Abstract
Potentially useful naturally occurring genetic variation is often difficult to identify as the effects of individual genes are subtle and difficult to observe. In this study, a novel genetic technique called Mutant-Assisted Gene Identification and Characterization is used to identify naturally occurring loci modulating the hypersensitive defense response (HR) in maize. Mutant-Assisted Gene Identification and Characterization facilitates the identification of naturally occurring alleles underlying phenotypic variation from diverse germplasm, using a mutant phenotype as a “reporter.” In this study the reporter phenotype was caused by a partially dominant autoactive disease resistance gene, Rp1-D21, which caused HR lesions to form spontaneously all over the plant. Here it is demonstrated that the Rp1-D21 phenotype is profoundly affected by genetic background. By crossing the Rp1-D21 gene into the IBM mapping population, it was possible to map and identify Hrml1 on chromosome 10, a locus responsible for modulating the HR phenotype conferred by Rp1-D21. Other loci with smaller effects were identified on chromosomes 1 and 9. These results demonstrate that Mutant-Assisted Gene Identification and Characterization is a viable approach for identifying naturally occurring useful genetic variation.
POTENTIALLY useful naturally occurring genetic variation is often difficult to identify as the effects of individual genes are subtle and difficult to observe. Furthermore, so many different alleles are available that it is a major challenge just to sift through the enormous diversity available. To this end, we recently conceptualized a simple yet effective method to discover and characterize variation present naturally in plant germplasm (Johal et al. 2008). This method, Mutant-Assisted Gene Identification and Characterization, makes use of a mutant phenotype for a gene affecting the trait of interest as a reporter to discover and analyze relevant, interacting genes present naturally in diverse germplasm. Mutant-Assisted Gene Identification and Characterization involves crossing a mutant to diverse germplasm and then evaluating the mutant progeny for transgressive changes (both suppressed and severe) in the mutant phenotype(s). If the mutation is recessive, the population needs to be advanced to the F2 generation to be able to detect and analyze such variation. However, for a dominant or partially dominant mutant, evaluations can be made immediately in the F1 to discover lines that contain suppressors or enhancers of the trait (mutation) under study. Mutant F1 progenies from such crosses can then be propagated further to identify, map, and clone genes/QTL that affect the trait positively or negatively. In the case of maize and other species for which genetically characterized mapping populations are available, modifying loci can be rapidly mapped by crossing a mutant line to each member of a mapping population and evaluating the resulting F1 families. In this study we provide a proof-of-concept for the Mutant-Assisted Gene Identification and Characterization technique, using it to identify loci involved in the defense response of maize.
Plants are constantly exposed to numerous potential pathogens with diverse modes of attack. Nevertheless, it is rather rare to see plants succumbing to disease. One key reason for this is the presence of a highly effective and inducible defense system, a major component of which is the hypersensitive response (HR). HR is usually associated with a specific recognition event and is activated after other nonspecific resistance mechanisms have been overcome or evaded (see Bent and Mackey 2007). Although it was initially coined to refer to the rapid collapse of cells at the site of infection, over the years the term HR has been used to refer to both cell death and the associated induction of a number of other defense responses, including the accumulation of phytoalexins and pathogenesis-related (PR) proteins at the site of infection, to name a few (Mur et al. 2007). Reactive oxygen species such as superoxide and H2O2 appear to be causally involved in cell death underlying the HR response (Jones and Dangl 2006).
HR is under the control of a subset of disease-resistance genes, commonly referred to as R genes. These R genes specifically recognize matching avirulence (Avr) effectors from the pathogen. Many R genes encode products containing a nucleotide-binding site (NBS) domain in the middle of the protein and a leucine-rich repeat (LRR) domain at the C-terminal end (Bent and Mackey 2007). R proteins are involved both in the recognition of the pathogen and the subsequent induction of the HR response. How R proteins remain in a quiescent but “vigilant” state remains to be established. Certain mutations in R genes have been found that abolish their dependence on AVR proteins for activation. Such aberrant R genes mostly behave as dominant or partially dominant alleles and trigger the HR constitutively in the absence of the pathogen (Hu et al. 1996; Zhang et al. 2003; Dodds et al. 2006). Two consequences of such “autoactive” or “ectopically active” R genes are a massive induction of cell death and the consequential stunting of the organism (Dodds et al. 2006). Although autoactive R genes have been found to exist in many plant species, the first few examples came from the maize Rp1 locus, which confers race-specific resistance to common rust, caused by Puccinia sorghi (Hu et al. 1996). Such autoactive R genes can be used to investigate HR genetics and etiology in the absence of confounding effects from the pathogen and constitute an excellent candidate for analysis using Mutant-Assisted Gene Identification and Characterization.
The details of the HR cell death reaction as well as the pathway(s) that link R gene activation with the HR remain unclear (Mur et al. 2007). Despite considerable research over the past decade, only a few components have been found thus far. Some of these, Ndr1, Eds1, Pad4, Rar1, and Sgt1, were identified in mutagenesis screens conducted to identify mutants that failed to undergo an HR reaction in response to infection by an avirulent pathogen (reviewed in Bent and Mackey 2007). A few others, RIN4, for example, were identified in yeast two-hybrid assays using an NBS–LRR protein as bait (Mackey et al. 2003). Recently, an Arabidopsis gain-of-function mutant that carries a point mutation in an R gene analog (a gene with the structure of an R gene but not known to be involved in resistance to any pathogen) was used to isolate a few more potential genes in the HR pathway in a second site suppressor approach following mutagenesis with ethane methyl sulfonate (EMS) (Palma et al. 2005; Zhang and Li 2005; Goritschnig et al. 2007). A problem with approaches based on intentional mutagenesis is that they fail to uncover genes that have either redundant or essential functions. One way to avoid this problem would be to seek naturally occurring allelic variants affecting HR. Such natural variation is pervasive in all species, being generated and selected for over millions of years of evolution.
Although natural variation has served as a constant provider of the R genes in all plant species, natural variability has not been tapped as a tool for understanding other aspects of the disease-resistance response (Holub 2007). The Rp1-D21 gene is an autoactive allele from the maize Rp1 disease-resistance locus that initiates HR randomly all over the plant (Pryor 1993; Collins et al. 1999; Sun et al. 2001). Our objective for this study was to use the Rp1-D21 gene phenotype as a test case for the Mutant-Assisted Gene Identification and Characterization approach. We show here that enormous variation exists in the maize germplasm that is capable of affecting the HR response positively or negatively and we identify loci that modulate expression of the HR phenotype segregating in the well-known Intermated B73 × Mo17 (IBM) advanced intercross line (AIL) population (Coe et al. 2002; Lee et al. 2002). This constitutes the first demonstration of the utility of the Mutant-Assisted Gene Identification and Characterization approach—an approach that is likely to prove widely applicable.
MATERIALS AND METHODS
Plant materials:
The Rp1-D21-H95 line was generated by crossing the Rp1-D21 variant to the maize inbred line H95 and backcrossing to the H95 parent four times, while selecting for the HR phenotype indicated by the spontaneous formation of cell death lesions. Since Rp1-D21 homozygotes in the H95 background are often unable to produce and sustain a viable ear, this stock is maintained in heterozygous condition by repeatedly crossing it to the H95 inbred.
The Rp1-D21-H95 heterozygote was crossed both to a collection of maize inbred lines (see Table 1) and to 233 recombinant inbred lines (RILs) from the maize IBM population (Lee et al. 2002). As expected, in each case, the resulting F1 families segregated in a 1:1 ratio for mutant (with Rp1-D21 HR lesions) to wild-type individuals. The IBM mapping population itself is composed of 302 F7:8 RILs derived from the cross of maize inbred lines B73 and Mo17. This population was intermated four times following the F2 stage before inbred lines were derived (Lee et al. 2002). Seed of IBM lines was received from the Maize Genetic Stock Center and also as gifts from Drs. A. Stapleton and O. Hoekenga.
TABLE 1.
The diversity of the Rp1-D21-mediated HR in maize
| Indiana scores |
NC scores |
|||
|---|---|---|---|---|
| Cross | Height ratio | Necrosis score | Height ratio | Necrosis score |
| A632 × Rp1-D21-H95 | 0.8 | 1.5 | 0.67 | 2.3 |
| B73 × Rp1-D21-H95 | 0.64 | 4 | 0.74 | 2.2 |
| B97 × Rp1-D21-H95 | 0.82 | 2 | 0.73 | 3.4 |
| CM103 × Rp1-D21-H95 | 0.23 | 7 | 0.50 | 5.6 |
| CML228 × Rp1-D21-H95 | 0.73 | 3.5 | 0.89 | 1.9 |
| CML322 × Rp1-D21-H95 | 0 | 10 | 0.32 | 7.2 |
| CML333 × Rp1-D21-H95 | 0.4 | 4 | 0.69 | 3.1 |
| CML69 × Rp1-D21-H95 | 0.48 | 6 | — | — |
| CML277 × Rp1-D21-H95 | 0.19 | 7 | 0.62 | 3.6 |
| CML247 × Rp1-D21-H95 | 0.73 | 3 | — | — |
| IL14H × Rp1-D21-H95 | 0.22 | 8 | 0.60 | 5.7 |
| Ki3 × Rp1-D21-H95 | 0 | 10 | 0.35 | 6.4 |
| Ki11 × Rp1-D21-H95 | 0.73 | 3 | — | — |
| Ky21 × Rp1-D21-H95 | 0.18 | 9 | 0.37 | 6.6 |
| M162W × Rp1-D21-H95 | 0 | 10 | 0.30 | 7.8 |
| M37W × Rp1-D21-H95 | 0 | 10 | 0 | 10 |
| Mo17 × Rp1-D21-H95 | 0.39 | 6 | 0.48 | 6.3 |
| Mo18w × Rp1-D21-H95 | 0.68 | 3 | 0.66 | 4.3 |
| MS-71 × Rp1-D21-H95 | 0.35 | 6.5 | 0.82 | 5.2 |
| NC350 × Rp1-D21-H95 | 0 | 10 | 0 | 10 |
| NC358 × Rp1-D21-H95 | 0.44 | 6 | 0.72 | 5.1 |
| Oh43 × Rp1-D21-H95 | 0.76 | 3.5 | 0.89 | 2.7 |
| Oh7B × Rp1-D21-H95 | 0.68 | 3 | — | — |
| P39 × Rp1-D21-H95 | 0 | 10 | — | — |
| Tx303 × Rp1-D21-H95 | 0 | 10 | 0.34 | 6.3 |
| Tzi8 × Rp1-D21-H95 | 0.51 | 4.5 | 0.81 | 3.9 |
F1 families were derived from crosses between a number of inbreds and the Rp1-D21-H95 line, which was heterozygous for the Rp1-D21 gene. The resulting F1 families therefore segregated 1:1 for the necrotic spotting phenotype associated with Rp1-D21. The height ratio was the ratio between the average height of the mutant and wild-type plants within a family. If all the mutant plants died as seedlings the height ratio was reported as 0. This was measured in Clayton, North Carolina, in 2006. The same families were planted in West Lafayette, Indiana, in 2009. In this case severity scores were taken on a 0–10 scale with 0 being no lesions at all and 10 being dead.
Field trials:
Experiments were performed at the North Carolina State University Central Crops Research Station located at Clayton, North Carolina, in the summers of 2006, 2007, and 2008 and at the Purdue Agronomy Center for Research and Education (ACRE) in West Lafayette, Indiana, in the summers of 2006 and 2007.
In Clayton 10 seeds per plot were planted in each plot and rows were not thinned. One plot of inbred border was planted on all sides of the experiment. Overhead irrigation was applied as needed to ensure satisfactory plant growth. Standard fertilizer and herbicide regimes for central North Carolina were used. Plots were 2 m in length with a 0.6-m alley at the end of each plot. Interrow spacing was 0.97 m. In 2009 F1 crosses between the Rp1-D21-H95 heterozygote and a set of diverse lines were planted in two replications. For the QTL mapping experiment, two replicates of the F1 families derived from crosses of the Rp1-D21-H95 heterozygote with individual IBM lines and the parental lines (B73 and Mo17) were planted in 2007 and 2008. In 2007, 184 F1 families were evaluated and in 2008, 233 F1 families were evaluated. Experimental units in each case consisted of single-row plots arranged in randomized complete blocks with two replications.
In West Lafayette, Indiana, 20 seeds per row were planted in 6-m rows spaced 0.76 m apart. As in Clayton, progenies of crosses of Rp1-D21-H95 with diverse inbreds were planted in 2009, while the test-cross progenies of 184 IBM RILs with Rp1-D21 were planted in 2007. Plants were drip irrigated as needed.
Scores were assigned on a row basis, only considering the mutant Rp1-D21 individuals within each row. They were scored on a 1–5 quantitative scale for HR lesion severity with 1 being complete absence of lesions and 5 being complete coverage of the ear leaf with lesions. In North Carolina, the plants were scored seven times in 2007 and 10 times in 2008 at approximately 1-week intervals during June and July. In Indiana the plants were scored twice, once on June 29 and once on July 9, 2007. At the end of the season the heights of all the plants were measured. In North Carolina in 2007, the time to anthesis of each plant was also measured.
Greenhouse trials:
F1 families from 184 different Rp1-D21-H95 heterozygote × IBM lines were examined in the greenhouse in Raleigh, North Carolina, in January 2006. Six complete randomized blocks were planted using single plants in 3-in.-diameter clay pots as experimental units. Pots were filled with a 50:50 mixture of Metromix (Scott's Inc., Marysville, OH) and sterilized soil. Supplemental lighting was used for 14hr/day to maintain a ∼16-hr day length. The plants were maintained at 26° during the day and 22° during the night. The actual temperature rarely deviated more than 4° from these target temperatures.
Each F1 family segregated 1:1 for the mutant to wild-type individuals so on average, only three Rp1-D21 individuals were grown for each family. By chance, for 10 families only wild-type plants and no mutant plants were planted while 5 families consisted solely of mutant Rp1-D21 plants. Plants were scored every 4 or 5 days from 17 days after planting to 44 days after planting on a 1–5 scale similarly to the field experiments. At 44 days, the height of all the plants was measured. For each family the average score of the Rp1-D21 phenotype (regardless of how many plants this was) was used as the score for subsequent analyses.
Marker-assisted analysis of Hrml1 in A632:
To check the status of Hrml1 in the maize line A632 a “pseudo-F2 population” was generated by crossing A632 and Rp1-D21-H95 to generate an F1 population segregating 1:1 for wild-type to mutant plants. Wild-type F1 plants were then crossed with mutant F1 plants to generate the pseudo-F2, which again segregated 1:1 for wild-type to mutant plants. A number of SSR markers located in bins 10.2 and 10.3 were examined for polymorphism between A632 and Rp1-D21-H95. The marker umc1962 was selected and used to genotype extreme segregants from the pseudo-F2. DNA was extracted by a fast extract protocol as described by Xin et al. (2003). PCR was performed in a total volume of 20 μl containing 2.5 mm MgCl2, 0.4 mm each dNTPs, 50 ng each of forward and reverse primers, and 0.3 units of Taq polymerase (Promega, Madison, WI). The PCR conditions were 30 sec each at 94°, 57°, and 72° for 35 cycles.
Expression analyses:
Expression analysis of maize defense response genes was conducted using semiquantitative reverse transcriptase polymerase chain reaction (RT–PCR). Total RNA was extracted from maize leaf tissue using TRIZOL (Invitrogen, Carlsbad, CA) and was treated with RNase-free DNase I (Promega Corp., Madison, WI). For RT–PCR, 200 ng of total RNA was reverse transcribed using oligo(dT) primers and AMV reverse transcriptase (Promega) to synthesize first-strand cDNA. In accordance with the manufacturer's instructions, 0.5 μg oligo(dT) primer/μg RNA was mixed in nuclease-free water and incubated at 70° for 5 min and then chilled on ice for 5 min. The AMV RT reaction mix (Promega) was then added to a final volume of 25 μl and the sample was incubated at 42° for 1 hr followed by 15 min at 70° to deactivate the reverse transcriptase. The resulting cDNA was then used to quantify transcript levels of several maize defense genes using the following PCR conditions: 94° for 30 sec, 57° for 30 sec, and 72° for 30 sec (32 cycles for the defense response genes and 28 cycles for the 18S ribosomal RNA [18S rRNA] control). The primers 18S-F (5′-TCCTGAGTAACGAACGAGACC-3′) and 18S-R (5′-CACGATGAAATTTCCCAAGAT-3′) were used to amplify the 18S rRNA control. The primers PR1-F (5′-AGGCTCGCGTGCCTCCTAGCTCTGG-3′) and PR1-R (5′-GGAGTCGCGCCACACCACCTGCGTG-3′) were used to amplify the maize PR1 defense response gene. The primers PR5-F (5′-AACAACTGCGGTTCACCGTG-3′) and PR5-R (5′-ACCGAGATGTCGTAGAAGTCC-3′) were used to amplify the PR5 defense response gene. The primer pair PRms-F (5′-ACCTGGAGCACGAAGCTGCAG-3′) and PRms-R (5′-GCAGCCGATGCTTGTAGTGGC-3′) was used to amplify the maize defense response gene PRms. The primers WIP1-F (5′-TGCTGATCCTGTGCCTCCAG-3′) and WIP1-R (5′-CTCTCTGATCTAGCACTTGGGG-3′) were used to amplify the WIP1 gene. All primers were obtained from Integrated DNA Technologies (Coralville, IA). Reaction products were visualized via gel electrophoresis (1% agarose) using a Gel-Doc imaging and documentation system (Bio-Rad, Hercules, CA).
Detection of H2O2 and superoxide ions:
The in situ presence of H2O2 and superoxide in Rp1-D21 leaves was visually detected with 3,3-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT), respectively, using the procedures described by Thordal-Christensen et al. (1997). The leaves undergoing Rp1-D21 lesion initiation were excised with a razor blade and allowed to take up NBT (1 mg/ml) or DAB (1 mg/ml) through the cut ends under high light intensity at room temperature. After 3 hr incubation, leaves were decolorized by boiling in 96% ethanol to remove chlorophyll before examining under a bright light transmission microscope.
Statistical analysis and QTL mapping:
F1 families derived from the cross between the Rp1-D21-H95 heterozygote line and 233 IBM lines were assessed in the field in both Clayton, North Carolina, in 2007 and 2008 and West Lafayette, Indiana, in 2007 (henceforth called NC ′07 and NC ′08 and IN, respectively). The population was also assessed in the greenhouse as juvenile plants in the winter of 2006. The population was scored for HR lesion intensity and severity (“necrosis”) and mutant:wild type height ratio within an F1 family (“height”) in all four environments and for differential between anthesis date of mutant and wild-type plants within an F1 family (“anthesis) in NC ′07 (see materials and methods for details of trait measurement). Least-squares means were calculated for the height and necrosis sAUDPC (standardized area under disease progress curve) traits measured in the three field trials. The traits were called “overall necrosis” and “overall height.” All correlation calculations were made using the PROC CORR procedure of SAS. The Windows QTL cartographer software package (Department of Statistics, North Carolina State University, Raleigh, NC) was used to detect the QTL. Composite interval mapping was used with a walk speed of 0.5 cM, window size 10 cM. Model 6 was used with five control markers and threshold values determined by permutation analysis with a significance level of 0.05. The following eleven traits were used for QTL analysis:
GH necrosis: The average standardized area under disease progress curve (sAUDPC) score was determined for all of the lesion mimic individuals for each Rp1-D21-H95 heterozygote × IBM lines F1 family grown on the greenhouse. sAUDPC ratings were calculated in the following way: The average value of two consecutive ratings was obtained and multiplied by the number of days between the ratings. Values were then summed over all intervals and then divided by the number of days of evaluation to determine the weighted average. This method is a standard way of measuring disease (or in this case lesion) development over time (Shaner and Finney 1977; Campbell and Madden 1990).
GH height: The ratio of the average height of the disease mimic plants divided by the average height of the wild-type plants within each Rp1-D21-H95 heterozygote × IBM line F1 family grown in the greenhouse at 44 days after planting.
IN necrosis: This is the average of the two scores for each Rp1-D21-H95 heterozygote × IBM lines F1 family grown in West Lafayette, Indiana, scored on the 1–5 scale described. In this case a single replication was assessed.
IN height: The ratio of the average height of the disease mimic plants divided by the average height of the wild-type plants within each Rp1-D21-H95 heterozygote × IBM line F1 family grown in West Lafayette, Indiana, in 2007, measured at the end of the season. By using the mutant:wild type height ratio rather than just the average height of the mutants, we could account for the variable levels of heterosis seen in each F1 family.
NC '07 necrosis and NC '08 necrosis: The average sAUDPC score for each Rp1-D21-H95 heterozygote × IBM lines F1 family grown in Clayton, North Carolina, in 2007 and 2008. Two replications were grown in complete randomized blocks. The sAUDPC was calculated for each F1 family for each replication and the average of the two sAUDPC scores for each F1 family was used.
NC height '07 and NC height '08: The ratio of the average height of the disease mimic plants divided by the average height of the wild-type plants within each Rp1-D21-H95 heterozygote × IBM lines F1 family grown in Clayton, North Carolina, 2007 and in 2008. Two replications were grown in complete randomized blocks. The height ratio was calculated for each F1 family in each replication and the average of the ratios for each F1 family was used.
NC anthesis 07: The average difference in days to anthesis of the disease mimic plants compared to the time to anthesis of the wild-type plants within each Rp1-D21-H95 heterozygote × IBM lines F1 family grown in Clayton, North Carolina. Two replications were grown in complete randomized blocks and the average of the anthesis differentials for each F1 family was used.
Overall necrosis: The least-squares means sAUDPC score for each F1 family over the three environments, NC '07, NC '08, and IN.
Overall height: The least-squares means height ratio for each F1 family over the three environments, NC '07, NC '08, and IN.
RESULTS
Rp1-D21 lesions form spontaneously in a developmentally programmed fashion:
Like many disease lesion mimic mutations, the HR lesions on Rp1-D21 mutants followed a developmental progression for initiation and expansion and were significantly affected by the environmental conditions. In the H95 background, cell death lesions first initiated on the oldest leaf at around week 2 after planting in the field (the three-leaf stage) but around week 3 in the glasshouse (the four-leaf stage). These lesions enlarged slightly and the new ones formed down the leaf blade in a basipetal fashion (i.e., progressing from the tip of the leaf to the base) (Figure 1). By the time they covered most of the leaf, new HR lesions initiated near the tip of the second leaf. This pattern of lesion initiation and expansion was repeated progressively as the plant grew and all the leaves were covered with Rp1-D21 lesions by anthesis with the lowermost leaves becoming entirely necrotic (Figure 1). The lesion initiation/formation was uniform either in a GH or field setting and their progression up the plant was gradual, suggesting that the lesions form spontaneously and did not need a stimulus for initiation.
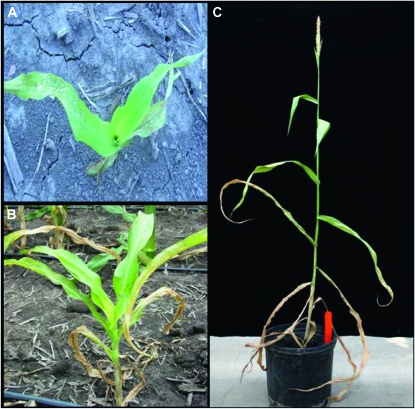
Figure 1.—
Manifestation of the Rp1-D21 phenotype in the heterozygous state in the Rp1-D21-H95 line. Phenotype of field-grown Rp1-D21-H95 plants 3 weeks after planting (A) and 8 weeks after planting (B). (C) An adult Rp1-D21-H95 mutant with a relatively normal tassel but no ear.
The growth and vigor of the Rp1-D21 mutants was significantly curtailed compared to their wild-type siblings. In the H95 background, Rp1-D21 mutants were about half the size of wild-type siblings. Rp1-D21-H95 plants were able to produce a small tassel that shed pollen normally, but they were never able to sustain a viable ear. As a result, the Rp1-D21-H95 line was maintained as heterozygotes by fertilizing H95 females with pollen from a Rp1-D21-H95 plant heterozygous for the Rp1-D21 gene.
The Rp1-D21 disease lesion phenotype has typical hallmarks of the hypersensitive response:
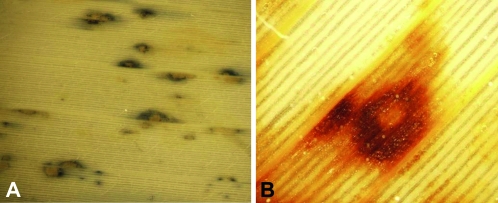
Genetic and molecular studies have clearly demonstrated that the Rp1-D21 gene is a structurally aberrant allele of functional R genes at the complex Rp1 disease-resistance locus (Collins et al. 1999; Sun et al. 2001). However, the question remained whether lesions associated with the Rp1-D21 mutation truly represented cell death typical of the HR response induced in response to pathogen attack. To determine this, we looked for two key hallmarks diagnostic of the bona fide HR response in mutants expressing the Rp1-D21 lesions in the Rp1-D21-H95 line. The first was the accumulation of reactive oxygen species superoxide (O2−) and hydrogen peroxide (H2O2), both of which have been shown to be causally involved in the HR response (Levine et al. 1994). We examined the in situ formation of these biochemical markers at developing Rp1-D21 lesions using the reagent NBT for O2− and DAB for H2O2. Both of these reactive oxygen species were detected uniquely around Rp1-D21 lesions (Figure 2), indicating their similarity to the HR cell death response.
Figure 2.—
In situ staining of developing Rp1-D21 lesions with nitroblue tetrazolium (A) and di-amino benzadine (B), showing the production of superoxide and H2O2, respectively.
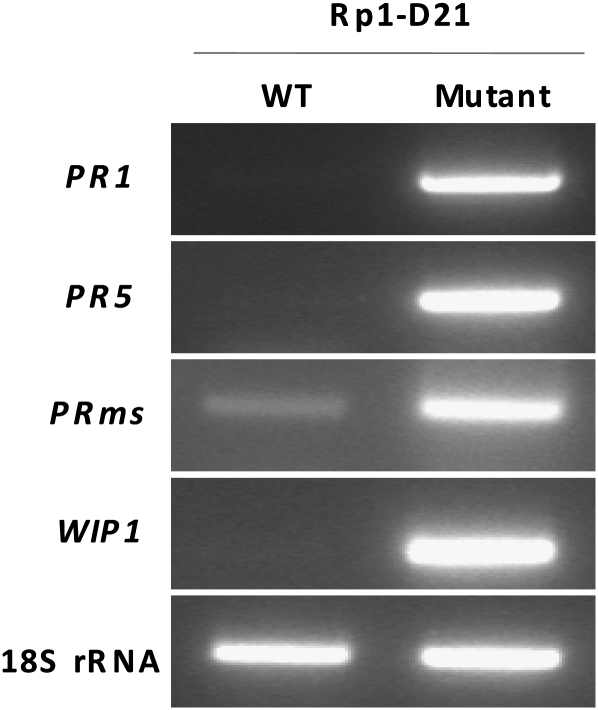
The second hallmark tested was the induction of defense response genes. In maize, the induction of four genes, PR1, PR5, PRms, and Wip1, has been associated with the HR defense response (Casacuberta et al. 1992; Rohrmeier and Lehle 1993; Morris et al. 1998; Simmons et al. 2002). We examined their expression in relation to developing Rp1-D21 lesions by RT–PCR. All four of the genes were markedly induced in plants exhibiting the Rp1-D21 phenotype compared to the wild-type siblings (Figure 3). These results support the conclusion that the cell death lesions that form on Rp1-D21 mutants are bona fide HR lesions and that the Rp1-D21 phenotype represents a spontaneous induction of the HR response.
Figure 3.—
RT-PCR assay showing constitutive induction of defense response genes in Rp1-D21 leaves. PR, Pathogenesis related; WIP, wound-inducible protein; 18S rRNA, control.
The phenotype conferred by Rp1-D21 is variable depending on the genetic background:
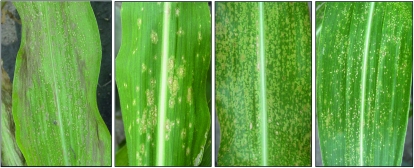
The Rp1-D21-H95 heterozygote line was crossed to a diverse set of 26 inbred lines comprising most of the founders of the maize nested association mapping (NAM) population (Yu et al. 2008). The resulting F1 families, segregating 1:1 for the mutant phenotype, were grown in the field in Indiana and North Carolina. A high degree of variation was observed in the Rp1-D21 phenotype, depending on the genetic background in both quantitative (Table 1 and Figure 4) and qualitative (Figure 5) terms. The HR phenotype conferred by Rp1-D21 was so strong in some F1 families that all the mutant plants died within 4 or 5 weeks after germination. In other F1 families the HR phenotype was extremely mild (Table 1 and Figure 4). It was noted that F1 families derived from crosses with Mo17 and B73, the two parents of the IBM population, differed substantially for the HR phenotype, the B73 cross being relatively mild and the Mo17 cross, relatively severe. Not surprisingly, the severity of the HR phenotype was well correlated with the degree of stunting observed when comparing the wild-type to the mutant individuals within each F1 family (Table 1). It should be noted that since different crosses produce different levels of heterosis, the appropriate measure of stunting is the ratio between wild-type and mutant individuals within a family rather than an absolute measure of height or yield. It was also evident that while the scores recorded in Indiana and North Carolina were highly correlated (>0.8 Person correlation coefficient for both traits), the phenotype was generally more severe in Indiana. This is likely due to the lower temperatures in the field in Indiana; the Rp1-D21 phenotype is temperature sensitive (Hu et al. 1996).
Figure 4.—
The progression of the Rp1-D21 phenotype in crosses between the Rp1-D21-H95 line and the lines indicated.
Figure 5.—
Examples of different morphology, size, and color of lesion of Rp1-D21 lesions in different backgrounds.
Identification of a QTL for suppression of the Rp1-D21 phenotype:
F1 families derived from the cross between the Rp1-D21-H95 heterozygote line and 233 IBM lines were assessed and scored as detailed in materials and methods. The correlations between the average sAUDPC scores for the two replications in NC '07 and NC '08 were 0.75 and 0.80, respectively. The correlations between all traits for all environments were moderate to high and all were highly significant (P < 0.0001; see Table 2). In the field tests, line and line-by-environment interactions were the main significant contributors to phenotypic variance for sAUDPC and height (Table 3). Environmental effects were large for both the phenotypes but they were not significant due to their large standard errors (Table 3).
TABLE 2.
Pearson correlation coefficients between different measured parameters affecting lesion mimic severity conferred by the Rp1-D21 gene
| GH necrosis | GH height | IN necrosis | IN height | NC '07 necrosis | NC '08 necrosis | NC '07 height | NC '08 height | |
|---|---|---|---|---|---|---|---|---|
| GH height | 0.58 | |||||||
| IN necrosis | 0.54 | 0.52 | ||||||
| IN height | 0.60 | 0.60 | 0.85 | |||||
| NC '07 necrosis | 0.69 | 0.50 | 0.73 | 0.72 | ||||
| NC '08 necrosis | 0.63 | 0.47 | 0.72 | 0.72 | 0.77 | |||
| NC '07 height | 0.52 | 0.43 | 0.68 | 0.66 | 0.71 | 0.71 | ||
| NC '08 height | 0.46 | 0.34 | 0.58 | 0.55 | 0.54 | 0.77 | 0.57 | |
| NC '07 anthesis | −0.39 | −0.46 | −0.56 | −0.54 | −0.56 | −0.48 | −0.61 | −0.28 |
Data were derived from a population from a cross between the Rp1-D21-H95 maize line heterozygous for Rp1-D21 and 233 different lines from the B73 × Mo17 advanced intercross IBM population. The population was assessed in four environments: in a greenhouse in winter 2006 in Raleigh, North Carolina (denoted GH), in the field in West Lafayette, Indiana, in summer 2007 (IN), and in the field in Clayton, North Carolina, in summer 2007 and 2008 (NC '07 and NC '08). Three different traits were measured: lesion severity measured on a 1–5 scale (with 1 being complete absence of lesions and 5 being complete coverage of the ear leaf with lesions—denoted as “necrosis” in the table), the ratio between the average height of lesion mimic and wild-type individuals within each F1 family (height), and the average divergence in time to anthesis in days between lesion mimic and wild-type individuals within each F1 family (anthesis). This last trait was measured only in Clayton, North Carolina, on 2007.
TABLE 3.
Variance component estimates and standard errors for standardized area under disease progress curve (sAUDPC) for the Rp1-D21 lesion phenotype and the mutant:wild type height ratio (Height) for a population consisting of F1 families from a cross between Rp1-D21-H95 heterozygote and 233 lines from the IBM population
| Variance component estimates (standard error) and P-values |
||||
|---|---|---|---|---|
| Parameter | sAUDPC | P-value | Height (×10−2) | P-value |
| Environment | 0.09 (0.09)a | NS | 1.37 (1.46) | NS |
| Replication within environment | 0.01 (0.01) | NS | 0.11 (0.11) | NS |
| Line | 0.18 (0.02) | <0.01 | 0.82 (0.11) | <0.01 |
| Environment by line | 0.03 (0.01) | <0.01 | 0.27 (0.07) | <0.01 |
| Residual | 0.06 (0.004) | <0.01 | 0.71 (0.06) | <0.01 |
The populations were scored in the field in Clayton, North Carolina, in the summers of 2007 and 2008 (two replications each) and in West Lafayette, Indiana, in 2007 (one replication). NS, not significant.
Standard error.
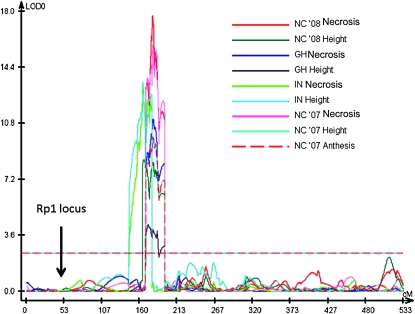
The strongest QTL identified for each trait and for each environment was on chromosome 10 in bin 10.03 (see Table 4). Although the detected QTL did not completely overlap in every case, they were so close that it is likely that they were all caused by the same underlying gene or genes (Figure 6). This QTL accounted for between 10 and 26% of the total variation, depending on the trait and the environment. For 6 of the 11 trait/environment combinations analyzed, the bin 10.03 QTL was the only significant QTL detected. We have termed this QTL Hrml1 for HR-modulating locus 1. A smaller effect QTL for GH Necrosis and NC Necrosis was also detected in bin 9.03 for 5 trait/environments including overall necrosis and overall height. A QTL of modest effect was identified in bin 9.02 for NC '08 necrosis. Effects were also detected at this locus in bin 9.02 for most of the necrosis traits, but they did not rise to the level of significance as defined by permutation analysis.
TABLE 4.
Chromosomal location in IBM map units and parameters associated with major quantitative trait loci (QTL), affecting lesion severity conferred by the Rp1-D21 gene
| Bin 1.05a |
Bin 9.02 |
Bin 9.03 |
Bin 10.03 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | 2 LOD intervalb | ac | LODd | R2(%)e | 2 LOD interval | a | LOD | R2(%) | 2 LOD interval | a | LOD | R2(%) | 2 LOD interval | a | LOD | R2(%) |
| GH necrosis | 463-473-482 | 0.21 | 8.9 | 14 | 241-249-257 | 0.16 | 5.2 | 8 | 174-179-185 | 0.23 | 11 | 19 | ||||
| IN necrosis | 164-165-178 | 0.58 | 13 | 24 | ||||||||||||
| NC '07 necrosis | 244-249-253 | 0.17 | 7.6 | 11 | 171-178-186 | 0.26 | 15.8 | 26 | ||||||||
| NC '08 necrosis | 122-133.6-142 | −0.14 | 5.4 | 7 | 235-248-253 | 0.12 | 5.4 | 6 | 176-180-183 | 0.24 | 17.8 | 23 | ||||
| Overall necrosis | 238-250-253 | 0.13 | 6.6 | 7 | 176-178-181 | 0.25 | 21.3 | 26 | ||||||||
| GH height | 168-173-188 | 0.05 | 4.3 | 1 | ||||||||||||
| IN height | 162-165-167 | 0.1 | 11.1 | 25 | ||||||||||||
| NC '07 height | 170-179-186 | 0.05 | 9.9 | 18 | ||||||||||||
| NC '08 height | 165-180-196 | 0.02 | 8.4 | 13 | ||||||||||||
| Overall height | 238-250-256 | 0.03 | 4.2 | 5 | 169-179-185 | 0.06 | 18 | 26 | ||||||||
| NC '07 anthesis | 169-184-196 | 0.96 | 5.2 | 11 | ||||||||||||
Data were derived from a population derived from a cross between the Rp1-D21-H95 maize line heterozygous for Rp1-D21 and 233 different lines from the B73 × Mo17 advanced intercross IBM population. The population was assessed in four environments: in a greenhouse in winter 2006 in Raleigh, North Carolina (denoted GH), in the field in West Lafayette, Indiana, in summer 2007 (IN), and in the field in Clayton, North Carolina, in summer 2007 and 2008 (NC '07 and NC '08). Three different traits were measured: lesion severity measured on a 1–5 scale (with 1 being complete absence of lesions and 5 being complete coverage of the ear leaf with lesions—denoted as “necrosis” in the table), the ratio between the average height of lesion mimic and wild-type individuals within each F1 family (height), and the average divergence in time to anthesis in days between lesion mimic and wild-type individuals within each F1 family (anthesis). Least-squares means for each F1 family over the three field environments (IN, NC '07, NC '08) were used to calculate QTL for “overall necrosis” and “overall height.” These traits are shown in bold.
Chromosome bin location of QTL peak on 1 of the 10 chromosomes of the maize genome. Bins divide the genetic map into 100 approximately equal segments. The segments are designated with the chromosome number followed by a two-digit decimal (e.g., 1.00, 1.01, 1.02, and so on).
The two outer numbers report the positions that define the two LOD interval around the position of peak likelihood for the QTL. The middle number defines the position of the peak. All values are in IBM map units (IcM; see Balint-Kurti et al. 2007) and are based on the IBM2 map (http://www.maizegdb.org/map.php).
The additive effect of the QTL. For necrosis ratings this is in terms of the 1–5 scale. For height ratings this is in terms of the ratio with “1” meaning a 1:1 ratio. For anthesis this is in terms of days. A positive number means the allele for decreased score (lower lesion level), increased ratio, or decreased anthesis differential derived from B73.
The log of odds (LOD) value at the position of peak likelihood of the QTL.
R2 estimates the proportion of phenotypic variance (%) explained by the detected QTL.
Figure 6.—
Chromosome 10 QTL likelihood plots for various Rp1-D21-associated traits in various environments: in a greenhouse in winter 2006 in Raleigh, North Carolina (denoted GH), in the field in West Lafayette, Indiana, in summer 2007 (IN), and in the field in Clayton, North Carolina, in summer 2007 and 2008 (NC '07 and NC '08). Three different traits were measured: lesion severity measured on a 1–5 scale (with 1 being complete absence of lesions and 5 being complete coverage of the ear leaf with lesions—denoted as Necrosis), the ratio between the average height of lesion mimic and wild-type individuals within each F1 family (Height), and the average divergence in time to anthesis in days between lesion mimic and wild-type individuals within each F1 family (Anthesis). Data were derived from a population derived from a cross between the Rp1-D21-H95 maize line heterozygous for Rp1-D21 and 233 different lines from the B73 × Mo17 advanced intercross IBM population. The y-axis shows the log of odds (LOD) likelihood ratio, and the x-axis denotes the position on the chromosome in IBM map units (Imu).
A QTL for GH Necrosis was detected in bin 1.05. There was no effect at this locus for any other trait. Since the GH traits were measured on young plants using scores taken between 17 days after planting to 44 days after planting, we thought this might be a juvenile-plant-specific QTL. To test this hypothesis we analyzed alone the earliest scores taken in NC '07 and NC '08 (which were taken at 24 and 37 days after planting respectively). We did not find an effect on these traits in bin 1.05 (data not shown).
The Hrml1 locus effects the RP1-D21 phenotype in other crosses:
To check if the Hrml1 locus modulated the effect of Rp1-D21 in other crosses, we analyzed the genetic basis for suppression of the Rp1-D21 HR phenotype in another inbred line A632, which, in crosses with Rp1-D21-H95, suppressed the Rp1-D21 phenotype even more strongly than B73. To accomplish this, a “pseudo-F2 population” was generated by crossing A632 and Rp1-D21-H95 to generate an F1 population segregating 1:1 for wild-type to mutant plants. Wild-type F1 plants were then crossed as females to their mutant siblings. The key reason for using this approach to generate an F2 mapping population was to keep the copy number of the Rp1-D21 allele constant in all mutant plants so that variation in the mutant phenotype was not due to different numbers of copies (one vs. two) of the Rp1-D21 allele. Because Rp1-D21 behaves in a partially dominant manner, plants containing two copies of the mutant allele (homozygous for Rp1-D21) would be more severe than plants carrying a single Rp1-D21 allele (Rp1-D21 heterozygotes). As expected, a range of variation of the Rp1-D21 phenotype was detected in the F2 population, including many plants that exhibited a highly suppressed Rp1-D21 phenotype. DNA was extracted from 35 of the most highly suppressed plants as well as from another 23 mutants randomly selected from a population of about 250 plants. These samples were evaluated for the segregation pattern of an Hrml1-linked SSR marker (umc1962), which was found to be polymorphic between A632 and H95. This marker exhibited no significant segregation distortion at the marker locus in the randomly selected Rp1-D21 plants (5 homozygous H95, 13 heterozygotes, 5 homozygous A632). However, all 35 Rp1-D21 plants having a highly suppressed phenotype were homozygous for the A632 allele at umc1962, suggesting that these suppressed plants were homozygous for the A632 Hrml1 allele and that the suppressive effect of the A632 Hrml1 allele was either recessive or partially dominant.
DISCUSSION
One standard way to identify components comprising genetic networks controlling biological processes is to do second-site mutagenesis (SSM) in lines that already possess mutations in genes affecting the phenotype of interest to reveal other genes that suppress or enhance the effect of the mutation (Page and Grossniklaus 2002). For instance, many of the genes known to be involved in the plant defense response were identified by this method (e.g., Li et al. 2001). Mutant-Assisted Gene Identification and Characterization provides a complimentary approach that allows one to tap into an additional, vast resource of genetic variation—natural variation, produced over millions of years of evolution (Johal et al. 2008). Results presented here show that Mutant-Assisted Gene Identification and Characterization is a viable approach for discovering natural variation underlying the HR response. Just as breeders have for many years exploited the high level of diversity in R genes produced by diversifying selection during evolution, so now Mutant-Assisted Gene Identification and Characterization provides a way in which diversity in the downstream components of the HR can be systematically identified. Mutant-Assisted Gene Identification and Characterization is conceptually similar to the SSM approach; however, instead of using induced variation, it relies on variation that is present naturally. For each locus, each different allele has presumably been selected for under specific environmental conditions and it is therefore likely that the natural variation unearthed by Mutant-Assisted Gene Identification and Characterization represents variation that could be adaptive and immediately valuable.
For the HR response conferred by Rp1-D21, which is a partially dominant mutant, another significant advantage of this approach is that it provides a highly quantitative and easy to measure parameter for the HR trait, which in the past could only be scored subjectively. We show that the severity of the HR response is positively correlated with the degree of stunting experienced by an Rp1-D21 mutant, such that the more severe the HR response, the more severe was stunting of the Rp1-D21 mutant. Thus using an Rp1-D21 mutant in heterozygous condition is beneficial, in that it allows the height ratio to be used as a highly sensitive and easy to record parameter of the HR response. This measure also mitigates against any bias that might arise as a result of different levels of heterotic vigor in different crosses. Height ratio obviously cannot be used with segregating population such as an F2 population in which every individual plant is genetically distinct. The remarkable genetic background-dependant variation in the phenotype conferred by Rp1-D21 is illustrated in Table1 and Figure 4. The fact that these comparisons were made between F1 hybrids, which shared 50% of their genome in common, makes the variation all the more noteworthy.
A major QTL, termed Hrml1, was detected in bin 10.03 in every trait and every environment assessed. Hrml1 represents a major modifier of the Rp1-D21 lesion mimic phenotype, with the B73 allele at this locus suppressing the phenotype and the Mo17 allele enhancing it. A recent review of the architecture of disease resistance in maize (Wisser et al. 2006), despite reporting coverage of 89% of the genome with disease resistance QTL, did not report a single major resistance gene or resistance QTL in bin 10.03, with the possible exception of a single QTL for gray leaf spot resistance, which had been mapped with poor precision.
Further work is required to identify the gene or genes underlying Hrml1. It is likely though that this locus is involved in the pathway controlling the elicitation or local spread of HR. We are currently performing detailed image analysis experiments designed to determine whether the B73 Hrml1 allele suppresses lesion initiation or lesion spread (or both). We are also currently working to construct B73 near isogenic lines differing for Hrml1 We will further cross them to lines carrying other Rp1 alleles to confirm that this locus also modifies the wild-type Rp1-mediated response. It also remains to be determined whether Hrml1 modulates cell-death phenotypes conferred by other major, HR-conferring, maize disease-resistance genes such as Rxo (Zhao et al. 2004), other Rp genes (e.g., Webb et al. 2002), and Rpp genes (Storey and Howland 1957; Futrell et al. 1975; Chen et al. 2004) and possibly also by non-HR causes of programmed cell death (Buckner et al. 2000).
It should be noted that, since we were assessing the effect of these genes in F1 hybrids between IBM lines and the Rp1-D21-H95 rather than in inbred lines, for a modifying locus to be detected, either the Mo17 or the B73 allele at that locus has to dominant or partially dominant with respect to the allele in the Rp1-D21-H95 line (i.e., they cannot both be completely recessive). If both the Mo17 and B73 alleles were recessive to the Rp1-D21-H95 allele, then no modifying effect could have been detected. It is therefore possible that some loss of function alleles in B73 or Mo17 with strong modifying effects when present in homozygous form could have been missed by this approach. This class of modifiers could in theory be identified by generating additional segregating populations, such as a backcross or an F2 population such as the population used in this article to show that Rp1-D21-suppressing alleles of Hrml1 also exits in A632. Conversely, the A632 Hrml1 allele identified here cannot be completely dominant as otherwise a mixture of A632 heterozygotes and homozygotes at the Hrml1 locus would have been identified among the most repressed individuals in the F2 population, rather than them all being A632 homozygotes. It is therefore quite likely that the Hrml1 suppressive allele is partially dominant in both cases. Further work using near-isogenic lines will clarify the allelic relationships at this locus.
It should also be noted that, in addition to Hrml1, the Rp1 locus itself is present on chromosome 10. However it is more than 100 IBM map units (approximately equivalent to 25 cM) away from the main QTL peak and therefore cannot be responsible for the QTL. To confirm this we divided up the IBM population into two groups, one homozygous B73 across the Rp1 locus and the other homozygous Mo17 (there was also a sizable group that was recombinant across the Rp1 locus—this group was ignored for this analysis). A highly significant effect at Hrml1 was detected by analysis of each group separately (data not shown). In other words, an Hrml1 effect could be detected in populations in which Rp1 was not segregating. Further evidence for the fact that Hrml1 is not an Rp1 allele comes from the analysis of the A632 × Rp1-D21-H95 F2 population. In this case, all the F2 plants that show a Rp1-D21 phenotype must be heterozygous at the Rp1 locus, with one copy of Rp1-D21 and one copy of either the wild-type A632 Rp1 allele or the wild-type H95 allele. In this population the profound effect of Hrml1 can still be detected as all the most suppressed plants are A632 homozygotes at the Hrml1 locus.
Although Mutant-Assisted Gene Identification and Characterization was conceived during genetic dissection of genetic background effects on les23, a recessive lesion mimic mutant of maize (Penning et al. 2004), use of an aberrant phenotype to identify genes involved in specifying specific phenotypes, one of the ideas underlying Mutant-Assisted Gene Identification and Characterization, has been used in the past in both plants and animals. Indeed, most restorers of fertility genes in all crops have been identified essentially by genetic schemes akin to Mutant-Assisted Gene Identification and Characterization (Duvick 1956). In Drosophila, many components of the sevens pathway that executes eye formation were identified using a genetically hypersensitive background generated by the ectopic expression of a component of the pathway (Gibson and Dworkin 2004). With Mutant-Assisted Gene Identification and Characterization we are able to additionally harness both the large amount of genetic diversity in maize (Liu et al. 2003) and the excellent genetic and genomic resources available for the crop such as RILs and NILs for the discovery of natural genes or gene variants. A key advantage here is that, in many cases, one must do the phenotyping only, the genotyping data of the resource being already available. The IBM mapping population used to uncover Hrml1 is an AIL maize population derived from a cross between the maize inbreds B73 and Mo17 with four generations of random mating following the formation of the F2 generation and prior to the development of inbred lines (Lee et al. 2002). The increased opportunity for recombination has had the effect of expanding the genetic map approximately fourfold compared to nonintermated, conventional RIL populations (Lee et al. 2002). The IBM population consists of a relatively large number of lines (302), which have been densely genotyped with more than 2000 molecular markers (Coe et al. 2002). Another advantage of using RILs for gene discovery is that the scores are derived from families and not individual segregants, as in an F2 population. So the same population can be evaluated for multiple traits, multiple times, and at multiple locations.
The maize nested association mapping (NAM) population (Yu et al. 2008) is a recently established 5200-line mapping population that consists of 26 RIL subpopulations, each of which was derived from a cross between B73 and 1 of 25 other diverse lines. Additional Hrml loci can almost certainly be identified using the NAM population with an approach identical to that used here. We demonstrate in this work that a great deal of diversity capable of modulating the HR response exists in the NAM founders (Table 1). A study is underway in our labs to conduct a Mutant-Assisted Gene Identification and Characterization screen on the NAM RILs.
One concern could be whether Rp1-D21 triggers a bona fide HR rather than causing cell death by some other mechanism. There are many reasons to suggest that it does. First, we know that Rp1-D21 is an autoactive allele of Rp1, a known disease resistance gene that confers an HR in response to specific P. sorghi isolates. In addition, Figures 2 and 3 show that the Rp1-D21 phenotype is associated with the production of superoxide and H2O2, together with the induction of the genes PR1, PR5, PRms, and WIP1, all hallmarks of a bona fide HR. It should be noted that these genes can also be induced by a defense response not including HR and by various other stresses including wounding (Casacuberta et al. 1991, 1992; Rohrmeier and Lehle 1993; Morris et al. 1998; Dunkle and Levy 2000). Even so, the preponderance of evidence suggests that Rp1-D21 triggers an exaggerated form of the normal maize hypersensitive response.
In 1983 it was proposed that mutants such as Rp1-D21 could be used as “a simplified system for the plant response to disease and stress” without the causative agent needing to be present (Walbot et al. 1983). In this study we have used this approach in conjunction with the Mutant-Assisted Gene Identification and Characterization concept and with modern mapping resources such as the IBM population. We have identified Hrml1 as a naturally occurring suppressor allele on chromosome 10 and have demonstrated that many more naturally occurring alleles likely effect the defense response and can be identified in a straightforward way. Most important, we have demonstrated the utility of the Mutant-Assisted Gene Identification and Characterization approach. Many mutants that could be exploited for Mutant-Assisted Gene Identification and Characterization already exist in maize and many other plant systems. For example, mutants that confer aluminum sensitivity (Pascholati et al. 1986; Sibov et al. 1999), alterations in starch accumulation (Braun et al. 2006), or sensitivity to drought (Postlethwait and Nelson 1957) are available and could be used in a conceptually very similar way to that demonstrated here to identify useful variation in the traits that they affect. In other cases, mutagenesis screens could be designed to specifically identify mutants for use in Mutant-Assisted Gene Identification and Characterization. Ultimately Mutant-Assisted Gene Identification and Characterization is an approach that could be harnessed to identify naturally occurring, useful alleles important for a large number of traits in many systems.
Acknowledgments
We are grateful to the following for assistance and valuable advice: Cliff Weil, Randy Wisser, Rebecca Nelson, Donna Stephens, Kristen Kump, Rahul Dhawan, and Hugh Young. This work was funded by USDA–ARS, Purdue University, and a National Science Foundation (NSF) Grant 0822495.
References
- Balint-Kurti, P. J., J. C. Zwonitzer, R. J. Wisser, M. L. Carson, M. Oropeza-Rosas et al., 2007. Precise mapping of quantitative trait loci for resistance to southern leaf blight, caused by Cochliobolus heterostrophus race O, and flowering time using advanced intercross maize lines. Genetics 176 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A. F., and D. Mackey, 2007. Elicitors, effectors, and R Genes: the new paradigm and a lifetime supply of questions. Ann. Rev. Phytopath. 45 399–436. [DOI] [PubMed] [Google Scholar]
- Braun, D. M., Y. Ma, N. Inada, M. G. Muszynski and R. F. Baker, 2006. Tie-dyed1 regulates carbohydrate accumulation in maize leaves. Plant Physiol. 142 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner, B., G. S. Johal and D. Janick-Buckner, 2000. Cell death in maize. Physiol. Plantarum 108 231–239. [Google Scholar]
- Campbell, C. L., and L. V. Madden, 1990. Introduction to Plant Disease Epidemiology, pp. 192–194. John Wiley & Sons, New York.
- Casacuberta, J. M., P. Puigdomenech and B. San Segundo, 1991. A gene coding for a basic pathogenesis-related pr-like protein from Zea mays molecular cloning and induction by a fungus Fusarium moniliforme in germinating maize Seeds. Plant Mol. Biol. 16 527–536. [DOI] [PubMed] [Google Scholar]
- Casacuberta, J. M., D. Raventos, P. Puigdomenech and B. San Segundo, 1992. Expression of the gene encoding the Pr-like protein Prms in germinating maize embryos. Mol. Gen. Genet. 234 97–104. [DOI] [PubMed] [Google Scholar]
- Chen, C. X., Z. L. Wang, D. E. Yang, C. J. Ye, Y. B. Zhao et al., 2004. Molecular tagging and genetic mapping of the disease resistance gene RppQ to southern corn rust. Theor. Appl. Genet. 108 945–950. [DOI] [PubMed] [Google Scholar]
- Coe, E., K. Cone, M. McMullen, S.-S. Chen, G. Davis et al., 2002. Access to the maize genome: an integrated physical and genetic map. Plant Physiol. 128 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N., J. Drake, M. Ayliffe, Q. Sun, J. Ellis et al., 1999. Molecular characterization of the maize Rp1-D rust resistance haplotype and its mutants. Plant Cell 11 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P. N., G. J. Lawrence, A. M. Catanzariti, T. Teh, C. I. Wang et al., 2006. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle, L. D., and M. Levy, 2000. Genetic relatedness of African and United States populations of Cercospora zeae-maydis. Phytopath. 90 486–490. [DOI] [PubMed] [Google Scholar]
- Duvick, D. N., 1956. Allelism and comparative genetics of fertility restoration of cytoplasmically pollen sterile maize. Genetics 41 544–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futrell, M. C., A. L. Hooker and G. E. Scott, 1975. Resistance in maize to corn rust, controlled by a single dominant gene. Crop Sci. 15 597–599. [Google Scholar]
- Gibson, G., and I. Dworkin, 2004. Uncovering cryptic genetic variation. Nature Rev. Genet. 5 681–690. [DOI] [PubMed] [Google Scholar]
- Goritschnig, S., Y. Zhang and X. Li, 2007. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 49 540–551. [DOI] [PubMed] [Google Scholar]
- Holub, E. B., 2007. Natural variation in innate immunity of a pioneer species. Curr. Op. Plant Biol. 10 415–424. [DOI] [PubMed] [Google Scholar]
- Hu, G., T. E. Richter, S. H. Hulbert and T. Pryor, 1996. Disease lesion mimicry caused by mutations in the rust resistance gene Rp1. Plant Cell 8 1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal, G. S., P. Balint-Kurti and C. F. Weil, 2008. Mining and harnessing natural variation: a little MAGIC. Crop Sci. 48 2066–2073. [Google Scholar]
- Jones, J. D. G., and J. L. Dangl, 2006. The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 X Mo17 (IBM) population. Plant Mol. Biol. 48 453–461. [DOI] [PubMed] [Google Scholar]
- Levine, A., R. Tenhaken, R. Dixon and C. Lamb, 1994. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79 583–593. [DOI] [PubMed] [Google Scholar]
- Li, X., J. D. Clarke, Y. Zhang and X. Dong, 2001. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant-Microbe Interact. 14 1131–1139. [DOI] [PubMed] [Google Scholar]
- Liu, K., M. Goodman, S. Muse, J.-S. Smith, E. Buckler et al., 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Y. Belkhadir, J.-M. Alonso, J.-R. Ecker and J.-L. Dangl, 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Morris, S. W., B. Vernooij, S. Titatarn, M. Starrett, S. Thomas et al., 1998. Induced resistance responses in maize. Mol. Plant Microbe Interact. 11 643–658. [DOI] [PubMed] [Google Scholar]
- Mur, L. A., P. Kenton, A. J. Lloyd, H. Ougham and E. Prats, 2007. The hypersensitive response: The centenary is upon us but how much do we know? J. Exp. Bot. 59 501–520. [DOI] [PubMed] [Google Scholar]
- Page, D. R., and U. Grossniklaus, 2002. The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 3 124–136. [DOI] [PubMed] [Google Scholar]
- Palma, K., Y. Zhang and X. Li, 2005. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15 1129–1135. [DOI] [PubMed] [Google Scholar]
- Pascholati, S. F., R. L. Nicholson and L. G. Butler, 1986. Phenylalanine ammonia-lyase activity and anthocyanin accumulation in wounded maize Zea–Mays mesocotyls. Phytopathol. Z.
- Penning, B. W., G. S. Johal and M. M. McMullen, 2004. A major suppressor of cell death, slm1, modifies the expression of the maize (Zea mays L.) lesion mimic mutation les23. Genome 47 961–969. [DOI] [PubMed] [Google Scholar]
- Postlethwait, S. N., and O. E. Nelson, 1957. A chronically wilted mutant of maize. Am. J. Bot. 44 628–633. [Google Scholar]
- Pryor, A. J., 1993. Transposon tagging of a rust resistance gene in maize, pp. 469–475 in Advances in Molecular Genetics of Plant–Microbe Interactions, edited by E. Nester and D. Verma. Kluwer Academic, Dordrecht, The Netherlands.
- Rohrmeier, T., and L. Lehle, 1993. WIP1, a wound-inducible gene from maize with homology to Bowman–Birk proteinase inhibitors. Plant Mol. Biol. 22 783–792. [DOI] [PubMed] [Google Scholar]
- Shaner, G., and P. E. Finney, 1977. The effect of nitrogen fertilizer on expression of slow mildewing resistance in Knox wheat. Phytopath. 67 1051–1056. [Google Scholar]
- Sibov, S. T., M. Gaspar, M. J. Silva, L. M. M. Ottoboni, P. Arruda et al., 1999. Two genes control aluminum tolerance in maize: genetic and molecular mapping analyses. Genome 42 475–482. [Google Scholar]
- Simmons, C. R., J. T. Tossberg, G. A. Sandahl, W. A. Marsh, P. F. Dowd et al., 2002. Maize pathogen defenses activated by avirulence gene avrRxv. Maize Genet. Coop. Newsl. 76 40–41. [Google Scholar]
- Storey, H. H., and A. K. Howland, 1957. Resistance in maize to the tropical American rust fungus, Puccinia polysora Underw. I. Genes Rpp1 and Rpp2. Heredity 11 289–301. [Google Scholar]
- Sun, Q., N. Collins, M. Ayliffe, S. M. Smith, J. Drake et al., 2001. Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H., Z. G. Zhang, Y. D. Wei and D. B. Collinge, 1997. Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11 1187–1194. [Google Scholar]
- Walbot, V., D. Hoisington and M. G. Neuffer, 1983. Disease lesion mimic mutations, pp. 431–432 in Genetic Engineering of Plants, edited by T. Kosuge, C. P. Meredith and A. Hollaender. Plenum Press, New York.
- Webb, C. A., T. E. Richter, N. C. Collins, M. Nicolas, H. N. Trick et al., 2002. Genetic and molecular characterization of the maize Rp3 rust resistance locus. Genetics 162 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisser, R. J., P. J. Balint-Kurti and R. J. Nelson, 2006. The genetic architecture of disease resistance in maize: a synthesis of published studies. Phytopath. 96 120–129. [DOI] [PubMed] [Google Scholar]
- Xin, Z., J. P. Velten, M. J. Oliver and J. J. Burke, 2003. High-throughput DNA extraction method suitable for PCR. BioTechniques 34 820–826. [DOI] [PubMed] [Google Scholar]
- Yu, J., J. B. Holland, M. D. McMullen and E. S. Buckler, 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., S. Goritschnig, X. Dong and X. Li, 2003. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1–1, constitutive 1. Plant Cell 15 2636–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and X. Li, 2005. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of Npr1–1, constitutive 1. Plant Cell 17 1306–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, B. Y., E. Ardales, E. Brasset, L. E. Claflin, J. E. Leach et al., 2004. The Rxo1/ Rba1 locus of maize controls resistance reactions to pathogenic and non-host bacteria. Theor. Appl. Genet. 109 71–79. [DOI] [PubMed] [Google Scholar]