Biosynthesis of the plant hormone auxin must be tightly controlled. This work shows that the STYLISH1 protein of the plant species Arabidopsis thaliana plays an important role in this process by directly binding to and activating at least one of the auxin biosynthesis genes.
Abstract
The establishment and maintenance of auxin maxima in vascular plants is regulated by auxin biosynthesis and polar intercellular auxin flow. The disruption of normal auxin biosynthesis in mouse-ear cress (Arabidopsis thaliana) leads to severe abnormalities, suggesting that spatiotemporal regulation of auxin biosynthesis is fundamental for normal growth and development. We have shown previously that the induction of the SHORT-INTERNODES/STYLISH (SHI/STY) family member STY1 results in increased transcript levels of the YUCCA (YUC) family member YUC4 and also higher auxin levels and auxin biosynthesis rates in Arabidopsis seedlings. We have also shown previously that SHI/STY family members redundantly affect development of flowers and leaves. Here, we further examine the function of STY1 by analyzing its DNA and protein binding properties. Our results suggest that STY1, and most likely other SHI/STY members, are DNA binding transcriptional activators that target genes encoding proteins mediating auxin biosynthesis. This suggests that the SHI/STY family members are essential regulators of auxin-mediated leaf and flower development. Furthermore, the lack of a shoot apical meristem in seedlings carrying a fusion construct between STY1 and a repressor domain, SRDX, suggests that STY1, and other SHI/STY members, has a role in the formation and/or maintenance of the shoot apical meristem, possibly by regulating auxin levels in the embryo.
INTRODUCTION
The plant hormone auxin plays critical roles during plant growth and development and has been proposed to act in pathways controlling embryo axis formation, organ phyllotaxis, organ and tissue differentiation, root meristem organization, and tropic responses (Went, 1974; Mattsson et al., 1999; Sabatini et al., 1999; Reinhardt et al., 2000; Benková et al., 2003; Friml et al., 2003). Auxin responses are largely dependent on the auxin concentration perceived by individual cells, resulting from tight homeostatic control of auxin levels (reviewed in Woodward and Bartel, 2005), and evidence for the involvement of local auxin gradients or peaks in several of the above-mentioned processes have been highlighted during the last few years (Friml et al., 2002, 2003; Benková et al., 2003; Heisler et al., 2005; Scarpella et al., 2006).
These local auxin peaks/gradients largely depend on tightly regulated timing and direction of polar auxin transport (PAT) mediated by, for example, members of the PIN family of auxin efflux facilitators, and disrupted functions of the PIN proteins result in defects in the establishment of embryo polarity, organ positioning, and organ development (Friml et al., 2002, 2003; Marchant et al., 2002; Benková et al., 2003; Reinhardt et al., 2003; Heisler et al., 2005). However, recent data also suggest that other aspects of auxin homeostasis play critical roles in determining the size and position of local auxin peaks/gradients. Auxin biosynthesis occurs both at distant sites as a source for PAT and at local auxin response sites. For example, in the root, auxin levels peak in the root tip, including the meristem (Sabatini et al., 1999). By measuring the auxin biosynthesis rate in young seedlings, Ljung et al. (2005) could demonstrate a peak in biosynthesis rate in the most apical 0.5 mm of the root tip, suggesting both auxin transport from the aerial part of the seedlings and local auxin biosynthesis contribute to the high auxin levels of Arabidopsis thaliana root tips. These authors could also show that the two auxin biosynthesis genes CYP79B2 and CYP79B3, which are rate limiting in a Trp-dependent pathway (Zhao et al., 2002), are active in the root meristem, further supporting that biosynthesis takes place in this high-auxin-response region. Members of the YUCCA (YUC) family of flavin monooxygenases have been suggested to act as rate-limiting enzymes in another Trp-dependent auxin biosynthesis pathway (Zhao et al., 2001) and have highly redundant functions in embryo and organ patterning (Cheng et al., 2006, 2007). Recently, Cheng et al. (2007) presented data suggesting YUC family–mediated auxin biosynthesis to partially commence at local auxin response sites (e.g., cotyledon tips), again implicating auxin asymmetries to be built up not solely by transport toward a specific site but also by local auxin biosynthesis.
We previously identified the SHORT INTERNODES/STYLISH (SHI/STY) gene family, which consists of highly redundant members that regulate the development of diverse plant organs (Fridborg et al., 1999; Kuusk et al., 2002, 2006). The nine active SHI/STY family members in Arabidopsis are very similar in two highly conserved regions, a 43–amino acid RING-like zinc finger domain and a more C-terminal companion domain of similar size, the IGGH domain (Fridborg et al., 2001; Kuusk et al., 2002, 2006). Zinc finger domains consist of one or several small protein motifs that each bind a zinc ion, allowing the formation of stable finger-like protrusions contacting their target molecules, which could be either DNA, RNA, protein, or lipids (Klug, 1999; Laity et al., 2001; Matthews and Sunde, 2002; Brown, 2005; Hall, 2005; Gamsjaeger et al., 2007). The zinc finger domain of SHI/STY proteins consists of two fingers that may form a cross-brace arrangement, and it has so far remained unclear if their targets are DNA, RNA, or proteins. Although this zinc finger domain is unique to the SHI/STY family proteins, an identical arrangement and spacing of Cys and His residues was found in an electron carrier/iron ion binding protein (Silverstein et al., 2005) otherwise showing only 35% similarity over the zinc finger motif. The IGGH domain is also unique to the SHI/STY family. In addition to these two domains, one or two Gln-rich regions and a nuclear localization signal are present in the SHI/STY proteins, suggesting that they might function as transcription factors.
At least six of the SHI/STY-related genes contribute to carpel fusion and marginal tissue formation in the apical part of the gynoecium. In addition, several of the SHI/STY-related genes appear to be important for proper apical-basal patterning of the gynoecium, vascular formation, and leaf polarity and organ identity in floral whorls two and three (Kuusk et al., 2006). Several of the SHI/STY family mutant phenotypes are reminiscent of defects related to reduced auxin responses, and sty1-1 confers hypersensitivity to chemical and genetic inhibition of PAT in the gynoecium (Sohlberg et al., 2006). PAT is essential for the apical-basal patterning of the gynoecium, and Nemhauser et al. (2000) proposed a model where an auxin gradient promotes regional patterning in the gynoecium; high levels define the style region, intermediate the ovule enclosing carpels, and low levels the basal part of the gynoecium, the gynophore. Reduced PAT results in apical shifts in the boundaries between the different tissues, based, according to the model, on apical reallocation of auxin threshold boundaries as auxin synthesized in the apical end of the gynoecium cannot efficiently be transported in the basipetal direction. The SHI/STY family mutants respond, just like the auxin response mutant ettin-1 (ett-1), with a more dramatic apical boundary shift upon PAT inhibition compared with the wild type, suggesting that auxin levels or responses are affected (Nemhauser et al., 2000; Sohlberg et al., 2006). Using a 35Spro:STY1-GR construct, we could show that dexamethasone (DEX)-mediated nuclear translocation of the STY1-GR protein induces an increased auxin biosynthesis rate (Ståldal et al., 2008), and elevated auxin accumulation preceded by increased expression of at least one of the YUC family members, YUC4 (Sohlberg et al., 2006), suggesting that auxin mediates some of the SHI/STY family functions. Overlapping expression domains between SHI/STY family members and YUC4 suggested the expression of YUC4 to be either directly or indirectly regulated by the SHI/STY proteins.
In this work, we aimed at further analyzing the function and regulatory role of STY1 in auxin biosynthesis. We demonstrate that STY1-regulated activation of YUC4 is independent of protein intermediates and that STY1 directly interacts with a short sequence proximal to a TATA-box in the YUC4 promoter. Furthermore, constitutive expression of a STY1-SRDX fusion protein represses transcription of YUC4 and phenocopies SHI/STY multiple loss-of-function mutants, suggesting that several SHI/STY family members participate in activating transcription of common downstream targets (e.g., YUC4). Thus, our data point out the SHI/STY family members as essential spatiotemporal regulators of auxin biosynthesis, thus revealing a pathway regulating auxin biosynthesis in a developmental manner.
RESULTS
STY1 Is a Nuclear Protein
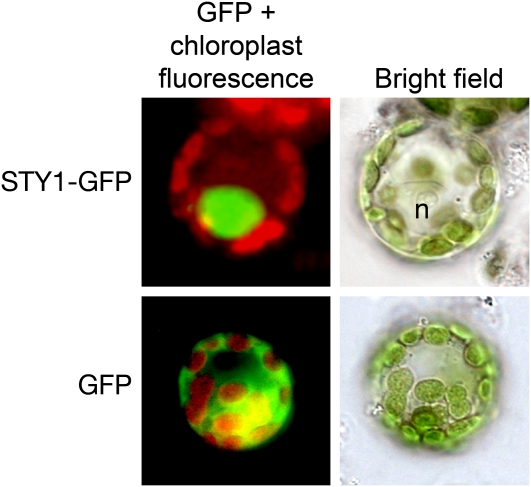
We previously described the 35Spro:STY1-GR line expressing a fusion protein between STY1 and the rat glucocorticoid receptor domain, capable of restoring the sty1-1 phenotype in a ligand (DEX)-dependent manner (Kuusk et al., 2006; Sohlberg et al., 2006). The ability of the STY1-GR fusion protein to rescue the sty1-1 mutant phenotype suggested STY1 to be active in the nucleus (Kuusk et al., 2006). To confirm the nuclear localization of STY1, we made a green fluorescent protein (GFP) fusion protein construct (35Spro:STY1-GFP) and expressed it transiently in onion epidermal cells and protoplasts of the moss Physcomitrella patens. The STY1-GFP fusion protein was localized to the nucleus in both systems, confirming STY1 to be a nuclear protein (Figure 1).
Figure 1.
Nuclear Localization of STY1-GFP.
Fluorescence microscopy pictures show the merged fluorescence signals from chloroplasts (red) and GFP (green) to the left. STY1-GFP fusion protein is localized only to the nucleus (n; top left). Expression of GFP without a fusion partner results mainly in a cytoplasmic localization (bottom left). To the right are bright-field images of the protoplasts shown in the left panels.
The STY1-Dependent Activation of YUC4 Does Not Require Protein Translation
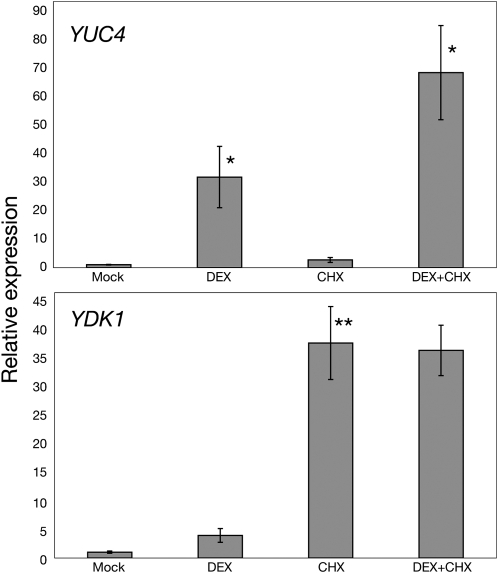
Using a 35Spro:STY1-GR construct, we could show that transcription of the YUC4 gene was induced 30 min after DEX treatment in 35Spro:STY1-GR sty1-1 sty2-1 seedlings subsequently resulting in increased levels of active auxin (Sohlberg et al., 2006). To investigate if STY1-GR–mediated induction of YUC4 expression requires de novo protein synthesis, we performed quantitative RT-PCR (qRT-PCR) on DEX and cycloheximide (CHX; an inhibitor of protein translation)-treated seedlings. Our data clearly suggest the STY1-GR induced activation of YUC4 to be independent of protein translation as would be expected for a direct downstream target (Figure 2). The auxin-induced gene YDK1/GH3-2, encoding a protein involved in auxin inactivation by conjugation (Staswick et al., 2005), was also induced in DEX-treated 35Spro:STY1-GR sty1-1 sty2-1 seedlings, although several hours later than YUC4 (Sohlberg et al., 2006). Because auxin responses require degradation of repressor proteins with high turnover rates, auxin-inducible genes are generally upregulated by CHX treatment (Ballas et al., 1993; Abel et al., 1995). As expected, YDK1 mRNA levels were increased by the CHX treatment, and no further activation was found by combined DEX and CHX treatment compared with that of CHX alone (Figure 2), suggesting that protein intermediates may be required for the STY1-GR–mediated induction of YDK1.
Figure 2.
STY1-Dependent Activation of YUC4 Does Not Require Protein Synthesis.
qRT-PCR analysis of 35Spro:STY1-GR sty1-1 sty2-1 seedling mRNA 2 h after treatment with mock, DEX, CHX, or DEX and CHX as indicated. The YUC4 gene is activated by STY1 in the presence of CHX as indicated in the top section. YDK1 is not activated by STY1 in the presence of CHX. The data represent the average of three biological replicates (three technical replicates for each biological sample). Error bars represent se. Asterisks denote significance according to a t test, *P < 0.05 and **P < 0.01, where DEX and CHX are compared with mock and DEX+CHX is compared with CHX. Primers for ACT2 were used to normalize the expression data.
STY1 Has Several Functional Transcriptional Activation Domains
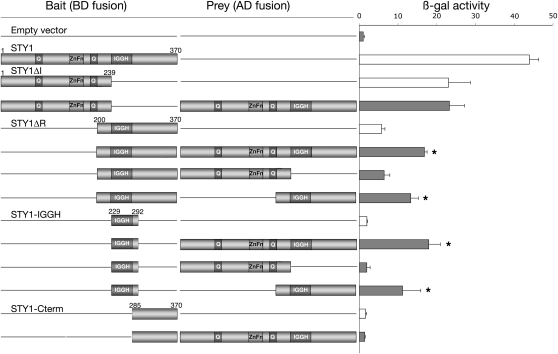
The activation of YUC4 transcription in 35Spro:STY1-GR sty1-1 sty2-1 seedlings upon DEX treatment suggests that STY1 acts as a transcriptional activator. The N-terminal part of STY1 contains a 17–amino acid region rich in Gln (8 glu), as does a region close to the nuclear localization signal (12 amino acids, 6 glu). In addition, a stretch of five acidic amino acids resides in the IGGH domain. These small domains have the characteristic features of transcriptional activation domains (Mitchell and Tijan, 1989; Triezenberg, 1995). We made a series of deletions in the STY1 cDNA and fused the partial sequences to the DNA binding domain of the GAL4 gene from yeast to elucidate if any specific region in STY1 was responsible for this activity. The full-length STY1 protein was, as previously described (Kuusk et al., 2006), able to activate transcription in yeast as shown by the activation of the LacZ reporter gene (Figure 3). The truncated versions of STY1 revealed that all regions except the C-terminal domain were able to induce transcription to some extent (Figure 3, white bars). Furthermore, the C-terminal half (STY1ΔR), which alone was found to be a rather poor activator, caused an almost 50% reduction of β-gal activity when removed from STY1 (STY1ΔI), compared with full-length STY1, suggesting that the different transcriptional activation domains have a synergistic effect on transcription. The result from the quantitative LacZ assay was confirmed by the growth of the different transformants on media selective for the ADE2 and HIS3 reporters.
Figure 3.
Dissecting the Transactivation and Homodimerization Domains of STY1 Using Truncated Fusion Proteins in Yeast.
Full-length STY1 (1 to 370) and the truncated versions STY1ΔI (1 to 239), STY1ΔR (200 to 370), STY1-IGGH (229 to 292), and STY1-Cterm (285 to 370) were analyzed by yeast two-hybrid assays. Results from a quantitative β-gal assay, measuring the activity of the LacZ reporter product in three independent transformants, are shown to the right (error bars indicate 1 sd). The level of autoactivation in cotransformants with the empty prey plasmid is indicated by white bars. Cotransformants displaying interaction are indicated by an asterisk. Q, Gln-rich region; ZnFn, the RING-like zinc finger domain; IGGH, the IGGH domain.
Fusion of STY1 to a Repressor Domain Demonstrates the Role of STY1 as a Transcriptional Activator
Because our data suggest that STY1 acts as a transcriptional activator, we hypothesized that a STY1 repressor fusion expressed by a constitutive promoter in planta would cause dominant-negative effects. Consequently, lines expressing this construct would have phenotypes resembling multiple mutants of SHI/STY family members, due to a reduction in transcription of genes targeted by SHI/STY family members.
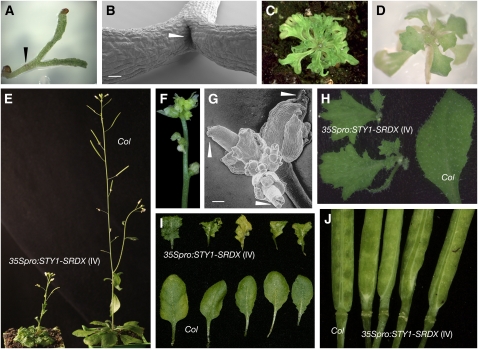
Transformation of accession Columbia (Col) with a 35Spro:STY1-SRDX construct resulted in 183 primary transformants classified into five groups (classes I to V). The majority (class I, n = 124) arrested in development after the emergence of two very narrow cotyledons (Figure 4A) and completely lacked a shoot apical meristem (SAM; Figure 4B). Because STY1 is expressed early during embryo development (Kuusk et al., 2002), SHI/STY genes could potentially be required for the formation of the SAM. Class II (n = 37) transformants never bolted but developed many tiny and heavily serrated rosette leaves (Figure 4C). A third class represented by a single transformant was phenotypically similar to class II, except that it bolted with a short thin stem producing severely defective flowers but no cauline leaves (Figure 4F). Interestingly, all analyzed gynoecia from this individual had a split in the stigma and style (Figure 4G), a phenotype associated with SHI/STY family multiple loss-of-function mutants (Kuusk et al., 2006). However, this individual did not produce any seeds and was not useful for further studies. The rosette leaves of the 12 class IV transformants resembled those of class II, although leaf serration was less severe and similar to that of the sty1-1 sty2-1 shi-3 lrp1 srs5-1 quintuple or yuc1 yuc2 yuc4 yuc6 quadruple mutants (Figures 4D, 4E, and 4I; Cheng et al., 2006; Kuusk et al., 2006). These lines often produced two or more highly serrated cauline leaves at each node and frequently produced aerial rosettes (Figure 4H). Class IV transformants flowered, although later compared with the wild type and produced gynoecia with elongated gynophore, resembling those of SHI/STY family quintuple mutants (Figure 4J). Some of these plants produced viable seeds and were used for further analysis. Class V (n = 9) lines were morphologically identical to the wild type (data not shown). In conclusion, class I to IV plants showed a variety of phenotypes associated with SHI/STY family multiple mutants and some new defects in additional regions where STY1 normally is expressed. Taken together, these data indicate that the STY1 function is reversed when fused to a strong repressor.
Figure 4.
The 35Spro:STY1-SRDX Transgene Acts in a Dominant-Negative Manner.
(A) 35Spro:STY1-SRDX seedling of class I. The arrowhead points out the hypocotyl.
(B) Scanning electron micrograph of 35Spro:STY1-SRDX (class I) seedling. The arrowhead points to the empty position where the SAM should have been. Bar =100 μm.
(C) 35Spro:STY1-SRDX plant of class II.
(D) 35Spro:STY1-SRDX plantlet of class IV.
(E) Col and 35Spro:STY1-SRDX (class IV) plants.
(F) 35Spro:STY1-SRDX (class III) inflorescence, displaying severe floral organ defects.
(G) Scanning electron micrograph of 35Spro:STY1-SRDX (class III) inflorescence. White arrowheads indicate shi/sty multiple mutant-like apical gynoecia defects. Bar = 100 μm.
(H) Cauline leaves from Col and 35Spro:STY1-SRDX (class IV).
(I) Rosette leaves from Col and 35Spro:STY1-SRDX (class IV) plants.
(J) Gynophores from Col and 35Spro:STY1-SRDX (class IV) siliques.
STY1-SRDX Is a Repressor of YUC4
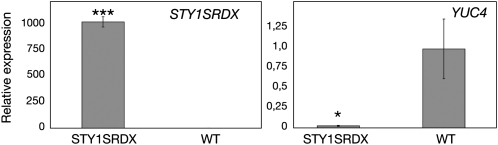
The STY1 and YUC4 promoters are both active in hydathodes of young leaves (Kuusk et al., 2002; Cheng et al., 2006). To elucidate the effect of STY1-SRDX on YUC4 expression in leaves, we extracted RNA from rosette leaves of wild-type (Col) and class IV transformants and analyzed STY1-SRDX and YUC4 mRNA levels. The STY1-SRDX transcript was ∼1000 times more abundant in class IV relative to the background noise of the wild type, suggesting that STY1-SRDX caused the class IV phenotypical abnormalities (Figure 5A). The YUC4 transcript level was reduced by ∼95% in class IV compared with the wild type (Figure 5B) (i.e., a much more severe reduction of YUC4 transcripts than that observed in floral buds of sty1-1 sty2-1 double mutants compared with wild type, which was ∼50%) (Sohlberg et al., 2006).
Figure 5.
STY1-SRDX Negatively Regulates Transcription of YUC4.
qRT-PCR measuring YUC4 and STY1-SRDX transcripts in rosette leaves of 35Spro:STY1-SRDX (class IV) and wild-type (Col) plants. Values on the y axis represent relative expression, where the wild-type expression levels are set to 1. The data represent the average of three and four biological replicates (three technical replicates for each biological sample). Error bars represent se. Stars denote significance according to a t test: *P < 0.05 and ***P < 0.001.
STY1 Interacts with the YUC4 Promoter
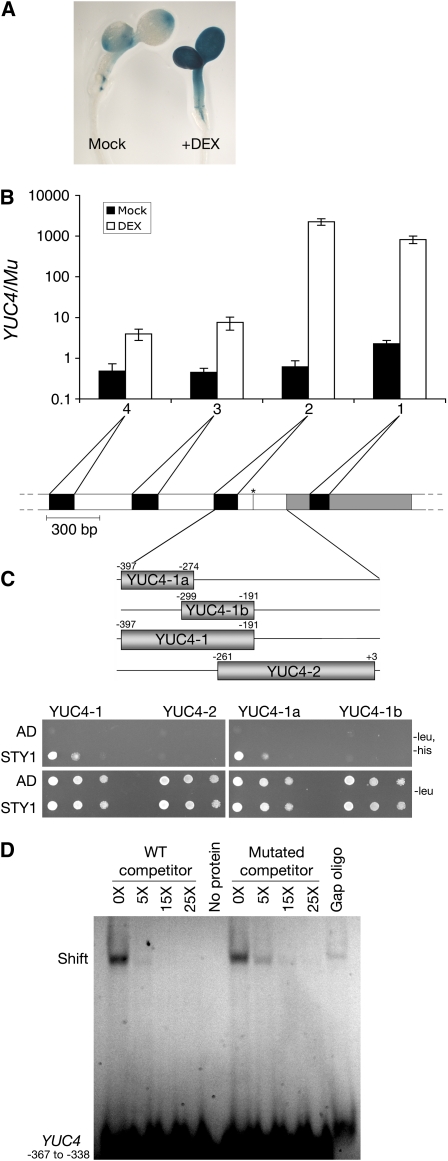
Because STY1 activates YUC4 transcription independently of protein synthesis, it seemed plausible that STY1 directly acts on the YUC4 promoter. By analyzing YUC4 promoter activity after DEX-mediated activation of STY1-GR in YUC4pro:GUS (for β-glucuronidase) 35Spro:STY1-GR seedlings, we showed that 3 h post DEX treatment, at the peak of STY1-GR–induced YUC4 transcription (Sohlberg et al., 2006), the entire aerial part demonstrated ectopic YUC4 promoter activity (Figure 6A; Cheng et al., 2006). This implies that STY1-mediated activation of YUC4 is independent of the YUC4 genomic context and only requires parts of the YUC4 promoter present in the YUC4pro:GUS construct. In an attempt to test whether STY1 is recruited to this region of the YUC4 promoter, chromatin immunoprecipitation (ChIP) experiments were performed using the 35Spro:STY1-GR sty1-1 sty2-1 line. Extracts of mock- and DEX-treated seedlings were used in combination with anti-GR antibodies as well as four primer pairs targeting different regions of the YUC4 promoter. While no selective amplification of YUC4 could be detected in mock-treated samples, a significant increase in amplification of the YUC4 promoter compared with control DNA was detected after DEX treatment (Figure 6B). Primer pairs 1 and 2 gave a stronger signal compared with 3 and 4, suggesting that STY1 interacts within the first 400 bp upstream of the YUC4 open reading frame.
Figure 6.
STY1 Interacts with the YUC4 Promoter.
(A) GUS histochemical analysis of mock- or DEX-treated 35Spro:STY1-GR YUC4pro:GUS seedlings.
(B) qRT-PCR showing the selective amplification of the YUC4 promoter relative to the Mu transposon from DNA coimmunoprecipitated with STY1-GR. 1, 2, 3, and 4 represent different primer pairs with different amplicons in the YUC4 promoter. White represents noncoding sequence, gray the first exon, and the asterisk denotes a putative TATA box. The graph shows the average of two technical repeats on one biological sample. Error bars represent 1 sd.
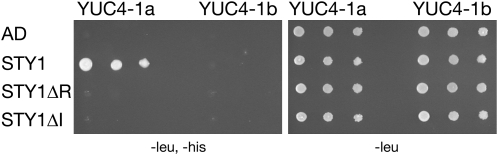
(C) STY1 interacts with the YUC4 promoter in a yeast one-hybrid assay. Size and location of the PCR-amplified fragments YUC4-1, -2, -1a, and -1b on the YUC4 promoter. Numbers indicates position in relation to initiation of translation. Bottom panels show yeast strain Y187, containing the different promoter fragments in front of a minimal promoter and the HIS3 reporter, transformed with a STY1-AD fusion (STY1) or empty vector control (AD). Yeast cultures were diluted (1:10 successive dilution series), spotted onto plates without Leu, and without Leu and His, and subsequently incubated 2 d at 30°C.
(D) A FAM-labeled wild-type double-stranded oligo corresponding to −367 to −338 of the YUC4 promoter was competing for STY1 binding with 5 to 25 times higher concentrations of unlabeled wild-type oligo (wild-type competitor) or an unlabeled oligo corresponding to the same promoter region but carrying a 6-bp mutation at the putative STY1 binding site (mutant competitor). Although the STY1 protein shows an unspecific binding to an unrelated oligo (gap oligo) by causing a band shift, a specific interaction at the CTCTAC site is also indicated, as the mutant competitor was less efficient than the wild-type competitor at 5× concentration.
[See online article for color version of this figure.]
Although both the timing of the STY1-GR–induced expression of YDK1 as well as the CHX experiments suggest activation of YDK1 to be a secondary effect of STY1 action, we studied the ability of the STY1 protein to interact with the YDK1 promoter. However, in a ChIP analysis, we could not detect any selective amplification of the YDK1 promoter region, targeted by our YDK1 promoter primers, in neither mock- nor DEX-treated seedlings (data not shown). This indicates that STY1 does not bind to the YDK1 promoter but affects its activity indirectly, supporting our previous data suggesting that YDK1 is activated by STY1-mediated elevation of auxin levels (Sohlberg et al., 2006).
We also assayed the ability of STY1 to interact with YUC4 promoter fragments in yeast cells. The promoter fragments were selected based on the ChIP results and covered the most probable region of interaction as indicated by primer pairs 1 and 2 (Figures 6B and 6C). The yeast one-hybrid analysis showed the ability of STY1 to bind a 207-bp region directly upstream of a predicted TATA box (YUC4-1; TSSP at http://www.softberry.com/berry.phtml) but not to DNA from the more proximal promoter region or the presumptive 5′ untranslated region of YUC4 (YUC4-2; Figure 6C). To narrow down the region targeted by STY1, we preformed another round of one-hybrid analysis on two fragments (YUC4-1a and -1b), covering regions −397 to −274 and −299 to −191, respectively. This showed that STY1 interacts with a 124-bp fragment (YUC4-1a) located 91 bp upstream of the presumptive TATA box. Because the zinc finger of STY1 is identical to that found in the most closely related SHI/STY family member, SHI (Kuusk et al., 2006), we analyzed the ability of SHI to interact with YUC4 in yeast. We found that SHI also can interact with fragment YUC4-1a (see Supplemental Figure 1 online). These data suggest that the two SHI/STY family members potentially could activate YUC4 transcription by direct binding to the YUC4 promoter. Alternatively, the SHI/STY1–YUC4 interaction could be indirect via another protein, or proteins, expressed both in plants and in yeast.
An STY1 Homolog from Madagascar periwinkle Interacts with the Promoter of the AP2/ERF-Encoding Gene OCTADECANOID-RESPONSIVE CATHARANTHUS AP2 3
vom Endt et al. (2007) describe an in vivo interaction between a putative Catharanthus roseus SHI/STY ortholog and the OCTADECANOID-RESPONSIVE CATHARANTHUS AP2 3 (ORCA3) promoter. They could also show that the C. roseus SHI/STY ortholog, when expressed and purified from Escherichia coli, could interact with the ORCA3 promoter in vitro, showing that SHI/STY family members indeed possess DNA binding activity in vitro. In a phylogenetic analysis, the potential C. roseus SHI/STY ortholog forms a clade together with SRS3, SRS5, and SRS7, supported by a bootstrap value of 71 (see Supplemental Figure 2 online), but also displays high sequence identity and similarity with STY1 (Figure 7A). Interestingly, the ORCA3 promoter fragment used in the yeast one-hybrid and in vitro assays performed by vom Endt et al. (2007) contains three regions highly similar to the YUC4-1a fragment used in our one-hybrid assay, as depicted by a MEME analysis (MEME 4.0.0; Bailey and Elkan, 1994; Figure 7B). vom Endt et al. (2007) mutated several regions in the ORCA3 promoter fragment, including two of the regions identified as similar to YUC4-1a, without affecting the affinity to the C. roseus SHI/STY ortholog. However, the authors never mutated the region carrying the ACTCTAC sequence (Figure 7B), suggesting that the C. roseus SHI/STY ortholog, and also STY1, interact with this region in the ORCA3 and YUC4 promoters, respectively. This region is located 173 to 167 bp upstream of a putative TATA box in the YUC4 promoter, covered by the YUC4-1a fragment, but not the YUC4-1b fragment (Figure 6C), and could thus serve as an important cis-regulatory element. Interestingly, ORCA3 appears to positively affect transcription of a Trp decarboxylase (TDC; van der Fits and Memelink, 2000), mediating the formation of tryptamine, the substrate for the YUC family monooxygenases (Zhao et al., 2001).
Figure 7.
Promoter Fragments Targeted by SHI/STY Family Proteins Show Sequence Similarities.
(A) and (B) Asterisks indicate identical residues; one and two dots indicate different degrees of similarity.
(A) Protein alignment of STY1 and a C. roseus ortholog (ABL63114.1). A gray box marks the RING-like zinc finger domain.
(B) Promoter fragments with affinity to SHI/STY family members. A black box marks the mutated residues in the YUC4mut2pro:HIS3 construct. Dark gray boxes mark the base pairs mutated in the ORCA3 promoter (vom Endt et al., 2007).
An Arabidopsis Gene Closely Related to ORCA3 Is Activated by STY1
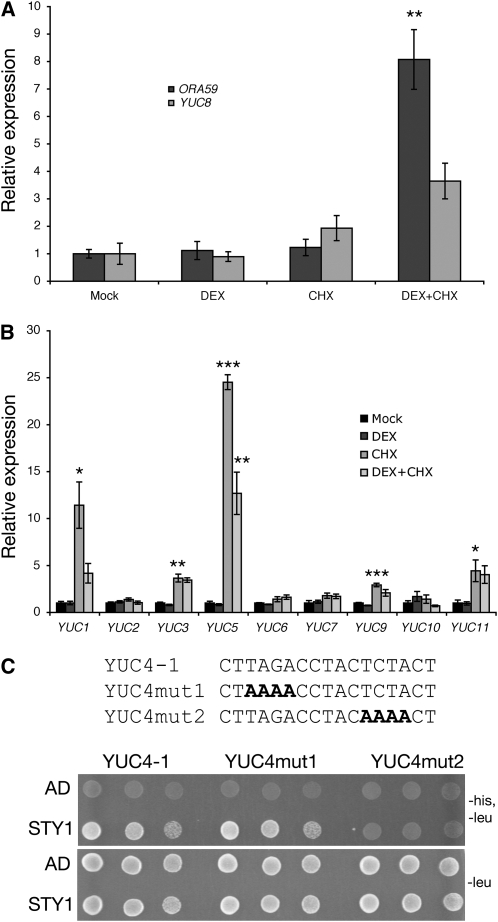
Because a SHI/STY family member appears to directly bind to the promoter of an AP2/ERF gene in C. roseus, we wanted to investigate if any closely related AP2/ERFs in Arabidopsis also are regulated by SHI/STY proteins. According to Nakano et al. (2006), ORCA3 is an AP2/ERF of subgroup IX. We analyzed the promoters of all 17 Arabidopsis genes in subgroup IX and found only two genes with ACTCTAC elements. In ERF15, we found the element 1000 bp upstream of the transcriptional start site. The other gene, OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF 59 (ORA59; Pré et al., 2008), had the putative STY1 binding site 489 bp upstream of the transcriptional start site, 455 bp upstream of a putative TATA box (see Supplemental Figure 3 online). Interestingly, ORA59 was upregulated approximately twofold in both biological replicates at 3 h after DEX treatment of 35Spro:STY1-GR sty1-1 sty2-1 seedlings in a microarray experiment (V. Ståldal and E. Sundberg, unpublished data). In the same microarray, no other AP2/ERF IX genes showed increased transcript levels. To confirm that ORA59 is activated by STY1, we performed DEX/CHX-mediated induction of STY1-GR in seedlings and measured ORA59 transcript levels by qRT-PCR. ORA59 is not activated by STY1 after 2 h of DEX treatment (Figure 8A). However, when seedlings were treated with CHX and DEX simultaneously, STY1 could activate ORA59 transcription, resulting in an eightfold upregulation of ORA59 (Figure 8A). This suggests that a repressor of STY1-mediated activation of ORA59 is removed by CHX.
Figure 8.
STY1 Induces the Activity of ORA59 and YUC8 and Binds to a Conserved Element in YUC4.
(A) and (B) qRT-PCR analysis of transcripts from 35Spro:STY1-GR sty1-1 sty2-1 seedling 2 h after treatment with mock, DEX, CHX, or DEX and CHX as indicated. The data represent the average of three biological replicates (three technical replicates for each biological sample). Error bars represent se. Stars denote significance according to a t test, *P < 0.05, **P < 0.01, and ***P < 0.001, where DEX and CHX values are compared with mock and CHX+DEX is compared with CHX. Primers for ACT2 were used to normalize the expression data.
(A) ORA59 and YUC8.
(B) YUC1-3, 5-7 and 9-11.
(C) Mutations in the YUC4 promoter abolishes its interaction to STY1. Yeast strains of Y187 harboring the YUC4mut1pro:HIS3 or YUC4mut2pro:HIS3 constructs were transformed with pGAD424 (AD) and pADSTY1 (STY1). The YUC4-1pro:HIS3 line served as a control. Yeast cultures were diluted (1:10 successive dilution series), spotted onto plates without Leu, and without Leu and His, and subsequently incubated 3 d at 30°C.
STY1 Interacts with a Short Sequence in the YUC4 Promoter
To confirm that the ACTCTAC sequence in the YUC4 promoter is targeted by STY1, we mutated parts of this sequence in the YUC4-1a promoter fragment and analyzed STY1 affinity to the mutated fragment in yeast. In fragment YUC4mut2, we mutated ACTCTAC to ACAAAAC, resulting in the complete inability of STY1 to interact with the DNA fragment (Figure 8C). In YUC4, the ACTCTAC sequence is part of a larger, partially palindromic, sequence (TAGA CCTACTCTAC). The YUC4mut2 fragment is mutated in the second part of this palindrome and because our data suggest STY1 to be able to homo- or heterodimerize with other STY/SHI family members (below), we assayed whether mutations in the first part of the palindrome (YUC4mut1; AAAACCTACTCTAC) could abolish STY1-YUC4 interaction. We found that STY1 interacts with YUC4mut1 with identical affinity as for YUC4-1a (Figure 8C).
STY1 Binds to the ACTCTAC Element in Vitro
To determine if the interaction between STY1 and the ACTCTAC promoter element is direct or if it requires a protein also present in yeast, we produced the STY1 protein by in vitro transcription/translation (see Supplemental Figure 4 online) and used it in electrophoretic mobility shift assays. Using a 30-bp oligo covering the putative binding site, we could detect a clear STY1-dependent mobility shift (Figure 6D; see Supplemental Figure 5 online). A faint shift was also seen with a completely unrelated control oligo, indicating some degree of unspecific DNA binding (the gap oligo; Figure 6D). However, a specific preference for binding to the ACTCTAC element was shown since wild-type unlabeled oligos could compete for binding to STY1 more efficiently than oligos carrying mutations in the identified element (Figure 6D). From these results, we conclude that STY1 binds directly to the identified element of the YUC4 promoter.
Two Members of the YUC Gene Family Are Activated by STY1
We searched for the STY1 binding site in proximal promoter regions (650 bp upstream of the transcriptional start site) of all 11 YUC genes in Arabidopsis but could only find the ACTCTAC sequence in YUC4. However, we found a similar sequence, ACTCTAA, 190 bp upstream of the annotated transcriptional start site, 142 bp upstream of a putative TATA box, in YUC8 (see Supplemental Figure 3 online). This prompted us to analyze STY1-GR–regulated expression of YUC8 in 35Spro:STY1-GR sty1-1 sty2-1 seedlings. We found that YUC8 is activated by STY1-GR in the presence of CHX, although not to the same extent as YUC4 (Figure 8A). The ACTCTAA sequence can also be found in the proximal promoter regions of YUC1, 5, and 9, although not as close to the TATA box as in YUC8 (see Supplemental Figure 3 online). To examine the effect of DEX-mediated nuclear translocation of STY1-GR on the remaining YUC genes, we measured transcripts of YUC1-3, 5-7, and 9-11. No YUC genes, except YUC4 and YUC8, were found to be induced by STY1-GR (Figure 8B). The activation of only two YUC genes suggests that other sequences or factors, in addition to the putative ACTCTA(C/A) promoter element, are required for STY1-mediated transcriptional activation.
The IGGH Domain Is Responsible for Intrafamily Homo-/Heterodimerization
We have previously shown that the SHI family member LRP1 is capable of dimerizing with itself and other SHI family members, suggesting that SHI/STY proteins exert at least some of their functions as homo- or heterodimers (Kuusk et al., 2006). To elucidate how STY1 interacts with the YUC4 promoter, we used yeast two-hybrid assays to identify the regions involved in the putative STY1 homodimerization. Truncated STY1-BD fusions were screened against truncated STY1-AD fusions (Figure 3). Full-length STY1 and STY1ΔR, lacking the N-terminal half including the zinc finger domain, showed similar interaction patterns when fused to the GAL4 activation domain and assayed for interaction with STY1ΔR-BD. However, the STY1ΔI-BD fusion, lacking the IGGH domain, showed no significant activation of the interaction reporters HIS3, ADE2, and LacZ when cotransformed with full-length STY1-AD (Figure 3). This indicates that the C-terminal half of STY1 harbors the dimerization domain. To further map the interaction site, IGGH domain and C-terminal domain fusions were made; these clearly show that the IGGH domain alone is sufficient for STY1 homodimerization in yeast (Figure 3).
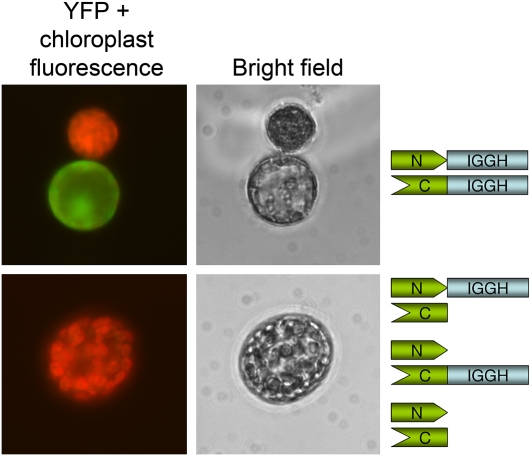
To test the capacity of the STY1 IGGH domain to homodimerize in planta, we used a bimolecular fluorescence complementation approach (Bracha-Drori et al., 2004). The IGGH domain was fused with the N- and C-terminal halves of yellow fluorescent protein (YFP), and IGGH dimerization-mediated YFP reconstitution was demonstrated by YFP signals only in protoplasts transformed with the two different IGGH domain-YFP fusions (Figure 9). The negative controls lacked YFP signals and were identical to untransformed protoplasts (Figure 9).
Figure 9.
The IGGH Domain Mediates Homodimerization in Planta.
Bimolecular fluorescence complementation analysis of the ability of IGGH domains to form homodimers. Fluorescence microscopy pictures, showing merged fluorescence signals from chloroplasts (red) and YFP (green) are shown to the left. A protoplast transformed with the two different YFP-IGGH constructs and an untransformed protoplast are shown in the top left picture. The bottom left picture shows a representative protoplast from one of the three negative controls. Bright-field microscopy images of the protoplasts are shown in the middle. The construct combination analyzing IGGH-mediated reconstitution of functional YFP (N-term YFP:IGGH + C-term YFP:IGGH) and the three negative controls (N-term YFP:IGGH + C-term YFP, N-term YFP + C-term YFP:IGGH, and N-term YFP + C-term YFP) are shown to the right.
Full-Length STY1 Is Required for Interaction with the YUC4 Promoter
The zinc finger in STY1 might serve as a DNA binding domain. In general, the zinc finger is an independently folded globular domain, and isolated zinc finger peptides have been shown to fold accurately and interact with DNA in solution (Parraga et al., 1988). If STY1 can interact with the YUC4 promoter as a monomer, then the putative DNA binding zinc finger domain of STY1ΔI could be enough for DNA interaction. Our one-hybrid data clearly show that the truncated fusions, either lacking the dimerization domain (STY1ΔI) or the putative DNA binding domain (STY1ΔR), are unable to interact with the YUC4 promoter as opposed to full-length STY1 (Figure 10). The requirement of a full-length STY1 protein suggests that several regions in STY1 might cooperate in protein-DNA complex formation and indicates that dimer formation might be essential for STY1 interaction with the YUC4 promoter in vivo.
Figure 10.
Both the Zinc Finger and IGGH Domains Are Required for STY1 Binding to the YUC4 Promoter.
Yeast strain Y187 harboring YUC4-1apro:HIS3 and YUC4-1bpro:HIS3 constructs was transformed with pGAD424 (AD), full-length STY1 (STY1), and the two truncated versions of STY1 containing amino acids 1 to 239 (STY1ΔI) or 200 to 370 (STY1ΔR). Yeast cultures were diluted (1:10 successive dilution series), spotted onto plates without Leu, and without Leu and His, and subsequently incubated 3 d at 30°C.
DISCUSSION
We previously characterized SHI and its nine paralogs in Arabidopsis and demonstrated that the SHI/STY genes share highly redundant functions, have overlapping spatial and temporal expression patterns, are predominantly expressed in apical regions of plant organs, and together exert essential functions during plant development (Kuusk et al., 2002, 2006). Recently, we showed that the SHI/STY gene family most likely controls developmental processes through regulation of auxin biosynthesis (Sohlberg et al., 2006). However, not much information regarding the properties of STY/SHI family proteins has been available. In this report, we reveal that STY1 is a transcriptional activator regulating auxin biosynthesis by binding to, and activating, YUC4 and probably also YUC8 and ORA59. These genes encode two closely related enzymes that are suggested to be rate limiting in auxin biosynthesis as well as a transcription factor putatively regulating Trp formation, respectively.
SHI/STY Family Members Can Act as Transcriptional Regulators
Here, we provide evidence that STY1 is a nuclear protein and that DEX-mediated nuclear import of STY1-GR activates transcription of the previously described downstream target YUC4 (Sohlberg et al., 2006) in the presence of CHX, an inhibitor of protein synthesis. We also demonstrate that DEX induction of nuclear STY1-GR localization leads to increased expression levels of YUC4pro:GUS. As protein intermediates do not appear to be required for the upregulation of YUC4 and because the promoter region used in the YUC4pro:GUS construct is enough for STY1 to activate GUS expression, we speculated that STY1 exerts its function directly or in concert with other proteins on one or several elements in the YUC4 promoter. To assess this hypothesis, we analyzed whether STY1 is associated with the promoter of YUC4 by ChIP, electrophoretic mobility shift assay, and yeast one-hybrid assays. Indeed, STY1 interacts with a small region located close to a putative TATA box, just as expected for a transcription factor binding a cis-regulatory element.
SHI/STY proteins have a conserved RING-like zinc finger domain, C3HC3H, in the N-terminal half of the protein (Fridborg et al., 2001). This is the only domain present in the family members that resembles a DNA binding domain, suggesting that it could be responsible for their potential protein–DNA interaction. Our in vitro STY1–YUC4 interaction study suggests a direct physical interaction, and in a recent study, vom Endt et al. (2007) describes an in vitro interaction between the ORCA3 promoter and a C. roseus SHI/STY ortholog, clearly suggesting that SHI/STY family members are capable of binding DNA in vitro.
The amino acids in the zinc finger domain loops are highly conserved in the nine Arabidopsis proteins (Fridborg et al., 2001) and in orthologs from other divergent species, such as the previously mentioned C. roseus protein (Figure 7A). Generally, residues at the loop tip of DNA binding C2H2 zinc finger interact with the targeted DNA sequence (reviewed in Laity et al., 2001). STY1 and its closest paralog SHI have identical zinc fingers, suggesting that they could bind the same DNA sequences; indeed, both proteins are able to interact with the YUC4 promoter fragments in yeast. However, the affinity of SHI to the YUC4 promoter is lower compared with that of STY1, indicating slight functional differences between the two proteins, at least in the yeast system. Alternatively, the two proteins might reach different steady state levels in the yeast system. The YUC4 promoter fragment targeted carries an ACTCTAC sequence, which when partially mutated completely abolishes STY1 binding in our yeast system. Other SHI/STY family members might bind slightly different DNA sequences in different promoters, but the high amino acid sequence identity/similarity in the zinc finger loops of STY1, STY2, and SHI, together with their high level of functional redundancy and the reduced YUC4 transcription in double mutants (sty1-1 sty2-1) and 35Spro:STY1-SRDX dominant-negative lines, suggest that at least STY1, STY2, and SHI target the same promoters.
The STY1-GR–mediated upregulation of YUC4 and YUC4pro:GUS activity suggest that STY1 in itself is a transcriptional activator or that it interacts with a coactivator. In Kuusk et al. (2006), we revealed that STY1 and SHI activate transcription of marker genes in yeast, and here we show that STY1 has at least three domains, two Gln-rich regions and the IGGH domain, capable of activating transcription in yeast. Gln-rich and acidic domains are known to recruit transcription factors initiating transcription of the targeted gene (Mitchell and Tijan, 1989; Triezenberg, 1995). The IGGH domain carries several acidic residues conserved among Arabidopsis SHI/STY family members (Fridborg et al., 2001). The acidic region is also conserved in moss (P. patens) orthologs, and an analysis of the ability of the acidic region to self-activate transcription in yeast, using a mutated Pp SHI1-GAL4BD fusion protein, showed that the IGGH domain loses most of its self-activating function when the acidic region is mutated (see Supplemental Figure 6 online). This suggests that the acidic residues are important for the transcriptional activator function of the IGGH domain. We could also demonstrate that when STY1 is fused to a strong transcriptional repressor (SRDX), its biological function is reversed, further strengthening the hypothesis of STY1 acting as a transcription factor activating target genes.
In an attempt to elucidate whether the STY1 monomer has affinity for the YUC4 promoter, we analyzed truncated forms of STY1, either lacking the DNA binding or the dimerization domain. We found that only full-length STY1 can interact with DNA in our yeast assay, indicating that a single isolated zinc finger lacks affinity for the promoter fragment. This suggests that SHI/STY family proteins homo- or heterodimerize to form functional units, that an additional region in the C-terminal half of STY1 is essential for DNA recognition and/or DNA binding, or that the truncated protein misfolds and thus loses its DNA binding activity. Many SHI/STY genes have overlapping spatial and temporal expression patterns, indicating that the proteins are active in the same cells at the same time. In Kuusk et al. (2006), we showed that STY1, SHI, and LRP1 can dimerize with LRP1, suggesting that the SHI/STY family members may form homo- and/or heterodimers and are active in protein complexes. Here, we show that homo- and heterodimerization between SHI/STY proteins is mediated by the IGGH domain. It remains to be established whether these proteins form dimers in planta and how this affects DNA sequence recognition and transcriptional activation properties.
STY1 Directly Regulates Auxin Biosynthesis
The data presented here support our hypothesis that transcriptional activators of the SHI/STY family can recognize and bind the whole or at least parts of the sequence ACTCTA(C/A) in the YUC4, YUC8, ORCA3, and ORA59 promoters.
It has been suggested that YUC proteins are responsible for catalyzing hydroxylation of the amino group in tryptamine, a rate-limiting step in one of the Trp-dependent indole-3-acetic acid biosynthesis pathways (Zhao et al., 2001), and multiple loss of YUC proteins demonstrates their importance in various auxin-regulated developmental processes (Cheng et al., 2006, 2007).
LEAFY COTYLEDON2 (LEC2) has previously been shown to directly activate an auxin biosynthesis gene (Stone et al., 2008). Apart from this recent finding, the temporal and spatial regulation of auxin biosynthesis is poorly understood. Because the YUC and SHI/STY family members share overlapping temporal and spatial expression patterns (e.g., in the embryo, cotyledon tips, vasculature, hydatodes, style, stigma, and shoot apex) (Kuusk et al., 2002, 2006; Cheng et al., 2006, 2007), it is likely that SHI/STY proteins contribute to determining the expression domains of YUC4 and YUC8.
Another SHI/STY-regulated gene carrying the ACTCTAC element that potentially is involved in auxin biosynthesis is the AP2/ERF IX family member ORA59 (Pré et al., 2008). Pré et al. (2008) found that 16 h of estradiol-induced expression of XVEpro:ORA59 resulted in accumulation of mRNA from TRYPTOPHAN SYNTHASE BETA SUBUNIT1 (TSB1), TSB2, and INDOLE-3-GLYCEROL PHOSPHATATE SYNTHASE (IGPS), all involved in biosynthesis of Trp (Ouyang et al., 2000). IGPS catalyzes the formation of indole-3-glycerol phosphate, which can be used for Trp-independent indole-3-acetic acid biosynthesis or be converted to Trp by TRYPTOPHAN SYNTHASE ALFA SUBUNIT1 and the TSB1/TSB2 heterodimer (Ouyang et al., 2000).
The SHI/STY family target ORCA3 positively regulates transcription of TDC, thereby mediating the formation of tryptamine from Trp (van der Fits and Memelink, 2000), which suggest ORCA3 to have a role in YUC-mediated auxin biosynthesis. ORCA3 and ORA59 are both AP2/ERF members of subgroup IX (Nakano et al., 2006). This suggests that ORA59 and ORCA3 might perform similar functions and that several AP2/ERF subgroup IX members might function as transcriptional regulators of local auxin biosynthesis in Arabidopsis.
35Spro:STY1-SRDX Transformants Reveal a Role for SHI/STY Family Members in SAM Formation/Maintenance during Embryo Development
The chimeric repressor silencing technology (CRES-T) is based on a modified EAR motif named SRDX, a 12–amino acid peptide acting as a strong repressor (Hiratsu et al., 2003). A STY1-SRDX fusion was ectopically expressed in Arabidopsis using the cauliflower mosaic virus 35S promoter, resulting in primary transformants with varying degree of phenotypic defects in organs where SHI/STY and YUC family members normally are expressed. Many transformants showed SHI/STY quintuple mutant-like flower and leaf phenotypes, suggesting that the STY1-SRDX proteins function in a dominant-negative manner, repressing the target genes normally activated by STY1. Consistent with this idea, the expression of YUC4 was reduced in these lines. Because the yuc4 mutant is morphologically identical to the wild type (Cheng et al., 2006), the STY1-SRDX fusion protein must affect the expression of additional genes, most likely YUC8, ORA59, and other yet unidentified direct targets. However, a majority of the primary transformants arrest in development at seedling stage, after the emergence of two cotyledons, and lack a functional SAM. This suggests a role for STY1 and related proteins in SAM formation and/or maintenance during embryo development. Although no comprehensive studies of the SHI/STY family expression pattern during embryo development have been made, we know that at least STY1 is expressed in early globular embryos prior to SAM formation, mainly at the sites of cotyledon initiation (Kuusk et al., 2002). The SHI/STY-mediated effect on SAM formation/maintenance might therefore be non-cell autonomous, and it is tempting to speculate that the mobile signal involved is auxin. Although the SAM and the boundary between the SAM and lateral organs appear to be low auxin zones, correct auxin gradients across the early globular embryo may be required for the formation/maintenance of the SAM.
In the future, it will be interesting to put SHI/STY-regulated biosynthesis of auxin in relation to LEC2-mediated activation of YUC family members, and other pathways controlling similar processes, to build a comprehensive view of the requirement of developmentally regulated auxin homeostasis, including biosynthesis, transport, and inactivation. It will also be interesting to identify additional downstream targets of the SHI/STY gene family to fully understand their DNA binding preferences and biological functions.
METHODS
Subcellular Localization
The vector psmRS-GFP (Davis and Vierstra, 1996) was used for construction of plasmid pSTY1-GFP, which contains a STY1-GFP translational fusion with STY1 in the N terminus. pSTY1-GFP was constructed by amplifying full-length STY1 cDNA using primers STY1BamHI5′ II and STY1BamHI3′ II. The amplified fragment was digested with BamHI and subsequently ligated into the BamHI site of psmRS-GFP. Biolistic transformation of onion epidermal cells was performed essentially according to Edqvist et al. (2004). Polyethylene glycol–mediated transformation of Physcomitrella patens protoplasts was performed as previously described (Schaefer et al., 1991). To visualize GFP and chloroplast autoflourescence signals in P. patens protoplasts, a Zeiss Axioskop 2 MOT fluorescence light microscope equipped with fluorescein isothiocyanate and tetramethylrhodamine isothiocyanate filters was used.
Gene Expression Analysis
Quantitative real-time PCR was preformed essentially as previously described (Sohlberg et al., 2006). Ten-day-old 35Spro:STY1-GR sty1-1 sty2-1 shi-3 or 35Spro:STY1-GR sty1-1 sty2-1 seedlings were treated with mock, DEX (10 μM), and CHX (10 μM) for 2 h. Rosette leaves from 35Spro:STY1-SRDX(IV) lines and Col were collected from adult plants. Total RNA was extracted using the RNeasy plant mini kit (Qiagen). Equal amounts of total RNA from each sample were reverse transcribed to cDNA with oligo (dT)20 primer using Superscript III reverse transcriptase (Invitrogen). The cDNA was diluted to 2 ng/ μL, and 5 μL of the diluted cDNA was used as template for amplification using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7000 thermocycler (Applied Biosystems). Primers targeting ACTIN2 (ACT2) were used to normalize the expression data for each gene. The PCR conditions were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15°C and 60°C for 1 min. At the end of the experiment, a dissociation kinetics analysis was performed to check the specificity of annealing. Three biological replicates were used, and three technical replicates were performed for each biological replicate. Primers targeting YUC4, YDK1, and ACT2 are described by Sohlberg et al. (2006). Primers to quantify YUC1-3 and 5-11 transcripts can be found in Supplemental Table 1 online.
STY1SRDXRTF and STY1SRDXRTR were used to target STY1-SRDX, and primers ORA59F and ORA59R were used to target ORA59.
Histochemical GUS staining was preformed according to Jefferson (1987).
Yeast Two-Hybrid Analysis
Full-length STY1 cDNA was PCR amplified using primers STY1pBDEcoRI5′ and STY1pBDEcoRI3′ and subsequently cloned into plasmid pBD GAL4 cam (Stratagene), creating plasmid pBD-STY1. To dissect the origin of the autoactivation observed for the pBD-STY1 construct, four truncated cDNAs (ΔR,ΔI, IGGH, and C-term) were amplified using primers STY1ΔR5′, STY1ΔR3′, STY1ΔI5′, STY1ΔI3′, STY1IGGH5′, STY1IGGH3′, and STY1c-term5′. Amplified fragments were then ligated into the Eco RI site of pBD GAL4 cam, creating plasmids pBD-STY1ΔR, pBD-STY1ΔI, pBD-STY1IGGH, and pBD-STY1c-term. Two truncated cDNAs were PCR amplified by primers STY1ΔRPstI5′, STY1ΔRPstI3′, STY1ΔIPstI5′, and STY1ΔIPstI3′ and ligated into the Pst I site of plasmid pGAD 424 (Clontech) to create plasmids pAD-STY1ΔR and pAD-STY1ΔI. pAD-STY1 is described by Kuusk et al. (2006). Yeast strain PJ69-4A harbors the reporters HIS3, ADE2, and LacZ under the influence of different GAL4-responsive promoters (James et al., 1996). PJ69-4A was cotransformed with BD and AD constructs (Gietz and Woods, 2002). Primary transformants were isolated on selective plates incubated 3 d at 30°C. Isolated transformants were subsequently analyzed on plates selective for the reporters ADE2 and HIS3. Transformants were also subjected to a liquid β-gal assay (http://130.15.90.245/lac_z_liquid_assay_for_yeast.htm) to quantify the expression levels of the LacZ reporter.
Generation of 35Spro:STY1-SRDX Plants
Primers STY1SRDXF and STY1SRDXR were used to amplify full-length STY1 cDNA. The PCR product was digested and subsequently ligated into the Sma I site of vector p35SSRDXG to create a 35Spro:STY1-SRDX-NosT-cassette flanked by attL sites. The cassette was recombined into the Arabidopsis thaliana transformation vector pBCKH using the gateway system (Invitrogen), creating plasmid p35Spro:STY1-SRDX. The construct was transformed into Agrobacterium tumefaciens strain GV3101, containing the helper plasmid pMP90, and introduced into Arabidopsis Col by A. tumefaciens–mediated transformation. T1 and T2 lines were analyzed and crossed to YUC4pro:GUS lines.
Scanning Electron Microscopy
Scanning electron microscopy was performed on fixed tissues as previously described (Fridborg et al., 1999) or on fresh tissue in low vacuum using a Zeiss Supra35VP as previously described (Sundström et al., 2006).
ChIP
The nuclear extracts were prepared following the procedure described by Ito et al. (1997), with some modifications. The immunoprecipitation of bound chromatin was performed using a ChIP kit (Upstate). Fourteen-day-old 35Spro:STY1-GR sty1-1 sty2-1 seedlings were treated with DEX (10 μM) or mock solution (Sohlberg et al., 2006) for 3 h and then ground in liquid nitrogen. The material was then cross-linked for 10 min in 1% formaldehyde. Chromatin was isolated from the tissues, resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH 8.1) with protease inhibitors (Complete Mini; Roche), diluted with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl) and sonicated to achieve an average DNA size of 0.5 to 1 kb. The chromatin extract was cleared by centrifugation and once again diluted, followed by a 2-h incubation with salmon sperm DNA/protein-A agarose beads (Upstate) at 4°C. The supernatants were then incubated with affinity-purified polyclonal anti-GR antibodies [GR (P-20); Santa Cruz Biotechnology] at 4°C overnight. The chromatin antibody complex was precipitated with salmon sperm DNA/protein-A agarose beads at 4°C for 1 h. The beads were then washed once with low salt buffer (20 mM Tris-HCl, pH 8.1, 2 mM EDTA, and 150 mM NaCl), once with high salt wash buffer (50 mM Tris-HCl, pH 8.0, 2 mM EDTA, and 500 mM NaCl), once with LiCl buffer (10 mM Tris-HCl, pH 8,0, 1 mM EDTA, 0.25 M LiCl, 1% IGEPAL-CA630, and 1% deoxycholic acid), and twice with TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA). The immunoprecipitated DNA and protein were eluted using 1% SDS and 0.1 M NaHCO3, and then the cross-link was reversed by incubation at 65°C for 5 h in 0.2 M NaCl. DNA was purified using the Qiagen PCR purification kit and eluted in 100 μL TE. DNA was analyzed by quantitative real-time PCR according to Sohlberg et al. (2006). Primers used for amplification of YUC4 and YDK1 promoter DNA were as follows: YUC4f1 and YUC4r1, YUC4f2 and YUC4r2, YUC4f3 and YUC4r3, YUC4f4 and YUC4r4, YDK1f1 and YDK1r1, YDK1f2 and YDK1r2, YDK1f3 and YDK1r3, and YDK1f4 and YDK1r4. To normalize the amplification signal by the YUC4/YDK1-specific primer pairs, Mu-2 transposon primers (Sundström et al., 2006) were used.
Yeast One-Hybrid Analysis
To amplify regions of the YUC4 promoter, primers 1HYUC4-F1 and 1HYUC4-R1, and primers 1HYUC4-F2 and 1HYUC4-R2 were used to produce fragment YUC4-1 and YUC4-2, respectively. To amplify fragments YUC4-1a and YUC4-1b, primers 1HYUC4-F1 and 1HYUC4-R3, and 1HYUC4-F3 and 1HYUC4-R1 were used, respectively. Amplified fragments were cloned into the NotI/BcuI sites of plasmid pINT1/pHIS3NB (Meijer et al., 1998), creating plasmids pINTYUC4-1, pINTYUC4-2, pINTYUC4-1a, and pINTYUC4-1b. Plasmid pINTYUC4-1a was mutated with the QuickChange site-directed mutagenesis kit (Stratagene) using primers YUC4mut1sense, YUC4mut1antisense, YUC4mut2sense, and YUC4mut2antisense to produce plasmids pINTYUC4-mut1 and pINTYUC4-mut2. All plasmids were linerized by NcoI and SacI before transformation of yeast strain Y187 (Harper et al., 1993). Positive clones were selected on G418 and subsequently transformed with vectors pAD-STY1 (Kuusk et al., 2006), pAD-STY1ΔI, pAD-STY1ΔR, or pGAD 424 (Clontech). Activation of the HIS3 reporter was observed after 3 d on plates without histidine.
Expression of Strep-Tagged STY1 Protein
In vitro transcription and translation of Strep-tagged STY1 protein was performed with the EasyXpress Insect Kit II (Qiagen) with the following modifications: In the translation step, 12 μL of column purified transcription reaction, 0.05 mM ZnCl2 and 2 μL 35S Met were added. The protein was concentrated 2.5 times and dialyzed in 25 mM HEPES, pH 8, 2 mM DTT, 0.05 mM ZnCl2, and 17% glycerol. Protein expression was verified by phosphor imager detection (Bio-Rad) following separation by SDS-PAGE.
Electrophoretic Mobility Shift Assay
Wild-type, mutated, or control gap oligonucleotides (oligos) were commercially synthesized (Eurogentec) as single stranded (ss) DNA. The wild-type oligo sequence corresponds to the stretch −367 to −338 in the YUC4 promoter. The mutated oligo differs from the wild-type one in that CTCTAC (−355 to −350) has been changed to TCTGCA. The gap oligo is completely unrelated and 40 bp long (Dobson and Allinson, 2006). To make double stranded (ds) oligos, equal amounts of complementary ss oligos were mixed, boiled for 5 min, and slowly cooled down to 25°C.
For the binding reaction, 9 μM of 5′ FAM (6′ carboxy-fluorescein group) labeled wild type or gap ds oligos was incubated with 8 μL of strep-tagged STY1 protein in binding buffer (20 mM HEPES, pH 8, 50 mM NaCl, 0.05 mM ZnCl2, 2 mM MgCl2, and 1 μg ss salmon sperm) at 4°C for 90 min. Five percent of glycerol was then added to each reaction before loading on a 6% native polyacrylamide gel in 0.25×TBE buffer run at 4°C and 9 V/cm. Band fluorescence was detected using a CCD camera system (Fujifilm LAS 3000). For competition experiments, different amounts of nonlabeled wild-type and mutated ds oligos were added to the binding reaction.
Bimolecular Fluorescence Complementation
A cDNA corresponding to the STY1 IGGH domain (amino acids 229 to 292) was PCR amplified using primers IGGHBiFCF and IGGHBiFCR. The PCR product was digested with SalI and BamHI and cloned into vectors pSY735 and pSY736 (Bracha-Drori et al., 2004), creating plasmids p35IGGH and p36IGGH. Plasmids were cotransformed into protoplasts of P. patens (Schaefer et al., 1991) and analyzed as for the GFP assay described above.
PCR Primers
All primers used are listed in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: STY1, At3g51060; STY2, At4g36260; SHI, At5g66350; LRP1, At5g12330; SRS3, At2g21400; SRS4, At2g18120; SRS5, At1g75520; SRS6, At3g54430; SRS7, At1g19790; YUC1, At4g32540; YUC2, At4g13260; YUC3, At1g04610; YUC4, At5g11320; YUC5, At5g43890; YUC6, At5g25620; YUC7, At2g33230; YUC8, At4g28720; YUC9, At1g04180; YUC10, At1g48910; YUC11, At1g21430; YDK1/GH3-2, At4g37390; ERF15, At2g31230; ORA59, At1g06160; ACT2, At5g09810; ORCA3, EU072424; Pp SHI1, AY953419; and C. roseus STY1 homolog, ABL63114.1
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. SHI Interacts with the YUC4 Promoter in Yeast.
Supplemental Figure 2. ABL63114.1 Is a Typical Member of the SHI/STY Family.
Supplemental Figure 3. The Putative STY1 Binding Site ACTCTA(C/A) Can Be Found in Proximal Promoter Regions of ORA59 and YUC1,4,5,8,9.
Supplemental Figure 4. In Vitro Transcription/Translation of Strep-Tagged STY1 Yields a Single Product of Expected Molecular Weight.
Supplemental Figure 5. STY1-Dependent Shift of a YUC4 Promoter Sequence.
Supplemental Figure 6. Acidic Residues in the IGGH Domain of the Physcomitrella patens SHI1 Protein Are Important for Its Function as a Transcriptional Activator.
Supplemental Table 1. PCR Primers.
Supplemental Data Set 1. GCG MSF Alignment of ABL63114.1 and SHI/STY Members from Arabidopsis.
Supplemental Methods and References.
Supplementary Material
Acknowledgments
We thank Gary Wife and Stefan Gunnarsson for scanning electron microscopy facilities, Gun Rönnqvist, Gunilla Swärd, and Agneta Ottosson for Arabidopsis transformation, Pieter Ouwerkerk for the pINT yeast one-hybrid system, and Yunde Zhao for YUC4pro:GUS seeds and plasmid. The technical assistance of Anders Nilsson and Jon Ramsell is gratefully acknowledged.
References
- Abel S., Nguyen M.S., Theologis A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology (Menlo Park, CA: AAAI Press; ), pp. 28–36 [PubMed] [Google Scholar]
- Ballas N., Wong L.M., Theologis A. (1993). Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J. Mol. Biol. 233: 580–596 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Brown R.S. (2005). Zinc finger proteins: Getting a grip on RNA. Curr. Opin. Struct. Biol. 15: 94–98 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Vierstra R.D. (1996). Soluble derivatives of green fluorescent protein (GFP) for use in Arabidopsis thaliana. Weeds World 3: 43–48 [Google Scholar]
- Dobson C.J., Allinson S.L. (2006). The phosphate activity of mammalian polynucleotide kinase takes precedence over its kinase activity in repair of single strand break. Nucleic Acids Res. 34: 2230–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist J., Ronnberg E., Rosenquist S., Blomqvist K., Viitanen L., Salminen T.A., Nylund M., Tuuf J., Mattjus P. (2004). Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein-2. J. Biol. Chem. 279: 53544–53553 [DOI] [PubMed] [Google Scholar]
- Fridborg I., Kuusk S., Moritz T., Sundberg E. (1999). The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg I., Kuusk S., Robertson M., Sundberg E. (2001). The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol. 127: 937–948 [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J., Wisniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Gamsjaeger R., Liew C.K., Loughlin F.E., Crossley M., Mackay J.P. (2007). Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32: 63–70 [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Hall T.M. (2005). Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 15: 367–373 [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75: 805–816 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Ito T., Takahashi N., Shimura Y., Okada K. (1997). A serine/threonine protein kinase gene isolated by an in vivo binding procedure using the Arabidopsis floral homeotic gene product, AGAMOUS. Plant Cell Physiol. 38: 248–258 [DOI] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Klug A. (1999). Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293: 215–218 [DOI] [PubMed] [Google Scholar]
- Kuusk S., Sohlberg J.J., Eklund D.M., Sundberg E. (2006). Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 47: 99–111 [DOI] [PubMed] [Google Scholar]
- Kuusk S., Sohlberg J.J., Long J.A., Fridborg I., Sundberg E. (2002). STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129: 4707–4717 [DOI] [PubMed] [Google Scholar]
- Laity J.H., Lee B.M., Wright P.E. (2001). Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11: 39–46 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Celenza J., Yamada M., Estelle M., Normanly J., Sandberg G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A., Bhalerao R., Casimiro I., Eklöf J., Casero P.J., Bennett M., Sandberg G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews J.M., Sunde M. (2002). Zinc fingers–Folds for many occasions. IUBMB Life 54: 351–355 [DOI] [PubMed] [Google Scholar]
- Mattsson J., Sung Z.R., Berleth T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126: 2979–2991 [DOI] [PubMed] [Google Scholar]
- Meijer A.H., Ouwerkerk P.B., Hoge J.H. (1998). Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast 14: 1407–1415 [DOI] [PubMed] [Google Scholar]
- Mitchell P.J., Tijan R. (1989). Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245: 371–378 [DOI] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Feldman L.J., Zambryski C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888 [DOI] [PubMed] [Google Scholar]
- Ouyang J., Shao X., Li J. (2000). Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 24: 327–333 [DOI] [PubMed] [Google Scholar]
- Parraga G., Horvath S.J., Eisen A., Taylor W.E., Hood L., Young E.T., Klevit R.E. (1988). Zinc-dependent structure of a single-finger domain of yeast ADR1. Science 241: 1489–1492 [DOI] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C.M., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Mandel T., Kuhlemeier C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern an polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer D., Zryd J.P., Knight C.D., Cove D.J. (1991). Stable transformation of the moss Physcomitrella patens. Mol. Gen. Genet. 226: 418–424 [DOI] [PubMed] [Google Scholar]
- Silverstein K.A., Graham M.A., Paape T.D., VandenBosch K.A. (2005). Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol. 138: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg J.J., Myrenås M., Kuusk S., Lagerkrantz U., Kowalczyk M., Sandberg G., Sundberg E. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 47: 112–123 [DOI] [PubMed] [Google Scholar]
- Ståldal V., Sohlberg J.J., Eklund D.M., Ljung K., Sundberg E. (2008). Auxin can act independently of CRC, LUG, SEU, SPT and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytol. 180: 798–808 [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Braybrook S.A., Paula S.L., Kwong L.W., Meuser J., Pelletier J., Hsieh T.-F., Fischer R.L., Goldberg R.B., Harada J.J. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: Implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 105: 3151–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström J.F., Nakayama N., Glimelius K., Irish V.F. (2006). Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J. 46: 593–600 [DOI] [PubMed] [Google Scholar]
- Triezenberg S.J. (1995). Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5: 190–196 [DOI] [PubMed] [Google Scholar]
- van der Fits L., Memelink J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289: 295–297 [DOI] [PubMed] [Google Scholar]
- vom Endt D., Soares e Silva M., Kijne J.W., Pasquali G., Memelink J. (2007). Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 144: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went F.W. (1974). Reflections and speculations. Annu. Rev. Plant Physiol. 25: 1–26 [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.J., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenases in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hull A.K., Gupta N.R., Goss K.A., Alonso J., Ecker J.R., Normanly J., Chory J., Celenza J.L. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.