Abstract
Comparative studies of Caenorhabditis briggsae and C. elegans have provided insights into gene function and developmental control in both organisms. C. elegans is a well developed model organism with a variety of molecular and genetic tools to study gene functions. In contrast, there are only very limited tools available for its closest relative, C. briggsae. To take advantage of the full potential of this comparative approach, we have developed several genetic and molecular tools to facilitate functional analysis in C. briggsae. First, we designed and implemented an SNP-based oligonucleotide microarray for rapid mapping of genetic mutants in C. briggsae. Second, we generated a mutagenized frozen library to permit the isolation of targeted deletions and used the library to recover a deletion mutant of cbr-unc-119 for use as a transgenic marker. Third, we used the cbr-unc-119 mutant in ballistic transformation and generated fluorescently labeled strains that allow automated lineaging and cellular resolution expression analysis. Finally, we demonstrated the potential of automated lineaging by profiling expression of egl-5, hlh-1, and pha-4 at cellular resolution and by detailed phenotyping of the perturbations on the Wnt signaling pathway. These additions to the experimental toolkit for C. briggsae should greatly increase its utility in comparative studies with C. elegans. With the emerging sequence of nematode species more closely related to C. briggsae, these tools may open novel avenues of experimentation in C. briggsae itself.
Keywords: Caenorhabditis briggsae, single-nucleotide polymorphism (SNP) mapping, knockout screening, cell lineage, gene expression
COMPARATIVE analysis of genes between related species is a powerful experimental approach, allowing one to exploit nature's experiments to illuminate the sequence basis of function. Commonly, conservation of sequence is used to delineate functional elements in a genome and is particularly effective with protein-coding sequence. However, in many instances differences in sequence may also be informative. For example, the small size of transcription factor binding sites allows for generation of new sites and loss of old sites, masking the conservation of functions (Andolfatto 2005). In other cases, a change in sequence is associated with a change in function that sheds light on the underlying mechanisms of gene action. In these instances, simple sequence comparisons are inadequate; they need to be supplemented with functional analysis.
For comparative functional analysis of Caenorhabditis elegans, C. briggsae remains the preferred species. Although it shows an estimated 1.6 substitutions per neutral site with C. elegans (Stein et al. 2003), it remains the closest identified living relative. Both are self-fertilizing hermaphrodites and have very similar morphology. Recent evidence suggests that the developmental cell lineage is remarkably constant between the two species (Zhao et al. 2008). However, changes in cellular signaling underlying these patterns have been revealed by quantitative study of development after cell ablation or manipulation of the signaling molecules (Felix 2007). Conservation of sequence has been used in detecting new candidate protein-coding genes (Stein et al. 2003) and in the discovery of large regulatory regions (Kuntz et al. 2008; Sleumer et al. 2009). In other cases, however, the differences have been informative for functional divergence. For example, changes in lin-48 expression alter the placement of the excretory duct opening (Wang and Chamberlin 2004). Genes in the sex determination pathway have diverged substantially, providing insights into alternative mechanisms for sex determination (Nayak et al. 2005; Hill et al. 2006). These and a growing list of examples illustrate the potential power of the comparative approach for these two organisms.
This potential has spurred the development of an increasing battery of resources in C. briggsae with which one may attack fundamental biological questions. For example, its genome has been sequenced using a shotgun strategy and assembled into contigs (Stein et al. 2003) and then been ordered along chromosomes (Hillier et al. 2007). RNAi by injection allows the knockdown of gene activity and an engineered strain containing the C. elegans sid-2 gene extends the use of RNAi to feeding (Winston et al. 2007). Mutants can be isolated either through conventional means (D. Baillie, personal communication; B. Gupta, unpublished results; Inoue et al. 2007) or by PCR-based screening methods (Hill et al. 2006). However, a frozen mutagenized library that allows routine PCR-based gene knockout is still lacking in C. briggsae. A rudimentary genetic map has been constructed through conventional mapping of mutants (B. Gupta, unpublished results) and this has been supplemented with a SNP-based map assayed through PCR-based methods (Hillier et al. 2007). Unfortunately, there are only a limited number of mutants mapped on chromosome, making it difficult to map a newly isolated genetic mutant in C. briggsae. Transgenic arrays can be generated through injection, but in many cases, an alternative transformation method such as ballistic bombardment is required to generate low- or single-copy chromosomally integrated transgenes (Praitis et al. 2001). We have recently reported the embryonic cell lineage of C. briggsae and demonstrated its remarkable similarity to that of C. elegans using a GFP-labeled strain (Zhao et al. 2008). However, the strain carries an extrachromosomal array and is unable to stably transmit and express the transgene, preventing the full use of the strain for other lineage-based analysis.
Here we added to the C. briggsae toolkit in several important ways. We applied to C. briggsae the methods recently developed in C. elegans (Flibotte et al. 2009) for using array comparative genomic hybridization (CGH) to map mutants. This allows rapid, inexpensive localization of new mutants into a region with only a few hundred kilobase pairs in a single step. We created a frozen mutagenized library to facilitate the isolation of deletion mutations and used this library to isolate an allele of cbr-unc-119. In turn we used the deletion allele as a selection marker for transformation with biolistic bombardment. With this ability we created strains that allow the ready determination of the C. briggsae embryonic cell lineage and in turn the high-resolution mapping of gene expression. Using transgenes as fate markers, we used RNAi and automated lineaging to study the effects of cbr-pop-1 loss of function on cell fates, confirming and extending the recent results (Lin et al. 2009).
MATERIALS AND METHODS
Strains used:
All the strains were maintained at room temperature for genetic manipulations. The strains used for this study were AF16, HK104, PS9329 (cbr-lin-11 I), DY214 (lev I), DY93 (cbr-pry-1 I), PS9148 (cbr-sma-6 II), RW10426 (unc ?), DY63 (rol ?), DY144 (cby-1 III), JT10250 (csp-1 III), PS9022 (cbr-dpy-1 III), PS9482 (unc IV), RW10417 (unc ?), PS9454 (unc V), BC6031 (unc V), Dy175 (unc V), RW10421 (unc ?), PS9001 (rot-1 X), RW20000 (cbr-unc-119), RW20022 (cbr-unc-119, stIs20022[cbr-HIS-72∷GFP, unc-119 (+)]), RW20025 (cbr-unc-119, stIs20025[cbr-HIS-72∷mCherry, unc-119 (+)]), RW20027 (cbr-unc-119, stIs20025[cbr-HIS-72∷mCherry], stIs20027[HLH-1∷GFP, unc-119 (+)]), RW20045 (cbr-unc-119, stIs20025[cbr-HIS-72∷mCherry], stIs20045[PHA-4∷GFP, unc-119 (+)]), RW20046 (cbr-unc-119, stIs20025[cbr-HIS-72∷mCherry, unc-119 (+)], stIs20046[EGL-5∷GFP, unc-119 (+)]), and RW20047 (cbr-unc-119, stIs20047[cbr-myo-2∷GFP, unc-119 (+)]).
SNP-based genetic mapping:
We adapted the methods of (Flibotte et al. 2009) for use in C. briggsae, including genetic crosses, DNA preparation, microarray design, and hybridization and analysis. A total of 9701 SNPs were selected among several hundred thousand detected by comparing the genomic sequences of HK104 obtained by Illumina sequencing methods with the AF16 sequence (Stein et al. 2003; D. Spencer, Z. Zhao, R. H. Waterston, M. Olson and L. Hillier, unpublished results). We avoided SNPs with other nearby SNPs that might overlap the oligonucleotides. The selected SNPs (supporting information, Table S2) spanned the sequences that were assigned to specific positions on chromosomes but do not include regions of the sequence not assigned to chromosomes or those assigned to chromosomes but not positioned along them. The microarrays were manufactured by Roche NimbleGen, with oligonucleotides synthesized at random positions on the arrays. Each SNP was represented on the microarray by an average of almost 40 50-mer oligonucleotides targeting both the AF16 (cb3 assembly) and the HK104 sequences. The typical spacing between adjacent oligonucleotides was 2 bases and the targeted strand was selected to ensure the SNPs were as far as possible from the slide to maximize the SNP signal (Flibotte et al. 2009). The current microarray design is available to the community under the name “080211_Moerman_cbriggsae_CGH” and researchers are encouraged to contact the authors to inquire about potential future releases. For all the experiments, normalization of fluorescence intensity ratios was performed with a LOESS regression as previously described (Maydan et al. 2007).
The C. briggsae mutants were generated using 25 mm EMS (see below; B. Gupta, unpublished data). They were mated with HK104 male animals and from the wild-type F1 progeny, a total of 10 F2 mutant worms were picked onto 12 15-cm plates seeded with OP50 (120 worms total) and allowed to propagate until starvation. Worms were pooled and genomic DNAs were extracted using the QIAGEN (Valencia, CA) Blood and Tissue DNeasy kit. The genomic DNAs from AF16 and the mutant worms were differentially labeled and hybridized to the array, either by Nimblegen or locally using standard protocols. For the initial test, the genomic DNAs from both AF16 and HK104 were differentially labeled and hybridized on the microarray to test the ability of the oligonucleotides to distinguish the two DNAs (Figure S1).
For rescue of the putative cbr-unc-22 mutation, we co-injected BAC clones 18K09 and 25D10 using myo-2∷GFP as a selection marker with concentrations of 50, 50, and 20 ng/μl, respectively. The rescued animals were judged by the presence of the active moving gravid adults on the bacterial lawn. The adult mutants were settled on the bacterial lawn with little crawling (data not shown).
BAC preparation and microinjection:
BAC preparations were performed essentially as described (Marra et al. 1997). On the basis of end-sequencing data in Wormbase (WS203), each of the two BACs (plate 18 K09 and plate 25 D20) partially covers the genomic regions of cbr-unc-22 but together they cover the entire gene. The two clones were cultured overnight in the presence of 10 μM chloramphenical of LB. The 5′ and 3′ ends of each BAC were verified by PCR. The purified BACs from the two clones were combined and co-injected into the cbr-unc-22 animals with AF16 genomic DNA, using the concentrations of 50 and 100 ng/μl, respectively. Rescue was scored as gravid adult progeny moving actively on the bacterial lawn.
Constructing a deletion library:
We built a mutagenized library for deletion screening using methods similar to those described for C. elegans (Ahringer 2006; H. Hutter, personal communication). Our detailed methods are described below to facilitate their use by others.
Synchronized L4 P0 worms were mutagenized with 50 mm EMS for 4 hr in a 15-ml Falcon tube on a shaker. The worms were washed four times with M9 buffer and grown overnight on large peptone-rich plates seeded with Escherichia coli OP50. The gravid adults were used for egg preparation followed by starvation on unseeded plates to synchronize the worms at the L1 stage. To measure the efficiency of mutagenesis, we examined the occurrence of putative mutations in the cbr-unc-22 locus by checking the twitching animals in 0.1 mm levamasole. The presence of at least 1 twitching animal of 1000 mutagenized F1 progeny was required to proceed to the next step.
The starved worms were mixed with aliquots from 2 liters of E. coli DH5α grown in LB at 37° overnight so that ∼20 animals were present in 50 μl medium in the presence of appropriate antibiotics and antifungal agents. The worms were cultured in flat-bottomed 96-well tissue culture plates with 50 μl culture per well for 5 days in a moist chamber at room temperature until the wells became “clear” due to the exhaustion of the bacteria; each well contained ∼1500 worms.
To harvest the library, one-fourth of the culture was pooled for genomic DNA preparation, one-fourth was left on the plates for the on-plate lysis with proteinase K, and the remaining one-half was used for freezing as described (Ahringer 2006). We used the QIAGEN Blood and Tissue DNeasy kit for the genomic DNA preparation with the DNA yield sufficient for ∼800 PCR screenings.
Recovery of a cbr-unc-119 deletion:
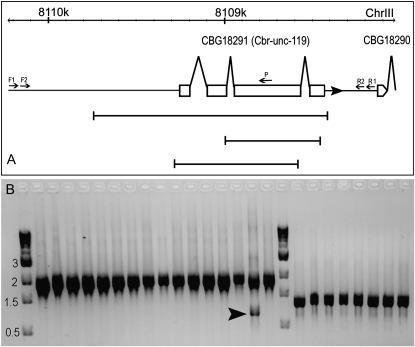
To recover a cbr-unc-119 mutant from the library, we designed a nested primer set with a single poisoning primer (Figure 3, Figure S2). Given the relatively small size of its genomic region as well as the fact that most of the recovered alleles in similar screens carried a deletion with a size ranging from 500 bp to 3 kb (Liu et al. 1999), we picked primers that amplified a fragment twice as big as its genomic region.
Figure 3.—
Deletion alleles detected for cbr-unc-119. (A) Schematic representation of a cbr-unc-119 gene model (exons depicted as boxes and transcription orientation indicated by a solid arrowhead). Three deletion alleles are shown below the gene model at the same scale. The primers used for the nested PCR screening are indicated as small arrows. The chromosomal position in kilobases is listed above the gene model. (B) A gel picture for the two rounds of PCR products with the second round on the left (∼1.9 kb) and the (partial) first round on the right (∼1.4 kb in the presence of poisoning primer). A 1-kb ladder was loaded both on the leftmost lane and in between the two rounds of PCR products with the sizes indicated in kilobases. An arrowhead indicates a shifted deletion band.
The sequences for the primers are as follows: external left (F1), ATATTTGACAGAAACCGGGATG; external right (R1), TGGGTACAATCGTATCCTCCTC; internal left (F2), ATGAATTGGCTCAAAGTTAGGG; internal right (R2), TCCTCGCTGGACATTCTCTAAT; poisoning primer, CACGTATCTTGCCGACTCCT.
For PCR, we used Fermentas 2× PCR Mastermix with 5 pmol individual primers and 10 ng genomic DNA as a template in a 25-μl volume reaction. One poison primer was included for the first round of the PCR screening (Figure 3). A total of 0.2 μl of the PCR product was used as a template for the second round of PCR.
Once a deletion band was identified, its sibling lysate plate was thawed (Figure S2) and PCR was performed as described above with the following modifications. Given the impurities and presence of PCR inhibitors like EDTA in the lysate, we diluted 1 μl of the lysate 10-fold with ddH2O. The diluted samples were pooled along row and column of each candidate microtiter plate, respectively, to make 20 samples and 1 μl was used as the template for the PCR screening as described above. The presence of deletion bands in both a row and a column identified a single well that contained the deletion allele. The lysate from the positive well was used as the template to perform the same two rounds of PCRs to verify the presence of the deletion allele.
The well corresponding to the positive one from the lysate plate was thawed as described (Ahringer 2006). For 3 consecutive days or until all the surviving worms were recovered, single L3 or L4 animals were transferred into individual wells of a 96-well culture plate containing ∼20 times less food than that used for the library construction. To synchronize the analysis, the worms picked on the first 2 days were put at 15° and moved to room temperature on the third day. After culturing for 1 week, the cultures were split into two sibling copies and on-well lysis was performed using one of the copies for one round of PCR, using the internal left and right primer pairs. Once a deletion was located to a single well, the culture from its sibling well was transferred onto a seeded NGM plate and allowed to grow for 3–5 hr followed by picking at least 20 single L3 or L4 worms into individual NGM plates. After overnight growth, single-worm PCR with the presence of poison primer was performed to genotype each worm to identify a homozygous strain. The isolated homozygous mutant was then backcrossed 5 times before being used for a rescuing test. The mutant demonstrates the expected Uncoordinated (Unc) and Dauer formation Defective (Daf-D) phenotypes as seen for its orthologous allele in C. elegans (Figure S3). Daf-D phenotypes were examined in 1% SDS solution for 0.5 hr.
Transformation rescue of cbr-unc-119 mutants:
We tested the utility of the cbr-unc-119 deletion allele as a transgenic selection marker by ballistic bombardment using constructs containing either the C. elegans or the C. briggsae version of wild-type genomic DNA as described (Praitis et al. 2001). Successful transformation was judged by the presence of actively moving dauer larvae. The number of the rescued lines varied among different bombardments, but both types of constructs were able to rescue the allele and produced integrated lines (Table S1).
Construction of lineaging strains:
For ubiquitous labeling of nuclei required for lineage tracing, we built a construct carrying a protein fusion between cbr-his-72 and a fluorescent reporter (GFP or mCherry). For the GFP fusion, vector pAZ132 (Praitis et al. 2001) was cut with XmaI and blunt ended and then cut with SacII. The vector backbone containing the pie-1 3′-UTR and unc-119 rescuing fragment was gel purified. A PCR-amplified cbr-his-72 genomic fragment was fused in frame with GFP coding sequences to generate a fragment flanked by SacII and XmaI sites by fusion PCR (Hobert 2002). The fused fragment was cut with XmaI and blunt ended and then cut with SacII followed by gel purification. The resulting fragment was ligated with the above vector backbone to give rise to pZZ22. For mCherry fusion, vector pAZ132 was cut with SpeI and blunt ended and then cut with SacII followed by gel purification. A PCR-amplified cbr-his-72 genomic fragment was fused in frame with mCherry coding sequences to generate a fragment flanked by SacII and EcoRV sites by fusion PCR. The fragment was first cut with EcoRV and blunt ended and then cut with SacII followed by gel purification. The fragment was ligated with the above vector backbone cut with SpeI and SacII to give rise to pZZ27. The two constructs were used to bombard the cbr-unc-119 mutant allele to give rise to multiple lines that are suitable for automatic lineaging. One line with bright and ubiquitous expression was selected for lineaging. RW20022 and RW20025 were used for lineaging using GFP and red fluorescent protein (RFP), respectively.
Single-cell gene expression profiling:
To examine expression patterns of three C. elegans genes, pha-4, egl-5, and hlh-1 in C. briggsae, we generated multiple chromosomal integrated transgenic lines by ballistic bombardment of protein:reporter fusion constructs into cbr-unc-119. The ORFs for these genes were tagged with GFP in the context of fosmids, using the recombineering technique (Sarov et al. 2006). The resulting transgenic strains were crossed into the RFP-based lineaging strain RW20025. Both lineaging marker and GFP-tagged loci were rendered homozygous before imaging using a Zeiss LSM510 confocal microscope. Due to the relatively weak expression of the red lineaging marker compared to that used in C. elegans, we increased the laser power to a range of 12–80% from the top to the bottom focal plane on the RFP channel after the 100-cell stage without impact on the survival of the imaged embryo. For the GFP channel, we used the same laser power as that for C. elegans, i.e., from 0.2 to 1% from the top to the bottom focal plane. We imaged one stack (31 planes at 1-μm spacing) every 1.5 min continuously for ∼6 hr at room temperature. Expression analysis was performed as previously described (Murray et al. 2008).
Ethyl methanesulfonate mutagenesis for mutant isolation:
Mutagenesis was performed as described (Brenner 1974) except that the P0 worms were mutagenized in 25 mm ethyl methanesulfonate (EMS) for 4 hr. F1 animals were singled out and animals with Unc, Dpy, Rol, and Sma were picked from F2 populations and crossed three times with AF16 before being used for mapping experiments.
RESULTS
A SNP-based oligonucleotide array for rapid and high-resolution genetic mapping in C. briggsae:
The isolation and characterization of mutants plays a central role in delineating gene functions and dissecting genetic pathways in C. elegans and other model organisms. Mutants are readily isolated in C. briggsae after mutagenesis, and, as in C. elegans, the availability of RNAi does not obviate the need for genetic mutants with stable defined effects. However, the lack of a detailed genetic map has slowed genetic analysis in C. briggsae. Precise genetic map localization simplifies assignment of mutations to complementation groups and in particular can greatly facilitate the molecular identification of the gene affected by the mutation. To advance genetic analysis in C. briggsae, we developed a SNP-based microarray for rapid mapping of genetic mutants by a bulk-segregant assay based on a newly published method (Flibotte et al. 2009).
The method utilizes SNPs between different isolates of the same species. For this purpose we used SNPs identified between AF16 [the sequenced strain (Stein et al. 2003)] and HK104 [a strain showing ∼0.4% divergence (Hillier et al. 2007)]. We obtained HK104 genomic sequence using next-generation sequencing to call SNPs across the genome (D. Spencer, Z. Zhao, M. Olson, R. H. Waterston and L. Hillier, unpublished data). From these data, we selected a total of 9701 SNPs well spaced across the chromosomally positioned sequences and used them to manufacture a SNP-based oligonucleotide array by NimbleGen, applying an algorithm similar to that developed for C. elegans (Flibotte et al. 2009).
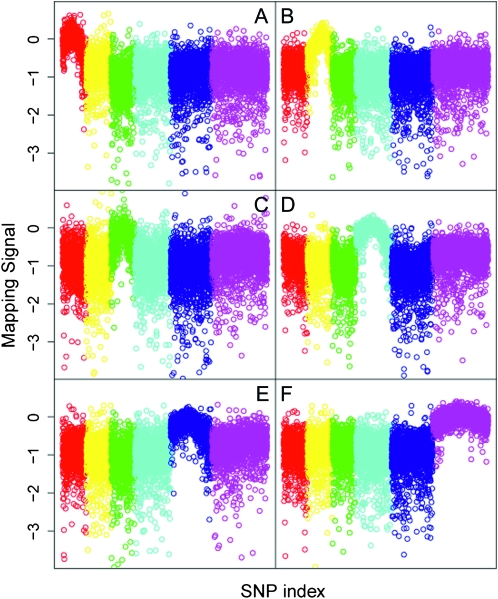
We validated the array in several experiments. We first examined the ability of the oligonucleotides to distinguish the sequences derived from each strain and found that most of them can robustly distinguish the two variants as expected (Figure S1). We next attempted to use the chip to map 14 mutations with both known and unknown map locations, including some newly recovered mutations from our EMS-based screen (Table 1, Figure 1). Generally, the analysis of the signal intensities yielded unambiguous assignment of the mutations to a chromosome and the arc of the signal provided an approximate position along the chromosome. For most mutations with genetic map positions, both the chromosomal assignment and the approximate position along the chromosome as determined by the array agreed with the conventional map location. However, for two mutations, cbr-lin-11(sy5336) and unc-(sy997), difficulty in scoring the phenotype led to a weak chromosome signal (cbr-lin-11) (Figure 2) or no localization [unc-(sy997)] (Table 1 and data not shown). In a third exception, cbr-dpy-1(sy5022), in addition to the expected chromosome III signal, there was also an increase in signal for chromosome I (Figure S1). Further, while most of the signal on chromosome III was located on the left arm, there was a separate sharp spike of signal on the right arm (Figure 2 and Figure S1). However, the other mutation mapping to chromosome III did not show this correlated increase in signal for chromosome I (Figure 1 and data not shown), suggesting the chromosome I signal for cbr-dpy-1 was specific for that cross.
TABLE 1.
Strains used for testing the SNP-based oligonucleotide microarray
| Strain | Allele | Gene name | Linkagea | Positionb | Homolog | Physical positionc |
|---|---|---|---|---|---|---|
| PS9349 | sy5336 | cbr-lin-11 | I | I: 7945234 | lin-11 | I: 6224361..6218662 |
| DY214 | sy5440 | lev | I | I: 6994809 | Unknown | Unknown |
| DY93 | sy5353 | cbr-pry-1d | I | I: 10047372 | pry-1 | I_random: 2174403..2183496 |
| PS9148 | sy5148 | cbr-sma-6 | II | II: 7983665 | sma-6 | II: 8576600..8573397 |
| RW10426 | st10426 | unc | Unknown | II: 5292517 | Unknown | Unknown |
| DY63 | sy5412 | rol | Unknown | III: 6544265 | Unknown | Unknown |
| PS9022 | sy5022 | cbr-dpy-1e | III | III: 241980 | dpy-1 | III: 12355146..12345594 |
| PS9482 | sy5422 | unc | IV | IV: 6731162 | Unknown | Unknown |
| RW10417 | st10417 | cbr-unc-22 | Unknown | IV: 6089638 | unc-22 | IV: 4927059..4962138 |
| PS9454 | sy5415 | unc | V | V: 15108872 | Unknown | Unknown |
| BC6031 | sy5094 | unc | V | V: 6905777 | Unknown | Unknown |
| DY175 | sa997 | unc | V | V: 7243478 | Unknown | Unknown |
| RW10421 | st10421 | unc | Unknown | V: 7441578 | Unknown | Unknown |
| PS9001 | sy5001 | rot-1 | X | X: 7200502 | Unknown | Unknown |
known linkage group before this work.
estimated positions from current mapping.
physical positions annotated in Wormbase (WS205)
The contig carrying cbr-pry-1 is not placed into the context of chromosome I.
The discrepancy between our estimated position and the Wormbase (WS205) annotated position is likely caused by a misassembly artifact.
Figure 1.—
Genomic views of the mapping signals for six C. briggsae mutations. For each SNP represented on the array the differences between the median log2 ratio (mapping signal) for probes with AF16 and HK104 sequences are plotted against the SNP index distributed along the six chromosomes and differentially color coded from left to right for I, II, III, IV, V, and X, respectively. The alleles used are listed as follows: (A) I, sy5440; (B) II, sy5148; (C) III, sy5142; (D) IV, st10417; (E) V, sy5094; and (F) X, sy5001.
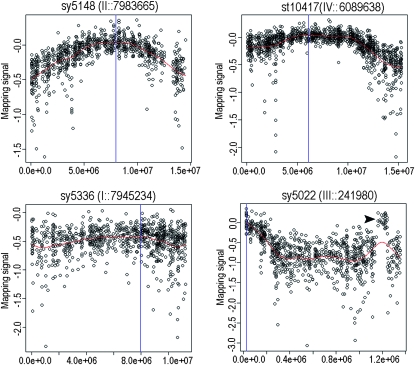
Figure 2.—
Linkage maximum estimation using spline fitting. Shown are the mapping signals plotted against the chromosome coordinates for four mutations located on four different chromosomes. The linkage maxima estimated by a cubic spline algorithm are indicated by blue lines. The chromosomal coordinates corresponding to the maxima are shown on the top of each panel along with allele names. Note a spike on the right arm of chromosome III (indicated by an arrowhead), which is likely caused by an error in the genome assembly.
To obtain a more refined position for the mutants we used a spline-fitting algorithm to estimate the site of maximal linkage (Figure 2). We compared the estimated positions with the sequence locations for the three genes with known genome locations (Table 1). For cbr-sma-6(sy5148) the oligonucleotides with the highest linkage value were only ∼600 kb from its actual position (WS205) (Table 1, Figure 2). On the other hand, the site of highest linkage to cbr-lin-11(sy5336), which had been difficult to score in the F2's, lay 1.7 Mb from its position in the assembly (WS205). The results from cby-4(sy5022), the C. briggsae ortholog of the C. elegans dpy-1 gene, were more complex. As indicated above, most probes with an increased signal lay on the left arm of III, suggesting a position ∼0.2 M from the left end. However, the sharp spike lay between 11.6 and 12.4 Mb, a region that includes the cby-4 gene (Figure 2, Figure S1). Because bulk segregant analysis is not expected to yield such a sharp spike, we investigated that region of the assembly in more detail. The region corresponds to a single supercontig positioned by a single sequence-tagged site (STS) (data not shown). Perhaps this region could be misassembled in the current sequence assembly and should instead be placed near the left end of the chromosome. On the basis of these findings we looked for other anomalous results associated with specific mutations and found only one other region that may represent a misassembly with a small segment of 125 kb currently located on chromosome V that may belong to chromosome IV (data not shown). Thus, overall our results validate the current genome assembly and the genetic recombination map used to provide long-range order for the physical map, but the systematic application of the method could reveal additional minor misassemblies or errors in marker order.
To test the utility of the information to clone unmapped mutations, we investigated the newly isolated unc-(st10417) allele in more detail. The SNP-array data positioned the mutation at 6.1 Mb on chromosome IV. In the 2 Mb surrounding this region there were three genes likely orthologous to an unc gene in C. elegans (WS205). On the basis of the described phenotypes of these Uncs, we deduced that the mutation most likely lay in the cbr-unc-22 gene [chrIV:4927059..4962138 (WS205)]. To test the hypothesis, two different, partially overlapping C. briggsae BAC clones, each carrying the partial genomic region for cbr-unc-22, were co-injected and together were able to rescue the Unc phenotype by microinjection, indicating that the observed Unc phenotype was produced by the mutation in this locus (see materials and methods, data not shown). Thus in this case, although the interval was somewhat large (∼1.2 Mb from the estimated position to its actual position within the genome), it was narrow enough that when combined with existing annotations from C. elegans mutants in the corresponding region, our mapping results were able to narrow the candidates to a few genes that could then be tested by transformation rescue. We expect that a combination of the SNP-mapping method with the use of C. elegans annotations should allow the rapid identification of many mutations in C. briggsae.
A mutagenized frozen library for deletion screening:
In addition to finding mutations through traditional methods, PCR-based screening has been widely used in recent years in C. elegans to isolate deletions in specific genes and has been applied to C. briggsae (Hill et al. 2006). To create an equivalent resource and test these methods in C. briggsae, we modified methods from those previously described for C. elegans (Liu et al. 1999; Edgley et al. 2002; Ahringer 2006) (Figure S2) and generated a mutagenized library with frozen aliquots to provide a resource for future use. EMS was the sole mutagen, as opposed to UV–trimethylpsoralen (TMP) (see materials and methods), to avoid the variability associated with the latter (D. Moerman and M. Edgley, unpublished observations). On the basis of the frequency of putative cbr-unc-22 “twitching” mutants (see materials and methods), the efficiency of the mutagenesis is comparable to that observed for C. elegans (Moerman et al. 1986). This is further supported by the frequency of recovery of cbr-unc-119 mutants (see below). The entire library consists of a total of 160 96-well plates, containing a total of some 640,000 mutagenized genomes. Our frozen library can be used for up to 800 screenings for C. briggsae deletion alleles.
To test the library, we performed a PCR-based deletion screening to isolate the deletion allele for cbr-unc-119, an ortholog of C. elegans unc-119 (Maduro 2008). The C. elegans unc-119(ed3) mutant provides an efficient selection marker for ballistic bombardment transformation, which results in integrated, low copy number transgenes in C. elegans (Maduro and Pilgrim 1995; Praitis et al. 2001). The orthologous C. briggsae gene can also effectively rescue the C. elegans unc-119(ed3) mutant (Sarov et al. 2006), suggesting strongly that a loss-of-function allele of cbr-unc-119 would exhibit the same phenotypes as its C. elegans counterpart, namely the Unc and Daf-D phenotypes important for the selection of transformed animals.
We screened the EMS mutagenized library for deletions of the 0.9-kb cbr-unc-119 gene and detected a total of three deletion alleles from an initial screening of 96 pooled plates with deletion sizes ranging from 570 bp to 1.4 kb (Figure 3). All three deletions were confirmed in the secondary screening of the lysate plates (Figure S2), indicating that each had been transmitted through the germline. We thawed the well of the frozen library corresponding to the 1.4-kb deletion and isolated a homozygous viable mutant worm that stably propagated the Unc phenotype through 10 generations. Sequence analysis of the endpoints shows the deletion removes the entire coding region of cbr-unc-119 together with some of its upstream intergenic region (Figure 3). The backcrossed cbr-unc-119(st20000) mutant exhibits the expected severe Unc and Daf-D phenotypes. However, cbr-unc-119(st20000) has a smaller body size and a weaker egg-laying defective (Egl) phenotype than the C. elegans mutant (Figure S3).
To demonstrate the utility of the cbr-unc-119 allele as a selection marker for DNA transformation, we attempted to rescue the mutant by both microinjection and ballistic bombardment. Plasmid constructs carrying the C. elegans wild-type unc-119 genomic regions and fosmids generated by the modENCODE project (www.modencode.org) carrying the C. briggsae unc-119 gene both rescued the mutant (Table S1, data not shown). Thus, the cbr-unc-119 allele can be used as an efficient selection marker for transformation in C. briggsae by bombardment or injection.
Automatic gene expression profiling at single-cell resolution in C. briggsae:
Precise knowledge of a gene's expression patterns is valuable information in deciphering its role in the animal. We recently developed methods that allow automated determination of expression patterns in embryogenesis with cellular resolution in C. elegans (Bao et al. 2006; Murray et al. 2008). These methods rely on a ubiquitously expressed fluorescent histone label to determine the lineage, which in turn provides the basis for identifying each cell.
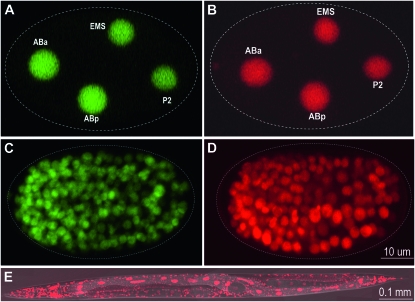
To extend these methods to C. briggsae we generated integrated transgenic lines expressing nuclear localized GFP or mCherry (RFP) in all embryonic cells. We used the cbr-his-72 promoter to drive expression in all cells, a cbr-HIS-72∷G/RFP protein fusion to localize the signal in the nucleus, and the C. elegans pie-1 3′-UTR to enhance expression in the germline with resultant labeling of early nuclei (see materials and methods). This construct was placed on a plasmid carrying a wild-type unc-119 gene and multiple lines carrying the integrated transgene were recovered after biolistic bombardment. The worms have brightly labeled nuclei throughout embryogenesis (Figure 4) and are satisfactory for automatic lineaging and expression analysis (see below). In addition, with the red-shifted mCherry-labeled strain we were able to obtain an improved signal-to-noise ratio compared to that in the GFP lines. This resulted in improved performance of the StarryNite algorithm (Bao et al. 2006) and allowed us to more readily extend the curated lineage to the point when there are 450 cells in the embryo (Figure S4).
Figure 4.—
Micrographs of transgene expression in C. briggsae lineaging strains. (A and C) Expression of histone GFP fusion in a four-cell embryo (A) and a bean stage embryo (C). (B and D) Expression of histone mCherry fusion in a four-cell embryo (B) and a bean stage embryo (D). The embryo boundary is indicated with dashed lines. (E) Expression of the same histone mCherry fusion in an adult C. briggsae animal merged with a DIC image. All the embryos and the adult animal are oriented so that anterior is to the left.
To test these strains for automatic gene expression profiling in C. briggsae, we generated stable transgenic C. briggsae strains expressing several tissue-specific protein∷reporter fusions for C. elegans genes and analyzed their expression by lineage analysis. The reporter constructs were derived from C. elegans genomic fosmid clones containing an entire transcription factor gene plus its flanking sequences (J. Perkins and D. Moerman, unpublished data). These fosmids were modified by recombineering to include a GFP∷FLAG(3×) fused in frame at the C terminus of the targeted protein and a cbr-unc-119 rescuing sequence (Sarov et al. 2006) and introduced into C. briggsae by bombardment. The resulting integrated strains were crossed into the mCherry-labeled lineaging strain to generate double homozygote lines for expression analysis [see materials and methods (Murray et al. 2008)].
We produced cellular resolution embryonic gene expression profiles in this way for three C. elegans genes: pha-4, hlh-1, and egl-5. For each gene, we determined the lineage out to 350 cells and extracted the expression signal for each cell. In turn we compared the pattern of expression with that seen for the same construct in C. elegans (Figure 5, Figure S5). This pairwise comparison of expression patterns between the two species revealed that the same cells expressed the reporter in the two organisms. For egl-5, a posterior hox gene required for normal specification of the tail and HSN neuron (Desai et al. 1988; Ferreira et al. 1999), both the onset and level of expression were similar between the two species. However, for pha-4, a forkhead transcription factor required for pharynx development (Mango et al. 1994), and hlh-1, a MyoD ortholog (Chen et al. 1994), the intensity of the signal was less with the C. elegans construct in the C. briggsae background than in its native background. This was often correlated with a later onset of detection in expressing lineages (Figure 5). Although these differences might result from reduced efficacy of the C. elegans regulatory sequences in the C. briggsae background, the differences are minor and will require additional experiments to establish the significance of the difference and to rule out other causes such as insertion site and construct broken point. The fact that these key cell fate regulators have the same lineal expression patterns further confirmed the conserved fate map between the two species as revealed by lineage analysis (Zhao et al. 2008).
Figure 5.—
Automatic gene expression profiling at single-cell resolution in C. briggsae. (A) An embryonic lineage tree showing the expression profile of pha-4 (red branches) in a 350-cell C. briggsae embryo. The major tissue fates are color coded at the bottom. PHA-4∷GFP is present at higher levels in the precursors of pharynx than in those of intestine. Also labeled are the major founder cells. (B) Pairwise comparisons of the expression profile of a Hox gene, egl-5 (left) and a MyoD homolog, hlh-1 (right) between C. briggsae and C. elegans. Both spatial and temporal patterns of the expressing cells are well preserved for EGL-5 between the two species but expression of HLH-1 in C. briggsae appears delayed and intensity decreased as opposed to those in C. elegans despite the well preserved spatial patterns between the two species. All of the DNA constructs used here are protein fusions in C. elegans fosmids with GFP at the C-terminal ends. These were bombarded into both species to generate stable reporter-expressing strains (see materials and methods).
In addition to the strains generated for automatic lineaging, we produced multiple stable lines expressing tissue-specific markers that should improve the utility of C. briggsae as a model animal in studies of molecular, cellular, and developmental biology (Table S1).
Comparison of lineage/cell fate transformation after gene perturbation between C. elegans and C. briggsae:
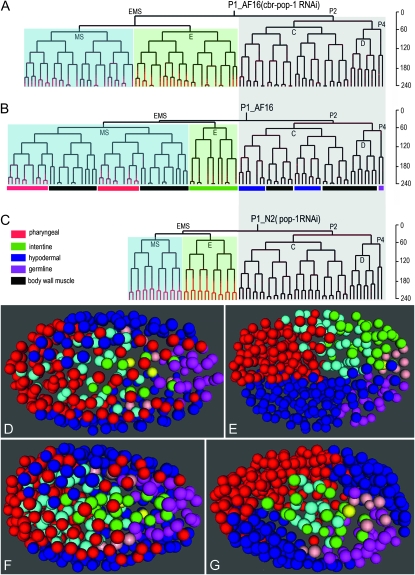
The methods used to generate the lineages do not depend on knowledge of the wild-type lineage and can thus be used to assay experimentally altered lineages. In addition, the expression reporters we have generated also provide markers for cell fate. To evaluate the utility of automated lineaging combined with cell fate markers in C. briggsae we investigated the recent report that reduction of the activity of a Wnt signaling pathway TCF-like effector, pop-1, by RNAi produced the opposite homeotic lineage transformation between E and MS in C. briggsae to that produced in C. elegans (Lin et al. 2009). We confirmed the results by parallel application of the lineaging technology to the two species; i.e., RNAi against pop-1 produced homeotic transformation from MS to E in C. elegans but from E to MS in C. briggsae (Figure 6). Indeed, the transformation from E to MS is nearly complete by the RNAi treatment as judged by the lineage transformation.
Figure 6.—
Comparison of cell lineages (A–C) and migrations (D–G) after RNAi against a Wnt signaling pathway component, POP-1 in both C. elegans and C. briggsae. The lineage trees are aligned with the P2 lineage with the P2-derived lineages shaded as light gray, the MS-derived lineages as light blue, and the E-derived lineages as light green. A homeotic lineage transformation from E to MS is observed in C. briggsae (A) while a transformation from MS to E is observed in C. elegans (C) as judged by cell division patterns. Note that the transformation from MSxp to MSxa is also likely in C. briggsae as evidenced by the ectopic expression of PHA-4 (red tree branches). The major tissue types are color coded in the wild-type P1 lineage (B). The developmental time in minutes at room temperature is shown on the right normalized to Sulston's time (Sulston et al. 1983). Differences in cell migrations after the RNAi against pop-1 are shown in a color-coded space-filling model. Note the similarity in cell positions of the wild-type embryos between C. briggsae (D) and C. elegans (F) but the differences in position for the progeny of ABp (dark blue), E (green), and MS (light blue) progeny between the pop-1-treated embryos of C. briggsae (E) and C. elegans (G). In particular, both MS and E cells remain on the surface of the pop-1 RNAi C. briggsae embryo (E) whereas the MS and E cells have taken up central, interior positions in the pop-1 RNAi-treated C. elegans embryo (G). The color keys for the models are red (ABa), dark blue (ABp), light blue (MS), green (E), magenta (C), light red (D), and yellow (P4).
By including the cell fate-specific marker pha-4 in the background we gained further insights into the transformations. We observed ectopic expression of PHA-4 in what would normally be MS derived body wall muscle precursors, supporting a role for pop-1 in the specification of the MSxp fate in C. briggsae (Figure 6). This extra expression was also seen in the transformed E lineage. Thus these lineages likely contributed to the extra pharynx-like cells previously reported (Lin et al. 2009). By contrast, the expression of PHA-4 in the AB lineage was similar to wild type after the RNAi treatment (data not shown). Furthermore, the cell migrations are drastically different between the two species after the RNAi treatment. EMS progeny migrated into the center of the embryos in C. elegans while those of in C. briggsae remained on the surface. Whether this is the primary effect of the RNAi or the secondary effect caused by extra cell divisions in E remains to be determined. In addition, ABp daughters failed to migrate along the anterior–posterior (a–p) axis in C. briggsae while those of C. elegans did but failed to spread evenly as seen in the wild-type embryos (Figure 6).
DISCUSSION
To increase the utility of C. briggsae as an experimental system, we have built a series of tools and resources that have been very useful in C. elegans but heretofore lacking in C. briggsae. These include efficient genetic mapping methods, a mutagenized frozen library for deletion screening, an unc-119 mutant for use in biolistic-based transformation, and automated gene expression profiling. We discuss aspects of each of these advances below.
The microarray SNP-based mapping method recently developed in C. elegans (Flibotte et al. 2009) can now be applied routinely in C. briggsae. With a single cross and hybridization an unknown mutant can be located to a fraction of a chromosome. The calculated map position relative to the sequence assembly location of the gene for the few cases we could study varied from just a few hundred kilobases to ∼2 Mb. More rigorous estimation of the mapping precision will depend on the molecular identification of additional C. briggsae genes that can be used in calibration. Of course the precision will depend on the frequency of recombination in the region, with arms of chromosomes in C. briggsae having much higher rates of recombination per megabase than in the centers, just as in C. elegans (Hillier et al. 2007). The precision of the positional estimates can also be influenced by the crossing scheme employed and these may be refined with experience. In any case, the current estimates are already sufficient to greatly limit the likely gene assignments and for mutants with distinctive phenotypes, such as the cbr-unc-22 used here, the position may suggest one or two specific genes. A next generation chip can replace poorly performing probes and could be adapted to allow multiplexing, thus substantially reducing costs. A priori there seems to be no reason why this positional accuracy of the array data should not be just as good as it is in C. elegans.
The mapping array also had the unanticipated benefit of detecting two likely misassemblies in the C. briggsae assembly. As additional mutations in other locations are mapped with the array, a few additional misassemblies may be detected, further improving the quality of the C. briggsae sequence. Also, future chips might incorporate SNPs from the portions of the genome not yet positioned along the chromosomes so that mapping experiments might suggest their position. In turn, with more directed crosses the arrays could be used to position the sequence more precisely.
The generation of a mutagenized resource for deletion screening will facilitate the comparison of specific systems and pathways between the two organisms. For example, sequence analysis might suggest differences between the two organisms, such as was seen for Notch pathway genes (Zhao et al. 2008). Alternatively, the distinct evolutionary histories or phenotypes of the two organisms might be investigated as was done for fem-2 homologs in C. briggsae (Hill et al. 2006). In such cases the ability to recover deletion alleles in C. briggsae will advance the study of the distinct functions in the two organisms. Unfortunately the library is not readily distributed, but our success combined with that of others (Hill et al. 2006) should encourage others to undertake similar studies.
Our initial use of the library to recover an unc-119 deletion allele allowed us to perform biolistic-based transformation methods readily. The resultant lines in C. briggsae, as in C. elegans, are stable and allow expression of germline-expressed genes. We expect that the trangenes are present in low copy number as they are in C. elegans and thus are more likely to faithfully represent endogenous gene expression than large extrachromosomal arrays obtained by injection. The marker may be able to be readily transferred to the newly identified sister species between which viable progeny can be produced with C. briggsae (M. Felix and E. Haag, personal communication).
The extension of automated gene expression profiling to C. briggsae now allows the detailed comparison of expression patterns between these two species. Our initial experiments demonstrated remarkable conservation of the regulatory program between these different organisms. We did note subtle differences in the onset and levels of expression of the C. elegans genes in the C. briggsae background for two of the three genes, which might reflect accumulated differences in the species. Without the ability to follow gene expression in such detail such differences would be difficult to appreciate. The methods open up this intriguing avenue for further study.
The application of expression profiling to examine pop-1 function in C. briggsae allowed us to confirm and extend recent findings (Lin et al. 2009). Instead of the cell lineage transformation of MS to E as is found in C. elegans after pop-1 loss of function, we saw an E to MS transformation based on the pattern of cell divisions, the expression of fate-dependent markers, and cell movements (Figure 6). We also noted, on the basis of hlh-1 expression patterns, the transformation of Cxa lineages to Cxp cells seen in C. elegans, which was not seen in C. briggsae (data not shown). Use of a marker of CxA (hypodermal) fate could help determine if Cxx fate transformation is also reversed by pop-1 loss of function in C. briggsae compared to C. elegans. Availability of these tools opens up this and similar questions for comparative study.
The tools and resources for C. briggsae that we have developed in this work combined with those previously developed make C. briggsae a powerful experimental system and its pairing with C. elegans will allow scientists to use the power of nature's experiments to decipher the information encoded in their genomes. C. briggsae has one advantage over C. elegans in comparative analysis: four species now exist with varying evolutionary distance from C. briggsae, ranging from the very closely related species 9, which can produce fertile offspring in some crosses, to species 11, which apparently branched off from C. briggsae shortly after the branch from C. elegans (K. Kiontke and D. Fitch, unpublished results). The experimental manipulations now possible in C. briggsae combined with this fortuitous evolutionary tree make C. briggsae attractive to study in its own right.
Acknowledgments
We thank Harold Hutter and the C. elegans Knockout Consortium for helpful discussion during the deletion library setup and James Thomas for helpful discussion in the SNP mapping setup. We also thank David Baillie and Paul Sternberg for providing C. briggsae mutant strains and Rick Zapf, Jon Taylor, and Iasha Chaudhry for assistance in preparing samples for CGH and performing microarray experiments. We are grateful to Mihail Sarov for generating GFP-tagged fosmid constructs. We thank the two reviewers for their helpful comments. We are grateful to members of the Waterston lab for helpful discussion and technical support. This work was supported by National Institutes of Health grant GM072675 to R. Waterston.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.110270/DC1.
References
- Ahringer, J., ed. Reverse Genetics (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook. 1.47.1, http://www.wormbook.org.
- Andolfatto, P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437 1149–1152. [DOI] [PubMed] [Google Scholar]
- Bao, Z., J. I. Murray, T. Boyle, S. L. Ooi, M. J. Sandel et al., 2006. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103 2707–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., M. Krause, M. Sepanski and A. Fire, 1994. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120 1631–1641. [DOI] [PubMed] [Google Scholar]
- Desai, C., G. Garriga, S. L. McIntire and H. R. Horvitz, 1988. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature 336 638–646. [DOI] [PubMed] [Google Scholar]
- Edgley, M., A. D'Souza, G. Moulder, S. McKay, B. Shen et al., 2002. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 30 e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, M. A., 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr. Biol. 17 103–114. [DOI] [PubMed] [Google Scholar]
- Ferreira, H. B., Y. Zhang, C. Zhao and S. W. Emmons, 1999. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev. Biol. 207 215–228. [DOI] [PubMed] [Google Scholar]
- Flibotte, S., M. L. Edgley, J. Maydan, J. Taylor, R. Zapf et al., 2009. Rapid high resolution single nucleotide polymorphism-comparative genome hybridization mapping in Caenorhabditis elegans. Genetics 181 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, R. C., C. E. de Carvalho, J. Salogiannis, B. Schlager, D. Pilgrim et al., 2006. Genetic flexibility in the convergent evolution of hermaphroditism in Caenorhabditis nematodes. Dev. Cell 10 531–538. [DOI] [PubMed] [Google Scholar]
- Hillier, L. W., R. D. Miller, S. E. Baird, A. Chinwalla, L. A. Fulton et al., 2007. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 5 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32 728–730. [DOI] [PubMed] [Google Scholar]
- Inoue, T., M. Ailion, S. Poon, H. K. Kim, J. H. Thomas et al., 2007. Genetic analysis of dauer formation in Caenorhabditis briggsae. Genetics 177 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz, S. G., E. M. Schwarz, J. A. DeModena, T. De Buysscher, D. Trout et al., 2008. Multigenome DNA sequence conservation identifies Hox cis-regulatory elements. Genome Res. 18 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, K. T., G. Broitman-Maduro, W. W. Hung, S. Cervantes and M. F. Maduro, 2009. Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev. Biol. 325 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. X., J. M. Spoerke, E. L. Mulligan, J. Chen, B. Reardon et al., 1999. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 9 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro, M., and D. Pilgrim, 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro, M. F., 2008. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim. Biophys. Acta 1789 250–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango, S. E., E. J. Lambie and J. Kimble, 1994. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development 120 3019–3031. [DOI] [PubMed] [Google Scholar]
- Marra, M. A., T. A. Kucaba, N. L. Dietrich, E. D. Green, B. Brownstein et al., 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan, J. S., S. Flibotte, M. L. Edgley, J. Lau, R. R. Selzer et al., 2007. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 17 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman, D. G., G. M. Benian and R. H. Waterston, 1986. Molecular cloning of the muscle gene unc-22 in Caenorhabditis elegans by Tc1 transposon tagging. Proc. Natl. Acad. Sci. USA 83 2579–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J. I., Z. Bao, T. J. Boyle, M. E. Boeck, B. L. Mericle et al., 2008. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods 5 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak, S., J. Goree and T. Schedl, 2005. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov, M., S. Schneider, A. Pozniakovski, A. Roguev, S. Ernst et al., 2006. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods 3 839–844. [DOI] [PubMed] [Google Scholar]
- Sleumer, M. C., M. Bilenky, A. He, G. Robertson, N. Thiessen et al., 2009. Caenorhabditis elegans cisRED: a catalogue of conserved genomic elements. Nucleic Acids Res. 37 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L. D., Z. Bao, D. Blasiar, T. Blumenthal, M. R. Brent et al., 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1 E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J. E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100 64–119. [DOI] [PubMed] [Google Scholar]
- Wang, X., and H. M. Chamberlin, 2004. Evolutionary innovation of the excretory system in Caenorhabditis elegans. Nat. Genet. 36 231–232. [DOI] [PubMed] [Google Scholar]
- Winston, W. M., M. Sutherlin, A. J. Wright, E. H. Feinberg and C. P. Hunter, 2007. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 104 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., T. J. Boyle, Z. Bao, J. I. Murray, B. Mericle et al., 2008. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev. Biol. 314 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]