This work identifies ZEP1, a rice protein required for the assembly of the synaptonemal complex during meiosis. It finds that, in rice, the number of crossovers between homologous chromosomes is increased in the absence of synapsis, emphasizing the idea that the role of synapsis in crossover formation has diverged among different organisms.
Abstract
ZEP1, a transverse filament (TF) protein, is the rice (Oryza sativa) homolog of Arabidopsis thaliana ZYP1. In the Tos17-insertional zep1 mutants, homologous chromosomes align along the entire length of the chromosome, but the synaptonemal complex is not assembled in early prophase I. Crossovers are well formed, and 12 bivalents could be detected from diakinesis to metaphase I, which leads to equal chromosomal segregation in anaphase I. Moreover, the number of crossovers has a tendency to be increased compared with that in the wild type. These phenomena are different from the TF mutants identified so far in other organisms. Chiasma terminalization of the bivalent, which occurs frequently in the wild type, seldom occurred in zep1. Transmission electron micrographs and immunodetection using an antibody against ZEP1 showed that ZEP1 is the central element of the synaptonemal complex. Although PAIR2 and MER3 were loaded normally in zep1, their dissociation was delayed severely compared with the wild type. In addition, ZEP1 is reloaded onto chromosomes in early microspores as the chromosome decondense, suggesting that ZEP1 might have other biological functions during this process.
INTRODUCTION
For all sexually propagating eukaryotes, meiosis is the crucial process of producing haploid gametes and consists of two rounds of chromosome division following a single round of DNA replication. The first division (meiosis I) is a process in which homologous chromosomes pair, synapse, recombine, and segregate; in the second division (meiosis II), which resembles mitosis, sister chromatids segregate toward opposite poles.
The synaptonemal complexes (SCs) are meiosis-specific proteinaceous structures that enhance the close attachment of homologous chromosomes in prophase I. SC assembly starts from early prophase I with the formation of the axial elements, which elongate along the chromosome arms and connect the sister chromatids (de Boer and Heyting, 2006). Transverse filament (TF) proteins are in the center of SCs and perpendicularly connect the lateral elements. In budding yeast, TFs first assemble at the sites of synapsis initiation complexes and then stretch along the homologs. TF proteins have been reported in many organisms, including budding yeast, Drosophila melanogaster, Caenorhabditis elegans, mouse, and Arabidopsis thaliana (Meuwissen et al., 1992; Sym et al., 1993; Heyting, 1996; Page and Hawley, 2001; MacQueen et al., 2002; Colaiácovo et al., 2003; de Vries et al., 2005; Higgins et al., 2005). Interestingly, these TF proteins have poor homology at the amino acid level but exhibit significantly similar structures. They all have a coiled-coil domain in the central region with globular domains at both ends (Page and Hawley, 2004). The C termini of the TFs have S/TPXX motifs, which are reported to interact with DNA. When TF proteins are assembled, they form parallel homodimers with the N termini overlapping in the center of the SCs and the C termini connected to the lateral elements.
ZIP1 in budding yeast was the first TF protein identified (Sym et al., 1993; Storlazzi et al., 1996). The assembly of SCs in budding yeast is closely coordinated with the initiation and maturation of homologous recombination events. ZMM complexes, which are required to implement interference-sensitive (class I) crossovers (COs), contain seven collaborating members, including ZIP1, ZIP2, ZIP3, ZIP4, MSH4, MSH5, and MER3. These proteins colocalize and are always present at the sites where SC polymerization initiates, so the ZMM proteins are referred to as the synapsis initiation complex and are markers of class I COs (Fung et al., 2004; Tsubouchi et al., 2006; Lynn et al., 2007). In mutants of non-ZIP1 ZMM components, ZIP1 always localizes to chromosomes as dots at the early stage of prophase I; while at pachytene, it forms polycomplexes that are never associated with chromosomes rather than the string-like signals along the entire chromosomes in the wild type (Agarwal and Roeder, 2000; Novak et al., 2001; Borner et al., 2004; Cheng et al., 2006). In the zip1 mutant, the immunosignals of ZIP2, ZIP3, and RAD51/DMC1 resemble those in wild-type nuclei, while those of Msh4 and Msh5 in zip1 become fainter (Chua and Roeder, 1998; Agarwal and Roeder, 2000; Shinohara et al., 2000; Novak et al., 2001; Shinohara et al., 2008).
c(3)G encodes the 744–amino acid TF protein in Drosophila females. Mutation of c(3)G results in the loss of all COs, indicating that all COs in Drosophila females depend on C(3)G (Page and Hawley, 2001; Bogdanov Iu et al., 2002). In C. elegans, two components of the central element of SCs, SYP-1 and SYP-2, have been reported (MacQueen et al., 2002; Colaiácovo et al., 2003). Both SYP-1 and SYP-2 are coiled-coil proteins that may cooperate to span the width of synaptic axial elements. In both syp-1 and syp-2 mutants, 12 univalents are present in the diakinesis nuclei, reflecting a failure during chiasma formation. Therefore, TFs are essential for the formation of COs in C. elegans, just like that in Drosophila (MacQueen et al., 2002; Colaiácovo et al., 2003; Hillers, 2004). In the syp-2 mutant, the Rad51 foci in early prophase I are similar to those of the wild type, but these foci fail to decrease at late pachytene and persist until early diakinesis. A similar phenomenon was also observed in syp-1 (Colaiácovo et al., 2003). Sycp1 has been identified as a TF protein in mouse. Homologous chromosomes in sycp1 spermatocytes can form normal axial elements but do not synapse, and >90% of COs disappear in sycp1 (de Vries et al., 2005). In the sycp1 mutant, the number and distribution of MSH4 foci are similar to those observed in the wild type, but they do not disappear at the appropriate time compared with the wild type. The same behavior was also observed for RAD51/DMC1 in sycp1.
Higgins et al. (2005) used an in silico method as described by Bogdanov et al. (2003) to overcome the lack of primary sequence homology and identified a TF protein in Arabidopsis. There are two redundant TF proteins in Arabidopsis, ZYP1a and ZYP1b, together referred to as ZYP1. In Arabidopsis plants treated with small interfering RNA for ZYP1, the number of foci of MLH1 (a marker of late recombination events destined to be COs) declines only slightly, which indicates that the formation of COs in Arabidopsis is less dependent on TF (Higgins et al., 2005; Jackson et al., 2006). Interestingly, nonhomologous chromosomes assembled to form bivalents and/or multivalents in the ZYP1 RNA interference (RNAi) plants, which may imply a new function of the TF proteins in Arabidopsis.
In both mitosis and meiosis, chromosomes must condense to facilitate chromosome segregation, and the condensin complex plays an important role in this process. The condensin complex (13S condensin) comprises two structural maintenance of chromosome (SMC) subunits (SMC2/CAP-E and SMC4/CAP-C), which form a heterodimer in a V-shape, as well as three non-SMC subunits (CAPD2, CAP-G, and CAP-H) (Hirano, 2002; Yu and Koshland, 2003; Nasmyth and Haering, 2005). The SMC superfamily proteins have ATP binding domains at both the N and C termini and two extended coiled-coil domains separated by a hinge in the middle. However, a condensin protein specific to meiosis has not been reported.
In this study, we identified the homolog of ZYP1 in rice (Oryza sativa), ZEP1, and characterized its function using both Tos17-insertion mutants and RNAi plants. Our data suggest that ZEP1 is the central element of SCs in rice. In the mutants of zep1, the SCs do not assemble normally, but COs are always formed between homologous chromosomes, which led to equal chromosome segregation in meiosis I. In addition, the chromosomes tended to condense in early microspores separated from the tetrad in zep1, which impeded the initiation of the first pollen mitosis and finally led to defects in pollen development.
RESULTS
Identification of ZEP1
To identify a putative ZYP1 gene in rice, a BLAST search was performed using the Arabidopsis ZYP1a and ZYP1b amino acid sequences. The two homology searches produced the same candidate with significant similarity at locus Os04g0452500. By means of RT-PCR and rapid amplification of cDNA ends (RACE) PCR with gene-specific primers, we redefined the cDNA sequence, which differed remarkably from the predicted sequence, and named the gene ZEP1 for its functional homology with ZIP1 in Saccharomyces cerevisiae and ZYP1 in Arabidopsis. Nucleotide sequence analysis of the ZEP1 cDNA revealed that it is comprised of 3391 bp with an open reading frame of 2610 bp. ZEP1 has 21 exons and 20 introns, which differs from the predicted structure (see Supplemental Figure 1 online). The corresponding protein sequence of ZEP1 is most similar to ZYP1b (362/874 residues identical and 555/874 residues positive). It also shows high similarity to ZYP1a (357/880 residues identical and 567/880 residues positive; see Supplemental Figure 2 online). By means of RT-PCR, we revealed that ZEP1 is expressed mainly in young panicles and roots, and to a lesser extent in leaves and stems (see Supplemental Figure 3A online). Although the expression of ZEP1 is not meiosis specific, ZEP1 protein was present only in the panicle and could not be detected in vegetative organs (see Supplemental Figure 3B online).
The ZEP1 protein contains 869 amino acid residues, with a central coiled-coil region of 650 residues (amino acids 64 to 713; COILS-Prediction of Protein Coiled-Coil Regions available at http://www.ch.embnet.org/software/COILS_form.html) (Bogdanov et al., 2003). The N-terminal globular domain of 63 residues (amino acids 1 to 63) is basic, which is different from the acidic N termini of the mouse and budding yeast TF proteins but consistent with the Arabidopsis protein. The C-terminal globular domain of 156 residues (amino acids 714 to 869) has a high pI (9.44) and putatively binds DNA (ProtParam tool available at http://www.expasy.ch/tools/protparam.html) (Bogdanov et al., 2003). Additionally, the C terminus of ZEP1 contains two S/TPXX motifs, SPET (amino acids 751 to 754) and SPIT (amino acids 767 to 770), which are possibly involved in DNA binding (Suzuki, 1989).
Isolation of the Tos17-Insertion Mutant of ZEP1
Four single Tos17-insertion mutant lines of the ZEP1 gene were identified by screening the public insertion line collections. Sequence analysis of their PCR products confirmed that Tos17 was inserted in exon 8 of two independent lines and in exon 12 and intron 12 of the other two lines, which were referred to as zep1-1, zep1-2, zep1-3, and zep1-4, respectively (see Supplemental Figure 1 online). The four homozygous zep1 mutants showed normal vegetative growth and development, but with decreased seed sets of 32.68, 34.55, 48.35, and 56.67%, respectively. Their pollen fertilities were 59.18, 57.63, 71.59, and 91.28%, respectively, when evaluated by 1% I2-KI solution staining (see Supplemental Table 1 online). We also checked both seed setting and pollen grain fertility evaluated with I2-KI staining of the ZEP1/zep1 heterozygote plants. The seed setting and pollen fertility were normal (see Supplemental Figure 4 online), indicating that the ZEP1 gene function is sporophytic. In the progeny of self-fertilized heterozygous plants of zep1 mutants, plants with normal fertility and lower fertility segregated in a ratio of 3:1 (89:26 for zep1-1), implying that the lower fertility mutation in zep1 is a monogenic recessive mutation. By conducting immunoblotting, we found that both zep1-1 and zep1-3 expressed truncated ZEP1 proteins (see Supplemental Figure 3B online). Because of the differences in pollen fertilities and seed sets, we proposed that zep1-1 and zep1-2 are strong alleles, whereas zep1-3 and zep1-4 are weak alleles. Thus, we mainly focused on zep1-1 to characterize the phenotype of zep1 and study ZEP1 function.
The zep1 Mutant Shows Abnormal Synaptic Behaviors and More COs during Prophase I
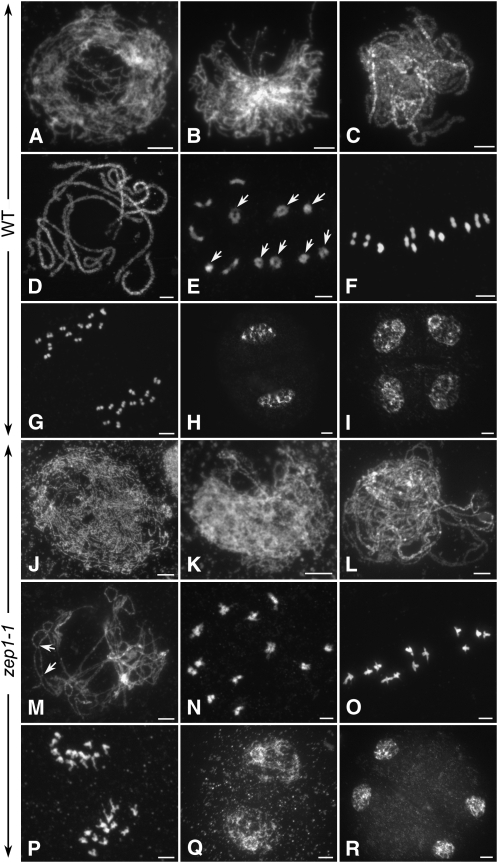
To determine whether the low fertility was a result of meiosis defects, the meiotic chromosomes of pollen mother cells at different stages from both the wild type and zep1-1 were investigated. During early prophase I of the wild type, the homologous chromosomes began to pair (Figures 1A and 1B). The SCs were fully assembled at pachytene (Figures 1C and 1D) and fell apart at diplotene. At diakinesis, several ring-shaped bivalents could always be found in the same nucleus, indicating that the chiasmata of these bivalents had already been terminated (Figure 1E). All 12 of the extremely condensed bivalents lined up on the equatorial plate at metaphase I (Figure 1F). From anaphase I to telophase I homologous chromosomes segregated away from each other and migrated to the opposite poles (Figures 1G and 1H). Meiosis II is a relatively short process compared with meiosis I, and the sister chromatids of each chromosome segregate just like in mitosis. After meiosis II, four microspores were produced (Figure 1I).
Figure 1.
Meiotic Chromosomes Stained with 4',6-Diamidino-2-Phenylindole in Both Wild-Type and zep1-1 Pollen Mother Cells.
(A) to (I) The wild type.
(J) to (R) in zep1-1. Bars = 5 μm.
(A) and (J) Leptotene.
(B) and (K) Zygotene.
(C) and (L) Early pachytene.
(D) and (M) Pachytene. In zep1-1, the COs are visible as indicated by arrows in (M).
(E) and (N) Diakinesis. The ring-shaped bivalents in the wild type are indicated by arrows in (E). In zep1-1, homologous chromosomes connect with each other tightly and ring-shaped bivalents are seldom observed (N).
(F) and (O) Metaphase I. In zep1-1, the two centromeres of each bivalent were dragged out by spindle fibers (O).
(G) and (P) Anaphase I.
(H) and (Q) Telophase I.
(I) and (R) Telophase II.
In zep1-1, the chromosome behavior was almost the same as in the wild type from leptotene to zygotene (Figures 1J and 1K). At early pachytene, the two threads of the bivalents were always visible, although the homologous chromosomes could align perfectly along the entire chromosome length (Figure 1L). At late pachytene, all of the chromosomes were separated into single threads, while the two homologous chromosomes of each bivalent were still connected by several chiasmata, indicating that COs had occurred during the early stages (Figure 1M). At diakinesis, 12 bivalents could be detected in every nucleus (Figure 1N). We found that the two homologous chromosomes of the bivalents were tightly connected side-by-side and ring-shaped bivalents were seldom detected, implying that the chiasmata were not terminated efficiently at diakinesis in zep1-1. At metaphase I, 12 bivalents in zep1-1 were lined up on the equatorial plate (Figure 1O), but the two centromeres of each bivalent were dragged out far away in opposite directions by spindle fibers. At anaphase I, the homologous chromosomes segregated normally and moved toward the opposite poles (Figure 1P). During meiosis II, no significant defects could be detected in zep1-1, and tetrads were normally formed at the end of meiosis (Figures 1Q and 1R).
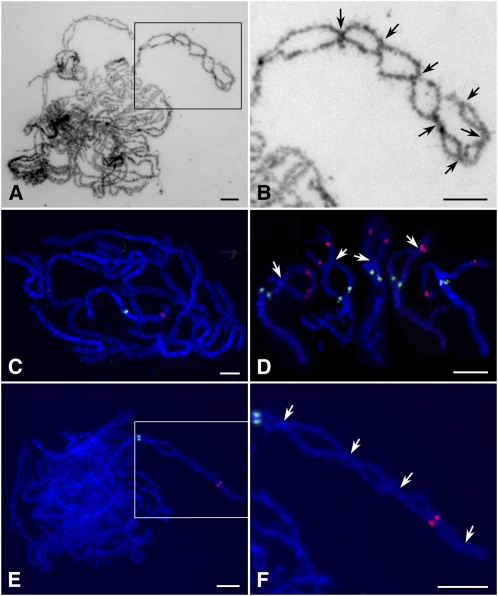
As indicated above, the two homologous chromosomes in zep1-1 could align well at pachytene stage, and the COs were obvious and easily counted (Figures 2A and 2B). In the wild type, by contrast, homologs connected to each other closely at pachytene and the COs were not visible until diplotene (Figures 2C and 2D). We found that each chromosome pair in zep1-1 formed several COs, implying that COs had a tendency to increase compared with the wild type, in which only ∼1.73 COs per pair are formed (Wang et al., 2009). Because pachytene chromosomes were relatively longer and difficult to trace along their length, the total number of COs in an entire pollen mother cell of zep1-1 was hard to estimate. Thus, we counted only the COs occurring between two cytological markers, OSJNBa0088I16 and 5S rDNA, near the end and close to the centromere, respectively, of the short arm of chromosome 11 (Figures 2C to 2F). Using this measure, the average number of COs on the short arm of chromosome 11 was 3.47 (n = 17) in zep1-1 and 0.92 (n = 26) in the wild type, consistent with COs being increased to a certain degree in zep1-1.
Figure 2.
Detection of COs in Both the Wild Type and zep1-1.
(A) zep1-1 pachytene chromosomes stained with 4',6-diamidino-2-phenylindole (DAPI).
(B) Magnified view of the boxed region in (A); visible COs of one chromosome arm are indicated by arrows.
(C) Pachytene chromosome in the wild type probed with OSJNBa0088I16 (red) and 5S rDNA (green) located near the end and close to the centromere, respectively, of the short arm of chromosome 11.
(D) Selected early diplotene chromosomes in the wild type probed with the same markers as in (C); arrows indicate COs.
(E) zep1-1 pachytene chromosomes probed with the same two markers.
(F) Magnified view of the boxed region in (E); arrows indicate COs.
Bars = 5 μm.
We performed interference to confirm that the abnormal chromosome behavior and low fertility were caused by the loss of ZEP1 gene function in zep1-1. Among the 73 ZEP1RNAi transgenic plants, 56 showed decreased seed sets ranging from 3.33 to 68.45%. We selected the lowest fertility transgenic plant, named ZEP1RNAi-3, for further cytological analysis. The chromosome behavior of ZEP1RNAi-3 also resembled that of zep1-1 (see Supplemental Figures 5A to 5D online).
We also observed meiosis of the other three mutants, zep1-2, zep1-3, and zep1-4, and found that they exhibited the same meiotic characters as those of zep1-1 (see Supplemental Figures 6A to 6C online). Even though some abnormal chromosome behavior was observed in zep1, chromosome segregation was still equal during meiosis. Therefore, the decreased pollen fertility of zep1 does not appear to be caused by chromosome segregation defects.
ZEP1 Is the TF Protein of SCs in Rice
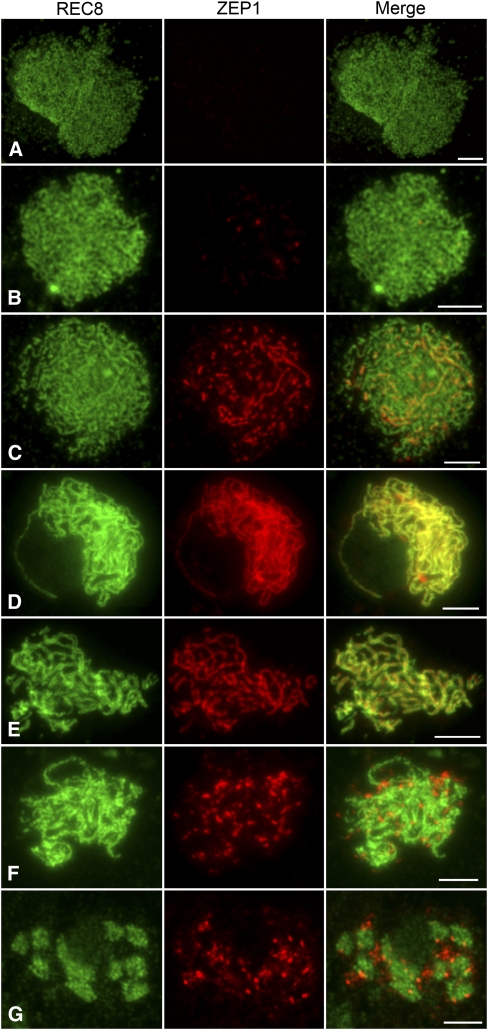
To investigate the precise spatial and temporal distribution of ZEP1 as meiosis progresses, dual immunolocalization was performed using polyclonal antibodies against REC8 and ZEP1(also known as RAD21-4) raised in mouse and rabbit, respectively. REC8 is a component of the cohesion complex and is required for sister chromatid cohesion, axial element formation, and homolog pairing (Molnar et al., 1995; Bhatt et al., 1999; Klein et al., 1999; Cai et al., 2003; Zhang et al., 2006). Thus, REC8 can be used as a marker to monitor early meiotic events during prophase I. REC8 was first present as punctuate foci at premeiotic interphase. At late leptotene and zygotene, these foci assembled to form linear signals and became thicker thereafter, corresponding to the condensing chromosomes (Figures 3A to 3C). At pachytene, REC8 was distributed over all the paired chromosomes (Figure 3D). After diakinesis, REC8 gradually disappeared (Figure 3G).
Figure 3.
Dual Immunolocalization of REC8 and ZEP1 in the Wild-Type Prophase I.
(A) Early leptotene.
(B) Late leptotene.
(C) Zygotene.
(D) Early pachytene. ZEP1 signals are narrower than the REC8 signals.
(E) Late pachytene. The double threads of REC8 signals of each bivalent align in parallel, while the single thread of ZEP1 situates in the center between REC8 signals, indicating that ZEP1 is the central element of SCs.
(F) Diplotene.
(G) Diakinesis.
Bars = 5 μm.
ZEP1 was first detected as punctuate foci at early leptotene (Figure 3B). From middle leptotene to zygotene, ZEP1 gradually elongated and formed short linear signals (Figure 3C). We found that all of the ZEP1 signals colocalized with the REC8 linear signals at this stage. Once the nucleus entered early pachytene, all ZEP1 signals aligned perfectly along the entire chromosome (Figure 3D). During this stage, the ZEP1 signals overlapped very well with the REC8 signals in length, whereas they were narrower than the REC8 signals in width and almost situated in the central region of the REC8 signals, showing that ZEP1 is the central element of SCs in rice (Figure 3E). The contiguous linear ZEP1 signals were maintained until late pachytene. At diplotene, ZEP1 fragments rapidly dissociated from the SCs and scattered as aggregates in the nucleoplasm (Figure 3F). At diakinesis, all ZEP1 signals were completely separated from the chromosomes and gradually depleted from the nucleoplasm (Figure 3G).
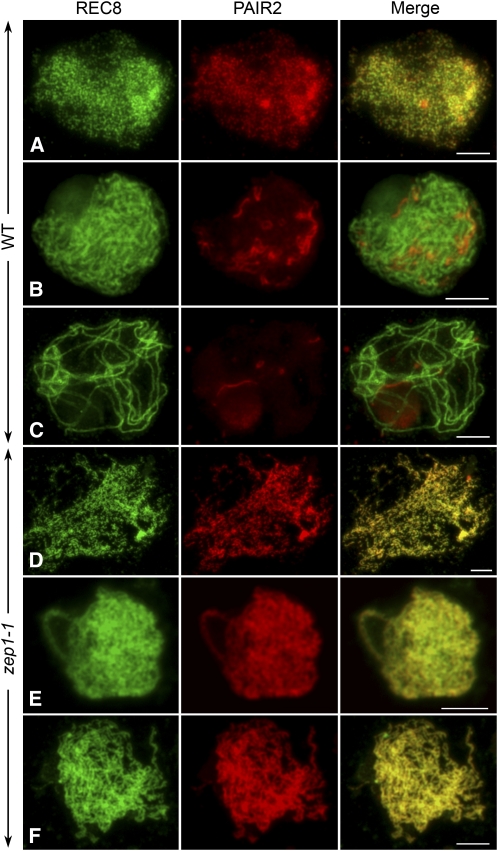
Immunolocalization studies were also performed using antibodies against REC8 and ZEP1 on meiocytes of the zep1-1 mutant. In the absence of ZEP1, the spatial and temporal distribution of REC8 was quite similar to that in the wild type. However, the two threads of REC8 on the homologous chromosomes at pachytene in zep1-1 were separated from each other, while those in the wild type were very close and always appeared to be a single thread (Figures 5 and 6). This phenomenon indicates that chromosome pairing in zep1-1 is not as intimate as that in the wild type. After diplotene, REC8 in zep1-1 started to dissociate from the chromosomes, so REC8 can still be used as a good marker to indicate chromosome axes in the zep1-1 mutant. In contrast with what was observed in the wild type, no ZEP1 signal was detected in zep1-1 (see Supplemental Figure 7A online). We also conducted the same immunodetection for the other three zep1 mutants and found besides some dot-like ZEP1 signals generated in zep1-3, very weak signals were detected in centromeric regions in zep1-2 and zep1-4 (see Supplemental Figure 7B online). However, no ZEP1 signal could be detected in ZEP1RNAi-3 (see Supplemental Figure 7C online), just like in zep1-1.
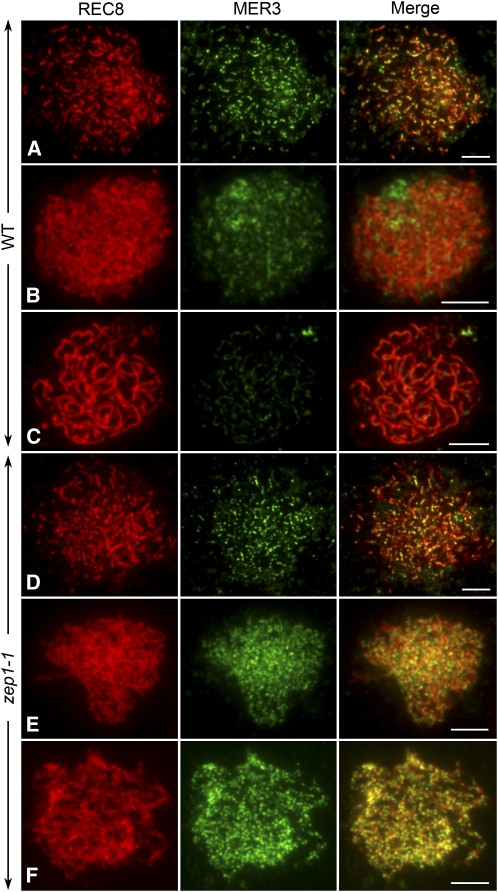
Figure 5.
Dual Immunolocalization of REC8 and MER3 in Both the Wild Type and zep1-1.
(A) to (C) The wild type.
(D) to (F) zep1-1. Bars = 5 μm.
(A) and (D) Leptotene. The MER3 foci in zep1-1 are more abundant than in the wild type.
(B) and (E) Zygotene.
(C) and (F) Pachytene. At pachytene, when MER3 foci in the wild type have almost disappeared, they are still abundant in zep1-1.
To elucidate further the deficient formation of SCs in zep1, we conducted transmission electron microscopy (TEM) analysis of zep1-1 and wild-type pollen mother cells. In the wild type, axial elements coaligned and the TF proteins, which connected axial elements to each other, formed obvious central elements at the pachytene stage (Figures 4A and 4B). In zep1-1, axial elements could still align, but the central elements were absent, showing that the formation of SCs was abolished in the mutant (Figures 4C and 4D). Thus, both immunolocalization and TEM observations confirmed that ZEP1 is the TF protein in rice and constitutes the central element of SCs.
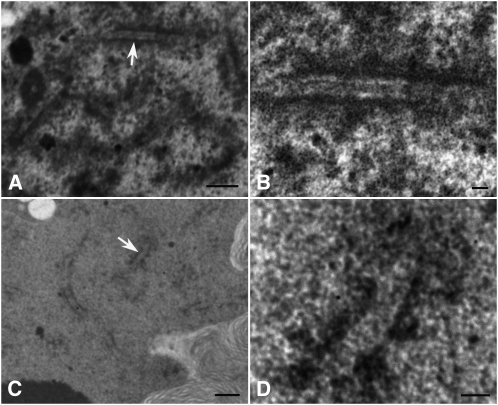
Figure 4.
Transmission Electron Micrographs Showing SCs in the Wild Type and zep1-1.
(A) and (B) The wild type.
(C) and (D) zep1-1. Bars = 5 μm in (A) and (C) and 1 μm in (B) and (D).
(A) and (C) Nucleus at pachytene stage with segments of SCs.
(B) and (D) Higher-magnification image of a stretch of SCs indicated by arrows in (A) and (C), respectively. In the wild type, the central elements of SCs are between the lateral elements and are less electron dense compared with lateral elements, whereas in zep1-1, no obvious central elements are visible.
The Number of MER3 Foci Increased and the Depletions of Both PAIR2 and MER3 Were Delayed in zep1-1
Dual immunolocalization experiments were also conducted in both the wild type and zep1-1 to investigate whether elimination of ZEP1 affected the distribution of meiotic proteins, including PAIR2 and MER3. PAIR2, the rice ortholog of HOP1 in S. cerevisiae and ASY1 in Arabidopsis, is first detected in premeiotic S/G2 cells and associates with axial elements through early prophase I. During synapsis, PAIR2 is removed from the chromosome and diffuses into the nucleoplasm (Nonomura et al., 2006). MER3 is a good marker to show most recombination events in rice (Wang et al., 2009). The distributions of MER3 and PAIR2 were investigated by immunodetection. In addition, the antibody against REC8 was also used to indicate the chromosome axes.
MER3 signals were first detected at early leptotene and began to decrease at zygotene in the wild type (Figures 5A to 5C). The stage at which MER3 appeared in zep1-1 resembled that in the wild type. However, the MER3 foci at late leptotene in zep1-1 (average 340, n = 5, range 299 to 370) were much more abundant than in the wild type (average 213, n = 5, range 182 to 241) (Figure 5D; see Supplemental Figure 8 online). When the homologous chromosomes paired, the number of MER3 foci decreased in both zep1-1 and the wild type, but the rate of decrease was much slower in zep1-1 (Figure 5E; see Supplemental Figure 8 online). At pachytene, when the MER3 foci in the wild type had almost disappeared (average 49, n = 5, range 38 to 67), there were still more foci in zep1-1 (average 224, n = 5, range 169 to 278), with only a small decrease compared with the number at zygotene (average 246, n = 5, range 180 to 320) (Figure 5F; see Supplemental Figure 8 online). Some MER3 foci even persisted until diakinesis in zep1-1, which was never observed in the wild type.
Additionally, the process of PAIR2 depletion was much different between the wild type and zep1-1. In the wild type, PAIR2 was first detected in premeiotic interphase and was always associated with axial elements through leptotene (Figure 6A). When synapsis occurred during zygotene, PAIR2 started to be removed from the chromosomes and released into the nucleoplasm (Figure 6B). At pachytene, only a few PAIR2 fragments were left on the chromosomes (Figure 6C). As in the wild type, the emergence of PAIR2 in zep1-1 was observed in premeiotic interphase. However, the signal of PAIR2 in zep1-1 totally overlapped with REC8 from leptotene to diakinesis (Figures 6D to 6F). After diakinesis, both PAIR2 and REC8 gradually disappeared in zep1-1.
Figure 6.
Dual Immunolocalization of REC8 and PAIR2 in Both the Wild Type and zep1-1.
(A) to (C) The wild type.
(D) to (F) zep1-1. Bars = 5 μm.
(A) and (D) Early leptotene.
(B) and (E) Zygotene.
(C) and (F) Pachytene. The PAIR2 signals in zep1-1 still overlap with REC8 at pachytene, when PAIR2 in the wild type is completely disassociated from the chromosome.
Taking these data together, although the loading of PAIR2 and MER3 occurred normally in zep1-1, depletion of both proteins was delayed severely compared with the wild type. In addition, the number of MER3 foci increased remarkably in absence of ZEP1.
The Loading of ZEP1 Requires both Homologous Chromosome Pairing and Recombination
To study the roles of homologous pairing and recombination in the process of SC formation, we selected two null mutants, pair2 and mer3, which represent defects in chromosome pairing and recombination (Nonomura et al., 2006; Wang et al., 2009), respectively, to investigate the loading of ZEP1. In addition, REC8 was also used as a marker to monitor the meiotic chromosomes in prophase I. We found that in both mutants, REC8 could be normally loaded onto prophase I chromosomes, while the localization of ZEP1 was defective (see Supplemental Figures 7D and 7E online). These results indicated that SC assembly requires both chromosome pairing and recombination. In the pair2 mutant, only a few ZEP1 foci could be detected in the prophase I nuclei (see Supplemental Figure 7D online). However, in the mer3 mutant, although the ZEP1 foci were abundant at the pachytene stage, the short linear signals were always distributed randomly in the nucleoplasm and were seldom loaded onto the chromosomes (see Supplemental Figure 7E online). Thus, normal loading of ZEP1 and further SC formation depend on both homologous pairing and recombination events.
Precocious Chromosome Condensation in Early Microspores Leads to Partial Abortion in zep1
As indicated above, chromosome segregation in zep1-1 was equal throughout meiosis. In order to find the real cause of the decreased fertility in zep1-1, chromosome behavior after meiosis in both the wild type and zep1-1 was further investigated. In the wild type, four early microspores were generated from a single pollen mother cell after meiosis (Figure 7A). As the early microspore developed, the pollen wall was gradually formed (Figure 7B). Meanwhile, the nucleus was moved to the peripheral region by the enlarging vacuole (Figure 7C), and each uninucleate microspore underwent an asymmetric mitotic division (pollen mitosis I) to produce two unequal daughter cells: the vegetative cell and the generative cell (Figure 7D). After nuclear migration, the generative nucleus completed the second mitotic division near the germinal pole, giving rise to the tricellular pollen (Figures 7E and 7F).
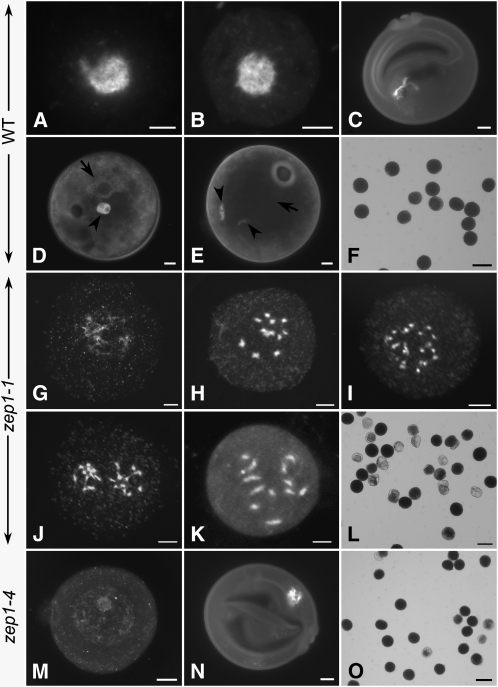
Figure 7.
Microspore Development in the Wild Type, zep1-1, and zep1-4.
(A) to (F) The wild type.
(G) to (L) zep1-1.
(M) to (O) zep1-4. Bars =50 μm in (F), (L), and (O) and 5 μm in the others.
(A) Early microspore just separated from tetrad.
(B) Early microspore with pollen wall formation.
(C) Uninucleate microspore with the nucleus migrating to the periphery of the cell.
(D) Bicellular pollen, with the arrowhead indicating the generative cell and the arrow indicating the vegetative nucleus.
(E) Tricellular pollen, with arrowheads indicating the generative cells and arrow indicating the vegetative nucleus.
(F) Mature pollen grains stained with 1% I2-KI solution.
(G) Early microspore in zep1-1 with less condensing chromosomes.
(H) Microspore in zep1-1 with 12 precocious condensed chromosomes in the central region.
(I) and (J) Two different microspores of zep1-1 with the condensed chromosomes tended to conduct equal segregation. Before the nucleus division (I) and after the nucleus division (J).
(K) Uninucleate microspore in zep1-1.
(L) Mature pollen grains stained with I2-KI in zep1-1.
(M) Early microspore in zep1-4 with decondensed chromosomes locating centrally within the cell, just like that in the wild type (B).
(N) Uninucleate microspore in zep1-4 resembling that observed in the wild type (C).
(O) Mature pollens stained with I2-KI in zep1-4.
[See online article for color version of this figure.]
Interestingly, in contrast with the decondensed chromatin in the early microspores in the wild type, severely condensed chromosomes were observed in ∼60% of the early microspores in the zep1-1 mutant (Figures 7G and 7H). We also observed that in a few early microspores, the chromosomes underwent equal segregation (Figures 7I and 7J). After pollen wall formation, the condensed chromosomes were still located in the center of the uninucleate pollen (Figure 7K). The abnormal chromosome condensation and nuclear location caused the microspore to be arrested at the uninucleate stage, finally leading to sterile pollen formation (Figure 7L). To test this hypothesis, we checked the microspore chromosome condensation status for the other three zep1 mutants and found that chromosome condensation events could be detected in 54.35, 12.67, and 5.38% of the microspores, respectively, in zep1-2, zep1-3, and zep1-4 (see Supplemental Table 1 online). In the weak allele zep1-4, the pollen formation was very similar to that in the wild type (Figures 7M to 7O). Nevertheless, the chromosome condensation of the uninucleate microspores in ZEP1RNAi-3 was much more severe than that in zep1-1 (see Supplemental Figures 5E and 5F online). It seems that the more chromosome condensation occurred in the microspores, the lower the fertility of the resulting pollen.
ZEP1 Reloads onto Chromosomes in Early Microspores When Chromosomes Are Decondensed
The defect in maintaining chromosome decondensation in the absence of ZEP1 indicates that ZEP1 might be involved in chromosome decondensation. We reanalyzed the behavior of ZEP1 though the entire meiotic process in both the wild type and the zep1 mutants. To our surprise, we found that ZEP1 could be reloaded onto chromosomes for two more rounds after diakinesis in the wild type. The first round was at prophase II when the chromosomes decondensed into chromatins (Figure 8A), and the second round was from telophase II until the first mitosis of the microspore, during which time the chromosomes were also in a decondensed chromatin state (Figures 8D and 8E). However, the chromosomes were highly condensed between the two stages, such as in metaphase II (Figure 8B) or anaphase II (Figure 8C). We found that all of the ZEP1 signals were localized in the nucleoplasm as the chromosomes condensed from metaphase II to anaphase II. However, after the first pollen mitosis, ZEP1 totally disappeared and could not be detected thereafter (Figure 8F). In contrast with the wild type, no ZEP1 immunosignal was detected in the corresponding stages in zep1-1 (Figures 8G to 8I) or zep1-2; however, those signals could constantly be detected in zep1-3 and zep1-4 (Figures 8J to 8L) at the same stages. We suspect that the truncated ZEP1 proteins in zep1-3 and zep1-4 might still perform their functions, so the chromosome decondensation status was maintained, just like that in the wild type.
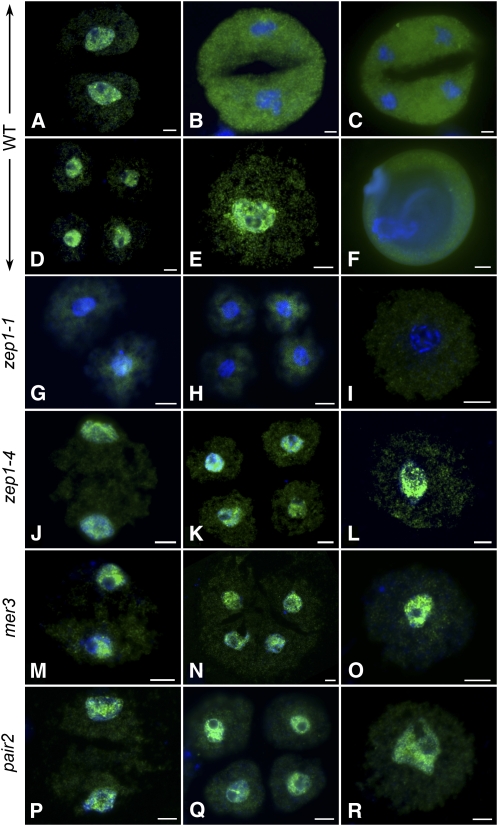
Figure 8.
The Localization of ZEP1 during Meiosis II and Microspore Development in the Wild Type, zep1-1, zep1-4, mer3, and pair2.
(A) to (F) The wild type.
(G) to (I) zep1-1.
(J) to (L) zep1-4.
(M) to (O) mer3.
(P) to (R) pair2. Bars = 5 μm.
(A) Prophase II. ZEP1 (green signal) reloads on chromosomes after its previous disassociation at diakinesis.
(B) Metaphase II with condensed chromosome; ZEP1 disassociates from chromosome and moves into the nuclearplasm.
(C) Telophase II.
(D) Tetrad. As chromosomes decondense, ZEP1 is reloaded onto the chromosome again.
(E) Early microspore.
(F) Uninucleate pollen.
(G), (J), (M), and (P) Prophase II in zep1-1, zep1-4, mer3, and pair2, respectively.
(H), (K), (N), and (Q) Tetrad in zep1-1, zep1-4, mer3, and pair2, respectively.
(I), (L), (O), and (R) Early microspore in zep1-1, zep1-4, mer3, and pair2, respectively. No ZEP1 is detected from meiosis II to early microspore in zep1-1, but the loading of ZEP1 in the corresponding stages in zep1-4, mer3, and pair2 resembles that in the wild type. Chromosomes are stained with DAPI (blue).
As described before, the defects in chromosome pairing or recombination would lead to failure of SC assembly in mer3 or pair2, respectively. However, in both the mer3 and pair2 mutants, chromosome decondensation was not obviously affected in the uninucleate microspores, though the chromosomes were unequally segregated in meiosis. Immunodetection was also performed to show whether ZEP1 localized normally at the corresponding stages in the mutants. We found that ZEP1 could still localize to chromosomes at prophase II, telophase II, and the early microspore stages in both mer3 (Figures 8M to 8O) and pair2 (Figures 8P to 8R), just like in the wild type. Thus, the results also support the idea that ZEP1 might be important for maintenance of chromosome decondensation during early microspore development.
DISCUSSION
Synapsis Is Dispensable for CO Formation in Rice
The role of synapsis in CO formation has diverged among different organisms. In animals, such as C. elegans, mouse, and Drosophilafemales, SCs have been shown to be required for CO formation. In C. elegans, the inappropriate repair of double strand breaks in two synapsis-deficient mutants, syp-1 and syp-2, induces apoptosis in most of the meiocytes. In the remaining meiocytes, only univalents can be observed at late prophase I (MacQueen et al., 2002; Colaiácovo et al., 2003). C(3)G is a TF protein that has been identified in Drosophila females. In the null mutant of c(3)G, double strand breaks seem to be inhibited and COs are accordingly unable to form (Page and Hawley, 2001). In fact, the initiation of recombination is not necessary for synapsis in Drosophila females and C. elegans, and they use specific pairing sites to achieve initial homolog recognition upon entering meiosis (McKim et al., 1993; McKee et al., 2000; de Boer and Heyting, 2006). In mouse, 90% of chiasmata in the wild type are dependent on SCYP1, and only univalents can be observed in sycp1 at metaphase I.
The situation is different in S. cerevisiae and Arabidopsis. In S. cerevisiae, the effect of the TF protein on CO formation varies according to the strain background, growth temperature, sporulation conditions, and so on. The zip1 mutant from the strain BR arrests during prophase I, and COs are reduced severely during growth at 30 or 34°C (Sym et al., 1993; Borner et al., 2004). However, in the strain SK1, the role of the TF protein in CO formation varies with different growth temperatures. At 33°C, defects in converting double strand breaks to single-end invasions result in most of the zip1 cells being arrested in meiosis and having severely altered sporulation. However, at 23°C, single-end invasions of the zip1 mutant cells in the SK1 strain can reach the wild-type level after a substantial delay, and most of the zip1 cells can go through meiosis II and produce spores (Borner et al., 2004). In Arabidopsis ZYP1RNAi plants, 70 to 80% of the wild-type MLH1 foci (the markers of chiasmata) can be assembled on the chromosomes, even though the multivalents are formed by nonhomologous chromosome association at metaphase I. Moreover, CO interference may still be maintained in zyp1 (Higgins et al., 2005).
Although the SC is often considered to be a universal feature of meiosis, it is absent from several organisms, including Schizosaccharomyces pombe, Aspergillus nidulans, and Drosophilamales (Rasmussen, 1973; Egel-Mitani et al., 1982; Molnar et al., 2003; Page and Hawley, 2004). In the absence of SCs, Drosophila males achieve homologous pairing and segregation in a special way rather than by meiotic recombination and chiasma formation (Page and Hawley, 2004). By contrast, in S. pombe and A. nidulans, large numbers of COs occur and homologs successfully disjoin without benefit of an SC (Kleckner, 2006). This lack of SCs in the two fungi makes it difficult to elucidate why the SC has evolved as a very common feature of the meiotic program (Kleckner, 2006). Here, the rice zep1 mutant, which retains CO formation and successful homolog segregation, resembles the phenotype of S. pombe and A. nidulans.
While the sequence and biological functions of most meiosis-specific proteins are highly conserved among organisms, the lower level of sequence conservation among TF proteins may cause divergent functions in homologous recombination among different organisms.
The Role of ZEP1 in Early Recombination Events
TF proteins have been reported to be required for programmed depletion of recombination-related proteins in many organisms. In mouse sycp1, 50 to 70% of the RAD51/DMC1, RPA, and MSH4 foci found in zygotene continued to persist into early diplotene, while most or all of these foci disappeared at the same stage in the wild type (de Vries et al., 2005). Additionally, the persistence of RAD51 foci into late prophase I was also observed in the mutants syp-1 and syp-2 in C. elegans (Colaiácovo et al., 2003). The delayed depletion of recombination-related proteins implies that meiotic recombination might be impeded at the step of single-end invasion.
In S. cerevisiae, MER3 can unwind duplex DNA from 3′to 5′as a helicase (Nakagawa and Ogawa, 1999; Nakagawa et al., 2001). MER3 is a component of the ZMM complex, which corresponds to the interference-sensitive CO (Lynn et al., 2007). The rice MER3 ortholog also is involved in interference-sensitive CO formation and may directly participate in double strand break repair (Wang et al., 2009). There are ∼12-fold more MER3 foci at late leptotene compared with the prospective COs, suggesting that the abundant ZMM foci are probably required for full synapsis in plants. Our data showed that there were many more MER3 foci in zep1 compared with those in the wild type at the same stage and that these foci persisted until diplotene, or even until diakinesis, while those in the wild type gradually disappeared as cells were entering into pachytene. As a result of later depletion of MER3, the recombination events may be accordingly delayed at the step in which MER3 is involved.
One model of CO interference has been proposed in which synapsis initiates at prospective CO sites and further prevents nearby COs. According to this model, COs are free to initiate in regions where synapsis does not occur and are excluded from synapsed regions (Egel, 1978; Sym and Roeder, 1994). Although the sites of synapsis initiation complexes display interference before SC formation in budding yeast, the prevention of CO initiation near synapsis initiation complexes by synapsis still quite likely exists (Fung et al., 2004). Additionally, our data show that CO is increased in the absence of ZEP1, providing the direct evidence that synapsis can inhibit the formation of excessive COs. This may occur through preventing the initiation of recombination events to some extent by limiting the number of MER3 foci in each nucleus. Thus, we infer that ZEP1 is quite important for regulating the process of meiotic recombination, although it does not participate directly in the progress of recombination events.
Chiasma Terminalization Relies on Regular Assembly of ZEP1
We found that ZEP1 played an important role in chiasma terminalization. At diakinesis in zep1, the condensed homologs associated with each other intimately and ring-like bivalents were seldom detected. Additionally, the terminal chromatin of zep1 bivalents was less condensed than that in the wild type. Therefore, we infer that this phenomenon is caused by defective chiasma terminalization in the absence of ZEP1. Chiasma terminalization was first described by Darlington (1929) and now is defined as shifting the position of chiasmata to or toward the ends of bivalents before metaphase I. The phenomenon of chiasma terminalization varies remarkably in different organisms, and the degree of terminalization may change from complete terminalization to the complete absence of terminalization (Egel, 1979; Baptista-Giacomelli et al., 2000). Chiasma terminalization might be affected by many factors, such as heterochromatic regions and structural rearrangement. In grasses, chiasma terminalization has been well characterized among different species, such as maize (Zea mays) and oat (Avena sativa; Maguire, 1981; Baptista-Giacomelli et al., 2000). Yet, the molecular mechanism of chiasma terminalization is still unknown. We suspect that the defects of chiasma terminalization in zep1 might be related to two aspects: one is the increased number of COs, and the other is the later depletion of PAIR2 and other meiotic elements.
ZEP1 Might Be Involved in Maintaining Chromosome Decondensation during the First Mitotic Division in Rice Pollen Genesis
During pollen development, which is specific to plants, a series of nuclear migration and cell division events are elaborately regulated (Twell et al., 1998; Zhang et al., 2005b). In this study, we found that precocious chromosome condensation prevented polar nuclear migration in absence of ZEP1 and therefore resulted in pollen abortion in zep1. There are many mutants defective in development from microspore to mature pollen in Arabidopsis, such as scp, gem1, duo1, duo2, etc. (Chen and McCormick, 1996; Park et al., 1998; Durbarry et al., 2005). By characterizing those mutants, several proteins specific to microspore development were identified. However, none of them have been shown to function during meiosis.
In Arabidopsis, REC8 can relocalize to the interphase II nucleus, but its function following meiosis I has not been reported (Cai et al., 2003). To our surprise, unlike anything reported in other organisms, ZEP1 can also reload onto chromosomes for two more rounds from prophase II to the early microspore stage as the chromosomes decondense in the wild type. In addition, we found that ZEP1 could reload normally onto chromosomes in dyads, tetrads, and in early microspores in mutants of both pair2 and mer3. This shows that the function of ZEP1 after meiosis I is independent of both PAIR2 and MER3. To a certain degree, the mechanism of ZEP1 reloading onto the chromosome during meiosis may be similar to INCENP, a component of the chromosomal passenger complex, which regulates key mitotic and meiotic processes by changing its position during cell division (Parra et al., 2003; Resnick et al., 2006; Ruchaud et al., 2007). In this respect, ZEP1 may have a parallel function like chromosomal passenger complex in normalizing chromosome behavior, but the exact function of ZEP1 after meiosis is still obscure.
The condensin complex plays a crucial role in chromosome condensation. In the presence of ATP, the condensin complex can produce supercoils in the protein-free region of closed circular DNA (Bazett-Jones et al., 2002). ZEP1 protein contains a domain (amino acids 61 to 530, domain A) with limited similarity to the prokaryote SMC proteins (conserved domains search in NCBI; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), which often form homodimers and are involved in condensing chromatin (Soppa et al., 2002). In this study, the Tos17 elements of zep1-1 and zep1-2 were inserted into domain A, while those of zep1-3 and zep1-4 were inserted downstream of this domain. The aberrant chromosome condensation in early microspores was observed mainly in zep1-1 and zep1-2, rather than in zep1-3 or zep1-4, although all of them were defective in SC formation. Thus, we suspect that the entire ZEP1 protein is required for SC formation, while domain A of ZEP1 might be important for maintaining chromosome decondensation in early microspore development. However, how ZEP1 prevents previous chromosome condensation in early microspores is still enigmatic. Although it is probable that ZEP1 might be involved in maintaining chromosome decondensation in early microspore development, our observations do not rule out the possibility that the defects of pollen development in zep1 may be the indirect consequences of the abnormal meiosis or other undetected defects. These suggestions still need to be tested with more investigations.
METHODS
Plant Materials
The japonica rice (Oryza sativa) variety, Nipponbare, was used in the present study as the wild type. The seeds of Tos17 insertion lines NG6573, NE7025, NF0601, and NG4542 named as zep1-1, zep1-2, zep1-3, and zep1-4, respectively, were kindly provided by the Rice Genome Resource Center of the National Institute of Agrobiological Sciences. pair2 and mer3 were isolated previously (Wang et al., 2009). All plant materials were grown in the paddy fields.
Cloning the Full-Length ZEP1 cDNA
Total panicle RNA extraction and reverse transcription experiments were performed as previously described (Wang et al., 2009). The cDNA was subject to PCR using Phusion (NEB) with gene-specific primers F1 (5′-CCTAGGGTTTCGACGACAG-3′) and R1 (5′-GAATCTTCTGAAGCAACTCT -3′), and F2 (5′-TCTGACCTAGAAGG GAGGGT-3′) and R2 (5′-CAGCA CCTCAAATGTGAGCT-3′) designed to amplify the predicted coding regions of ZEP1 by two halves. The PCR products were cloned into pMD18-T vector (Takara) and sequenced. The 3′RACE and 5′RACE were performed according to the protocol of the kit (3′-Full RACE Core Set and 5′-Full RACE Core Set; Takara). For 3′RACE, the first and second PCRs were performed using 3′RACE-F1 (5′-CCAACACTCAGAAGTCATTG-3′) and 3′adaptor (5′-CTGATCTAGAGGT ACCGGATCC-3′), and 3′RACE nested-F (5′-CTTAGGCATCTGACCATCC-3′) and 3′RACE nested-R (5′-TTCAGTTGATGCTGTACGC-3′), respectively. For 5′RACE, the RNA was reverse transcribed using 5′phosphorylated primer 5′RACE-RT (5′-CAACTTGGTATGCGTCA-3′). The first and second PCRs were performed using 5′RACE-S1 (5′-TCCGATATTGGAGGGAGCA-3′) and 5′RACE-R1 (5′-AGGTGGATCCAGCGAGAGA-3′), and 5′RACE-S2 (5′-CTGGTTAAGGAGCAGGCTT-3′) and 5′RACE-R2 (5′-GATAAAC CCAGCTTCTGC-3′), respectively. The products of 3′RACE-PCR and 5′RACE-PCR were cloned and sequenced.
Real-Time PCR for Transcript Expression Assay
Total RNA was extracted from the stem, leaf, root, and panicle of Nipponbare. Real-time PCR analysis was performed using the Chromo4 real-time PCR instrument (CFD-3240; Bio-Rad) and SYBR Green I (Invitrogen).RT-PCR was performed using ZEP1RT-F (5′-CAGCAGGATAATGAGCATAA-3′) and ZEP1RT-R (5′-GGTCTCAGGACTAACCAACT-3′) for ZEP1, and UBQF (5′-CAAGATGATCTGCCGCAAATGC-3′) and UBQR (5′-TTTAACCAGTCCATGAACCCG-3′) for Ubiquitin. The real-time PCR results were analyzed using Opticon Monitor analysis software 3.1 (Bio-Rad). The experiment had four replicates.
Tos17 Insertion Site Mapping
Primer pairs, TosP (5′-ATTGTTAGGTTGCAAGTTAGTTAAGA-3′) and 6573P (5′-TCTCCTTGCAGAGATTCCGT-3′), were used to amplify the Tos17 inserted regions of zep1-1 and zep1-2. Primer pairs, TosP and 0601P (5′-TTTGCAGGCA ATCAATGAAA -3′), as well as TosP and 4542P (5′-CCCTCAATTCATTCTCAGCC-3′), were used to amplify the Tos17 inserted regions of zep1-3 and zep1-4, respectively. The PCR products were cloned into pMD18-T vector (Takara) and sequenced.
Computational and Database Analysis
The methods of drawing the gene structure schematic and aligning amino acid sequences were previously described (Wang et al., 2009).
Construction of the ZEP1 RNAi Cassette and Rice Transformation
A 414-bp fragment of ZEP1 cDNA sequence was amplified with the following primers: RNAiF, 5′-ATGCTCGAGCACAAGCAAGAGTTGCTTCAG-3′(adding an XhoI site) and RNAiR, 5′-GCTAGATCTTTCATATTTCCTACGGATTTC-3′(adding a BglII site). The construction and transformation were performed as described (Wang et al., 2009).
Antibody Production and Protein Gel Blot Analysis
The antibodies against REC8, PAIR2, and MER3 were produced as described (Wang et al., 2009). To generate the antibody against ZEP1, a 561-bp fragment of ZEP1 cDNA (amino acids 426 to 612) was amplified with the following primers: ZEP1AbF, 5′-GCGGATCCCAAAAACTTCTGGAGGATTCT-3′(adding a BamHI site) and ZEP1AbR, 5′-GCGTCGACGCTCAGAATCTTGCCTGTTTTC-3′(adding a SalI site). The construction, fusion peptide expression, and purification were performed as described (Wang et al., 2009). Additionally, 20 residues (amino acids 97 to 116) of ZEP1 were synthesized to generate an antibody from mouse for immunoblotting.
For protein gel blot analysis, all protein samples (20 μg each) were separated by 10% SDS-PAGE and then electrophoretically transferred onto a polyvinylidene difluoride membrane. Hybridization was performed using polyclonal primary antibody against ZEP1 (diluted 1:2000). Goat anti-mouse antibody labeled by horseradish peroxidase (DingGuo) was used as the secondary antibody (diluted 1:500). The signal appeared after incubating in Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
Meiotic Chromosome Preparation and Fluorescence in Situ Hybridization
Young panicles of both the wild type and mutants were harvested and fixed in Carnoy's solution (ethanol:glacial acetic, 3:1). Microsporocytes undergoing meiosis were squashed in an acetocarmine solution. Slides with chromosomes were frozen in liquid nitrogen. After removing the cover slips, the slides were dehydrated through an ethanol series (70, 90, and 100%). Chromosomes were counterstained with DAPI in an antifade solution (Vector). Fluorescence in situ hybridization (FISH) analysis was conducted as described (Zhang et al., 2005a). The pTa794 clone containing the coding sequences for the 5S rRNA genes of wheat (Triticum aestivum; Cuadrado and Jouve, 1994) was used as one FISH probe, and the rice BAC OSJNBa0088I16 was used as the other FISH probe to monitor the short arm of chromosome 11. Chromosome images were captured under the Olympus BX51 fluorescence microscope with a microCCD camera.
TEM
For TEM of the meiocytes in the wild type and the zep1 mutant, the fixation and infiltration procedures were performed as described previously (Owen and Makaroff, 1995). Ultrathin sections were examined with a JEOL JEM-1400 transmission electron microscope at 80 kV. At least 50 ultrathin sections 100 nm thick were analyzed for pachytene pollen mother cells of both the wild type and the mutant individually.
Immunofluorescence
Fresh young panicles were fixed in 4% (w/v) paraformaldehyde for 30 min at room temperature. Anthers in the proper stage were squashed on a slide with PBS solution and covered with a cover slip. After soaking in liquid nitrogen and removing the cover slip, the slide was dehydrated through an ethanol series (70, 90, and 100%) prior to being used in immunostaining. Slides were then incubated in a humid chamber at 37°C for 4 h in different antibody combinations mentioned in the Results (diluted 1:500 in TNB buffer: 0.1 M Tris-HCl, pH 7.5, 0.15 M NaCl, and 0.5% blocking reagent). After three rounds of washing in PBS, Texas red–conjugated goat anti-rabbit antibody and fluorescein isothiocyanate-conjugated sheep anti-mouse antibody (1:1000) were added to the slides. The chromosomes were counterstained with DAPI in an antifade solution (Vector).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: ZEP1, GU479042; REC8, AY371049; MER3, FJ008126; PAIR2, AB109238; ZYP1a, At1g22260; and ZYP1b, At1g22275.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of the ZEP1 Gene.
Supplemental Figure 2. Alignment of ZEP1 with ZYP1a and ZYP1b from Arabidopsis thaliana.
Supplemental Figure 3. Expression Analysis of ZEP1.
Supplemental Figure 4. The Matured Panicle and Pollen Grains Stained with 1% I2-KI Solution in the ZEP1/zep1-1 Heterozygote Plant.
Supplemental Figure 5. DAPI Staining of Meiotic Chromosomes in ZEP1RNAi-3 Pollen Mother Cells.
Supplemental Figure 6. Pachytene Chromosomes Stained with DAPI in zep1-2, zep1-3, and zep1-4 Pollen Mother Cells.
Supplemental Figure 7. Dual Immunolocalization of REC8 and ZEP1 in Different Meiotic Mutants.
Supplemental Figure 8. Average Number of MER3 Foci at Successive Stages of Meiotic Prophase I in Both the Wild Type and zep1-1.
Supplemental Table 1. Fertility Investigation of the zep1 Homozygous Mutants.
Acknowledgments
This work was supported by grants from the Ministry of Sciences and Technology of China (2005CB120805 and 2006AA10A101) and the National Natural Science Foundation of China (30530070, 30671285, and 30621001).
References
- Agarwal S., Roeder G.S. (2000). Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255 [DOI] [PubMed] [Google Scholar]
- Baptista-Giacomelli F.R., Pagliarini M.S., de Almeida J.L. (2000). Chiasma frequency, distribution and terminalization in hexaploid oats (Avena sativa L.). Acta Sci. 22: 269–273 [Google Scholar]
- Bazett-Jones D.P., Kimura K., Hirano T. (2002). Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell 9: 1183–1190 [DOI] [PubMed] [Google Scholar]
- Bhatt A.M., Lister C., Page T., Fransz P., Findlay K., Jones G.H., Dickinson H.G., Dean C. (1999). The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 19: 463–472 [DOI] [PubMed] [Google Scholar]
- Bogdanov Y.F., Dadashev S.Y., Grishaeva T.M. (2003). In silico search for functionally similar proteins involved in meiosis and recombination in evolutionarily distant organisms. In Silico Biol. 3: 173–185 [PubMed] [Google Scholar]
- Bogdanov I.F., Grishaeva T.M., Dadashev S. (2002). CG17604 gene from Drosophila melanogaster–Possible functional homolog of the yeast ZIP1 and SCP1 (SYCP1) mammalian genes, coding for synaptonemal complex proteins. Genetika 38: 108–112 [PubMed] [Google Scholar]
- Borner G.V., Kleckner N., Hunter N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Cai X., Dong F., Edelmann R.E., Makaroff C.A. (2003). The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116: 2999–3007 [DOI] [PubMed] [Google Scholar]
- Chen Y.C., McCormick S. (1996). sidecar pollen, an Arabidopsis thaliana male gametophytic mutant with aberrant cell divisions during pollen development. Development 122: 3243–3253 [DOI] [PubMed] [Google Scholar]
- Cheng C.H., Lo Y.H., Liang S.S., Ti S.C., Lin F.M., Yeh C.H., Huang H.Y., Wang T.F. (2006). SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 20: 2067–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P.R., Roeder G.S. (1998). Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359 [DOI] [PubMed] [Google Scholar]
- Colaiácovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M. (2003). Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474 [DOI] [PubMed] [Google Scholar]
- Cuadrado A., Jouve N. (1994). Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6x-triticale. Chromosome Res. 2: 331–338 [DOI] [PubMed] [Google Scholar]
- Darlington C.D. (1929). Chromosome behaviour and structural hybridity in the Tradescantiae. J. Genet. 21: 207–286 [Google Scholar]
- de Boer E., Heyting C. (2006). The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115: 220–234 [DOI] [PubMed] [Google Scholar]
- de Vries F.A.T., de Boer E., van den Bosch M., Baarends W.M., Ooms M., Yuan L., Liu J.G., van Zeeland A.A., Heyting C., Pastink A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19: 1376–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbarry A., Vizir I., Twell D. (2005). Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 137: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. (1978). Synaptonemal complex and crossing-over: Structural support or interference. Heredity 41: 233–237 [DOI] [PubMed] [Google Scholar]
- Egel R. (1979). Telomeres and chiasma terminalization. Hereditas 91: 138–140 [Google Scholar]
- Egel-Mitani M., Olson L.W., Egel R. (1982). Meiosis in Aspergillus nidulans: Another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas 97: 179–187 [DOI] [PubMed] [Google Scholar]
- Fung J.C., Rockmill B., Odell M., Roeder G.S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802 [DOI] [PubMed] [Google Scholar]
- Heyting C. (1996). Synaptonemal complexes: Structure and function. Curr. Opin. Cell Biol. 8: 389–396 [DOI] [PubMed] [Google Scholar]
- Higgins J.D., Sanchez-Moran E., Armstrong S.J., Jones G.H., Franklin F.C. (2005). The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19: 2488–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers K.J. (2004). Crossover interference. Curr. Biol. 14: R1036–R1037 [DOI] [PubMed] [Google Scholar]
- Hirano T. (2002). The ABCs of SMC proteins: Two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16: 399–414 [DOI] [PubMed] [Google Scholar]
- Jackson N., Sanchez-Moran E., Buckling E., Armstrong S.J., Jones G.H., Franklin F.C. (2006). Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25: 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. (2006). Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194 [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S.B., Michaelis C., Nairz K., Nasmyth K. (1999). A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103 [DOI] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Borner G.V. (2007). ZMM proteins during meiosis: Crossover artists at work. Chromosome Res. 15: 591–605 [DOI] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiacovo M.P., McDonald K., Villeneuve A.M. (2002). Synapsis-dependent and-independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire M.P. (1981). Chiasma maintenance and terminalization across a homoeologous bivalent segment. Chromosoma 82: 107–112 [Google Scholar]
- McKee B.D., Hong C., Das S. (2000). On the roles of heterochromatin and euchromatin in meiosis in Drosophila: Mapping chromosomal pairing sites and testing candidate mutations for effects on X–Y nondisjunction and meiotic drive in male meiosis. Genetica 109: 77–93 [DOI] [PubMed] [Google Scholar]
- McKim K.S., Peters K., Rose A.M. (1993). Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R.L., Offenberg H.H., Dietrich A.J., Riesewijk A., Van Iersel M., Heyting C. (1992). A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M., Bahler J., Sipiczki M., Kohli J. (1995). The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141: 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M., Doll E., Yamamoto A., Hiraoka Y., Kohli J. (2003). Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 116: 1719–1731 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Flores-Rozas H., Kolodner R.D. (2001). The MER3 helicase involved in meiotic crossing over is stimulated by single-stranded DNA-binding proteins and unwinds DNA in the 3′to 5′direction. J. Biol. Chem. 276: 31487–31493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Ogawa H. (1999). The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 18: 5714–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. (2005). The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Nonomura K., Nakano M., Eiguchi M., Suzuki T., Kurata N. (2006). PAIR2 is essential for homologous chromosome synapsis in rice meiosis I. J. Cell Sci. 119: 217–225 [DOI] [PubMed] [Google Scholar]
- Novak J.E., Ross-Macdonald P.B., Roeder G.S. (2001). The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen H.A., Makaroff C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185: 7–21 [Google Scholar]
- Page S.L., Hawley R.S. (2001). c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15: 3130–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20: 525–558 [DOI] [PubMed] [Google Scholar]
- Park S.K., Howden R., Twell D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Parra M.T., Viera A., Gomez R., Page J., Carmena M., Earnshaw W.C., Rufas J.S., Suja J.A. (2003). Dynamic relocalization of the chromosomal passenger complex proteins inner centromere protein (INCENP) and aurora-B kinase during male mouse meiosis. J. Cell Sci. 116: 961–974 [DOI] [PubMed] [Google Scholar]
- Rasmussen S.W. (1973). Ultrastructural studies of spermatogenesis in Drosophila melanogaster Meigen. Cell Tissue Res. 140: 125–144 [DOI] [PubMed] [Google Scholar]
- Resnick T.D., Satinover D.L., MacIsaac F., Stukenberg P.T., Earnshaw W.C., Orr-Weaver T.L., Carmena M. (2006). INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev. Cell 11: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. (2007). Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 8: 798–812 [DOI] [PubMed] [Google Scholar]
- Shinohara M., Gasior S.L., Bishop D.K., Shinohara A. (2000). Tid1/Rdh54 promotes colocalization of rad51 and dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. USA 97: 10814–10819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M., Oh S.D., Hunter N., Shinohara A. (2008). Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40: 299–309 [DOI] [PubMed] [Google Scholar]
- Soppa J., Kobayashi K., Noirot-Gros M.F., Oesterhelt D., Ehrlich S.D., Dervyn E., Ogasawara N., Moriya S. (2002). Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 45: 59–71 [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Xu L., Schwacha A., Kleckner N. (1996). Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc. Natl. Acad. Sci. USA 93: 9043–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. (1989). SPXX, a frequent sequence motif in gene regulatory proteins. J. Mol. Biol. 207: 61–84 [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht J.A., Roeder G.S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378 [DOI] [PubMed] [Google Scholar]
- Sym M., Roeder G.S. (1994). Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292 [DOI] [PubMed] [Google Scholar]
- Tsubouchi T., Zhao H., Roeder G.S. (2006). The meiosis-specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev. Cell 10: 809–819 [DOI] [PubMed] [Google Scholar]
- Twell D., Park S.K., Lalanne E. (1998). Asymmetric division and cell-fate determination in developing pollen. Trends Plant Sci. 3: 305–310 [Google Scholar]
- Wang K., Tang D., Wang M., Lu J., Yu H., Liu J., Qian B., Gong Z., Wang X., Chen J., Gu M., Cheng Z. (2009). MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sci. 122: 2055–2063 [DOI] [PubMed] [Google Scholar]
- Yu H.G., Koshland D.E. (2003). Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 163: 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Tao J., Wang S., Chong K., Wang T. (2006). The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol. Biol. 60: 533–554 [DOI] [PubMed] [Google Scholar]
- Zhang W., Yi C., Bao W., Liu B., Cui J., Yu H., Cao X., Gu M., Liu M., Cheng Z. (2005a). The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol. 139: 306–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lu Y., Liu X., Feng J. (2005b). Nuclear and cell migration during pollen development in rice (Oryza sativa L.). Sex. Plant Reprod. 17: 297–302 [Google Scholar]