. 2001; Colasanti et al. 2006; Muszynski et al. 2006; Salvi et al. 2007; Colasanti and Coneva 2009). Variation at or near D8 and VGT1 is related to latitudinal adaptation, but these genes do not appear to regulate photoperiod responses and account for only a limited proportion of the standing flowering time variation in maize (Camus-Kulandaivelu et al. 2006, 2008; Ducrocq et al. 2008; Buckler et al. 2009).
Quantitative trait loci (QTL) mapping was a key first step to identifying the genes underlying natural variation for flowering time in Arabidopsis (Koornneef et al. 2004). Photoperiodic QTL have been mapped previously in individual biparental maize mapping populations (Koester et al. 1993; Moutiq et al. 2002; Wang et al. 2008; Ducrocq et al. 2009). Such studies are informative with respect to the parents from which the populations were derived, but often do not reflect the genetic heterogeneity of broader genetic reference populations (Holland 2007).
Association mapping (Thornsberry et al. 2001; Ersoz et al. 2007) and combined analysis of multiple biparental crosses (Rebaï et al. 1997; Rebaï and Goffinet 2000; Blanc et al. 2006; Verhoeven et al. 2006; Yu et al. 2008) represent alternative approaches to understanding the variation in genetic control for complex traits among diverse germplasm. Association mapping has limited power to identify genes that affect traits closely associated with population structure, such as flowering time in maize (Camus-Kulandaivelu et al. 2006; Ersoz et al. 2007). In contrast, joint QTL analysis of multiple populations is not hindered by the associations between causal genes and population structure. Combined QTL analysis of multiple mapping populations provides improved power to detect QTL, more precise estimation of their effects and positions, and better understanding of their functional allelic variation and distribution across more diverse germplasm compared to single-population mapping (Rebaï et al. 1997; Wu and Jannink 2004; Jourjon et al. 2005; Blanc et al. 2006; Verhoeven et al. 2006; Yu et al. 2008; Buckler et al. 2009). Joint analysis also provides a direct test of the importance of higher-order epistatic interactions between founder alleles at individual loci with genetic backgrounds (Jannink and Jansen 2001; Blanc et al. 2006). In this study, joint analysis of multiple populations was used to test directly the hypothesis that diverse tropical maize lines carry functionally similar alleles at key photoperiod loci, which would imply genetic homogeneity for a common set of mutations and a shared evolutionary pathway for photoperiod insensitivity.
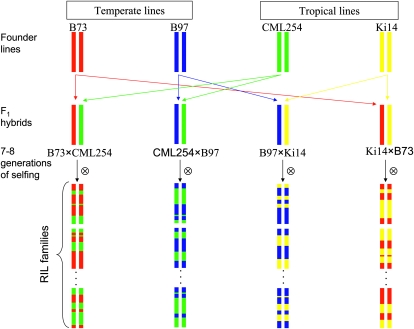
The objective of this study was to integrate candidate gene analyses with photoperiod QTL mapping across multiple maize populations. We tested candidate floral regulators known from other species for associations with natural variation for photoperiod response in maize. We analyzed flowering time in four interrelated recombinant inbred line (RIL) populations, each derived from crosses between temperate and tropical maize parents (Figure 1), in both long- and short-day environments to characterize their responses to distinct photoperiods. Joint population analysis provided high resolution of many QTL positions, permitting robust testing of underlying candidate genes. We directly and indirectly mapped homologs of flowering time candidates genes from Arabidopsis, rice, and barley on a dense consensus genetic map of these four populations, permitting identification of homologs that colocalize with genome regions associated with variation for photoperiod response. These mapping families are being integrated into the maize nested association mapping (NAM) population (Buckler et al. 2009; McMullen et al. 2009) because they were genotyped with the maize NAM map SNP markers, they involve the common parent B73, and their seed and genotypic information (File S1 cont.) are publicly available. Their availability further expands the genetic diversity represented by the maize NAM population and enhances this valuable public community resource.
Figure 1.—
Factorial mating of two temperate (B73 and B97) and two tropical (CML254 and Ki14) inbred maize lines to create four related recombinant inbred line mapping populations.
MATERIALS AND METHODS
Plant materials for recombinant inbred line populations:
We studied maize inbred lines B73, B97, CML254, and Ki14 and recombinant inbred lines derived from crosses among them. Inbred parents for this study were chosen as a sample of maize germplasm diversity from both tropical and temperate groups and on the basis of previously observed photoperiodic responses in summer vs. winter nurseries (http://www.panzea.org). B73 and B97 were both developed at Iowa State University (Russell 1972; Hallauer et al. 1994) and represent two distinct maize germplasm groups for U. S. Corn Belt Dent maize: Stiff Stalk temperate (B73), and non-Stiff Stalk temperate (B97; Liu et al. 2003). CML254 was developed by the International Maize and Wheat Improvement Center (CIMMYT) in Mexico (Srinivasan 2001) and Ki14 by Kasetsart University in Thailand (Chutkaew et al. 1997). CML254 and Ki14 are both tropical, but they represent distinct subgroups of tropical germplasm (Liu et al. 2003).
Recombinant inbred line population development:
We developed four populations of maize RILs by making all possible crosses between temperate (B73 or B97) and tropical (CML254 or Ki14) parental lines (B73 × CML254, CML254 × B97, Ki14 × B73, and B97 × Ki14; Figure 1). The Ki14 × B73 population was developed at The Ohio State University, whereas the other three were developed at North Carolina State University. From F2 progeny of each cross, single seed descent was employed to produce RILs at either the F5:6 or F6:7 generations. Selfing generations alternated between long-day summer nurseries in Clayton, NC or Wooster, OH and short-day winter nurseries in Homestead, FL or Puerto Rico. Substantial losses of lines due to large anthesis–silking intervals (ASI), male and female sterility, and overall poor vigor were observed in all populations, which limited sample sizes of RILs in later generations. RIL seed was increased via full-sib mating and bulking of seed from up to 20 plants per line. In total, after eliminating several lines in each population that carried nonparental marker alleles, we studied 120, 126, 214, and 109 RILs from the B73 × CML254, CML254 × B97, B97 × Ki14, and Ki14 × B73 populations, respectively. Small seed lots of RILs are freely available from J. B. Holland upon request.
RIL experimental design and phenotyping:
Phenotypes were measured under long-day conditions during four summer seasons from 2004 to 2007 at Clayton, NC (latitude 35.7° N, average day length from May 1 to July 31—the typical period from planting to flowering—is 14 hr 10 min from sunrise to sunset). The number of replications per environment is explained in supporting information, Table S1. In all years, locations, and populations, we employed an augmented alpha lattice design with an average of 9% of plots planted to repeated parental checks, which permitted the adjustment of phenotypic data for incomplete block effects (Wolfinger et al. 1997), even for the environment with only one complete replication. An additional long-day measurement of days to anthesis (DTA) was taken in 2006 at Andrews, NC—a location with a similar latitude and day length as Clayton, but at a higher elevation (latitude 35.2° N, 518 vs. 134 meters above sea level). Short-day phenotyping was carried out during three winter seasons from 2005 to 2007 at Homestead, FL (25.4° N, average day length from October 1 to December 31—the typical period from planting to seed filling—is 11 hr and 9 min from sunrise to sunset).
We measured five traits in each plot, including DTA, days to silking (DTS), plant height (PH), ear height (EH), and total leaf number (TLN). We defined DTA and DTS as the number of days between planting and when 50% of the plants in a plot were shedding pollen or exerting silks, respectively. Because of the strong influence of temperature on DTA and DTS, flowering time measurements were converted to growing degree days (GDD) following McMaster and Wilhelm (1997), with 10° as TBASE and 30° as Tmax. GDDTA and GDDTS represent the cumulative average daily heat units that a row of plants received from the time that seeds were sown in the field until anthesis or silking occurred, respectively. In replications where both GDDTA and GDDTS were evaluated, the anthesis–silking interval was also calculated as the difference in GDD between DTA and DTS (GDDASI). PH and EH were measured in centimeters as the distance between the ground and the last leaf node or the primary ear branch, respectively. The earliest emerging leaves naturally senesce before flowering in maize, which prevents measuring total leaf number (TLN) by simply counting visible leaves at the end of the growing season. Therefore, we marked fifth, tenth, and occasionally fifteenth leaves of each measured plant soon after they emerged. The marked tenth leaf was almost always identifiable at the end of the growing season, permitting counts of the total number of leaves on that same plant at the end of the growing season. We measured PH, EH, and TLN on two plants per plot in summer 2006, and on three plants per plot in all subsequent replications.
Phenotypic data analysis:
Data from the RIL populations were analyzed using SAS version 9.1 Proc Mixed (SAS Institute, 2002–2004). Data from summer long-day environments were analyzed separately from data from winter short-day environments. Parents were considered fixed genotypes and RILs were considered random effects, permitting estimation of best linear unbiased predictors (BLUPs) for each RIL, following Piepho et al. (2006). Complete replications, incomplete blocks, environments, and year-by-genotype interactions were also considered random effects. Columns and rows representing the physical layouts of the summer environments were also included as random effects to account for spatial variation. Each combination of the six measured characters and environmental day length (short- or long-day length) was considered a distinct trait for genetic analysis. We also estimated the photoperiodic response of each trait by subtracting the BLUPs for the trait measured in short-day environments from the BLUPs for the trait measured in long-day environments for each RIL within each year and analyzing the response across years. Thus, for each measured characteristic we obtained three distinct BLUPs, representing the genotypic value in short-day environments, in long-day environments, and for the photoperiod response. In total, BLUPs for 18 traits were obtained for each mapping line.
Genotypic data:
Genomic DNA was extracted from the bulked leaf tissue of 10 plants representing each of the RILs using the Invitrogen Charge Switch Genomic DNA extraction kit (Invitrogen, Carlsbad, CA). We used simple sequence repeat (SSR) markers from the maize genetic database (http://www.maizegdb.org) and SNP markers from the Panzea project (http://www.panzea.org) to define linkage groups. SSR markers were genotyped using gel electrophoresis on SFR agarose (Amresco, Solon, OH). SNP markers were genotyped using the Illumina Golden Gate SNP genotyping assay (Fan et al. 2003) with a 1536-locus array. The context sequences for the successful assays from this array can be obtained from http://www.panzea.org/lit/data_sets.html#SNPs, file NAM_snp-080715.xls. This same set of SNP markers was used to create the maize NAM map (www.maizegdb.org; McMullen et al. 2009). Thus, these populations can be incorporated into the overall NAM platform in future genetic analyses to expand the genetic diversity sampled in that mapping platform.
Four of the SNP markers were developed from sequences identical or homologous to flowering time or morphology genes known from maize: D8, ZFL2, FEA2, and HD1. We also developed several markers from maize DNA sequences with high homology to photoperiodic and flowering time genes of Arabidopsis and rice (DNA sequences for these genes from B73 and the other 25 founders of the maize NAM population obtained from Edward Buckler, USDA-ARS), including CHLOROPHYLL A/B BINDING PROTEIN (CAB), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), CONSTANS-LIKE (CO), CRYPTOCHROME 2 (CRY2), HEADING DATE3A/FLOWERING LOCUS T (HD3A), INDETERMINATE-RELATED PROTEIN 7 (ID7), ZEA MAYS FLORICAULA1 (LEAFY), and PHYTOCHROME C1 (PHYC). Sequences were aligned and indels and SNPs called. PCR primer pairs were designed to flank targeted indels or SNPs (Table S3). SNPs were assayed as cleaved amplified polymorphic sequences (CAPS) or derived cleaved amplified polymorphic sequences (dCAPS; Neff et al. 1998). Because the sequencing panel did not include CML254 or Ki14, several CAPS and dCAPS markers were developed for each gene and screened on the founders of our populations to identify polymorphic sequences. In addition, we sequenced alleles of two LATE ELONGATED HYPOCOTYL homologs (LHY and LHYC8) from the four founder lines of the populations studied here and developed CAPS or dCAPS markers for these genes.
All RILs were genotyped at polymorphic SSR and candidate gene sequences, and all RILs in the B73 × CML254, CML254 × B97, and Ki14 × B73 populations were also genotyped for all 1536 SNP markers. Because of limited resources, only a subset consisting of the 15 earliest, 15 latest, and a random sample of 98 RILs out of the remaining 184 total RILs from the B97 × Ki14 population were genotyped with SNPs.
A combined genetic map with the data from the four RIL populations was created. Preliminary genetic order of all loci was established with JoinMap 3.0 (Van Ooijen and Voorrips 2001). The COMPARE feature of Mapmaker/exp 3.0 (Lincoln et al. 1992) was then used to clarify regions where the JoinMap order differed greatly from the order of either the maize NAM or intermated B73 × Mo17 (IBM2) 2008 neighbors maps (http://www.panzea.org; http://www.maizegdb.org). Using Mapmaker, a combined map of the four RIL mapping populations was produced with 1339 (1088 SNP, 241 SSR, and 10 candidate gene) markers in the revised marker order. Individual maps for each of the four populations were then produced utilizing a similar method. A total of 866 (767 SNP, 97 SSR, and 2 candidate gene), 546 (459 SNP, 83 SSR, 4 candidate gene), 561 (476 SNP, 79 SSR, and 4 candidate gene), and 839 (765 SNP and 74 SSR) markers were placed on the genetic maps of B73 × CML254, CML254 × B97, B97 × Ki14, and Ki14 × B73, respectively. Seventy-three markers were mapped on all four populations, and each pair of populations had 221–315 common polymorphic markers.
QTL mapping:
QTL mapping in individual populations was undertaken with MCQTL 4.0 software (Jourjon et al. 2005) on the basis of the RIL BLUPs. Permutation analysis (Churchill and Doerge 1994), based on 1000 permutations for each trait, was used to estimate empirically the genomewide α = 0.05 LOD threshold rates for each trait in each of the RIL populations. The automated iterative QTL mapping (iQTLm) procedure of Charcosset et al. (2000) was used to search for QTL for each trait. In each population, cofactors were selected using 0.9 times the LOD threshold estimated for each specific combination of model and trait as recommended by Delannoy et al. (2006). Significant QTL were declared when LOD scores exceeded the genomewide α = 0.05 threshold.
Two combined population QTL mapping models (connected and disconnected models) were tested for each trait with MCQTL. The connected analysis assumes that the founder parental allele effects are consistent across populations, so that four allele effects were estimated (with three degrees of freedom) at each QTL, whereas the disconnected analysis assumes that founder parent allele effects vary across populations, resulting in eight allele-by-population effect estimates (with four degrees of freedom) at each QTL (Blanc et al. 2006; Mangin et al. 2007). Permutation analyses and iQTLm procedures were used to search for QTL for each trait under both connected and disconnected models in the same way as for individual populations. The final connected and disconnected models for each trait were compared using Schwarz's Bayesian information criterion (BIC), and the model with the better BIC value was selected. QTL effects and 2-LOD support intervals were estimated from the final model with best BIC for each trait. Allelic effects at each locus were compared with approximate t-tests (α = 0.05).
We tested for QTL × QTL epistasis using the epistasy function of MCQTL, which searches for any genome positions that interact with the QTL previously declared with significant effects. When epistatic interactions were detected, the BIC of the QTL models with and without epistasis were compared to judge their significance.
Inferring candidate gene locations:
Candidate gene positions were inferred on the combined map if they could not be mapped directly due to lack of a polymorphic gene marker. First, a list of the key candidate photoperiod/flowering genes from Arabidopsis, rice, maize, and barley was created (Table S4). For genes previously identified in other species, we searched for their maize homologs using BLAST-X at the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/). DNA sequences originating from maize were considered to be homologous to candidate genes from other plant species if their BLAST-X homologies were smaller than E = 1.0 × e−15. If no homolog was identified with the BLAST-X search, then a second search was conducted with BLAST-N directly on the MaizeGDB.org database utilizing the maize reference BAC library. In addition to BLAST searches, predicted genes homologous to a subset of Arabidopsis and rice genes were identified by searching the Gramene.org database for the Arabidopsis or rice gene, then using the “gene trees” tool to identify predicted orthologs in the maize reference genome sequence (Liang et al. 2007). Candidate genes and candidate gene homologs were positioned on our map using the maize genome browser at MaizeGDB.org to identify their location on the physical map of sequenced and ordered BAC clones (obtained from MaizeGDB, September 2009). The SNP markers forming the backbone of the genetic map also have known BAC and contig addresses (Table S4). Mapped SNP loci flanking the candidate gene-containing BACs defined map intervals containing the candidate gene. In cases where local physical sequence order disagreed with map order, we extended the candidate gene interval to flanking markers that displayed ordering consistent with the physical map to reflect the uncertainty of these positions.
RESULTS
Phenotypic variation under short- and long-day lengths:
Tropical inbred parent lines Ki14 and CML254 had significant photoperiod responses for all traits measured, with increased time to flower, height, and total leaf number in long-day environments (Table 1). Temperate parents B73 and B97 had significant but smaller photoperiod responses for height and leaf number and no significant photoperiod responses for flowering time (Table 1). These results are congruent with a phytotron study of inbred responses to different photoperiods where other environmental conditions were maintained constant (File S1; Table S2; Figure S1; Figure S4), demonstrating that the phenotypic differences between long- and short-day length field environments primarily represent photoperiodic responses.
TABLE 1.
Parental mean phenotypic values from long- and short-day environments and for photoperiod response (long-day−short-day means)
| GDDTA |
GDDTS |
GDDASI |
PH |
EH |
TLN |
|
|---|---|---|---|---|---|---|
| Inbred line | Growing-degree days (GDD) | cm | No. of leaves | |||
| Short-day environments | ||||||
| B73 | 873 (31) | 881 (36) | 9 (6.8) | 142 (12.3) | 63 (5.0) | 18 (0.37) |
| B97 | 809 (38) | 841 (43) | 31 (9.2) | 120 (14.0) | 50 (6.8) | 15 (0.53) |
| CML254 | 1003 (35) | 1005 (39) | 1 (8.0) | 126 (13.4) | 71 (6.3) | 20 (0.45) |
| Ki14 | 936 (36) | 994 (40) | 58 (8.4) | 109 (13.1) | 63 (6.1) | 19 (0.45) |
| Long-day environments | ||||||
| B73 | 879 (19) | 902 (24) | 18 (5.8) | 173 (7.2) | 76 (3.0) | 20 (0.26) |
| B97 | 867 (21) | 910 (28) | 40 (6.6) | 159 (7.7) | 74 (3.6) | 19 (0.34) |
| CML254 | 1377 (26) | 1440 (40) | 58 (9.9) | 162 (9.2) | 99 (5.0) | 29 (0.37) |
| Ki14 | 1107 (23) | 1302 (29) | 194 (7.1) | 164 (8.1) | 91 (4.0) | 24 (0.39) |
| Photoperiodic responses (long-day minus short-day responses) | ||||||
| B73 | 5 | 22 | 9 | 31* | 12* | 2*** |
| B97 | 58 | 69 | 9 | 38* | 23** | 3*** |
| CML254 | 373*** | 433*** | 57*** | 37* | 28** | 8*** |
| Ki14 | 171*** | 308*** | 136*** | 55** | 29*** | 4*** |
Values in parentheses represent the standard error of each estimate. *Significantly different from zero at 0.05 < P < 0.01. **Significantly different from zero at 0.01 < P < 0.001. ***Significantly different from zero at P < 0.0001.
Trait heritabilities ranged from 42 to 96% within mapping families, and most traits had heritabilities >75% (Table S5), demonstrating the reliability of phenotypic data for mapping QTL. Under short day lengths, phenotypic variation was compressed and no genomic region was associated with more than one day difference in flowering time (Table 2). We focus here on the genomic regions associated with photoperiod response, which tended to be similar to those regions causing variation under long-day lengths (Table 2; Figure 2).
TABLE 2.
Peak LOD score positions, positions representing endpoints of 2-LOD support interval, proportion of phenotypic variation explained (r2), and allelic effects of QTL detected with combined connected population analysis model
| Short-day QTL | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| QTL 2-LOD support interval and peak |
Phenotypic r2 | Additive effect |
|||||||
| Trait | Chr | Peak − 2 LOD | LOD Peak | Peak + 2 LOD | B73 | B97 | CML254 | Ki14 | |
| GDDTA | 1 | 262 | 278 | 287 | 0.07 | 7.6a | −5.7b | −1.4bc | −0.5c |
| GDDTA | 4 | 5 | 11 | 21 | 0.04 | 0.8a | 1.7a | −5.4b | 2.9a |
| GDDTA | 5 | 46 | 52 | 133 | 0.05 | −2.5a | −0.5a | 5.8b | −2.9a |
| GDDTA | 8 | 65 | 66 | 76 | 0.08 | −3.5a | −0.9a | 7.7b | −3.4a |
| GDDTA | 9 | 99 | 103 | 147 | 0.06 | 4.3a | −7.2b | −0.5c | 3.3ac |
| GDDTS | 1 | 179 | 193 | 245 | 0.05 | 0.0ab | −5.6a | 2.9b | 2.8b |
| GDDTS | 1 | 296 | 300 | 313 | 0.05 | 7.4a | −2.6b | −2.4b | −2.4b |
| GDDTS | 5 | 48 | 54 | 60 | 0.06 | −3.3a | −0.2a | 7.8b | −4.3a |
| GDDTS | 6 | 25 | 35 | 43 | 0.05 | −1.1ab | −4.7a | 1.1ab | 4.6b |
| GDDTS | 8 | 65 | 74 | 98 | 0.08 | −4.5a | 3.1b | 8.3b | −6.9a |
| GDDTS | 9 | 99 | 103 | 110 | 0.08 | 6.6a | −7.8b | −1.1c | 2.4ac |
| GDDTS | 10 | 124 | 136 | 179 | 0.05 | 1.5ab | −4.3a | −2.5a | 5.4b |
| GDDASI | 1 | 49 | 59 | 89 | 0.06 | 0.1ab | 2.7b | −2.0a | −0.8a |
| GDDASI | 2 | 104 | 108 | 125 | 0.08 | −0.8ab | −2.6b | 0.6ac | 2.8c |
| GDDASI | 3 | 72 | 93 | 102 | 0.06 | −0.7a | 3.2b | −2.3a | −0.3a |
| GDDASI | 4 | 37 | 46 | 62 | 0.08 | 0.7ab | 2.8b | −1.1ac | −2.5c |
| GDDASI | 6 | 86 | 107 | 134 | 0.03 | −0.8a | −0.8a | −0.8a | 2.4b |
| GDDASI | 8 | 61 | 67 | 93 | 0.13 | −1.0a | 5.1b | −1.7a | −2.4a |
| GDDASI | 10 | 89 | 97 | 172 | 0.04 | 0.5a | 0.3a | −3.2b | 2.5a |
| PH | 1 | 227 | 236 | 242 | 0.05 | 1.8a | −1.4b | −1.0bc | 0.6ac |
| PH | 3 | 89 | 96 | 113 | 0.08 | 0.7a | −2.3b | 0.2a | 1.4a |
| PH | 8 | 46 | 55 | 61 | 0.15 | 1.1a | −3.6b | 2.0a | 0.5a |
| PH | 10 | 142 | 158 | 168 | 0.05 | 0.2ab | 1.5a | −1.4b | −0.3b |
| EH | 1 | 256 | 276 | 293 | 0.07 | 1.6a | −1.5b | −0.9b | 0.7a |
| EH | 1 | 324 | 331 | 340 | 0.05 | 1.4a | 0.2ab | −1.0b | −0.6b |
| EH | 3 | 95 | 98 | 102 | 0.09 | 0.6a | −2.0b | 0.6a | 0.8a |
| EH | 5 | 153 | 157 | 164 | 0.05 | −0.2a | −0.9a | 1.5b | −0.3a |
| EH | 6 | 30 | 40 | 44 | 0.07 | −2.0a | 0.5bc | 1.6b | −0.1c |
| EH | 6 | 61 | 69 | 76 | 0.05 | 1.8a | −0.3b | −0.1b | −1.3b |
| EH | 7 | 55 | 59 | 63 | 0.08 | −0.5ab | 1.4c | 0.8ac | −1.6b |
| EH | 8 | 56 | 59 | 64 | 0.10 | 0.8ab | −2.4c | 2.1a | −0.4b |
| EH | 8 | 117 | 123 | 128 | 0.07 | −0.9a | −1.2a | 1.5b | 0.5ab |
| EH | 10 | 48 | 60 | 66 | 0.06 | −0.5ab | 1.0ac | −1.8b | 1.4c |
| TLN | 1 | 115 | 134 | 153 | 0.05 | −0.1a | −0.1a | 0.0ab | 0.2b |
| TLN | 1 | 184 | 200 | 280 | 0.05 | 0.1a | −0.2b | 0.0a | 0.1a |
| TLN | 2 | 80 | 87 | 125 | 0.10 | 0.2a | 0.1a | −0.2b | 0.0a |
| TLN | 5 | 84 | 88 | 135 | 0.05 | −0.2a | 0.0ab | 0.1b | 0.0b |
| TLN | 5 | 155 | 156 | 158 | 0.11 | 0.0a | −0.2b | 0.2c | 0.1ac |
| TLN | 7 | 48 | 60 | 80 | 0.07 | 0.0ab | 0.2a | −0.0b | −0.2b |
| TLN | 8 | 124 | 130 | 133 | 0.08 | −0.1ab | −0.2a | 0.2c | 0.0b |
| TLN | 10 | 119 | 122 | 126 | 0.07 | −0.2a | 0.0b | 0.2c | −0.1ab |
| Long-day QTL | |||||||||
| GDDTA | 1 | 131 | 133 | 139 | 0.12 | −8.4a | −5.1ab | 11.3c | 2.2b |
| GDDTA | 2 | 180 | 184 | 187 | 0.09 | −6.6a | −4.2ab | 10.9c | −0.2bc |
| GDDTA | 3 | 163 | 181 | 181 | 0.07 | −8.0a | −2.2ab | 6.0c | 4.2bc |
| GDDTA | 8 | 70 | 74 | 76 | 0.15 | −7.4a | −6.2a | 15.6b | −1.9a |
| GDDTA | 9 | 106 | 115 | 120 | 0.09 | −8.8a | −2.8ab | 10.4c | 1.2b |
| GDDTA | 10 | 59 | 60 | 69 | 0.25 | −6.6a | −14.2a | 9.9b | 11.0b |
| GDDTA | 10 | 158 | 174 | 191 | 0.05 | 2.4a | −11.5b | 6.4a | 2.7a |
| GDDTS | 1 | 138 | 144 | 146 | 0.08 | −11.2a | −2.4b | 9.5c | 4.1bc |
| GDDTS | 1 | 200 | 214 | 239 | 0.06 | −3.7ab | −8.2a | 10.2c | 1.7c |
| GDDTS | 2 | 180 | 183 | 185 | 0.09 | −9.0a | −4.4a | 13.5b | 0.0a |
| GDDTS | 3 | 159 | 171 | 181 | 0.05 | −6.0ab | −5.7a | 3.0bc | 8.7c |
| GDDTS | 8 | 69 | 75 | 127 | 0.08 | −10.4a | −0.4a | 14.8b | −4.0a |
| GDDTS | 9 | 103 | 107 | 124 | 0.07 | −10.1a | −2.3ab | 10.1c | 2.3bc |
| GDDTS | 10 | 59 | 60 | 63 | 0.28 | −11.9a | −16.8a | 9.7b | 19.1b |
| GDDTS | 10 | 135 | 166 | 190 | 0.05 | 5.5a | −13.4b | 1.2a | 6.7a |
| GDDASI | 1 | 144 | 162 | 181 | 0.05 | −5.4a | 0.4b | 1.8b | 3.2b |
| GDDASI | 2 | 28 | 41 | 47 | 0.05 | −2.4a | −2.3a | −0.1a | 4.8b |
| GDDASI | 2 | 78 | 92 | 99 | 0.06 | 2.2a | −5.6b | 2.2a | 1.3a |
| GDDASI | 4 | 155 | 158 | 165 | 0.05 | 0.4ab | 3.3a | 0.8a | −4.5b |
| GDDASI | 6 | 106 | 115 | 135 | 0.04 | −1.5a | −2.3a | 0.3ab | 3.5b |
| GDDASI | 8 | 86 | 90 | 99 | 0.09 | −3.4a | 7.1b | −0.2a | −3.5a |
| GDDASI | 10 | 58 | 60 | 63 | 0.18 | −4.3a | −3.8a | −1.1a | 9.2b |
| PH | 1 | 224 | 229 | 236 | 0.09 | 3.7a | −5.2b | −1.4c | 2.9a |
| PH | 2 | 6 | 53 | 160 | 0.04 | −2.3a | 3.3b | 0.2a | −1.3a |
| PH | 3 | 97 | 105 | 114 | 0.09 | −0.8ab | −3.0a | 1.3bc | 2.5c |
| PH | 5 | 91 | 100 | 105 | 0.06 | 1.7a | 1.6a | −0.5ab | −2.8b |
| PH | 8 | 61 | 66 | 81 | 0.10 | 3.4a | −6.3b | 1.7a | 1.2a |
| PH | 8 | 118 | 123 | 126 | 0.06 | −2.9a | −1.4ab | 3.0c | 1.3bc |
| PH | 10 | 48 | 53 | 83 | 0.05 | −0.1ab | −2.0a | −0.8a | 3.0b |
| EH | 1 | 254 | 276 | 292 | 0.06 | 2.1a | −1.8b | −1.4b | 1.1a |
| EH | 2 | 151 | 155 | 164 | 0.06 | 0.2a | 0.3a | −2.6b | 2.1a |
| EH | 3 | 94 | 100 | 120 | 0.08 | 0.2a | −2.4b | 0.9a | 1.3a |
| EH | 4 | 18 | 31 | 80 | 0.04 | 1.5a | 0.4ab | −1.7b | −0.3ab |
| EH | 7 | 57 | 79 | 91 | 0.05 | −0.9ab | 1.6c | 0.8ac | −1.6b |
| EH | 8 | 65 | 70 | 73 | 0.16 | 0.9a | −5.0b | 1.9a | 2.2a |
| EH | 8 | 118 | 123 | 126 | 0.06 | −1.3a | −1.3a | 3.1b | −0.5a |
| EH | 9 | 114 | 118 | 134 | 0.05 | −2.2a | 0.5bc | −0.3ab | 1.9c |
| EH | 10 | 51 | 78 | 84 | 0.07 | −0.4a | −1.5a | −0.5a | 2.3b |
| TLN | 1 | 123 | 144 | 150 | 0.08 | −0.0ab | −0.2a | 0.0b | 0.2b |
| TLN | 2 | 92 | 103 | 108 | 0.12 | 0.1ab | 0.2a | −0.3c | −0.1bc |
| TLN | 3 | 114 | 117 | 122 | 0.08 | −0.0a | −0.2a | 0.3b | −0.1a |
| TLN | 5 | 155 | 156 | 171 | 0.07 | −0.1ab | −0.2a | 0.2c | 0.1bc |
| TLN | 6 | 0 | 5 | 37 | 0.04 | −0.1a | −0.1a | 0.2b | 0.0a |
| TLN | 7 | 46 | 54 | 63 | 0.07 | 0.0ab | 0.2a | −0.1b | −0.2b |
| TLN | 8 | 67 | 69 | 70 | 0.22 | −0.1a | −0.4b | 0.3c | 0.2c |
| TLN | 9 | 127 | 128 | 135 | 0.09 | −0.2a | −0.1ab | 0.2c | 0.1bc |
| TLN | 10 | 72 | 73 | 82 | 0.16 | −0.3a | −0.2a | 0.3b | 0.1b |
| Photoperiodic QTL | |||||||||
| GDDTA | 1 | 119 | 128 | 141 | 0.10 | −5.2a | −4.3a | 7.0b | 2.4b |
| GDDTA | 2 | 179 | 182 | 185 | 0.07 | −4.5a | −2.6a | 7.7b | −0.6a |
| GDDTA | 3 | 109 | 116 | 181 | 0.05 | −0.8ab | −5.4a | 5.3b | 0.9b |
| GDDTA | 4 | 142 | 150 | 153 | 0.06 | −2.2a | −1.8a | 8.8b | −4.9a |
| GDDTA | 8 | 70 | 73 | 75 | 0.12 | −4.4ab | −5.8a | 9.1c | 1.1b |
| GDDTA | 9 | 114 | 118 | 120 | 0.12 | −10.9a | 3.5bc | 8.5b | −1.1c |
| GDDTA | 10 | 60 | 60 | 62 | 0.39 | −6.0a | −16.2b | 13.7c | 8.6c |
| GDDTS | 1 | 119 | 128 | 145 | 0.09 | −7.8a | −4.8a | 8.6b | 4.1b |
| GDDTS | 2 | 180 | 183 | 185 | 0.09 | −7.2a | −3.3a | 12.4b | −1.9a |
| GDDTS | 3 | 77 | 94 | 98 | 0.07 | −0.3a | −9.1b | 3.8a | 5.5a |
| GDDTS | 8 | 70 | 73 | 76 | 0.07 | −6.4a | −4.9a | 8.1b | 3.3b |
| GDDTS | 9 | 83 | 87 | 90 | 0.11 | −14.3a | 3.7bc | 10.7b | −0.1c |
| GDDTS | 10 | 60 | 60 | 61 | 0.40 | −10.7a | −20.5b | 16.3c | 14.9c |
| GDDASI | 3 | 80 | 96 | 127 | 0.05 | 1.3a | −4.4b | −0.1ab | 3.2a |
| GDDASI | 10 | 59 | 60 | 63 | 0.16 | −3.7a | −4.7a | 1.6b | 6.8c |
| PH | 2 | 41 | 62 | 100 | 0.05 | −0.6ab | 2.1c | 0.6ac | −2.0b |
| PH | 3 | 97 | 104 | 134 | 0.04 | −0.9a | −1.3a | 0.4ab | 1.8b |
| PH | 4 | 27 | 72 | 112 | 0.04 | 1.2a | 1.3a | −0.9ab | −1.6b |
| PH | 8 | 66 | 70 | 106 | 0.10 | 0.2a | −3.4b | 1.9a | 1.4a |
| PH | 9 | 57 | 65 | 83 | 0.06 | −3.1a | 2.6b | 1.6b | −1.1a |
| PH | 10 | 76 | 78 | 84 | 0.10 | 0.3ab | −3.1c | 0.1a | 2.7b |
| EH | 8 | 69 | 70 | 78 | 0.13 | −0.1a | −2.6b | 1.5a | 1.2a |
| EH | 9 | 87 | 94 | 99 | 0.08 | −2.6a | 1.1b | 0.5b | 1.0b |
| EH | 10 | 77 | 78 | 83 | 0.11 | 0.4a | −2.3b | 0.7a | 1.3a |
| TLN | 1 | 117 | 144 | 165 | 0.04 | 0.0ab | −0.1a | 0.1b | 0.0a |
| TLN | 2 | 95 | 107 | 130 | 0.05 | 0.0ab | 0.1a | 0.0ab | −0.1b |
| TLN | 3 | 49 | 72 | 144 | 0.04 | −0.1a | −0.1a | 0.1b | 0.1ab |
| TLN | 5 | 90 | 94 | 105 | 0.04 | 0.1a | 0.3a | −0.2b | 0.0a |
| TLN | 8 | 68 | 70 | 72 | 0.17 | −0.1a | −0.2b | 0.1ac | 0.2c |
| TLN | 9 | 87 | 89 | 95 | 0.16 | −0.3a | 0.1b | 0.1b | 0.1b |
| TLN | 10 | 79 | 82 | 85 | 0.22 | −0.1a | −0.3b | 0.2c | 0.2c |
Allelic effects are in GDD for flowering time traits, cm for height traits, and number of leaves for TLN. Allelic effects at a QTL followed by the same letter are not significantly different at P = 0.05.
Figure 2.—
Linkage map of chromosomes 1, 8, 9, and 10 containing the four key photoperiod response QTL regions ZmPR1–ZmPR4 based on the combined analysis of four RIL mapping populations. For the sake of clarity, chromosome 1 is displayed in two parts and only a subset of marker loci spaced about every 10 cM or more is shown. The complete genetic map with all QTL positions is shown in Figure S3. Candidate genes directly mapped are indicated in red. Map intervals containing candidate genes localized by inference indicated as segments with blue diagonal filling, with the candidate gene name in blue positioned at interval midpoint. Map intervals exhibiting significant segregation distortion in the combined analysis indicated as magenta segments on the linkage group. The map interval on chromosome 10 identified as having undergone a selection sweep by Tian et al. (2009) is indicated by a pink segment on the linkage group. QTL bars represent the 2-LOD support interval of the QTL position; the middle hash mark of a bar represents the maximum likelihood position of the QTL. Trait names for QTL are abbreviated as ANTHESIS for GDDTA, SILK for GDDTS, ASI for GDDASI, EHT for ear height, and PHT for plant height. SD, LD, and PR refer to QTL identified under short-day length or long-day length environments, or for photoperiod response, respectively.
Single population and combined multiple population QTL mapping:
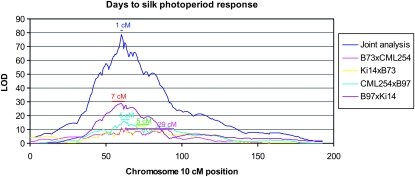
Analysis of each population separately suggested that many QTL differed between populations, but rarely did we find QTL that were unique to a single mapping population (Figure S2). Joint population analyses had much lower permutation-based significance thresholds and much higher QTL LOD scores than individual population analyses (Figure 3; Figure S2). Furthermore, the combined connected population analysis increased the precision of QTL position estimates over those of individual population analyses (Figure 3). Specifically, the 2-LOD support intervals of QTL mapped with the connected population analysis were significantly smaller on average (24 cM) than those mapped in individual populations (35 cM; P < 0.0001). The increased resolution of the joint analysis can be illustrated at the QTL with largest effect on the photoperiod response, centered at position 60.1 cM on chromosome 10, which explains 39–40% of the phenotypic variation for the anthesis and silking date photoperiod responses in the combined analysis (Table 2). The 2-LOD support interval around this QTL (expected to contain the causal gene with good probability with large population sizes; Van Ooijen 1992) ranged in size from 5 to 29 cM in the individual population analyses, but was narrowed to 1 cM in the joint analysis. The high resolution of QTL positions in the joint analysis greatly reduces the genome space in which to search for candidate genes, providing robust tests for candidate genes and simplifying the search for causal genes in forward genetic analyses. Because of the increased precision, power, and parsimony of the connected population analysis, we discuss further only the results of the joint multiple population QTL analyses.
Figure 3.—
Plots of LOD scores for the presence of a QTL affecting photoperiod response for days to silk along chromosome 10 of maize. Five separate analyses are plotted: the joint analysis (connected model) of four populations simultaneously and four individual population analyses (B73 × CML254, Ki14 × B73, CML254 × B97, and B97 × Ki14). Two-LOD support intervals around the peak QTL position are drawn with horizontal bars, with the length of the support interval presented above the bar.
Tests for epistasis:
Two forms of epistasis could be directly tested with the joint multiple population analysis. First, the overall significance of interactions between detected QTL and genetic backgrounds (i.e., different mapping families) was tested. This was done by comparing two multiple population analyses: connected (which assumes consistent founder allele effects across backgrounds) and disconnected analysis (which allows founder allele effects to vary across backgrounds). We found that the number, positions, and confidence intervals of the QTL detected by both analyses did not differ significantly. However, the BIC of the combined analysis was superior for the connected analysis for all traits (Table S6). Thus, QTL allele effects were consistent across genetic backgrounds and there was no evidence for epistatic interactions of QTL alleles with genetic backgrounds for any trait.
Second, we tested each QTL for epistatic interactions with all other positions in the genome, including epistasis between QTL pairs as well as between QTL and other genome regions that had no main effect. On the basis of the permutation thresholds computed specifically for this set of tests, we detected no significant epistatic interactions for any trait with the combined connected analysis.
Direct mapping of candidate genes:
We mapped 14 candidate gene markers with homology to known circadian rhythm/photoperiod response, floral transition, or floral morphology genes from maize, Arabidopsis, or rice: CHLOROPHYLL A/B BINDING PROTEIN (CAB), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), CONSTANS-LIKE (CO-2), CRYPTOCHROME 2 (CRY2), HEADING DATE1 (HD1), HEADING DATE 3A/FLOWERING LOCUS T (HD3A), and LATE ELONGATED HYPOCOTYL (LHY and LHYC8; Hayama and Coupland 2004); INDETERMINATE 1-LIKE (ID7; Colasanti et al. 2006); PHYTOCHROME C1 (PHYC; Sheehan et al. 2004), DWARF8 (D8; Thornsberry et al. 2001), ZEA MAYS FLORICAULA1 (LEAFY) and ZEA MAYS FLORICAULA2 (ZFL2; Bomblies and Doebley 2006); and FASCIATED EAR2 (FEA2; Bommert et al. 2005). Each candidate gene mapped to single genome positions distributed across all chromosomes except chromosome 3. Two of these genes control key regulatory steps in the photoperiod response pathways of other species: CO of Arabidopsis (closest match mapping to chromosome 2) and its homolog HD1 of rice (closest match mapping to chromosome 9), and FT of Arabidopsis (homologous to HD3A of rice, mapping to chromosome 6). CO, CRY2, FEA2, HD1, and LHYC8 homologs mapped within plant height photoperiod QTL intervals, but not within QTL intervals directly affecting flowering time photoperiod response (Table 2; Figure 2; Figure S3). None of the candidate genes that we directly mapped localized within flowering time photoperiod response QTL intervals.
Inferring positions of candidate genes on map:
A second approach to testing candidate genes for their effects on photoperiod response was to infer candidate gene positions on the genetic map by referencing the maize physical map and sequence. We identified predicted maize genes homologous to candidate genes from Arabidopsis, rice, and barley through the Gramene.org database browser and with direct sequence matching searches through the NCBI and MaizeGDB databases (Table S4). The positions of each gene were inferred on our combined map by reference to mapped SNP markers derived from flanking BACs on ordered BAC physical sequence. In total, we positioned 82 unique candidate gene sequences on the map (Figure S3; Table S4). Three homologs of TOC1/PRR7, two homologs each of CCA1/LHY, CRY2, ELF4, and HY1/SE5, and single homologs of CO/HD1, CRY1, EHD1/2, FCA, FKF1/ZTL, FLD, FY, LD, PIE1, PIF3, SNZ, ZCN1, ZCN2, ZCN8, ZCN10, ZCN12, ZCN19, ZCN21, and ZCN25 (together representing 37% of candidate gene positions inferred on the map) mapped with photoperiod response QTL intervals (Figure S3; Table S4), but all other candidate genes tested in this way did not map to genome regions with significant effects for photoperiod response traits.
DISCUSSION
QTL support intervals for all 18 traits covered 60% of the genetic map (Figure S3). QTL with smaller effects had the largest support intervals and comprised the majority of the map coverage (Table 2). Therefore, we focus discussion on the four most important QTL regions for photoperiod response, which were responsible for the majority of the photoperiodic responses in the six traits (each QTL explaining up to 40% of the phenotypic variation). Support intervals for all photoperiod trait QTL covered a combined 25% of the genetic map, but the four key QTL to be discussed here represent only 2% of the genetic map. These QTL also coincide with the four key flowering time QTL detected under long days on chromosomes 1, 8, 9, and 10 (Table 2; Figure 2) and also with the four most important flowering time QTL detected under long-day conditions in the maize NAM population (Buckler et al. 2009). To facilitate discussion, we named these four photoperiodic response QTL ZmPR1–4 (for Zea mays Photoperiodic Response). ZmPR1 is located between markers bnlg1811 and umc1754 on chromosome 1 (Figure S3), associated with ∼10% of the photoperiodic flowering time response in these populations. This QTL also had a significant effect on TLN, but was not a major contributor to PH or EH variation. ZmPR2 is located between the markers umc1130 and PHM3993.28 on chromosome 8 (Figure S3) and contributed 9% of the photoperiodic flowering time response in these populations. The chromosome 9 QTL, ZmPR3, is located between markers PZB00547.3 and PZA03057.3 (Figure S3). This locus contributed ∼11% of the photoperiodic flowering time variation. ZmPR4 is located on chromosome 10 between PZA00337.4 and umc1827 (Figure S3), explained 39% of the photoperiodic flowering time response in our study, and also had significant effects on PH, EH, and TLN. Genomic regions with significant segregation distortion across populations coincided with or were linked to all four of these QTL (Figure 2; Table S7), likely due to natural selection against very late flowering during RIL development, further highlighting their importance to adaptation.
Chromosome 1 QTL–ZmPR1:
An important flowering time QTL in the ZmPR1 region was previously detected in several populations (Beavis et al. 1994; Chardon et al. 2004; Blanc et al. 2006; Briggs et al. 2007; Buckler et al. 2009); however, neither of the two previous studies specifically addressing tropical maize photoperiod response detected a significant QTL in this region (Table S8; Moutiq et al. 2002; Wang et al. 2008), suggesting that ZmPR1 does not segregate in all temperate × tropical maize populations, as observed by Buckler et al. (2009). One flowering time candidate gene mapped to the ZmPR1 QTL interval: ELF4, which is involved in sensing day length and regulation of CCA1 in Arabidopsis (Doyle et al 2002). Buckler et al. (2009) also identified a homolog of rice Ghd7 (Xue et al. 2008) in this region.
Chromosome 8 QTL–ZmPR2:
ZmPR2 was associated with both the photoperiodic and lateness per se flowering time responses. Neither Moutiq et al. (2002), Wang et al. (2008), nor Briggs et al. (2007) detected a flowering time QTL in this region in other populations segregating for photoperiod response (Table S8). This region includes the major flowering time and height QTL Vgt1, which was originally identified genetically on the basis of an allelic difference between temperate maize lines (Vlăduţu et al. 1999; Salvi et al. 2007), although the position of Vgt1 on our map is uncertain because of disagreements in the maize genome database. The Vgt1 QTL affects flowering under short-day photoperiods (Salvi et al. 2002), similar to the effect that we observed at this QTL region. The noncoding functional polymorphism underlying Vgt1 regulates the expression of a Rap2.7, a maize homolog of an Arabidopsis floral identity gene APETELA 2 (AP2) (Salvi et al. 2007).
The known functional variants at Vgt1 and Rap2.7 do not segregrate in these populations, however (Buckler et al. 2009). Thus, the observed ZmPR2 QTL effects may be due to Vgt2, which is tightly linked to Vgt1 (Vlăduţu et al. 1999; Chardon et al. 2005). Danilevskaya et al. (2008) proposed that the gene underlying Vgt2 is Zea mays CENTRORADIALIS8 (ZCN8), which because of its expression patterns and sequence homology, bears the closest resemblance of any gene in the maize genome to the FT gene of Arabidopsis. FT functions in Arabidopsis as the long-distance floral promoter referred to as florigen (Corbesier et al. 2007). ZCN8 maps to the center of the QTL region.
Chromosome 9 QTL–ZmPR3:
ZmPR3 was also observed to affect flowering time and photoperiodic QTL in previous maize studies (Table S8; Moutiq et al. 2002; Chardon et al. 2004; Briggs et al. 2007; but not Wang et al. 2008). Several homologs of Arabidopsis and rice flowering time genes are located within the ZmPR3 interval, including CONSTANS of Zea mays1 (CONZ1), the putative hemeoxygenase HY1/SE5, and a homolog of circadian clock-associated genes TOC1/PRR3/PRR5/PRR7/PRR9 (Chardon et al. 2004; Cockram et al. 2007; Miller et al. 2008).
CONSTANS-like genes have been shown to play important roles in the photoperiodic responses of several plant species, including barley and rice (Griffiths et al. 2003), but to date, genetic data indicating that homologs of CO/HD1 function as activators of the floral transition in maize are lacking (Colasanti and Muszynski 2009). The inferred position of CONZ1 is inside the 2-LOD support intervals for regions affecting photoperiod response for TLN and EH, but just outside the interval affecting GDDTS photoperiod response. A gene family of CO/HD1 homologs appears to exist in maize; we also mapped a SNP marker on the basis of an additional HD1 homolog (ZHD1_1) at least 25 cM distant from Conz1 on chromosome 9, and a CAPS marker of a CO homolog to chromosome 2. ZHD1_1 mapped outside of the photoperiod response QTL intervals for all traits except plant height and the CO CAPS marker also mapped within a broad PH photoperiod response QTL interval on chromosome 2. CONZ1 is differentially expressed in response to photoperiodic stimuli, and the long-day expression of CONZ1 in B73 is different from that of photoperiod-sensitive teosinte and CML254 (Miller et al. 2008; T. Dlugi, personal communication). The gene expression results in combination with our genetic data suggest that CO-like genes in maize mediate height and leaf number responses to long-day lengths, but do not provide evidence of their effect on flowering time photoperiod responses.
Chromosome 10 QTL–ZmPR4:
The genome region with largest effect on all photoperiod response traits was ZmPR4 on chromosome 10, which was associated with up to 40% of the phenotypic variation for photoperiod response. The 2-LOD support intervals of the ZmPR4 QTL corresponding to GDDTA and GDDTS did not overlap with a tightly linked QTL for PH, EH, and TLN, suggesting that there are at least two distinct genes in the ZmPR4 region, each with important but unique effects on photoperiod response.
Previous studies have consistently detected major photoperiodic response, flowering time, PH, and TLN QTL in the ZmPR4 QTL region (Abler et al. 1991; Moutiq et al. 2002; Blanc et al. 2006; Briggs et al. 2007; Lauter et al. 2008; Wang et al. 2008; Ducrocq et al. 2009). ZmPR4 is syntenic to the major photoperiodic QTL of sorghum (Ma1; Ulanch 1999), which has undergone intense directional selection in temperate-adapted sorghum populations (Klein et al. 2008). Together, these results suggest that the gene or genes promoting photoperiod sensitivity at ZmPR4 are conserved in tropical maize, teosinte, and sorghum populations. Tian et al. (2009) reported a >1-Mb region that underwent a selection sweep during maize domestication or improvement which maps immediately adjacent to the ZmPR4 region (Figure 2). The QTL appears to be outside the region of the selection sweep, although map ordering in this region is difficult because it exhibits reduced recombination (Ducrocq et al. 2009; McMullen et al. 2009; Tian et al. 2009) and differences in SNP locus orders between the physical BAC sequence and both the NAM genetic map and the combined map from this study. Physical rearrangements of this region among maize lines may be associated with these phenomena.
Ducrocq et al. (2009) recently mapped a photoperiod response QTL to a 170-kb region inside the ZmPR4 region in a different maize cross. This interval included a noncoding sequence adjacent to a homolog of Ghd7, which encodes a CCT protein domain and regulates flowering time in rice (Xue et al. 2008). In addition to Ghd7, a number of other candidate gene homologs are more loosely associated with ZmPR4, including PHYTOCHROME INTERACTING FACTOR3 (PIF3; Oda et al. 2004), a cryptochrome gene (CRY2), a circadian clock-associated gene (CCA1/LHY), and one FT homolog, ZCN19 (Chardon et al. 2004; Danilevskaya et al. 2008).
Effects of maize genes defined by major mutations:
Aside from Vgt1, which has a quantitative effect on flowering time, the other maize flowering time genes cloned to date, D8, ID1, and DLF1, are associated with major effects on flowering time. None of these three latter genes coincided with photoperiod response QTL in this study. Despite previous studies indicating the importance of D8 to flowering time and latitudinal adaptation in maize (Thornsberry et al. 2001; Camus-Kulandaivelu et al. 2006), D8 colocalized only with QTL for ear height under long- and short-day lengths (7% of the variation) and TLN in short days (5% of the variation of a QTL with a very large support interval). D8 encodes a transcriptional regulator of the gibberellin flowering pathway, which controls flowering time independently of photoperiod in Arabidopsis (Peng et al. 1999; Simpson and Dean 2002), so it was not expected to have an effect on photoperiod response.
Our experimental approach may fail to identify causal genes as candidates if they are not well annotated in the genome databases, or if QTL interval position estimates are incorrect. Annotation of the maize genome sequence is ongoing, and many gene predictions are preliminary. An additional problem with the maize genome sequence is the possibility that some contigs were not placed correctly on the physical map on the basis of FPC alignments used here; we noted several cases where our SNP markers place a BAC on our genetic map in a different chromosome than they are currently positioned in the maize sequence. In addition, some candidate genes may affect photoperiod response in other crosses but could not be detected in this study because they were not segregating for functional nucleotide variants. Our approach also has the converse problem of potentially identifying incorrect candidate genes. The QTL intervals often encompass many genes, so any gene within the interval by coincidence may be considered a candidate until the interval can be narrowed by further genetic analysis. Many of the candidate genes we searched for appear to exist as gene families in maize, increasing the chance of a gene homologous to a known flowering regulator from another species residing within a genetic interval. Nevertheless, our results clearly indicate that at least some homologs of important photoperiod regulatory genes are not associated with significant photoperiod responses in these mapping families. There may be related genes that we did not detect in these genome regions or maize may have a unique regulatory pathway for photoperiod response that has not yet been observed in other species.
Genetic architecture of photoperiod sensitivity in maize:
We compared the results of this study with those of previous genetic analyses of photoperiod sensitivity in tropical maize (Moutiq et al. 2002; Wang et al. 2008; Table S8). The main photoperiodic QTL on chromosomes 8, 9, and 10 were observed to affect flowering time across the vastly different environments and with distinct genetic materials represented by these studies. Indeed, some of these photoperiodic alleles may be functionally equivalent to their ancestral teosinte ortholog, on the basis of their importance to photoperiod-dependent flowering time in a maize × teosinte cross (Briggs et al. 2007). Across each of these populations, the ZmPR4 QTL explained >30% of the variation for photoperiodic flowering time response. A study by Blanc et al. (2006) of temperate European maize lines also identified a large flowering time allele in the ZmPR4 region that affected flowering time in long-day environments, which might indicate that photoperiod-sensitive alleles of ZmPR4 can be found in temperate populations. On the basis of a meta-analysis of 22 maize flowering QTL mapping studies (representing both temperate and tropical germplasm and long- and short-day environments), Chardon et al. (2004) suggested that six QTL, including the four ZmPR QTL identified here, affect flowering time across many populations.
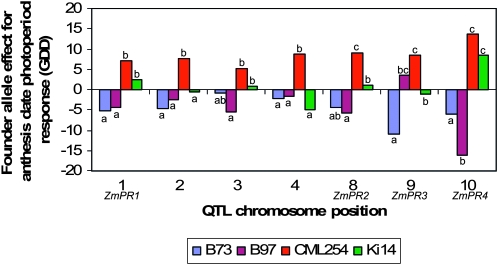
Comparisons of QTL across studies are generally confounded by differences in allelic composition, evaluation environments, and methodology. Thus, it can be difficult to determine whether the detection of a QTL in one population but not another is due to true differences in the QTL effects across populations, QTL-by-environment interactions, or statistical issues, such as limited power in small population sizes (Beavis 1998; Melchinger et al. 1998). The combined analysis of multiple mapping populations evaluated in common environments permits direct comparison of the effects of specific genome regions across genetic backgrounds. Our results indicate largely consistent QTL regions, but heterogeneity of allelic effects at the ZmPR loci within both tropical and temperate inbred line groups (Table 2). For example, CML254 alleles at many of the ZmPR loci delay flowering time more than the Ki14 alleles in the long-day environment, particularly at ZmPR2 and ZmPR3 (Table 2; Figure 4). Across all QTL detected, the temperate alleles differed significantly from each other at 41% of QTL, the pair of tropical alleles differed significantly at 39% of QTL, and temperate and tropical-derived allele effects differed at an average of 57% of QTL (Table 2). These results suggest the existence of allelic series at the ZmPR loci across diverse maize germplasm. Flowering time under long-day conditions in the maize NAM population also is controlled by QTL with allelic series, with significant variation among the allelic effects of tropical founder lines (Buckler et al. 2009). Our results suggest substantial standing genetic variation for photoperiod response in tropical maize (perhaps due to adaptation to environments with slight photoperiod differences and precipitation cycles) and multiple potential evolutionary trajectories to reduced photoperiod response. The apparently simpler genetic architecture of flowering time photoperiod response observed here compared to the more numerous genes with smaller effects on flowering time identified in the NAM population (Buckler et al. 2009) may be due in part to the smaller number of families derived from parents with very distinct photoperiod responses evaluated in this study.
Figure 4.—
Variation for functional allele effects at seven QTL detected for photoperiod response for anthesis date. At each QTL, four founder allele effects were estimated from the joint population analysis. Allele effects are estimated relative to recombinant inbred line population means: negative effects reduce the photoperiod response and positive effects increase the photoperiod response. Allelic effects within a QTL labeled with the same letter are not significantly different at P = 0.05.
Acknowledgments
We are grateful to Josie Bloom, Bob Baesman, David Rhyne, and Emily Silverman for technical assistance in SSR and candidate gene genotyping, and to Kate Guill for assistance with SNP genotyping. We thank Magen Eller, Brooke Peterson, Stella Salvo, Kathleen Starr, Scott Reed, and the crew of 27 farms for field assistance and help in phenotypic data collection; Arturo Garcia for data management assistance; Ed Buckler for candidate gene sequence information; and Janet Shurtleff and the staff of the North Carolina State University (NCSU) Phytotron. Sylvain Jasson generously provided technical assistance with MCQTL. GENOPLANTE MCQTL was generated within a GENOPLANTE program (Mangin et al. 2007) and improved thanks to a grant from Toulouse Genopole Midi-Pyrénées. MCQTL analyses were performed using the resources of the NCSU High Performance Computing Center with the assistance of Gary Howell. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture (USDA). This research was supported by a USDA–Initiative for Future Agricultural and Food Systems multidisciplinary training grant (award 2001-52101-11507), the National Science Foundation (DBI–0321467), and USDA–Agricultural Research Service.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.110304/DC1.
References
- Abler, B. S. B., M. D. Edwards and C. W. Stuber, 1991. Isoenzymatic identification of quantitative trait loci in crosses of elite maize inbreds. Crop Sci. 31 267–274. [Google Scholar]
- Allison, J. C. S., and T.B Daynard, 1979. Effect of change in time of flowering, induced by altering photoperiod or temperature, on attributes related to yield in maize. Crop Sci. 19 1–4. [Google Scholar]
- Beavis, W. D., O. S. Smith, D. Grant and R. Fincher, 1994. Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci. 34 882–896. [Google Scholar]
- Beavis, W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by A. H. Paterson. CRC Press, New York.
- Blanc, G., A. Charcosset, B. Mangin, A. Gallais and L. Moreau, 2006. Connected populations for detecting quantitative trait loci and testing for epistasis: an application in maize. Theor. Appl. Genet. 113 206–224. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., and J. F. Doebley, 2006. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert, P., N. Satoh, D. Jackson and H. Hirano, 2005. Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46 69–78. [DOI] [PubMed] [Google Scholar]
- Briggs, W. H., M. D. McMullen, B. S. Gaut and J. Doebley, 2007. Linkage mapping of domestication loci in a large maize–teosinte backcross resource. Genetics 177 1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S., J. B. Holland, P. J. Bradbury, C. Acharya, P. J. Brown et al., 2009. The genetic architecture of maize flowering time. Science 325 714–718. [DOI] [PubMed] [Google Scholar]
- Caicedo, A. L., J. R. Stinchcombe, K. M. Olsen, J. Schmitt and M. D. Purugganan, 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, H., M. Cooper, G. O. Edmeades, C. Loeffler, J. R. Schussler et al., 2006. Changes in drought tolerance in maize associated with fifty years of breeding for yield in the US Corn Belt. Maydica 51 369–381. [Google Scholar]
- Camus-Kulandaivelu, L., J. B. Veyrieras, D. Madur, V. Combes, M. Fourmann et al., 2006. Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172 2449–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus-Kulandaivelu, L., L.-M. Chevin, C. Tollon-Cordet, A. Charcosset, D. Manicacci et al., 2008. Patterns of molecular evolution associated with two selective sweeps in the Tb1-Dwarf8 region in maize. Genetics 180 1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charcosset, A., B. Mangin, L. Moreau, L. Combes, M.-F. Jourjon et al., 2000. Heterosis in maize investigated using connected RIL populations, pp. 89–98 in Quantitative Genetics and Breeding Methods: The Way Ahead. INRA Editions, Paris.
- Chardon, F., B. Virlon, L. Moreau, M. Falque, J. Joets et al., 2004. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics 168 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon, F., D. Hourcade, V. Combes and A. Charcosset, 2005. Mapping of a spontaneous mutation for early flowering time in maize highlights contrasting allelic series at two-linked QTL on chromosome 8. Theor. Appl. Genet. 112 1–11. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutkaew, C., W. Mekdum and C. Malumpong, 1997. Descriptors of Kasetsart University inbred lines. Available at http://www.ku.ac.th/AgrInfo/corn/; accessed November 24, 2008.
- Cockram, J., H. Jones, F. J. Leigh, D. O'Sullivan, W. Powell et al., 2007. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J. Exp. Bot. 58 1231–1244. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., and V. Coneva, 2009. Mechanisms of floral induction in grasses: something borrowed, something new. Plant Phys. 149 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti, J., and M. Muszynski, 2009. The maize floral transition, pp 41–55 in Handbook of Maize: Its Biology, edited by J. Bennetzen and S. Hake. Springer, New York.
- Colasanti, J., R. Tremblay, A. Y. Wong, V. Coneva, A. Kozaki et al., 2006. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7 158–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier, L., C. Vincent, S. Jang, F. Fornara, Q. Fan et al., 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. [DOI] [PubMed] [Google Scholar]
- Danilevskaya, O. N., X. Meng, Z. Hou, E. V. Ananiev and C. R. Simmons, 2008. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 146 250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy, D., R. Cathelin, M.-F. Jourjon, S. Jasson, J. Marcel et al., 2006. MCQTL: multi-allelic QTL mapping in multi-cross design. MCQTL Tutorial. INRA, Toulouse, France. [DOI] [PubMed]
- Doyle, M. R., S. J. Davis, R. M. Bastow, H. G. McWatters, L. Kozma-Bognár et al., 2002. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 74–77. [DOI] [PubMed] [Google Scholar]
- Ducrocq, S., D. Madur, J.-B. Veyrieras, L. Camus-Kulandaivelu, M. Kloiber-Maitz et al., 2008. Key impact of Vgt1 on flowering time adaptation in maize: evidence from association mapping and ecogeographical information. Genetics 178 2433–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrocq, S., C. Giauffret, D. Madur, V. Combes, F. Dumas, et al., 2009. Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics 183 1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersoz, E.S., J. Yu and E.S. Buckler, 2007. Applications of linkage disequilibrium and association mapping in crop plants, pp. 97–120 in Genomics-Assisted Crop Improvement, edited by R. Varshney and R. Tuberosa. Springer, Dordrecht, The Netherlands.
- Fan, J. B., A. Oliphant, R. Shen, B. G. Kermani, F. Garcia et al., 2003. Highly parallel SNP genotyping. Cold Spring Harbor Symp. Quant. Biol. 68 69–78. [DOI] [PubMed] [Google Scholar]
- Goodman, M. M., 1988. The history and evolution of maize. CRC Crit. Rev. Plant Sci. 7 197–220. [Google Scholar]
- Goodman, M. M., 1999. Broadening the genetic diversity in breeding by use of exotic germplasm, pp. 139–148 in Genetics and Exploitation of Heterosis in Crops, edited by J. G. Coors and S. Pandey. Crop Science Society of America, Madison, WI.
- Gouesnard, B., C. Rebourg, C. Welcker and A. Charcosset, 2002. Analysis of photoperiod sensitivity within a collection of tropical maize populations. Gen. Res. Crop Evol. 49 471–481. [Google Scholar]
- Griffiths, S., R. P. Dunford, G. Coupland and D. A. Laurie, 2003. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131 1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallauer, A. R., K. R. Lamkey, W. A. Russell and P. R. White, 1994. Registration of B97 and B98 parental lines of maize. Crop Sci. 34 318–319. [Google Scholar]
- Hayama, R., and G. Coupland, 2004. The molecular basis of diversity in the photoperiodic flowering response of Arabidopsis and rice. Plant Physiol. 135 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht, V., C. L. Knowles, J. K. Vander Schoor, L. C. Liew, S. E. Jones et al., 2007. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, J. B., and M. M. Goodman, 1995. Combining ability of tropical maize accessions with U.S. germplasm. Crop Sci. 35 767–773. [Google Scholar]
- Holland, J. B., 2007. Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 10 156–161. [DOI] [PubMed] [Google Scholar]
- Izawa, T., 2007. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58 3091–3097. [DOI] [PubMed] [Google Scholar]
- Jannink, J.-L., and R. C. Jansen, 2001. Mapping epistatic quantitative trait loci with one-dimensional genome searches. Genetics 157 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourjon, M.-F., S. Jasson, J. Marcel, B. Ngom and B. Mangin, 2005. MCQTL: multi-allelic QTL mapping in multi-cross design. Bioinformatics 21 128–130. [DOI] [PubMed] [Google Scholar]
- Klein, R. R., J. E. Mullet, D. R. Jordan, F. R. Miller, W. L. Rooney et al., 2008. The effect of tropical sorghum conversion and inbred development on genome diversity as revealed by high-resolution genotyping. Crop Sci. 48 S12–S26. [Google Scholar]
- Koester, R. P., P. H. Sisco and C. W. Stuber, 1993. Identification of quantitative trait loci controlling days to flowering and plant height in two near-isogenic lines of maize. Crop Sci. 33 1209–1216. [Google Scholar]
- Kojima, S., Y. Takahashi, Y. Kobayashi, L. Monna, T. Sasaki et al., 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., C. Alonso-Blanco and D. Vreugdenhil, 2004. Naturally occurring genetic variation in Arabidopsis thaliana. Ann. Rev. Plant Biol. 55 141–172. [DOI] [PubMed] [Google Scholar]
- Kwak, M., D. Velasco and P. Gepts, 2008. Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 99 283–291. [DOI] [PubMed] [Google Scholar]
- Lauter, N., M. J. Moscou, J. Habiger and S. P. Moose, 2008. Quantitative genetic dissection of shoot architecture traits in maize: towards a functional genomics approach. Plant Genome 1 99–110. [Google Scholar]
- Liang, C., P. Jaiswal, C. Hebbard, S. Avraham, E. S. Buckler et al., 2007. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 36 D947–D953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, S. E., M. J. Daly and E. S. Lander, 1992. Mapping genes controlling quantitative traits with MAPMAKER/QTL 1.1. pp. 1–73 in Whitehead Institute Technical Report, Cambridge, MA.
- Liu, K., M. M. Goodman, S. Muse, J. S. Smith, E. Buckler et al., 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin, B., R. Cathelin, D. Delannoy, S. Lambert, J. Marcel et al., 2007. MCQTL: A Reference Manual. Rapport UBIA Toulouse No. 2007/2. Département de Mathématiques et Informatique Appliquées, INRA, Toulouse, France. http://carlit.toulouse.inra.fr/MCQTL/images/UserManual_complete.pdf
- Matsuoka, Y., Y. Vigouroux, M. M. Goodman, J. Sanchez, E. S. Buckler et al., 2002. A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. USA 99 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster, G. S., and W. W. Wilhelm, 1997. Growing degree-days: one equation, two interpretations. Agric. For. Meteorol. 87 289–298. [Google Scholar]
- McMullen, M. M., S. Kresovich, H. Sanchez Villeda, P. Bradbury, H. Li et al., 2009. Genetic properties of the maize nested association mapping population. Science 325 737–740. [DOI] [PubMed] [Google Scholar]
- Melchinger, A. E., H. F. Utz and C. C. Schön, 1998. Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149 383–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T. A., E. H. Muslin and J. E. Dorweiler, 2008. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 227 1377–1388. [DOI] [PubMed] [Google Scholar]
- Moutiq, R., J. M. Ribaut, G. Edmeades, M. D. Krakowsky and M. Lee, 2002. Elements of genotype by environment interaction: genetic components of the photoperiod response in maize, pp. 257–267 in Quantitative Genetics, Genomics, and Plant Breeding, edited by M. Kang. CABI Publishing, Oxford, UK.
- Muszynski, M. G., T. Dam, B. Li, D. M. Shirbroun, Z. Hou et al., 2006. Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M. M., J. D. Neff, J. Chory and A. E. Pepper, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- Oda, A., S. Fujiwara, H. Kamada, G. Coupland and T. Mizoguchi, 2004. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett. 557 259–264. [DOI] [PubMed] [Google Scholar]
- Peng, J., D. E. Richards, N. M. Hartley, G. P. Murphy, K. N. Devos et al., 1999. ‘Green revolution’ genes encode mutant giberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Piepho, H. P., E. R. Williams and M. Fleck, 2006. A note on the analysis of designed experiments with complex treatment structure. HortScience 41 446–452. [Google Scholar]
- Rebaï, A., and B. Goffinet, 2000. More about quantitative trait locus mapping with diallel designs. Genetic. Res. 75 243–247. [DOI] [PubMed] [Google Scholar]
- Rebaï, A., P. Blanchard, D. Perret and P. Vincourt, 1997. Mapping quantitative trait loci controlling silking date in a diallel cross among four lines of maize. Theor. Appl. Genet. 95 451–459. [Google Scholar]
- Ribaut, J.-M., D. A. Hoisington, J. A. Deutsch, C. Jiang and D. Gonzalez-de-Leon, 1996. Identification of quantitative trait loci under drought conditions in tropical maize. 1. Flowering parameters and the anthesis-silking interval. Theor. Appl. Genet. 92 905–914. [DOI] [PubMed] [Google Scholar]
- Russell, W. A., 1972. Registration of B70 and B73 parental lines of maize. Crop Sci. 12 721. [Google Scholar]
- Salvi, S., R. Tuberosa, E. Chiapparino, M. Maccaferri, S. Veillet et al., 2002. Toward positional cloning of Vgt1, a QTL controlling the transition from the vegetative to the reproductive phase in maize. Plant Mol. Biol. 48 601–613. [DOI] [PubMed] [Google Scholar]
- Salvi, S., G. Sponza, M. Morgante, D. Tomes, X. Niu et al., 2007. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute, 2002. –2004 SAS 9.1.3 help and documentation. SAS Institute, Cary, NC.
- Sheehan, M. J., P. R. Farmer and T. P. Brutnell, 2004. Structure and expression of maize phytochrome family homeologs. Genetics 167 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C., M. J. Aranzana, C. Lister, C. Baxter, C. Nicholls et al., 2005. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Phys. 138 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G. G., and C. Dean, 2002. Arabidopsis, the Rosetta Stone of flowering time? Science 296 285–289. [DOI] [PubMed] [Google Scholar]
- Slotte, T., K. Holm, L. M. McIntyre, U. Lagercrantz and M. Lascoux, 2007. Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae). Plant Phys. 145 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, G., 2001. Maize inbred lines released by CIMMYT. Available at http://www.cimmyt.org/english/wps/obtain_seed/cimmytCMLS.htm; accessed November 24, 2008.
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Tian, F., N. M. Stevens and E. S. Buckler, 2009. Tracking footprints of maize domestication and evidence for a massive selection sweep on chromosome 10. Proc. Natl. Acad. Sci. USA 106 9979–9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A., J. Beales, S. Faure, R. P. Dunford and D. A. Laurie, 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034. [DOI] [PubMed] [Google Scholar]
- Ulanch, P. E., 1999. Steps towards map-based cloning Ma1, a gene controlling photoperiod sensitivity in sorghum. Ph.D. Thesis, Texas A&M University, College Station, Texas.
- Van Ooijen, J. W., 1992. Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 84 803–811. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap version 3.0: software for the calculation of genetic linkage maps. Wageningen: Plant Research International. http://www.kyazma.nl/index.php/mc.JoinMap/
- Verhoeven, K. J. F., J. L. Jannink and L. M. McIntyre, 2006. Using mating designs to uncover QTL and the genetic architecture of complex traits. Heredity 96 139–149. [DOI] [PubMed] [Google Scholar]
- Vlăduţu, C., J. McLaughlin and R. L. Phillips, 1999. Fine mapping and characterization of linked quantitative trait loci involved in the transition of the maize apical meristem from vegetative to generative structures. Genetics 153 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. L., F. F. Cheng, Z. H. Sun, J. H. Tang, L. C. Wu et al., 2008. Genetic analysis of photoperiod sensitivity in a tropical by temperate maize recombinant inbred population using molecular markers. Theor. Appl. Gen. 117 1129–1139. [DOI] [PubMed] [Google Scholar]
- Warrington, I. J., and E. T. Kanemasu, 1983. a Corn growth response to temperature and photoperiod I. Seedling emergence, tassel initiation, and anthesis. Agron. J. 75 749–754. [Google Scholar]
- Warrington, I. J., and E. T. Kanemasu, 1983. b Corn growth response to temperature and photoperiod. III. Leaf number. Agron. J. 75 762–766. [Google Scholar]
- Wolfinger, R. D., W. T. Federer and O. Cordero-Brana, 1997. Recovering information in augmented designs, using SAS PROC GLM and PROC MIXED. Agron. J. 89 856–859. [Google Scholar]
- Wu, X. L., and J. L. Jannink, 2004. Optimal sampling of a population to determine QTL location, variance, and allelic number. Theor. Appl. Genet. 108 1434–1442. [DOI] [PubMed] [Google Scholar]
- Xue, W., X. Yongzhong, X. Weng, Y. Zhao, W. Tang et al., 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40 761–767. [DOI] [PubMed] [Google Scholar]
- Yan, L., A. Loukoianov, A. Blechl, G. Tranquilli, W. Ramakrishna et al., 2004. Flowering repressor down-regulated by vernalization. Science 303 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., J. B. Holland, M. D. McMullen and E. S. Buckler, 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]