Posttranscriptional processes play a crucial role in plant mitochondrial gene expression. This work describes the identification and characterization of a pentatricopeptide repeat protein required for 5′ end processing of distinct mitochondrial transcripts. This protein is related to RESTORERS OF FERTILITY, which can suppress cytoplasmic male sterility, a mitochondrially inherited trait.
Abstract
In mitochondria of higher plants, the majority of 5′ termini of mature mRNAs are generated posttranscriptionally. To gain insight into this process, we analyzed a natural 5′ end polymorphism in the species Arabidopsis thaliana. This genetic approach identified the nuclear gene At1g62670, encoding a pentatricopeptide repeat protein. The functional importance of this mitochondrial restorer of fertility-like protein, designated RNA PROCESSING FACTOR2 (RPF2), is confirmed by the analysis of a respective T-DNA knockout mutant and its functional restoration by in vivo complementation. RPF2 fulfills two functions: it is required for the generation of a distinct 5′ terminus of transcripts of subunit 9 of the NADH DEHYDROGENASE complex (nad9) and it determines the efficiency of 5′ end formation of the mRNAs for subunit 3 of the CYTOCHROME C OXIDASE (cox3), the latter also being influenced by mitochondrial DNA sequences. Accordingly, recombinant RPF2 protein directly binds to a nad9 mRNA fragment in vitro. Two-dimensional gel electrophoresis and immunodetection analyses reveal that altered 5′ processing does not influence accumulation of the nad9 and cox3 polypeptides. In accessions C24, Oystese-1, and Yosemite-0, different inactive RPF2 alleles exist, demonstrating the variability of this gene in Arabidopsis. The identification of RPF2 is a major step toward the characterization of 5′ mRNA processing in mitochondria of higher plants.
INTRODUCTION
Plant mitochondrial genomes encode ∼60 genes (Kubo and Newton, 2008). This essential genetic information is expressed via a multistep process in which posttranscriptional events contribute to the complexity of the plant mitochondrial gene expression system. Alterations of the structure and primary sequence of transcripts are necessary for the formation of translatable mitochondrial mRNAs. Posttranscriptional processes include C-to-U RNA editing, splicing of cis and trans introns, and the formation of mature 5′ and 3′ ends (Gagliardi and Binder, 2007; Bonen, 2008; Takenaka et al., 2008).
The consequences of splicing and RNA editing are quite evident as they directly affect the sequence of the encoded protein, while the biological impact of posttranscriptional processing of mRNA extremities is less clear. Functional studies of mRNA 3′ maturation indicated the importance of this process. Two 3′ to 5′ exoribonucleases, the mitochondrial polynucleotide phosphorylase (PNPase) and the RNase R homolog 1, have crucial functions in the removal of extra sequences at the 3′ termini of mitochondrial RNA. The knockout or knockdown of these proteins has severe consequences for plant fitness, demonstrating the importance of mitochondrial mRNA 3′ processing (Perrin et al., 2004b). Trimming of mRNA 3′ ends is linked to mRNA stability, which is influenced also by polyadenylation. As in bacteria and chloroplasts, 3′ poly(A) tails enhance degradation of mRNAs in plant mitochondria.
In contrast to mRNA 3′ maturation, virtually nothing is known about the posttranscriptional generation of 5′ termini in plant mitochondria. A systematic analysis of major extremities of mitochondrial mRNAs in Arabidopsis thaliana showed that most 5′ termini of mature mRNAs are generated posttranscriptionally. Only a few mature 5′ transcript ends directly originate from transcription initiation (Forner et al., 2007). The prevalence of processed 5′ ends indicates that their generation is an important process, which might play an unidentified regulatory role in plant mitochondrial gene expression.
The inspection of mitochondrial mRNA extremities in Arabidopsis also revealed the frequent occurrence of multiple, abundant 5′ ends per gene. These can be processed ends or termini generated by transcription initiation as observed in other species. Thus, high variability of 5′ ends is a common feature of plant mitochondrial mRNAs. In addition, variation in 5′ mitochondrial transcript ends is seen between different Arabidopsis accessions. Comparative mapping of mRNA extremities in the three accessions Columbia (Col), C24, and Landsberg erecta (Ler) identified several 5′ end polymorphisms, while no difference was found in the 3′ ends of mitochondrial transcripts (Forner et al., 2008). This further demonstrates the variation and flexibility of 5′ ends of plant mitochondrial mRNAs, opposing the clearly defined nature of 3′ ends. The 5′ variation in different Arabidopsis accessions can be associated with differences in the mitochondrial DNA or with distinct factors encoded in the nucleus (Forner et al., 2005, 2008). The 5′ transcript end polymorphisms can now be used to identify the proteins that are required for posttranscriptional generation of 5′ termini of mitochondrial transcripts.
To date, only a few proteins implicated in posttranscriptional RNA metabolism have been identified and/or characterized in plant mitochondria. These include the two above-mentioned exoribonucleases (Perrin et al., 2004a, 2004b), two DEAD-box proteins (Matthes et al., 2007), and a small number of pentatricopeptide repeat (PPR) proteins. These PPR proteins almost exclusively represent restorers of male fertility (encoded by RF genes) in cytoplasmic male sterility (CMS) systems in different plant species (Andrés et al., 2007; Schmitz-Linneweber and Small, 2008). The RF proteins have particular roles in translation, RNA stabilization, and RNA cleavage of the CMS-specific mitochondrial transcripts, while their intrinsic normal functions are almost exclusively unknown. Apart from the RF genes, two other PPR proteins, ORGANELLAR TRANSCRIPT PROCESSING DEFECT43, involved in trans-splicing of the intron 1 of the mRNA for subunit 1 of the NADH DEHYDROGENASE (nad1) in mitochondria of Arabidopsis (de Longevialle et al., 2007), and MITOCHONDRIAL RNA EDITING FACTOR1, have been characterized (Zehrmann et al., 2009). However, no protein involved in 5′ end processing of plant mitochondrial mRNA has yet been described.
Here, we report the identification of a nuclear gene, RNA PROCESSING FACTOR2 (RPF2), required for the formation of 5′ ends 202 bp upstream of the nad9 reading frame in Arabidopsis mitochondria. The RPF2 locus is identified by map-based cloning, analysis of a respective T-DNA insertion mutant, and in vivo complementation studies. RPF2 encodes a PPR protein that is similar to the PPR proteins encoded by the restorer of fertility genes (Budar and Pelletier, 2001; Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003; Wang et al., 2006). Phenotyping of mitochondrial transcript extremities in several PPR knockout mutants reveals that RPF2 is also involved in the formation of distinct cox3 5′ transcript ends. In C24, cox3 5′ end formation is influenced both by RPF2 and a cis-element in the mitochondrial DNA.
RESULTS
The nad9 5′ Transcript End Polymorphism in Different Arabidopsis Accessions
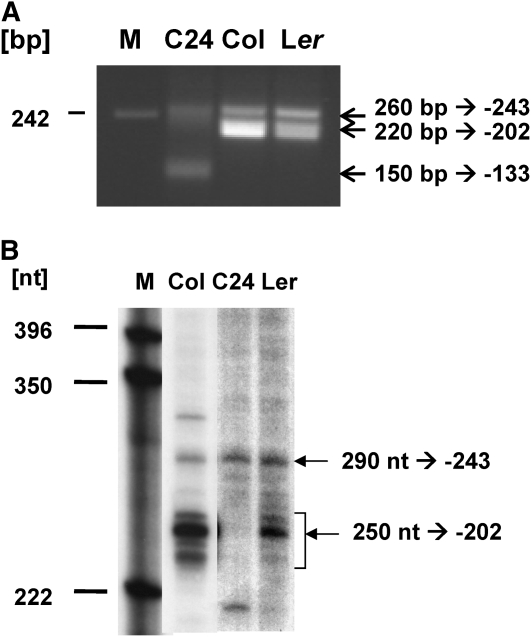
In Arabidopsis, the mitochondrial nad9 gene is transcribed into mRNAs of 0.85 and 0.9 kb in the accessions Col and Ler with distinct 5′ ends. In these accessions, circularized RNA (CR)-RT-PCR analysis identified two major 5′ ends at positions −202 and −243 with respect to the translation start codon (NATG, n = −1) (Forner et al., 2007). In C24, the latter end is present at levels comparable to Col and Ler, while the −202 5′ ends are hardly detectable (Figure 1A) (Forner et al., 2008). A reexamination of the nad9 mRNAs by primer extension confirmed the presence of the −243 end in these three accessions. In Col and Ler, a group of additional 5′ termini were found around position −202, in agreement with the CR-RT-PCR product pattern, which showed at least two products from this cluster of ends (Figure 1B). This analysis confirmed the absence of substantial amounts of the 5′ ends at −202 in C24. However, a second CR-RT-PCR approach amplified a faint cDNA product, indicating that minute amounts of −202 ends still exist in C24 (see Supplemental Figure 1 online). Also, in C24, a unique nad9 5′ transcript terminus was found 133 bp upstream of the ATG (Figure 1A).
Figure 1.
nad9 5′ Ends in Different Arabidopsis Accessions.
(A) CR-RT-PCR analysis (at least three biological and technical replicates) of nad9 mRNAs. PCR products of 260, 220, and 150 bp contain 5′ termini at positions −243, −202, and −133 relative to the ATG (A = +1).
(B) Primer extension analysis of nad9 mRNAs. Extension products of 290 nucleotides confirm the presence of the nad9 −243 5′ end in all accessions investigated. The cDNA strands of ∼250 nucleotides (nt) correspond to nad9 5′ ends around position −202 in Col und Ler. These results are consistent with the CR-RT-PCR analysis. Accession-specific products of 330 nucleotides (Col) and 210 nucleotides in C24 were not detected in respective CR-RT-PCR experiments.
The investigation of the nad9 −243 and −202 termini by primer extension analysis of terminator exonuclease-treated mtRNA confirmed these ends to be derived from posttranscriptional processing (see Supplemental Figure 2A online). This exonuclease specifically digests mRNAs containing 5′ monophosphate groups typically generated by posttranscriptional processing. By contrast, primary mRNAs with 5′ ends directly generated by transcription initiation contain 5′ tri- or diphosphate groups and are not degraded by this enzyme. Apart from our experimental data, a recent analysis of transcription initiation sites in Arabidopsis mitochondria identified four nad9 promoters more than 1 kb upstream of the −202 5′ ends (Kühn et al., 2009).
Delimiting the Genomic Region Containing the RPF2 Locus
Reciprocal crosses between Col and C24 have previously shown that the nad9 5′ end polymorphism is attributed to a nuclear locus, which we now designate RPF2 (Forner et al., 2008). In order to map this locus, an F2 mapping population was established from Col/C24 F1 hybrids. Phenotyping of nad9 transcript ends in 200 F2 plants revealed the C24 nad9 5′ end phenotype (i.e., the almost complete absence of the −202 ends) in 48 F2 individuals (see Supplemental Figure 3 online; Table 1). These plants correspond to 24% of the population, consistent with a 1:3 ratio of the C24 nad9 5′ end phenotype versus the nad9 mRNA 5′ end phenotype of Col. Therefore, a single genetic locus is responsible for this nad9 5′ end polymorphism, with the almost complete absence of the nad9 −202 mRNA end being a recessive trait. In addition, this analysis revealed that the nearly complete lack of the nad9 −202 5′ termini in C24 is independent from the occurrence of the nad9 −133 end (see Supplemental Figure 3 online; see below), demonstrating that another genetic locus is responsible for the generation of this terminus in C24.
Table 1.
CR-RT-PCR Analysis of Col/C24 F2 Hybrid Plants
| ♀ C24 × Col ♂ | ♀ Col × C24 ♂ | Total | |
| Number of plants | |||
| Without nad9 −202 | 25 | 23 | 48 |
| With nad9 −202 | 75 | 77 | 152 |
| Percentage without nad9 −202 | 25% | 23% | 24% |
| Ratio without:with nad9 −202 | 1:3.0 | 1:3.4 | 1:3.2 |
Using insertion/deletion markers distributed over the nuclear genome (see Supplemental Table 1 online), 47 plants with the C24 nad9 5′ end phenotype were genotyped (see Supplemental Figure 2 online). For all markers located on chromosomes 2, 3, 4, and 5, an approximately equal distribution of the Col and C24 genotypes was found. By contrast, the C24 genotype of marker CER 479928 on chromosome 1 is strongly linked to the recessive C24 nad9 5′ end phenotype. Therefore, further markers on the lower arm of chromosome 1 were tested to delimit the genetic interval containing the RPF2 locus (see Supplemental Figure 4 online). This mapping approach defined a genetic interval containing the RPF2 locus between 22.3 to 24.0 Mbp on the lower arm of chromosome 1. This region encodes 442 protein-coding genes, from At1g60470 to At1g64620 (Arabidopsis Genome Initiative, 2000).
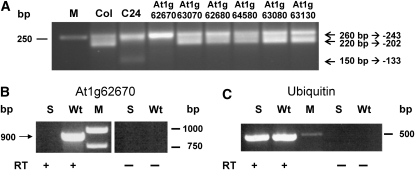
A T-DNA Insertion in At1g62670 Promotes the Absence of the nad9 −202 5′ Ends
RPF2 is expected to encode a protein targeted to mitochondria with a predicted function in nucleic acid metabolism. These criteria reduced the number of candidate genes in the defined interval to 23 genes encoding PPR proteins (Lurin et al., 2004). Thirteen of these PPR genes are very similar to restorers of fertility (RF) genes implicated in cytoplasmic male sterility systems in various plant species (Budar and Pelletier, 2001; Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003; Wang et al., 2006). Since fertility restoration correlates with altered expression of the CMS-related mitochondrial gene, it was investigated whether one of these genes is linked to the nad9 5′ end polymorphism. To this end, homozygous T-DNA insertion lines were established for nine candidate PPR genes and the 5′ termini of the nad9 mRNAs were inspected by CR-RT-PCR. No nad9 5′ end difference was observed between wild-type Col and the SALK lines carrying T-DNA insertions in At1g63070, At1g62680, At1g64580, At1g63080, At1g62590, At1g64100, At1g63230, and At1g63130 (Figure 2; see Supplemental Figure 5 online). However, in line SALK 146198, which harbors a T-DNA insertion in At1g62670, the nad9 −202 5′ ends were undetectable in a single round of CR-RT-PCR as in C24. An RT-PCR analysis of this mutant with primers flanking the T-DNA insertion demonstrated that the T-DNA causes a knockout of At1g62670 (Figure 2B). This correlates the inactivation of this gene with the nearly complete absence of the nad9 −202 5′ ends. Like C24, this knockout mutant contains minimal amounts of the 5′ ends around −202 (see Supplemental Figure 1 online). In addition, this CR-RT-PCR analysis revealed that the C24-specific nad9 −133 5′ end is not present in this T-DNA insertion line (Figure 2A), further confirming the abundance of the −202 5′ ends to be independent from the accumulation of nad9 transcripts with the −133 end.
Figure 2.
Phenotypes of nad9 5′ Termini in Different Arabidopsis Mutant Lines.
(A) Nad9 mRNA extremities were analyzed by CR-RT-PCR analysis as described in Methods. While the T-DNA insertions in At1g63070 (SALK 128068), At1g62680 (SALK 139736), At1g64580 (SALK 087235), At1g63080 (SALK 020638), and At1g63130 (SALK 020230) did not alter the 5′ end pattern of nad9 transcripts in comparison to Col wild type, an insertion in At1g62670 (SALK 146198) abolished the detection of the nad9 5′ ends at −202 as seen in the C24 wild type. In this mutant, the nad9 −133 5′ end is not detectable. CR-RT-PCR analysis of SALK 146198 was performed with three independent biological samples in at least three technical replicates. Lines with CR-RT-PCR product patterns identical to the wild type were investigated with single tests.
(B) RT-PCR analysis of SALK 146198 (S) mutant and Col wild type. An At1g62670 cDNA product of the expected size of 900 bp was amplified from wild-type RNA, while no such fragment is detectable in the mutant (RT +). No product is obtained without reverse transcriptase (RT −), demonstrating that the product derives from cDNA and not from DNA contaminations.
(C) The integrity and the use of equal amounts of the RNA and cDNA are indicated by the amplification of a 500-bp product from the ubiquitin 10 mRNA.
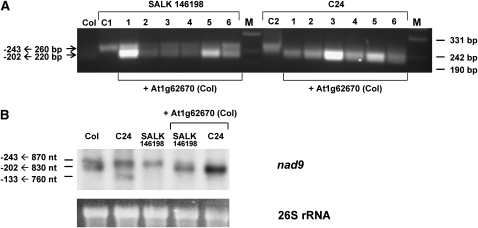
The Wild-Type Col Allele of At1g62670 Restores the Formation of Abundant nad9 -202 5′ Ends in Its T-DNA Insertion Line and in C24
The barely detectable levels of the nad9 -202 5′ ends in the T-DNA knockout line of At1g62670 are a strong indication that this gene is required for the formation of these termini. However, we could not exclude that another T-DNA insertion in line SALK 146198 might cause this phenotype. Thus, the At1g62670 gene from the Col wild type, including the regions ∼1.6 kb upstream and 1.3 kb downstream, was cloned into the pMDC123 vector and transformed into SALK 146198 and C24 plants. The nad9 transcript ends were then investigated by CR-RT-PCR analysis in several T1 plants. In all transformants (SALK 146198 and C24), the presence of the At1g62670 wild-type allele from Col restores the formation of the nad9 −202 5′ ends (Figure 3A). This result was confirmed by RNA gel blot analysis of total RNA from the different lines. In SALK 146198 and C24, the smaller nad9 transcript with the −202 5′ termini was only detected after introduction of the At1g62670 gene from Col (Figure 3B). In addition, both the CR-RT-PCR and the RNA gel blot analysis consistently showed that the presence of this gene in C24 leads to increased amounts of the nad9 mRNAs with the −202 5′ ends, while the transcripts with the −243 5′ ends were reduced in comparison to the C24 wild type. In C24, another mRNA of ∼740 nucleotides is detected. This nad9 mRNA species contains the −133 5′ end (Figure 1, panel C24). In summary, these data unambiguously demonstrate that At1g62670 encodes RPF2 required for the formation of considerable quantities of nad9 mRNAs with 5′ ends around nucleotide −202. The T-DNA insertion allele in SALK 146198 is accordingly designated rpf2-1.
Figure 3.
The At1g62670 Gene from Col Restores the Efficient Formation of the nad9 −202 5′ Ends in SALK Line 146198 and in C24.
(A) A 4.75-kb construct carrying the At1g62670 from Col was transformed into At1g62670 knockout line SALK 146198 and into the C24 wild type (+At1g62670). Nad9 mRNA of six individual transformants of each type was inspected by single CR-RT-PCRs. C1 (SALK 146198) and C2 (C24) are untransformed control plants.
(B) RNA gel blot analysis of total RNA from Col, C24, and SALK 146198, as well as SALK 146198 and C24 complemented with the At1g62670 gene from Col (+At1g62670). Loading of the gel was regulated by visualization of 26S rRNA.
To further explore the role of RPF2 in mRNA processing, we investigated whether this protein can bind to nad9 RNAs. To this end, electrophoretic mobility shift assays (EMSAs) were performed with recombinant RPF2 and a synthetic nad9 transcript covering the −202 5′ ends from –296 to –124 with respect to the translation initiation codon (NATG, n = −1). This experiment revealed that RPF2 indeed has the potential to bind to this RNA, further confirming its importance in 5′ end processing of nad9 transcripts (see Supplemental Figure 6 online).
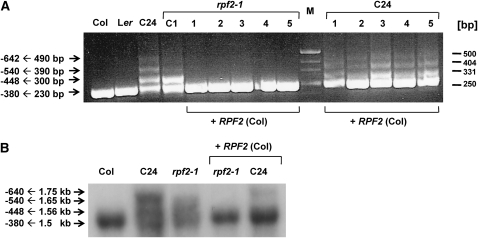
Knockout of RPF2 Affects the 5′ Ends of cox3 Transcripts
To investigate if the 5′ termini of other mitochondrial mRNAs are affected by the knockout of different Arabidopsis PPR genes with similarity to RF genes, we analyzed the 5′ and 3′ ends of all mitochondrial mRNAs in Col, Ler, C24, rpf2-1, and several additional lines with T-DNA insertions in RF-like PPR genes (Figure 4; see Supplemental Figure 5 online). In Col and Ler, a CR-RT-PCR product of 230 bp indicated a single cox3 5′ transcript end at position −380 relative to the ATG, while in C24, larger products (300, 390, and 490 bp) showed the presence of additional termini located upstream of the mature 5′ end at −448 (Figure 4A). These results are consistent with previous observations (Forner et al., 2005, 2008). The knockouts of the other RF-like PPR genes investigated did not exhibit any altered 5′ termini of mitochondrial transcripts (see Supplemental Figure 5 online); however, we found that cox3 mRNA ends are affected in the rpf2-1 mutant (Figure 4A). In addition to the single major cox3 mRNA 5′ end seen in Col wild type (−380), an additional CR-RT-PCR product was amplified from the rpf2-1 mutant. Sequence analysis of this product identified a cox3 5′ end at position −448 (Figure 4A; see Supplemental Figure 7 online). This observation suggested that RPF2 is also involved in 5′ end maturation of cox3 transcripts. To substantiate this result, we also inspected cox3 transcripts in rpf2-1 and C24 plants containing the Col RPF2 gene, which restored the nad9 −202 5′ end formation. In complemented rpf2-1 plants, a single major 5′ end of cox3 mRNA is detected by CR-RT-PCR analysis as in Col wild type (Figure 4A). By contrast, the complemented C24 plants still have additional cox3 5′ transcript ends due to a specific cis-element in the mitochondrial DNA of this accession (Forner et al., 2005, 2008). This result was confirmed by a RNA gel blot analysis of complemented rpf2-1 and C24 plants (Figure 4B). In both lines, a cox3 probe detected mature cox3 mRNAs of ∼1.5 kb and two additional cox3 RNA species of 1.56 and 1.65 kb. An additional cox3 RNA species of 1.75 kb was found in C24 consistent with previously mapped ends (Forner et al., 2005). These cox3 RNA patterns changed substantially after the introduction of the RPF2 gene into both lines. In the rpf2-1 knockout line, the efficient generation of the 1.5-kb mature cox3 mRNA was fully restored by the presence of RPF2. Likewise, the presence of the Col RPF2 gene in C24 plants enhanced the formation of the mature cox3 mRNA; however, in these plants, cox3 RNA species of 1.75 and 1.56 kb were still detectable.
Figure 4.
RPF2 and a C24-Specific Mitochondrial cis-Element Influence cox3 mRNA 5′ End Formation.
(A) The 5′ termini of the cox3 transcripts were determined by single CR-RT-PCRs in wild-type plants from accession Col, Ler, C24, and rpf2-1 (C2), as well as five rpf2-1 and five C24 plants transformed with the RPF2 complementation construct (+RPF2 Col).
(B) RNA gel blot analysis of total RNA from Col, C24, and rpf2-1, as well as C24 and rpf2-1 containing the RPF2 gene from Col (+RPF2 Col) using a cox3 probe.
Taken together, these results confirm that in both Col and C24, 5′ end formation of cox3 transcripts is influenced by RPF2. In C24, 5′ end maturation of cox3 mRNAs is additionally affected by a mitochondrially encoded cis-element (Forner et al., 2005).
Impaired 5′ Processing Does Not Interfere with Protein Accumulation
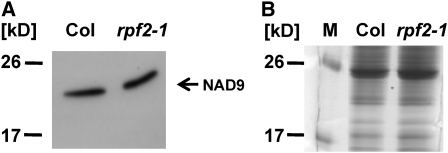
The defective RPF2 gene results in the loss of one of the nad9 mRNA species and a substantial reduction of mature cox3 mRNA (Figures 3 and 4). A potential impact of these deficiencies on accumulation of the nad9 polypeptide was investigated by an immunodetection analysis with an antiserum against the nad9 polypeptide from wheat (Triticum aestivum; Lamattina et al., 1993). In mitochondrial lysates from cell suspension cultures established from the Col wild-type and the rpf2-1 knockout lines, respectively, proteins with an apparent molecular mass of ∼21 kD were detected, which is consistent with the size expected for the nad9 polypeptide (Figure 5). In both lines, the nad9 subunits were present in identical amounts, demonstrating that the almost complete lack of the nad9 mRNA with the −202 ends in the rpf2-1 mutant has no influence on accumulation of this protein. This result was confirmed by two-dimensional blue native (BN)/SDS-PAGE of mitochondrial protein of the above-mentioned lines and a C24 wild-type culture. In addition, cox3 polypeptides were found in identical amounts in all lines investigated, and the total amount of protein complexes and their composition was indistinguishable between these lines (see Supplemental Figure 8 online). These results demonstrate that the altered nad9 and cox3 mRNA patterns in C24 and rpf2-1 do not interfere with accumulation of the respective polypeptides.
Figure 5.
Immunodetection Analysis of Mitochondrial Protein from Col Wild Type or rpf2-1.
(A) An antiserum against the nad9 polypeptide from wheat binds to a protein with an apparent size of ∼21 kD, consistent with the calculated size of nad9 in Arabidopsis (22.7 kD). No difference was observed between the amounts of the nad9 polypeptides in wild-type and rpf2-1 mutant plants.
(B) Loading of the gel was controlled by Coomassie blue staining.
Defective RPF2 Alleles in Different Arabidopsis Accessions
In C24, impaired nad9 and cox3 5′ end processing strongly suggests that this accession contains a defective RPF2 allele. To resolve the structure of the RPF2 gene in this and other accessions deficient in nad9 and cox3 5′ end processing (Oy-1 and Yo-0), the gene was amplified with primer pair At1g62670.H/At1g62670.R (see Supplemental Table 2 online) annealing to sequences at the 5′ and 3′ extremities of the RPF2 reading frame. Ler and Col were included as controls. In Col, Ler, and Yo-0, products of 2.0 kb were amplified from total DNA, as expected from the genomic sequence from Col-0 (see Supplemental Figure 9 online) (Arabidopsis Genome Initiative, 2000). By contrast, a 2.4-kb product was obtained in C24 while in Oy-1 two fragments of 3.7 and 0.4 kb were generated. These results suggest the presence of different types of RPF2 alleles defective in nad9 and cox3 5′ end processing. Two of these alleles seem to have alternative structures, as indicated by the sizes of the PCR products that differ from the expected size in Col (C24 and Oy-1), while another one has a size identical with Col (Yo-0). Sequencing of the PCR products obtained from C24 and Oy-1 confirmed the aberrant RPF2 gene structures and revealed that these alleles are chimeras composed of RPF2 and At1g62930 sequences (see Supplemental Figures 10 and 11 online). In contrast with these chimeric alleles, the RPF2 gene in Yo-0 contains 13 single nucleotide polymorphisms, which alter the amino acid sequence of RPF2 in comparison to Col.
To obtain information about the function of the RPF2 alleles from Oy-1 and Yo-0, these alleles were tested for the potential to complement impaired nad9 and cox3 5′ end processing in the rpf2-1 mutant. We found that both alleles of RPF2 were incapable of restoring efficient processing in rpf2-1 (data not shown). On the contrary, introduction of the Col RPF2 gene into these accessions led to an enhanced generation of a nad9 mRNA with −202 5′ ends as well as efficient and complete cox3 5′ end processing. These results demonstrate that the RPF2 alleles of Oy-1 and Yo-0 were nonfunctional with respect to their contribution to 5′ end processing of nad9 and cox3 mRNAs (see Supplemental Figure 12 online). In summary, these data indicate that the RPF2 gene in Arabidopsis does not encode a vital protein and that both changes in gene structure or alteration of amino acid identities can lead to the loss of function in 5′ end processing of nad9 and cox3 transcripts.
RPF2 Is a Mitochondrial Protein
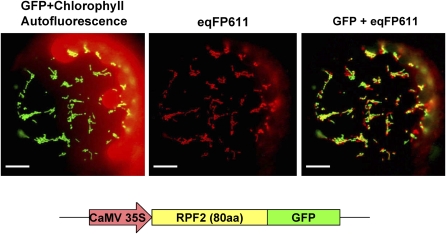
As outlined above, RPF2 is a nuclear-encoded factor expected to be imported into mitochondria. To investigate the subcellular localization of RPF2, DNA fragments representing the full-length RPF2 reading frame or 80 amino acids of its N terminus were fused in frame upstream of the gene encoding the green fluorescent protein (GFP; Davis and Vierstra, 1998). These constructs were transiently transformed into protoplasts prepared from transgenic tobacco (Nicotiana tabacum) stably expressing the isovaleryl-CoA dehydrogenase:red fluorescent fusion protein (eqFP611) as a mitochondrial marker (Forner and Binder, 2007). Inspection of transformed protoplasts by microscopy identified the GFP fluorescence exclusively in mitochondria, as indicated by the almost identical pattern of the green and the red fluorescence (Figure 6; see Supplemental Figure 13 online). This result suggests that the native RPF2 protein is imported into mitochondria in vivo.
Figure 6.
RPF2 Is a Mitochondrial Protein.
A fusion protein consisting of the RPF2 mitochondrial targeting sequence (80 amino acids) and GFP was transiently expressed in transgenic tobacco protoplasts. Fluorescence of GFP and chlorophyll and the red fluorescent protein from the sea anemone Entacmaea quadricolor (eqFP611) were visualized through different filter sets. eqFP611 C-terminally fused to the N-terminal part of the mitochondrial isovaleryl-CoA dehydrogenase has previously been established as a mitochondrial marker in transgenic tobacco (Forner and Binder, 2007). A merger (GFP + eqFP611) of both images demonstrates the match of both patterns (yellow represents colocalization). Small differences were due to the rapid movement of mitochondria in the protoplast. Bars = 10 μm.
DISCUSSION
RPF2 Is Involved in 5′ End Formation of the nad9 and cox3 mRNAs
We recently found several nuclear encoded natural polymorphisms affecting the 5′ termini of mitochondrial transcripts in distinct Arabidopsis accessions (Forner et al., 2008). Taking advantage of this natural genetic variation, we now identified At1g62670 as RPF2, a gene encoding a PPR protein. Several lines of evidence unambiguously demonstrate the function of the identified gene in 5′ end formation of mitochondrial nad9 and cox3 mRNAs. First, the knockout of At1g62670 in Col results in the absence of nad9 5′ ends around nucleotide −202 (Figures 2A and 3B) and in the accumulation of additional cox3 5′ transcript termini (Figure 4). Second, these phenotypes could be restored in vivo by introducing the RPF2 wild-type allele from Col into the rpf2-1 knockout line or into C24 (Figures 3 and 4). Third, C24, Oy-1, and Yo-0 contain aberrant RPF2 alleles. Accordingly, these accessions lack the nad9 mRNAs with −202 5′ ends and accumulate additional cox3 RNAs with 5′ termini located further upstream than in accessions with functional RPF2 alleles.
RPF2, an Efficiency Factor for the Maturation of cox3 mRNAs
As shown by our analysis, RPF2 has at least two functions. It is required for the generation of substantial amounts of nad9 mRNA with 5′ ends around −202, and it is involved in the formation of 5′ ends of the cox3 mRNAs. In the latter process, the inactivation of RPF2 provokes the accumulation of additional abundant cox3 RNAs with 5′ ends at position −448 (1.56-kb RNA species) and −540 (1.65-kb RNA species), upstream of the major cox3 5′ end (−380; 1.5-kb mature mRNA) (Figure 4). The relative amounts of these transcripts indicate that the occurrence of the larger cox3 mRNA species is accompanied by a decrease of the mature cox3 mRNA. This indicates that the 5′ extended RNA molecules are intermediates occurring during the generation of the mature cox3 mRNA, suggesting that the inactivation of RPF2 reduces the efficiency of this process. Thus, RPF2 is probably an efficiency factor for the generation of mature cox3 mRNA and is potentially required for different steps of this process, as indicated by the different 5′ extended cox3 precursor molecules. The accumulation of the cox3 processing intermediates argues against a function of this protein in modulating mRNA stability or a role of RPF2 as cox3 transcription factor. Furthermore, no difference in the efficiency in RNA editing of nad9 and cox3 mRNAs has been observed between rpf2-1 and wild-type plants, indicating that an involvement of RPF2 in RNA editing is highly unlikely.
The dual function of RPF2 in 5′ processing of nad9 and cox3 transcripts is reminiscent of the MMT (modifier of mitochondrial transcripts) locus in Brassica. The MMT locus also influences transcripts of two mitochondrial genes (ccmF and nad4); however, the changes of the transcript patterns of these genes might be linked to different genes within this nuclear locus (Singh et al., 1996).
Maturation of nad9 Transcripts
In contrast with the role of RPF2 in cox3 mRNA processing, the function of this protein in nad9 mRNA formation is less obvious. In C24 and rpf2-1 plants, the substantial reduction in the abundance of the short nad9 mRNAs with the 5′ ends around −202 is not accompanied by an apparent increase of the larger mRNA with the −243 5′ end or any other nad9 precursor molecules (Figure 3B; see Supplemental Figure 14 online). This observation suggests that the larger nad9 RNA might not serve as an intermediate or precursor for the formation of the short nad9 mRNA. On the other hand, the introduction of the intact RPF2 allele from Col into rpf2-1 knockout plants results in the generation of the nad9 −202 5′ ends, while the nad9 mRNA species with the −243 end is reduced. This effect is particularly strong when the Col RPF2 gene is brought into C24. In these complemented plants, the short nad9 mRNA accumulates in relatively high amounts, while the larger nad9 transcript almost completely disappears (Figure 3B). This result is confirmed by the absence of the 260-bp CR-RT-PCR product containing the −243 5′ end (Figure 3A). These observations suggest that the larger nad9 mRNA can indeed serve as precursor for the generation of the short nad9 transcript with −202 5′ termini. The particularly strong accumulation of the short nad9 mRNA in the complemented C24 plants might be explained with specific features of the nuclear background, but this phenomenon needs further investigation.
The apparent lack of accumulation of nad9 precursors in the rpf2-1 knockout line and in C24 might be explained by very moderate increases of precursor RNAs of different sizes originating from various upstream located promoters (Kühn et al., 2009).
Taken together, our data suggest that RPF2 is a factor that enhances 5′ processing of cox3 mRNAs and also of nad9 transcripts.
Mode of 5′ End Maturation of nad9 and cox3 mRNAs
The exact molecular function of RPF2 in 5′ processing of nad9 and cox3 transcripts is presently unclear. We could not detect a 5′ leader molecule with a 3′ end located directly upstream of the −202 5′ mRNA termini that would be indicative of endonucleolytic processing. Nevertheless, a 5′ to 3′ exonucleolytic generation of the different 5′ ends seems to be rather unlikely, since no such enzymatic activity has ever been reported in plant mitochondria (Gagliardi and Binder, 2007). Moreover, such an exonucleolytic formation of the nad9 −202 5′ ends would mean that RPF2 has to protect the −202 end from degradation, similar to what has been observed for the plastidal PPR10, which binds in a sequence-specific manner to different RNAs, protecting them from 5′ as well as 3′ exonucleolytic degradation (Pfalz et al., 2009). However, such a potential function of RPF2 in mRNA protection would be consistent with a function as stability factor, which is unlikely, as indicated by the increase of cox3 precursor RNAs in the rpf2-1 mutant, as mentioned above.
Considering all aspects, RPF2 is most likely a factor that enhances the endonucleolytic generation of the nad9 −202 and cox3 −380 5′ ends. A direct function as endonuclease is very unlikely, since the cox3 −380 5′ terminus and the nad9 −202 5′ ends are generated in the absence of RPF2. An endonucleolytic activity has been assigned to several DYW domain PPR proteins (Okuda et al., 2009), one of which (At2g02980) is targeted to mitochondria (Nakamura and Sugita, 2008). RPF2 does not contain a DYW domain required for endonucleolytic cleavage, supporting that RPF2 might interact with such PPR proteins or other endonucleases. In this scenario, RPF2 might interact with mitochondrial RNaseP and RNaseZ, enzymes normally involved in endonucleolytic tRNA processing. However, the latter has recently been found to cleave also some mitochondrial tRNA-like structures, so-called t-elements, which retained some features of real tRNAs but lost their original function (Canino et al., 2009). In plant mitochondria, such t-elements are present at 5′ or 3′ ends of several mRNAs, indicating that both RNaseP an RNaseZ are also involved in posttranscriptional generation of mRNA extremities. It seems conceivable that RPF2 might direct these endonucleases to higher-order structures that determine processing sites otherwise not or barely cleaved by these endonucleolytic enzymes. This model, which favors definition of the cleavage sites by secondary structure, is also compatible with the observation that the primary sequence at the nad9 −202 and cox3 −380 5′ ends is not conserved. The involvement of secondary structures is also implicated by the C24-specific mitochondrial DNA influencing the cox3 5′ end maturation process (Forner et al., 2005). Here, sequences located at least 200 bp upstream of −380 provoke the generation of alternative cox3 5′ termini. Such an effect across a long distance is best explained by the formation of alternative secondary structures, which interfere with 5′ processing of cox3 mRNAs. Further studies will be necessary to resolve potential processing complexes and to unambiguously define cis-elements.
RPF2 Is a Variable Restorer of Fertility-Like PPR Protein
In recent years, a number of PPR proteins have been identified as RF genes in CMS systems from various species (Bentolila et al., 2002; Brown et al., 2003; Desloire et al., 2003; Koizuka et al., 2003; Hanson and Bentolila, 2004; Wang et al., 2006; Chase, 2007). CMS is a maternally inherited deficiency to produce or release viable pollen (Budar and Pelletier, 2001; Chase, 2007). This trait, which is linked to chimeric, CMS-specific mitochondrial genes, can be used to generate F1 hybrids in many crops. Here, crosses between male sterile lines with fertile lines containing nuclear-encoded RF genes produce hybrid seeds. The presence of the RF genes restores male fertility in the F1 plants, most often by altering the expression of the CMS-specific mitochondrial genes at the posttranscriptional level. Most proteins encoded by the RF genes are highly similar to each other and to a group of PPR proteins from the autogamous plant species Arabidopsis, where CMS has not been found (Desloire et al., 2003; Schmitz-Linneweber and Small, 2008). RPF2, comprising 15 PPR repeats, groups with RF-like PPR proteins that form a small cluster of ∼20 genes within the huge family of 442 PPR genes identified in Arabidopsis (Lurin et al., 2004; O'Toole et al., 2008). Among the restorers of fertility, RPF2 is very similar to RF1B (32% identical amino acids) and RF1A (31%) from rice (Oryza sativa) as well as to RF592 (35%) from petunia (Petunia × hybrida; Bentolila et al., 2002; Wang et al., 2006). While the molecular function of RF592 in restoration of Petunia CMS is unclear, the two RF proteins from rice exhibit distinct fertility restoration functions in the Boro II CMS system. RF1A promotes the endonucleolytic cleavage of the discistronic B-atp6/orf79 transcript, which is consistent with the role of RPF2 in the nad9 and cox3 mRNA processing. Cleavage by RF1A generates two different mRNAs with the B-atp6 and the orf79 reading frame, respectively. This cleavage restores pollen fertility since the orf79 mRNA is inefficiently translated. RF1B enhances the degradation of the discistronic B-atp6/orf79 mRNA by an as yet unknown mechanism. The concerted action of RF1A and RF1B substantially reduces levels of the toxic orf79 protein, which allows the formation of fertile pollen (Wang et al., 2006; Kazama et al., 2008).
Interestingly, Arabidopsis RPF2 is a relatively variable gene with three distinct alleles being inactive in nad9 and cox3 5′ end processing in different accessions. It presently cannot be excluded that these alleles retained or even gained other yet unknown functions; however, their divergent structures suggest that another function common to these different alleles is rather unlikely. The diversity between these alleles seems only possible since the loss of the short nad9 mRNA and the impaired cox3 5′ end processing have no influence on protein accumulation and, thus, no detrimental effects on plant survival (Figure 5; see Supplemental Figure 4 online).
The sequences of the RPF2 loci from C24 and Oy-1 originate from different RF-like PPR genes (like At1g62930, At1g63130, or At1g62910), suggesting that chimeric gene structures frequently arise from recombinations between these highly similar genes. In line with this assumption, the RPF2 alleles from C24 and Oy-1 probably do not originate from a single recombination event, but are likely the outcome of independent recombinations, as indicated by the different parts of At1g62930, which are linked to the N-terminal portion of At1g62670 (see Supplemental Figures 6 and 7 online). This generation of new PPR genes is related to the Death-and-Birth model, which suggests either nonconservative or conservative transposition followed by rapid loss of a gene. This leads to a so-called nomadic character of PPR genes with highly variable positions in otherwise colinear segments of closely related genomes (Geddy and Brown, 2007).
In summary, our analysis reveals basic information about the intrinsic function of the RF-like PPR genes in Arabidopsis and allows insights into the evolution of such genes in this model species.
METHODS
Plants and Plant Cultivation
Arabidopsis thaliana and Nicotiana tabacum petit Havana plants and Arabidopsis cell suspension cultures were established and cultivated as described previously (Forner et al., 2007, 2008).
Analysis of RNA
Phenotypes (i.e., the 5′ ends of mitochondrial mRNAs) were determined by CR-RT-PCR analysis as described (Kuhn and Binder, 2002; Forner et al., 2007, 2008). Briefly, total RNA (up to 5 μg) was self-ligated by RNA ligase (10 units) in a bulk reaction (total volume 100 μL). After desalting on Microcon YM-10 microconcentrators, ligated RNA was used as template for first-strand cDNA synthesis using 200 units of M-MLV reverse transcriptase under conditions recommended by the manufacturer. One-tenth of the cDNA was used as template for a PCR across the ligated 3′ and 5′ ends of a given RNA species. PCR products were then separated by agarose gel electrophoresis, visualized by ethidium bromide staining, and recovered from the gel using the GFX Gel Band purification kit according to instructions given by the manufacturer (GE Healthcare). RNA extremities were determined by sequence analyses of the PCR products.
To determine nad9 5′ ends, cDNA synthesis was initiated with oligonucleotide Atnad9-1 followed by PCR with the primer pair Atnad9-3/Atnad9-7. For the 5′ end mapping of cox3 mRNAs, cDNA synthesis was initiated from oligonucleotide Atcox3-3′ PS followed by PCR with primers Atcox3-Mega3′ and Atcox3-Mega5′. Total RNA and mtRNA were isolated using an RNeasy plant mini kit (Qiagen). To discriminate between primary ends derived from transcription initiation (carrying 5′ triphosphate groups) and secondary ends generated by posttranscriptional processing (having 5′ monophosphate groups), 8 μg of mitochondrial RNA was digested with 1 unit of Terminator phosphate-dependant exonuclease according to the instructions given by the manufacturer (Epicentre Biotechnologies). Both exonuclease-treated and untreated RNAs were subsequently analyzed by primer extension analysis according to standard protocols (Sambrook and Russel, 2000). Oligonucleotides used in the primer extension experiments were nad9-2 and atp9-2. All primer sequences are given in Supplemental Table 2 online. RNA gel blot hybridization was performed with Duralon UV membranes according to instructions given by the manufacturer (Stratagene). Hybridization probes were generated by PCR with primer pairs cox3-5′ Primersonde/cox3-3′ Primersonde (cox3) and Atnad9-NS.H/Atnad9-NS.R (nad9).
Investigation of Nuclear and Mitochondrial DNAs
Crude and fine mapping of the genomic region containing RPF2 was done using insertion/deletion and single nucleotide polymorphism markers (Jander et al., 2002; Schmid et al., 2003). PCRs were performed with genomic DNA isolated with the DNeasy plant mini kit (Qiagen) from 14- to 18-d-old plants. A complete list of all markers used is given in Supplemental Table 1 online.
The RPF2 gene was amplified with oligonucleotide pair At1g62670.H/At1g62670.R from ∼200 ng total DNA from 14-d-old seedlings of the different accessions. The cox3 upstream DNA fragments were amplified with primers Atcox3-2 and Atcox3-23 for the Col/C24 configurations and primers Le-cox3-HindIII.H and Le-cox3-HindIII.R for the Ler configuration. These PCRs were performed with total DNA isolated as described above using Go-Taq DNA polymerase according to the instructions given by the manufacturer (Promega).
In Vivo Complementation
Complementation constructs were established in pMDC123 using standard procedures (Curtis and Grossniklaus, 2003). The RPF2 gene (At1g62670), including the 1.6-kb upstream and 1.3-kb downstream sequence, was amplified with primers At1g62670-Kompl.H and At1g62670-Kompl.R, digested with AscI and PacI, and cloned into the respective sites in pMDC123. Since we did not sequence the cloned PCR fragments, we transformed a mixture of 20 different clones per construct to avoid potential effects of PCR errors. Plant transformation was done by floral dip (Clough and Bent, 1998). Selection of transformed plants was done with Basta (120 mg/L; Bayer CropScience). Selected plants were analyzed as described above.
Miscellaneous Methods
Two-dimensional analysis of 1 mg of total mitochondrial protein obtained from Percoll purified organelles was done by BN/SDS-PAGE as described previously (Wittig et al., 2006; Matthes et al., 2007). Immunodetection assays were performed with protein extracted from mitochondria isolated and purified as described before (Matthes et al., 2007). The antiserum raised against nad9 from wheat (Triticum aestivum) was used at a 1/10,000 dilution.
EMSAs used gel-purified RNA directly transcribed from PCR products (amplified with primers Atnad9-EMSA.H + Atnad9-EMSA.R; see Supplemental Table 2 online) using a T7 RNA polymerase. The synthetic nad9 transcript corresponds to sequences in the 5′ -untranslated region ranging from position −296 to −124 with respect to the translation initiation codon (NATG, n = −1), which includes the −202 and −243 5′ ends. The binding reactions were essentially performed as described previously with slight modifications (Williams-Carrier et al., 2008). Two different buffer systems were used. Reactions performed with buffer B1 contained 20 mM Tris-HCl, pH 8.0, 20 mM NaCl, 0.2 mM EDTA, 4 mM DTT, 0.4 μg/ μL BSA, 5% (v/v) glycerol, 20 units Ribolock (Fermentas), 150 pM RNA, and 1 μg of purified RPF2 or control lysate (see below) in a total volume of 10 μL. Assays performed in buffer B2 contained 50 mM Na phosphate, pH 8.0, 12.5 mg/mL heparin, 20 mM NaCl, 0.2 mM EDTA, 4 mM DTT, 0.4 μg/mL BSA, 20 units Ribolock (Fermentas), 150 pM RNA, and 150 to 300 ng of purified protein or control protein extract.
To generate recombinant protein, the RPF2 reading frame was amplified without the putative mitochondrial targeting sequence (24 amino acids), with primers At1g62670UEx.H2 and At1g62670UEx.R. The PCR product was cloned into the BamHI/SacI sites in the expression vector pET32a (Novagen). Expression and affinity purification using S-protein agarose was performed according to the instructions of the manufacturer (Novagen), except that bacteria (Escherichia coli) were first grown at 37°C to an OD600 of 0.4 and then kept for 20 min at 20°C and, after induction with l-arabinose (0.2% [w/v]) and isopropyl β-d-1-thiogalactopyranoside (1.0 mM), grown for another 2 h at 20°C. A control lysate was generated under identical experimental conditions, except that an empty pET32a vector was used.
An RPF2 fragment corresponding to the first 80 amino acids was amplified with oligonucleotides At1g62670-Kompl.H3 and At1g62670-Kompl.R5 and cloned into the SmaI site of the psmGFP4 vector. The RPF2 full-length reading frame was amplified with primers At1g62670-Kompl.H3 and At1g62670-Kompl.R3 and analogously cloned into the psmGFP4 vector. Protoplast transformation and fluorescence microscopy were done as described previously (Matthes et al., 2007).
DNA sequencing was commercially performed (4baselab). All other basic methods of molecular biology and plant analysis were done following standard protocols (Weigel and Glazebrook, 2002). In silico sequence analyses were done at the National Center for Biotechnology Information server, applying various BLAST tools (McGinnis and Madden, 2004), and amino acid sequence alignments were done with ClustalW2 (Larkin et al., 2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RPF2, At1g62670; RPF2 allele in Oy-1, FN665655; RPF2 allele in Yo-0, FN665658; and RPF2 allele in C24, FM211647.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Detection of the −202 5′ End of nad9 mRNA in C24 and rpf2-1.
Supplemental Figure 2. The nad9 −243 and −202 Transcript Ends Are Generated by Posttranscriptional Processing.
Supplemental Figure 3. Phenotyping of nad9 mRNA Extremities in an F2 Mapping Population.
Supplemental Figure 4. Rough Mapping of the RPF2 Locus in Chromosome 1.
Supplemental Figure 5. CR-RT-PCR Analyses of Mitochondrial mRNA Extremities in PPR T-DNA Insertion Mutants.
Supplemental Figure 6. RPF2 Binds to a nad9 Transcript.
Supplemental Figure 7. Mapping of the cox3 −448 5′ Terminus.
Supplemental Figure 8. Accumulation of nad9 and cox3 Polypeptides Is Not Affected in the rpf2-1 Mutant.
Supplemental Figure 9. PCR Analysis of RPF2 Alleles in Different Accessions.
Supplemental Figure 10. RPF2 Alleles in Accessions Col, C24, Oy-1, and Yo-0.
Supplemental Figure 11. Alignment of Amino Acid Sequence Deduced from the RPF2 Alleles (62670) from Accessions Col, Yo-0, Oy-1, and C24 and from the At1g62930 Gene (62930) from Col.
Supplemental Figure 12. Complementation Studies with RPF2 Alleles from Oy-1 and Yo-0.
Supplemental Figure 13. RPF2 Is a Mitochondrial Protein.
Supplemental Figure 14. RNA Gel Blot Analysis of nad9 Transcripts in Col, C24, and rpf2-1 Plants.
Supplemental Table 1. List of Genetic Markers Used for Mapping of the RPF2 Locus.
Supplemental Table 2. Oligonucleotide Sequences.
Supplementary Material
Acknowledgments
We thank Gonny Guha and Uli Tengler Ulm University for excellent technical assistance and Anna Rapp Ulm University for her help in the mutant genotyping. We also thank Dagmar Lewejohann and Hans-Peter Braun from the Leibniz University in Hannover for their help with BN/SDS-PAGE analyses, the plant mitochondria group from the Institut de Biologie Moléculaire des Plantes in Strasbourg for the kind gift of the nad9 antiserum, and Solomon Stonebloom (Berkeley) for his help with the nad9 immunodetection assay and his valuable comments on the manuscript. J.F. was a fellow of the Studienstiftung des Deutschen Volkes. This work was supported by the Deutsche Forschungsgemeinschaft (Bi 590/10-1).
References
- Andrés C., Lurin C., Small I. (2007). The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 129: 14–22 [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bentolila S., Alfonso A.A., Hanson M.R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99: 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. (2008). Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8: 26–34 [DOI] [PubMed] [Google Scholar]
- Brown G.G., Formanova N., Jin H., Wargachuk R., Dendy C., Patil P., Laforest M., Zhang J., Cheung W.Y., Landry B.S. (2003). The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35: 262–272 [DOI] [PubMed] [Google Scholar]
- Budar F., Pelletier G. (2001). Male sterility in plants: Occurrence, determinism, significance and use. C. R. Acad. Sci. III 324: 543–550 [DOI] [PubMed] [Google Scholar]
- Canino G., Bocian E., Barbezier N., Echeverria M., Forner J., Binder S., Marchfelder A. (2009). Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 150: 1494–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase C.D. (2007). Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23: 81–90 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–742 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Vierstra R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36: 521–528 [DOI] [PubMed] [Google Scholar]
- de Longevialle A.F., Meyer E.H., Andres C., Taylor N.L., Lurin C., Millar A.H., Small I.D. (2007). The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desloire S., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4: 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J., Binder S. (2007). The red fluorescent protein eqFP611: Application in subcellular localization studies in higher plants. BMC Plant Biol. 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J., Hölzle A., Jonietz C., Thuss S., Weber B., Schwarzländer M., Meyer R.C., Binder S. (2008). Mitochondrial mRNA polymorphisms in different Arabidopsis thaliana accessions. Plant Physiol. 148: 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J., Weber B., Thuss S., Wildum S., Binder S. (2007). Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: T-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 35: 3676–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J., Weber B., Wietholter C., Meyer R.C., Binder S. (2005). Distant sequences determine 5′ end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res. 33: 4673–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi D., Binder S. (2007). Expression of the plant mitochondrial genome. In Plant Mitochondria, Logan D., (Ames, IA: Blackwell Publishing; ), pp. 50–95 [Google Scholar]
- Geddy R., Brown G.G. (2007). Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genomics 8: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.R., Bentolila S. (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16 (suppl.): S154–S169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T., Nakamura T., Watanabe M., Sugita M., Toriyama K. (2008). Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 55: 619–628 [DOI] [PubMed] [Google Scholar]
- Koizuka N., Imai R., Fujimoto H., Hayakawa T., Kimura Y., Kohno-Murase J., Sakai T., Kawasaki S., Imamura J. (2003). Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34: 407–415 [DOI] [PubMed] [Google Scholar]
- Kubo T., Newton K.J. (2008). Angiosperm mitochondrial genomes and mutations. Mitochondrion 8: 5–14 [DOI] [PubMed] [Google Scholar]
- Kuhn J., Binder S. (2002). RT-PCR analysis of 5′ to 3′ -end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res. 30: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K., Richter U., Meyer E.H., Delannoy E., Falcon de Longevialle A., O'Toole N., Börner T., Millar A.H., Small I.D., Whelan J. (2009). Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamattina L., Gonzalez D., Gualberto J., Grienenberger J.M. (1993). Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217: 831–838 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes A., Schmidt-Gattung S., Köhler D., Forner J., Wildum S., Raabe M., Urlaub H., Binder S. (2007). Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria. Plant Physiol. 145: 1637–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis S., Madden T.L. (2004). BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32: W20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Sugita M. (2008). A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett. 582: 4163–4168 [DOI] [PubMed] [Google Scholar]
- Okuda K., Chateigner-Boutin A.L., Nakamura T., Delannoy E., Sugita M., Myouga F., Motohashi R., Shinozaki K., Small I., Shikanai T. (2009). Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Perrin R., Lange H., Grienenberger J.M., Gagliardi D. (2004a). AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 32: 5174–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R., Meyer E.H., Zaepfel M., Kim Y.J., Mache R., Grienenberger J.M., Gualberto J.M., Gagliardi D. (2004b). Two exoribonucleases act sequentially to process mature 3′ -ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 279: 25440–25446 [DOI] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russel D.W. (2000). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Schmid K.J., Sörensen T.R., Stracke R., Törjek O., Altmann T., Mitchell-Olds T., Weisshaar B. (2003). Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13: 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Singh M., Hamel N., Menassa R., Li X.Q., Young B., Jean M., Landry B.S., Brown G.G. (1996). Nuclear genes associated with a single Brassica CMS restorer locus influence transcripts of three different mitochondrial gene regions. Genetics 143: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M., Verbitskiy D., van der Merwe J.A., Zehrmann A., Brennicke A. (2008). The process of RNA editing in plant mitochondria. Mitochondrion 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. (2006). Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Williams-Carrier R., Kroeger T., Barkan A. (2008). Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14: 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I., Braun H.P., Schagger H. (2006). Blue native PAGE. Nat. Protoc. 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Zehrmann A., Verbitskiy D., van der Merwe J.A., Brennicke A., Takenaka M. (2009). A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell 21: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.