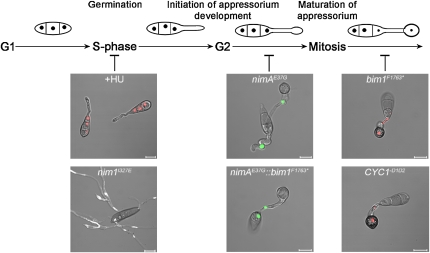
This study uses a combination of live-cell imaging and gene functional analysis to explore the relationship between the cell cycle and appressorium-mediated plant infection. It finds that there are as many as three distinct cell cycle checkpoints in the establishment of plant disease by M. oryzae.
Abstract
To gain entry to plants, many pathogenic fungi develop specialized infection structures called appressoria. Here, we demonstrate that appressorium morphogenesis in the rice blast fungus Magnaporthe oryzae is tightly regulated by the cell cycle. Shortly after a fungus spore lands on the rice (Oryza sativa) leaf surface, a single round of mitosis always occurs in the germ tube. We found that initiation of infection structure development is regulated by a DNA replication-dependent checkpoint. Genetic intervention in DNA synthesis, by conditional mutation of the Never-in-Mitosis 1 gene, prevented germ tubes from developing nascent infection structures. Cellular differentiation of appressoria, however, required entry into mitosis because nimA temperature-sensitive mutants, blocked at mitotic entry, were unable to develop functional appressoria. Arresting the cell cycle after mitotic entry, by conditional inactivation of the Blocked-in-Mitosis 1 gene or expression of stabilized cyclinB-encoding alleles, did not impair appressorium differentiation, but instead prevented these cells from invading plant tissue. When considered together, these data suggest that appressorium-mediated plant infection is coordinated by three distinct cell cycle checkpoints that are necessary for establishment of plant disease.
INTRODUCTION
Many of the most important plant diseases are caused by fungal pathogens that form specialized infection cells to breach the leaf surface. These infection structures are an especially common feature of cereal pathogens, including the causal agents of rust diseases, powdery mildews, and the devastating rice blast disease (Dean, 1997; Tucker and Talbot, 2001). The blast fungus Magnaporthe oryzae forms dome-shaped appressoria and uses a turgor-driven process to rupture the cuticle and send a narrow penetration hypha into an underlying plant epidermal cell (for reviews, see Ebbole 2007; Wilson and Talbot, 2009). The appressorium is a differentiated fungal cell with a specialized cell wall rich in chitin and an inner layer lined with melanin (Henson et al., 1999). The melanin layer is essential to M. oryzae appressoria because it reduces the porosity of the cell wall, which allows generation of osmotic pressure by accumulation of glycerol and the application of mechanical force at the leaf surface (Howard et al., 1991; De Jong et al., 1997). Interfering with initial plant infection offers one of the best means of controlling the most economically significant plant diseases, and an understanding of fungal infection-related development is therefore essential.
Formation of appressoria involves a switch from polarized hyphal growth to isotropic expansion of the germ tube tip and cellular differentiation of the appressorium. This is followed by turgor generation, reorientation of the axis of polarity, and resumption of polarized fungal growth at the base of the infection cell, rupturing the host cuticle. Following entry into the leaf, M. oryzae undergoes a further developmental switch to form specialized invasive hyphae, which secrete effector proteins and sequester nutrients from living host cells (Kankanala et al., 2007).
To initiate appressorium development, the fungus responds to a set of inductive cues, including surface hardness, hydrophobicity, cuticular wax, and the absence of exogenous nutrients (Ebbole, 2007). How each of these signals is perceived is not clear, but the process involves at least one G-protein–coupled receptor, Pth11, and cognate G-α and G-βγ-subunit proteins (reviewed in Wilson and Talbot, 2009). Adenylate cyclase-mediated generation of cAMP and protein kinase A are necessary for efficient appressorium morphogenesis (Choi and Dean, 1997; Xu et al., 1997; Adachi and Hamer, 1998; Thines et al., 2000), and a mitogen-activated protein kinase pathway composed of the Mst11, Mst7, and Pmk1 protein kinases is essential for appressorium formation and subsequent invasive growth (Zhao et al., 2005). However, how these signaling pathways are connected to the developmental biology of the fungus as it undergoes the morphogenetic transitions to form differentiated infection structures is unclear.
In this study, we set out to explore the relationship between cell cycle regulation and infection structure development in the rice blast fungus. The cell cycle is pivotal to cellular differentiation in multicellular eukaryotes, which must synchronize cell division to form specific tissues and organs effectively (Kipreos, 2005; Théry and Bornens, 2006; Cools and De Veylder, 2009). The switch between isotropic and polarized growth in fungi is also intimately linked to cell cycle regulation in yeasts and filamentous and dimorphic fungal species (Momany, 2002; Whiteway and Bachewich, 2007; Steinberg and Perez-Martin, 2008). We therefore hypothesized that cell cycle regulation would be likely to provide control points for infection structure development by M. oryzae. Previously, we showed that mitosis is necessary for appressorium formation and infection-associated autophagy during plant infection (Veneault-Fourrey et al., 2006), providing the first indication of cell cycle regulation of this process. In this study, we set out to use a combination of live-cell imaging and gene functional analysis to determine how such cell cycle regulation of infection-related development might operate and, in particular, to define the individual cell cycle checkpoints necessary for appressorium morphogenesis and subsequent plant infection. We report here that the key step in initiating infection structure development in M. oryzae is entry into S-phase, which causes formation of an incipient infection cell at the germ tube tip. Cellular differentiation of this terminal swelling requires mitosis to have occurred, with mitotic entry marking the regulatory commitment point for appressorium maturation. We further present evidence that subsequent progression through mitosis is dispensable for appressorium maturation but is essential for plant infection. When considered together, this suggests that there are three pivotal cell cycle checkpoints that operate in the rice blast fungus to allow it to gain entry to its host plant.
RESULTS
Mitosis Always Precedes Appressorium Development in M. oryzae
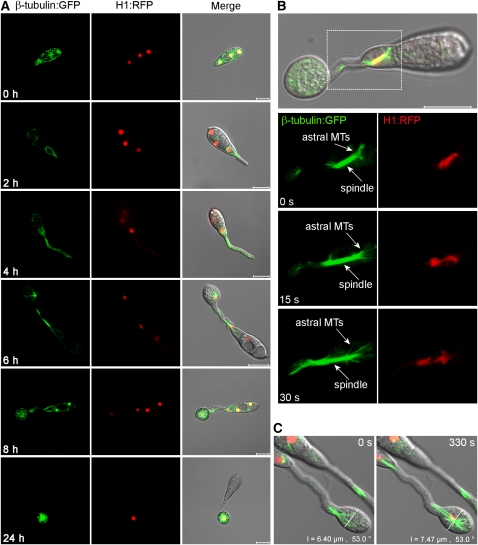
To investigate nuclear division and migration during appressorium formation, we performed live-cell imaging of M. oryzae expressing a histone H1-enhanced red fluorescent protein (RFP; tdTomato) fusion (Shaner et al., 2004) and β-tubulin-synthetic green fluorescent protein (sGFP) fusion, as shown in Figure 1 and Supplemental Movie 1 online. Following spore germination, a polarized germ tube was formed and microtubules aligned parallel to its longitudinal axis (Figure 1A). Mitosis occurred 4 to 6 h later with an influx of β-tubulin:sGFP into the nucleus as the spindle formed (see Supplemental Movie 1 online; Figure 1B). Astral microtubules were evident during spindle elongation and rapid separation of chromosomes (Figure 1B). Following mitosis, one daughter nucleus migrated to the swollen germ tube tip, while the other returned to the original conidial cell. The germ tube tip swelled prior to mitosis and continued to develop, increasing in diameter upon receipt of the daughter nucleus (Figure 1C). The three nuclei that remained in the spore were then broken down by autophagic cell death (Veneault-Fourrey et al., 2006) with a single nucleus remaining in the appressorium as shown in Figure 1A.
Figure 1.
Live-Cell Imaging of Microtubule Dynamics and Nuclear Division in M. oryzae during Germination and Appressorium Development.
(A) Time series of micrographs showing nuclear division during appressorium development in M. oryzae. The grg(p):H1:RFP and ccg1(p):bm1:sGFP gene fusion vectors were introduced into the M. oryzae wild-type strain Guy-11. Nuclei (red) and microtubules (green) were observed during conidial germination and appressorium development in M. oryzae.
(B) Astral microtubules were observed during mitotic spindle elongation and the concurrent rapid separation of chromosomes. MTs, microtubules.
(C) Continued expansion in diameter of the incipient appressorium during mitotic division. Bars = 10 μm.
Initiation of Appressorium Morphogenesis Is Regulated at S-Phase
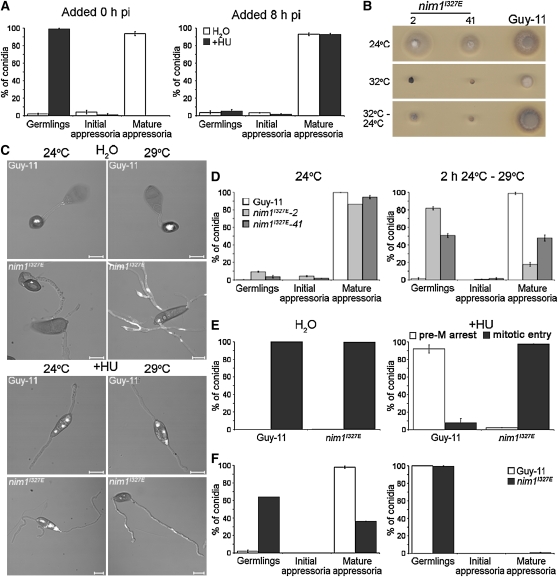
To investigate the importance of cell cycle progression to appressorium development, we applied the DNA synthesis inhibitor hydroxyurea (HU) to conidia of M. oryzae. Following addition of HU, germinating conidia arrested predominantly with undifferentiated germ tubes, rather than incipient appressoria, as shown in Figures 2A and 2C. This is in contrast with perturbation of the cell cycle in M. oryzae with either benomyl or a nimAE37G mutation, both of which cause a late G2 arrest but allow the formation of swollen germ tube tips (Veneault-Fourrey et al., 2006). We reasoned that DNA replication (S-phase) might be necessary for formation of the terminal swelling that later develops into an appressorium. To test this idea, we generated a mutant that was affected in DNA replication but that would still allow mitotic entry. In the filamentous fungus Aspergillus nidulans, the temperature-sensitive nimO18 mutant (for never in mitosis) is unable to replicate DNA, although aberrant segregation of chromatin can occur, leading to cells advancing into mitosis with unreplicated DNA at the restrictive temperature (James et al., 1999). NimO is functionally related to Saccharomyces cerevisiae Dbf4p, the regulatory subunit of the Cdc7p-Dbf4p kinase complex, which is required for Cdc7p kinase activity and initiation of DNA replication (Jackson et al., 1993). We identified an M. oryzae gene, NIM1, which putatively encodes a protein of 651 amino acids with 31% sequence identity to A. nidulans NimO (see Supplemental Figure 1 online). Analysis of the Nim1 sequence revealed a BRCT domain (residues 116 to 166 [e-value 0.32]) that is characteristic of cell cycle regulators involved in the DNA damage checkpoint (Bork et al., 1997), the Dbf4 M motif (residues 241 to 369 [e-value 9.1e-45]), and a zinc finger (residues 582 to 630 [e-value 2e-26]), all of which are conserved in Dbf4-related proteins (Ogino et al., 2001).
Figure 2.
DNA Replication in M. oryzae Is Necessary for Initiation of Appressorium Development.
(A) The effect of chemical inhibition of DNA replication on appressorium development in M. oryzae. Water (white bars) or the DNA synthesis inhibitor HU (+HU, black bars) was added (200 mM) to conidial suspensions of the H1:RFP strain, 0 or 8 h postinoculation (hpi). The percentage of conidia developing undifferentiated germ tubes (germlings), swollen germ tube tips (initial appressoria), and differentiated infection cells (mature appressoria) was recorded after 8 h.
(B) Temperature sensitivity of the nim1I327E mutant of M. oryzae; two putative nim1I327E transformants (2 and 41) and Guy-11 were incubated at a permissive temperature of 24°C or a restrictive temperature of 32°C for 4 d. Restoration of hyphal growth was assessed by incubation for a further 3 d at 24°C.
(C) Representative images of the nim1I327E mutant during appressorium development, in the presence (bottom panels) or absence (top panels) of HU; nim1I327E and Guy-11 germinating conidia were fixed and stained with the DNA-specific stain 4',6-diamidino-2-phenylindole 24 h postinoculation, following incubation at 24 or 29°C. Bars = 10 μm.
(D) Bar charts to show frequency of appressorium development in M. oryzae nim1I327E. Conidial suspensions of nim1I327E transformants 2 and 41 were incubated on hydrophobic surfaces at 24 or 29°C. The percentage of conidia developing undifferentiated germ tubes (germlings), swollen germ tube tips (initial appressoria), or differentiated infection cells (mature appressoria) was recorded after 24 h.
(E) Bar charts to show the frequency of germinating conidia displaying a premitotic arrest (pre-M arrest; white bars) or the ability to progress into mitosis uninhibited (mitotic entry; black bars).
(F) HU was added to nim1I327E and Guy-11 conidia at 0 h and nuclear division and appressorium development assessed 24 h later following incubation at 24°C (left bar chart) or 29°C (right bar chart). All graphs represent two biological and three technical replicate observations of 100 germinating conidia. Error bars are 1 se.
We generated a temperature-sensitive mutant of NIM1 by making a nim1I327E allele, identical to the mutation found in the thermo-labile nimO18 mutant of A. nidulans (James et al., 1999), and introduced this into the M. oryzae H1:RFP-expressing strain of Guy11 by targeted allelic replacement. Transformants were selected that showed a severe hyphal growth defect at the restrictive temperature of 32°C, and growth was only partially restored by subsequent incubation at 24°C (Figure 2B). Following DNA gel blot analysis, transformants with a single targeted replacement of the nim1I327E allele were identified, and two transformants (2 and 41) were selected each containing the I327E substitution.
We determined the frequency of nuclear division and appressorium development in the nim1I327E mutants and the isogenic H1:RFP strain. Appressorium development was significantly reduced in nim1I327E mutants (t test; P < 0.001) compared with the wild type, and conidia instead formed undifferentiated germlings when incubated at the semirestrictive temperature (Figures 2C and 2D). However, we still observed nuclear division in nim1I327E strains (Figure 2C), consistent with the reported phenotype of A. nidulans nimO18 mutants (James et al., 1999). The M. oryzae nim1I327E mutant was able to initiate numerous rounds of aberrant nuclear division, and we reasoned that this was likely to be due to the partial inactivation of the Nim1 protein at 29°C (higher temperatures affected appressorium morphogenesis, as reported previously [Veneault-Fourrey et al., 2006]). We interpreted the nim1I327E phenotype as being consistent with segregation of unreplicated, or partially replicated, chromatin. To test this idea, we added the DNA synthesis inhibitor HU to germinating nim1I327E conidia. We consistently observed a postmitotic growth arrest phenotype in which nuclei still divided in nim1I327E mutants exposed to HU. By contrast, the wild-type H1:RFP strain always arrested growth prior to mitosis (Figure 2E). This strongly indicates that nim1I327E mutants are capable of circumventing the DNA replication checkpoint, thereby enabling nuclear division to take place in the absence of DNA replication (James et al., 1999). High-resolution imaging of the chromatin status of nim1I327E cells in the presence of HU was consistent with this conclusion (see Supplemental Figure 2 online). Importantly, nim1I327E mutants were unable to initiate appressorium development or conidial autophagic cell death in the presence of HU (Figure 2F), demonstrating that successful execution of DNA replication is necessary for initiation of appressorium morphogenesis by the rice blast fungus.
Mitotic Entry Is Both Necessary and Sufficient for Appressorium Development
Previously, we showed that blocking the cell cycle at mitotic entry (G2-M) was sufficient to prevent appressorium development by M. oryzae (Veneault-Fourrey et al., 2006). This conclusion was based on the fact that temperature-sensitive nimAE37G mutants were unable to make mature appressoria at the restrictive temperature (Veneault-Fourrey et al., 2006). M. oryzae NimA encodes a functional homolog of the A. nidulans NimA protein kinase, which is essential for mitosis (Osmani et al., 1991). We noted that nimAE37G mutants were able to form swollen germ tube tips but could not develop mature, melanized infection cells, preventing nimAE37G mutants from causing plant disease (see Supplemental Figure 3A online). We therefore set out to determine at which stage M. oryzae germlings became committed to appressorium differentiation by attempting to block mitosis at a point subsequent to mitotic entry.
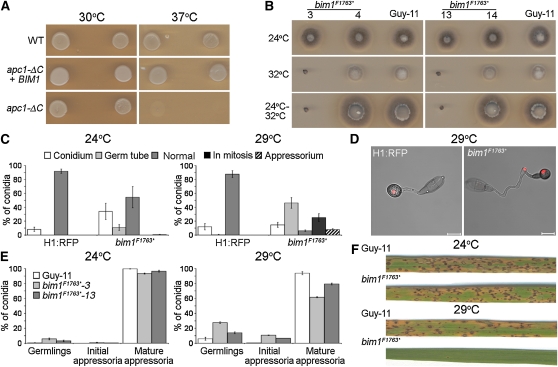
In A. nidulans, the blocked in mitosis bimE7 mutant has been described, which arrests cells in mitosis in a pre-anaphase state (Osmani et al., 1988). The A. nidulans BimE protein is homologous to the large subunit of the S. cerevisiae anaphase-promoting complex (APC; Peters et al., 1996). We identified an M. oryzae gene, BIM1, from the M. oryzae genome database (Dean et al., 2005), which showed 35% sequence identity with A. nidulans BimE. Sequence analysis of Bim1 revealed three proteasome/cyclosome repeat regions (residues 1140 to 1174 [e-value 0.78], residues 1185 to 1223 [e-value 0.37], and residues 1328 to 1362 [e-value 0.3]), consistent with the protein acting as a component of the multisubunit APC in M. oryzae (see Supplemental Figure 4 online). To test this relationship, we introduced BIM1 into the S. cerevisiae thermosensitive mutant, apc1- ΔC (Winzeler et al., 1999). Expression of M. oryzae BIM1 allowed the mutant to grow at the restrictive temperature, consistent with M. oryzae Bim1 protein acting as a component of the APC (Figure 3A). We then sequenced the previously uncharacterized A. nidulans bimE7 allele, which revealed a point mutation altering a Gln residue to a premature stop codon at position 1966. A corresponding bim1F1763* allele was constructed, introduced into M. oryzae by homologous recombination, and transformants selected that were unable to grow at 32°C (Figure 3B). Following DNA gel blot analysis, the allelic replacement of BIM1 was confirmed by amplification of the locus and DNA sequencing (data not shown).
Figure 3.
The M. oryzae bim1F1763* Mutant Forms Appressoria but Is Unable to Cause Plant Disease.
(A) M. oryzae BIM1 is a functional homolog of S. cerevisiae APC1. The M. oryzae BIM1 gene coding sequence under the control of the GAL1 promoter was introduced into the S. cerevisiae apc1- ΔC mutant. Induced expression of BIM1 restored growth of apc1- ΔC at 37°C.
(B) Temperature sensitivity of the bim1F1763* mutant of M. oryzae. Hyphal growth analysis of bim1F1763* mutant transformants (3 and 13), ectopic transformants (4 and 14), and Guy-11 after incubation at 24 or 32°C for 4 d. Restoration of hyphal growth was assessed by incubation for a further 3 d at 24°C.
(C) Bar charts to illustrate nuclear distribution during appressorium development in bim1F1763*. Nuclear position was recorded following appressorium development of the bim1F1763* mutant and the H1:RFP wild-type strain after 24 h at 29°C. Nuclei were scored as being both retained in the spore (Conidium), arrested in the germ tube (Germ tube), blocked in mitosis (Mitosis), both migrating to the developing infection cell (Appressorium), or progressing normally through mitosis, resulting in a single nucleus in the appressorium and degradation of the remaining three nuclei (Normal).
(D) Representative images to show nuclear distribution during appressorium formation of the bim1F1763* mutant and H1:RFP strain, after 24 h at 29°C.
(E) Quantitative analysis of appressorium development of the bim1F1763* mutant after incubation of germinating conidia at 24 or 29°C for 24 h. All graphs represent two biological and three technical replicate observations of 100 germinating conidia. Error bars are 1 se.
(F) M. oryzae bim1F1763* mutants are unable to cause rice blast disease. Leaves from rice cultivar CO-39 inoculated with 5 × 104 spores mL−1 of bim1F1763* and Guy 11. Rice plants were incubated initially for 24 h at either 24 or 29°C. All plants were then transferred to 24°C and leaves were harvested 5 d later.
To test the effect of the bim1F1763* mutation on appressorium development, we selected two independent transformants and first quantified their abilities to complete mitosis, as shown in Figure 3C. The wild-type H1:RFP strain progressed through mitosis normally at both temperatures, resulting in degradation of the three conidial nuclei and retention of a single nucleus in the appressorium (Figures 3C and 3D). By contrast, bim1F1763* mutants arrested in mitosis, or displayed a range of nuclear migration defects, consistent with impairment of mitosis (Figures 3C and 3D; Osmani et al., 1988). The phenotypes observed suggest that Bim1 acts as a regulator of mitotic progression, consistent with its role as a component of the APC, which catalyzes ubiquitination of anaphase-specific inhibitors (Cohen-Fix et al., 1996), targeting them for proteasome degradation and thereby facilitating mitotic progression. Transferring germinating conidia to 29°C at later times had a less dramatic effect on nuclear division and migration because the nonsynchronously germinating conidia had increasingly passed through mitosis before transfer to the semirestrictive temperature (for a full quantitative analysis, see Supplemental Figure 3B online).
Importantly, bim1F1763* mutants were consistently able to develop mature, melanized appressoria even at the restrictive temperature (Figure 3E), indicating that arrest of the cell cycle after mitotic entry has no effect on appressorium morphogenesis. We conclude that mitotic entry is therefore both necessary and sufficient for the induction of appressorium differentiation in M. oryzae.
To analyze further this phenotype, we decided to carry out epistasis analysis of the bim1F1763* and nimAE37G mutations by constructing a double mutant. In A. nidulans when the G2-specific nimA5 and bimE7 mutations are combined, nimA5 bimE7 double mutant cells enter mitosis and arrest in a pre-anaphase state, a phenotype indistinguishable from that of the bimE7 single mutant (Osmani et al., 1991). We quantified nuclear division in a M. oryzae nimAE37G bim1F1763* double mutant and observed that nuclear division always arrested at mitotic entry, as observed in nimAE37G single mutants (see Supplemental Figure 5 online). These results demonstrate that although the A. nidulans NimA and BimE proteins appear functionally conserved in M. oryzae, the epistatic relationship between them is clearly distinct.
Appressoria formed normally in bim1F1763* mutants, and we were therefore interested to find out whether these were functional infection cells capable of initiating rice blast disease. We therefore infected the rice (Oryza sativa) seedlings of cultivar CO-39 at permissive and semipermissive temperatures. We found that disease symptoms caused by bim1F1763* mutants were significantly reduced at the semirestrictive temperature of 29°C (Figure 3F; two-sample ttest assuming equal variances, t = 9.27, df = 10, P < 0.05), although no effect was observed at the permissive temperature of 24°C when compared with the wild-type strain, Guy 11 (Figure 3F; two-sample ttest assuming equal variances, t = 0.94, df = 16, P > 0.05). Cytological analysis on rice or sterile onion epidermal layers indicated that appressoria of bim1F1763* mutants were unable to elaborate penetration hyphae at the semipermissive temperature of 29°C. When considered together, our results suggest that mitotic entry is sufficient for appressorium morphogenesis but that plant infection depends on completion of mitosis during appressorium morphogenesis by the rice blast fungus.
Expression of Stabilized Forms of the M. oryzae B-Type Cyclin Genes Prevents Mitotic Exit
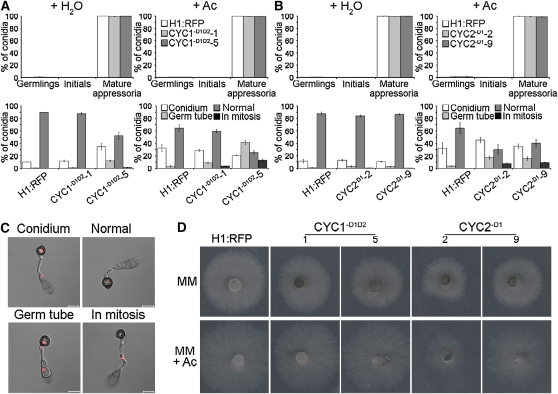
To test our hypothesis that mitotic entry was sufficient to enable appressorium development by M. oryzae, we decided to carry out an alternative strategy to block the cell cycle prior to mitotic exit. To do this, we identified two putative M. oryzae cyclin B-encoding genes, CYC1 and CYC2, which showed similarity to cyclin B homologs from other fungi (see Supplemental Figure 6 online). We analyzed the predicted amino acid sequences of CYC1 and CYC2 to identify destruction box consensus sequences (RXXLXXXXN) essential for targeting cyclin B for ubiquitin-mediated degradation during mitotic exit (reviewed in Hochstrasser, 1996). We reasoned that by removing these sequences and expressing the resulting alleles in M. oryzae, we would be able to prevent mitotic exit, thus providing an alternative means of establishing the cell cycle commitment point for appressorium development.
We identified and removed the destruction box consensus sequences from CYC1 and CYC2, to create the CYC1-D1D2 and CYC2-D2 alleles, respectively (see Supplemental Figure 6 online). The modified dominant alleles were then placed under the ICL1 promoter from the isocitrate lyase-encoding gene of M. oryzae (Wang et al., 2003), to enable induction of gene expression by acetate, and introduced into the fungus. We identified transformants carrying single copies of the CYC1-D1D2 and CYC2-D2 alleles, respectively, by DNA gel blot analysis and DNA sequencing. We then quantified the ability of these strains to undergo nuclear division in the presence or absence of acetate, as shown in Figure 4. The wild-type H1:RFP strain progressed through mitosis normally and, following autophagy of the spore, contained a single nucleus in each appressorium (Figures 4B and 4C). By contrast, in the presence of acetate, we found that nuclear division was disrupted during appressorium development in both the CYC1-D1D2 and CYC2-D1 strains, which also expressed H1:RFP, and they displayed either a complete block in mitosis or a range of nuclear distribution phenotypes in which nuclei failed to migrate to the appressorium or conidium or migrated together. The impairment of mitotic exit was most pronounced in the CYC1-D1D2-5 mutant, in which 41% of germinated conidia arrested nuclear migration with two daughter nuclei remaining very close together in the germ tube (Figure 4B). By contrast, we observed hyphal growth defects of CYC2-D1 strains when incubated in the presence of acetate (Figure 4D) compared with the wild type or CYC1-D1D2 strains. This indicates that CYC2 may be more important during vegetative hyphal growth, suggesting stage-specific functions for the two cyclins in M. oryzae.
Figure 4.
Expression of Stabilized Cyclin B Proteins in M. oryzae Inhibits Mitotic Exit but Does Not Prevent Appressorium Morphogenesis.
(A) Bar charts to show frequency of appressorium development (top panel) and nuclear distribution patterns (bottom panel) in two independent CYC1-D1D2 transformants (1 and 5). Appressorium development and nuclear distribution during appressorium morphogenesis were assessed in the presence or absence of 50 mM sodium acetate (Ac) 24 h postinoculation to induce expression of stabilized cyclins. For key, see legend to Figure 3.
(B) Bar charts to show frequency of appressorium development and nuclear distribution patterns in two CYC2-D1 transformants (2 and 9). Germinating conidia were incubated to allow appressorium development in the presence or absence of 50 mM Ac and observed 24 h later. All graphs represent two biological and three technical replicate observations of 100 germinating conidia. Error bars are 1 se.
(C) Representative images of CYC1-D1D2, CYC2-D1, and H1:RFP strains in the presence or absence of Ac during appressorium morphogenesis. Nuclei were scored as being both retained in the spore (Conidium), arrested in the germ tube (Germ tube), blocked in mitosis (in mitosis),or progressing normally through mitosis, resulting in a single nucleus in the appressorium and degradation of the remaining three nuclei (Normal).
(D) Phenotypic analysis of CYC1-D1D2 and CYC2-D1 expression on hyphal growth in M. oryzae. Plugs of mycelium (5 mm diameter) from ICL1(p):CYC1-D1D2, ICL1(p):CYC2-D1, and H1:RFP were incubated in the presence (MM + Ac) or absence (MM) of 50 mM Ac. Hyphal growth was assessed 4 d later. Bars = 10 μm.
The ability of CYC1-D1D2 and CYC2-D1 strains to develop appressoria was analyzed in the presence and absence of acetate. Appressoria developed normally in both CYC1-D1D2 and CYC2-D1 strains in the presence of acetate (Figure 4A), suggesting that mitotic exit is not necessary for appressorium differentiation and providing further evidence that mitotic entry is the commitment point for appressorium maturation by the rice blast fungus.
DISCUSSION
In this study, we set out to explore the relationship between cell cycle progression and infection structure formation by a plant pathogenic fungus. Previous evidence had suggested that mitosis was needed for appressorium formation by M. oryzae (Veneault-Fourrey et al., 2006) but did not reveal which stage of the cell cycle governed initiation and differentiation of infection structure formation. We reasoned that by constructing conditional mutants, impaired at each stage of the cell cycle, and combining these with quantitative cytological analysis of fluorescently tagged strains of M. oryzae, we would be able to determine the most likely regulation points for infection-related development.
S-Phase Regulation of Infection Structure
Our first conclusion is that entry into S-phase is critical for regulating initiation of appressorium development. The evidence to support this idea is based on analysis of the nim1I327E mutant, which is impaired in DNA replication but still undergoes nuclear division. Even when exposed to HU, which completely blocked DNA replication, nim1I327E mutants showed a postmitotic arrest phenotype, indicating that the mutation circumvents the DNA replication checkpoint, enabling nuclear division to take place in the absence of DNA replication. Because these mutants were unable to initiate appressorium development, the regulation point for initiating appressorium development must occur prior to mitosis and depend on a checkpoint operating at S-phase (Figure 5). Consistent with this, time-lapse microscopy of mitosis showed that the swollen germ tube tip always developed prior to mitosis, providing the destination for one of the daughter nuclei, which rapidly migrates into the germ tube apex (see Supplemental Movie 1 online).
Figure 5.
Model for the Regulation of Infection Structure Development by Cell Cycle Progression in M. oryzae.
Entry into S-phase leads to initiation of appressorium development in the rice blast fungus. Arresting S-phase with HU or nim1I327E mutation prevents germ tubes from swelling and initiating appressorium formation. Entry into mitosis is both necessary and sufficient for appressoria to fully differentiate, as demonstrated by the phenotypes of the nimAE37G and nimAE37Gbim1F1763* mutants. Mitotic progression and exit are not necessary for appressoria to mature but may regulate repolarization and subsequent disease development. Bars = 10 μm.
The inductive cues for appressorium development, such as surface hydrophobicity, cuticular wax, and the absence of nutrients, must therefore be perceived at a very early stage following spore germination on the rice leaf and stimulate the nucleus to pass rapidly from G1-S, as a necessary prerequisite for appressorium formation. By contrast, previous work has shown that when M. oryzae spores are incubated under noninductive conditions, they undergo mitosis much later (after 6 to 8 h), and this does not lead to terminal swelling of the germ tube or to degeneration of the nuclei within the spore (Veneault-Fourrey et al., 2006). Therefore, the inductive properties of the rice leaf cause a fundamental change in cell cycle regulation that is necessary for appressorium formation. How such an S-phase checkpoint operates is not clear, but one possibility is through the action of the Pmk1 mitogen-activated protein kinase (MAPK) cascade, which is essential for appressorium development (Zhao et al., 2005). Δpmk1 mutants do not form appressoria and instead develop undifferentiated germ tubes on rice leaves. Interestingly, successive rounds of mitosis occur in germ tubes of Δpmk1 mutants, suggesting that the MAPK may be involved in regulating S-phase and initiation of appressorium formation. Pmk1 is functionally related to the Fus3/Kss1 MAPKs in yeast, which are involved in the pheromone signaling pathway and also modulate the cell cycle through the action of Far1, which induces a cell cycle arrest that is required for yeast mating to occur (McKinney and Cross, 1995). The Pmk1 MAPK module consists of three protein kinases, Mst11, Mst7, and Pmk1, held together by a scaffold protein Mst50, which interacts with Mst11 and Mst7 (Park et al., 2006). Mst50 also interacts with the Rho GTPase, Cdc42, an essential polarity marker, providing evidence that the pathway regulates the switch between polarized and isotropic growth that is necessary for appressorium formation. Our results suggest that this process is likely to be integrated with cell cycle regulation. Cyclic AMP signaling has also been shown to be essential for the initiation of appressorium development in M. oryzae (Choi and Dean, 1997; Adachi and Hamer, 1998) and may also operate upstream of the cell cycle regulation points identified in this study. Consistent with this idea, we found that addition of cAMP (up to 10 mM) to germinating conidia on hydrophobic plastic surfaces did not rescue the phenoytpe of nim1I327E mutants or the ability of HU to suppress appressorium formation (data not shown).
Mitosis Is Necessary and Sufficient for Appressorium Maturation in M. oryzae
The second major finding from this study is that entry into mitosis represents a commitment point for appressorium morphogenesis by M. oryzae. Previous analysis of the nimAE37G mutant showed that blocking the cell cycle prior to mitosis prevented appressoria from developing properly and becoming fully differentiated, melanized infection structures (Veneault-Fourrey et al., 2006). This study shows that blocking the cell cycle at later stages, such as anaphase or mitotic exit, has no effect on appressorium formation; infection structures form normally and become melanized. This strongly suggests that the checkpoint regulating cellular differentiation operates at the G2-M boundary, marking the point at which M. oryzae becomes committed to making an appressorium.
Two lines of evidence support this conclusion. First, the bim1F1763* mutation caused a severe impairment in mitosis at a semipermissive temperature, which was evident by our observation of failure of nuclei to separate correctly or undergo nuclear migration. Strikingly, we observed that in spite of a clear mitotic block in bim1F1763* mutants, appressoria formed normally, consistent with mitotic entry being sufficient for their development.
Our second test of this idea was to block mitotic exit, which we performed by regulated expression of stabilized forms of the two M. oryzae cyclin B genes, CYC1 and CYC2, lacking their inherent destruction box consensus sequences. We found that CYC1-D1D2 and CYC2-D1 strains both displayed mitotic exit phenotypes, which is consistent with stabilized expression of truncated cyclin B genes in U. maydis (Garcia-Muse et al., 2003). There were, however, stage-specific differences in the phenotypes of the mutants, with CYC1-D1D2 strains showing more severe impairment in mitotic exit during infection-related development and CYC2-D1 strains severely affecting vegetative growth. In the human pathogenic fungus Candida albicans, the Clb2 cyclin has been shown to be essential for vegetative growth, while Clb4 negatively regulates pseudohyphal growth, consistent with differential expression of B-type cyclins being important in the regulation of polarized hyphal growth (Bensen et al., 2005). Similarly the U. maydis B-type cyclins fulfill distinct developmental roles, with Clb2 acting as a negative regulator of budding, independent of Clb1 (Garcia-Muse et al., 2003). Impairment of mitotic exit in CYC1-D1D2 and CYC2-D1 strains did not, however, affect appressorium formation in M. oryzae, providing independent support for the hypothesis that entry into mitosis is sufficient for appressorium formation.
Our final, and more tentative, conclusion is that completion of mitosis during appressorium morphogenesis is an essential prerequisite to plant infection. This is based on the fact that although bim1F1763* mutants form appressoria normally, they are unable to cause rice blast disease. This suggests that completion of mitosis is necessary for repolarization of the penetration hypha at the base of the appressorium to bring about cuticle rupture. We propose that the nucleus within the appressorium has to progress to G1 phase to allow the infection structure to be functionally competent. What we cannot tell at this time is whether a further round of DNA replication is also necessary to allow repolarization of the appressorium, with the nucleus passing into G2, prior to penetration peg development. Analysis of conidial germination, however, suggests that germ tube emergence occurs while the nucleus is still in G1 and progression through to G2 only occurs after polar growth has been established (Veneault-Fourrey et al., 2006). If the same holds true for emergence of the penetration peg at the base of the appressorium, then this would suggest that repolarization of the appressorium is also a G1-regulated event and, therefore, logically requires the previous mitosis to have been completed, although this will require further investigation.
In summary, this study has identified an interdependent relationship between the morphological transitions associated with plant infection and cell cycle progression exhibited by M. oryzae (presented in Figure 5). Initiation of appressorium morphogenesis is dependent on execution of DNA replication. Mitotic entry is both necessary and sufficient to initiate appressorium maturation in M. oryzae, and exit from mitosis appears to be required for plant infection.
METHODS
Fungal Strains, Growth Conditions, and Infection Structure Development Assays
All isolates of Magnaporthe oryzae (Couch and Kohn, 2002; formerly M. grisea) used in this study were stored in the laboratory of N.J.T. See Supplemental Table 1 online for strains generated in this work. Growth and maintenance of M. oryzae, media composition, nucleic acid extraction, transformation, and plant infection assays were all as described previously (Talbot et al., 1993). Conidial germination and development of appressoria were monitored over time on hydrophobic borosilicate glass cover slips (Fisher Scientific) using a method adapted from Hamer et al. (1988). Conidial suspensions at 5 × 104 conidia mL−1 were inoculated onto cover slips, incubated at 24°C, and the frequency of conidial germination and appressorium development recorded 24 h later, unless otherwise stated.
Live-Cell Imaging of Nuclei and Cytoskeletal Components of M. oryzae
Details of the construction of the H1:RFP (tdTomato) and β-tubulin:sGFP gene fusion vectors are given in Supplemental Methods online. Constructs were introduced into the M. oryzae wild-type strain Guy-11 (Leung et al., 1988) and transformants selected by DNA gel blots. At least two independent transformants were investigated for all experiments and were demonstrated to be fully pathogenic and grow normally (see Supplemental Figure 7 online). All images of conidial germination and appressorium development were recorded using a Zeiss LSM510 Meta confocal laser scanning microscope system. Slides were prepared for processing by sealing cover slip appressorium preparations to a slide with petroleum jelly (Vaseline; Unilever). Blue diode (405 nm), argon (458, 477, 488, and 504 nm), and helium-neon (543 and 633 nm) lasers were used to excite fluorochromes and all images recorded following examination under the × 63 oil objective. Offline image analysis was performed using the LSM image browser (Zeiss) or MetaMorph 7.5 (Molecular Devices). The confocal laser scanning microscope multitrack setting was used to enable synchronized image acquisition.
For nuclear staining, conidial suspensions of M. oryzae were prepared on cover slips and inverted onto 100- μL aliquots of fixative solution (3.7% formaldehyde, 50 mM phosphate buffer, pH 7, and 0.2% Triton X-100) and incubated at room temperature for 5 min. Cover slips were then submerged in 20 mL water for 5 min and stained by submersion in 1 μg mL−1 4',6-diamidino-2-phenylindole (Sigma-Aldrich) for 5 min. Cover slips were washed a second time in 20 mL water for 5 min and allowed to air dry. Germinating conidia were then mounted in 10 μL Na-PO4, pH 8, 50% glycerol (v/v), and 0.1% propyl-gallate (Fluka).
Identification of Putative Cell Cycle Regulator Genes in M. oryzae
Putative M. oryzae cell cycle regulators were identified by interrogation of the sixth release of the annotated genome sequence of M. oryzae from the Broad Institute (http://www.broad.mit.edu/annotation/fungi/magnaporthe) using the BLASTP algorithm (Altschul et al., 1990). Protein sequences were aligned using ClustalW (http://www.ebi.ac.uk/clustalw/) and shaded using Boxshade v 2.01 (http://www.ch.embnet.org/software/BOX_form.html). For detailed protein sequence analysis, we used the Pfam database (http://www.sanger.ac.uk/Software/Pfam/). Details of construction for the allelic replacement vectors for nim1I327E, bim1F1763*, and nimAE37Gbim1F1763* mutant generation are given in Supplemental Methods online.
M. oryzae BIM1 Complementation of the Saccharomyces cerevisiae apc1- ΔC Mutant
Complementation of the S. cerevisiae apc1 temperature-sensitive (ts) mutant, apc1- ΔC (Winzeler et al., 1999), was performed by amplifying the BIM1 coding sequence from M. oryzae RNA using the Titanium One-Step RT-PCR kit (BD Biosciences) and cloning into pYES2 (Invitrogen) using primers 5BimE-EcoR1 and 3BimE-HindII, as shown in Supplemental Table 2 online. The S. cerevisiae strain MATa apc1- ΔC::KanMX4 (YNL171c::KanMX4) was provided by Claus Zachariae (University of Copenhagen, Denmark) and used for yeast transformation using the Yeastmarker system (Clontech). Six putative transformants were selected and grown in 50 mL YPD with 2% glucose for 16 h in a shaking incubator at 30°C and 230 rpm with the apc1- ΔC mutant and an isogenic wild-type strain. The cultures were then used to inoculate two sets of YPD plates supplemented with 2% glucose and 2% galactose, respectively. Successful complementation of BIM1 in S. cerevisiae strain apc1- ΔC was assessed by observing the ability of putative transformants to restore growth of the apc1- ΔC strain at 37°C.
Identification of the Aspergillus nidulans bimE7 Mutation
The full-length 6.2-kb A. nidulans bimE gene was amplified from both the A. nidulans bimE7 strain (kindly provided by Steve Osmani, Ohio State University, Columbus, OH) and a wild-type strain A89 (Fungal Genetics Stock Center). Both bimE alleles were sequenced (Eurofins MWG Operon) and a single mutation identified in bimE7, which altered a Gln residue in the amino acid sequence to introduce a premature stop codon at position 1966. This was verified by separate amplification of the two alleles and further DNA sequence analysis.
Identification of Putative M. oryzae B-Type Cyclins
Two M. oryzae putative cyclin B-encoding genes, CYC1 and CYC2, were identified from the M. oryzae genome database, with the Ustilago maydis B-type cyclins, Clb1 and Clb2. The 1.5-kb M. oryzae cyclin B gene CYC1 and 1.9-kb cyclin B homolog CYC2 were amplified from RNA using the Titanium One-Step RT-PCR kit (BD Biosciences) and introduced into pCR 2.1-TOPO (primers 5CyclinB1, 3CyclinB1, 5CyclinB2, and 3CyclinB2). Modified CYC1 and CYC2 alleles in which the destruction box consensus sequences were removed, as described in the Supplemental Methods online, were introduced into the M. oryzae H1:RFP strain. Regulated expression was performed in the presence of acetate; for details, see Supplemental Methods online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: M. oryzae NIM1 (MG00597), M. oryzae BIM1 (MG03314), M. oryzae CYC1 (MG05646), M. oryzae CYC2 (MG07065), M. oryzae NIMA (MG03026), A. nidulans Histone H1 vector pMF280 (AY598429), A. nidulans BimE (AN2772), U. maydis Clb1 (AY260969), U. maydis Clb1 (AY260970), S. cerevisiae APC1 (CAA96060), C. albicans CLB2 (U40430), C. albicans CLB4 (AF094507), A. nidulans NimE (X64602), S. pombe Cig2 (S67490), S. pombe Cdc13 (X12557), S. cerevisiae CLB2 (X62319), S. cerevisiae CLB3 (X69425), S. cerevisiae CLB4 (X69426), S. cerevisiae CLB5 (X70435), and S. cerevisiae CLB6 (X70436).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the Predicted M. oryzae Nim1 Amino Acid Sequence with A. nidulans NimO and S. cerevisiae Dbf4.
Supplemental Figure 2. High-Resolution Micrographs of Chromatin Status of M. oryzae Arrested at Different Stages of the Cell Cycle.
Supplemental Figure 3. Mitotic Entry Is Sufficient for M. oryzae Appressorium Maturation, and bim1F1763* Inhibits Nuclear Division during Appressorium Morphogenesis.
Supplemental Figure 4. Alignment of the Predicted M. oryzae Bim1 Amino Acid Sequence with A. nidulans BimE and S. cerevisiae Apc1.
Supplemental Figure 5. M. oryzae nimAE37G:bim1F1763* Displays a Typical nimAE37G Phenotype during Appressorium Development.
Supplemental Figure 6. Alignment of the Predicted M. oryzae B-Type Cyclins with Characterized B-Type Cyclins from A. nidulans, U. maydis, C. albicans, S. cerevisiae, and S. pombe, Which Act Specifically at the G2-Mitotic Transition.
Supplemental Figure 7. Construction of the grg(p):H1:RFP and ccg1(p):bm1:sGFP Gene Fusion Vectors and Expression in M. oryzae.
Supplemental Table 1. M. oryzae Strains Generated in This Study.
Supplemental Table 2. Detailed Information of the Primers Used in This Study.
Supplemental Methods.
Supplemental Movie 1. Live-Cell Imaging of Mitotic Division during Appressorium Development in M. oryzae.
Supplementary Material
Acknowledgments
This work was supported by a grant to N.J.T. from the Biotechnology and Biological Sciences Research Council and a graduate fellowship for D.G.O.S from the University of Exeter. We thank Steve James (Gettysburg College, Gettysburg, PA) and Steve Osmani (Ohio State University, Columbus, OH) for kindly providing Aspergillus mutants and for valuable discussions, Gero Steinberg and M.J. Kershaw (University of Exeter) for help with figure and movie preparation, and Richard Wilson (University of Nebraska, Lincoln, NE) for helpful discussions.
References
- Adachi K., Hamer J.E. (1998). Divergent cAMP signaling pathways regulate growth and pathogenesis in the rice blast fungus Magnaporthe grisea. Plant Cell 10: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bensen E.S., Clemente-Blanco A., Finley K.R., Correa-Borders J., Berman J. (2005). The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16: 3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Hofmann K., Bucher P., Neuwald A.F., Altschul S.F., Koonin E.V. (1997). A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11: 68–76 [PubMed] [Google Scholar]
- Choi W., Dean R.A. (1997). The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9: 1973–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D. (1996). Anaphase inhibition in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10: 3081–3093 [DOI] [PubMed] [Google Scholar]
- Cools T., De Veylder L. (2009). DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12: 23–28 [DOI] [PubMed] [Google Scholar]
- Couch B.C., Kohn L.M. (2002). A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 94: 683–693 [DOI] [PubMed] [Google Scholar]
- Dean R.A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35: 211–234 [DOI] [PubMed] [Google Scholar]
- Dean R.A., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- De Jong J.C., McCormack B.J., Smirnoff N., Talbot N.J. (1997). Glycerol generates turgor in rice blast. Nature 389: 244–245 [Google Scholar]
- Ebbole D.J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45: 437–456 [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T., Steinberg G., Perez-Martin J. (2003). Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot. Cell 2: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer J.E., Howard R.J., Chumley F.G., Valent B. (1988). A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 239: 288–290 [DOI] [PubMed] [Google Scholar]
- Henson J.M., Butler M.J., Day A.W. (1999). The dark side of the mycelium: Melanins of phytopathogenic fungi. Annu. Rev. Phytopathol. 37: 447–471 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996). Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30: 405–439 [DOI] [PubMed] [Google Scholar]
- Howard R.J., Ferrari M.A., Roach D.H., Money N.P. (1991). Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 88: 11281–11284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Pahl P.M., Harrison K., Rosamond J., Sclafani R.A. (1993). Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell. Biol. 13: 2899–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James S.W., Bullock K.A., Gygax S.E., Kraynack B.A., Matura R.A., MacLeod J.A., McNeal K.K., Prasauckas K.A., Scacheri P.C., Shenefiel H.L., Tobin H.M., Wade S.D. (1999). nimO, an Aspergillus gene related to budding yeast Dbf4, is required for DNA synthesis and mitotic checkpoint control. J. Cell Sci. 112: 1313–1324 [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E.T. (2005). C. elegans cell cycles: Invariance and stem cell divisions. Nat. Rev. Mol. Cell Biol. 6: 766–776 [DOI] [PubMed] [Google Scholar]
- Leung H., Borromeo E.S., Bernardo M.A., Notteghem J.L. (1988). Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Genetics 78: 1227–1233 [Google Scholar]
- McKinney J.D., Cross F.R. (1995). FAR1 and the G1 specific specificity of cell cycle arrest by mating factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2509–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany M. (2002). Polarity in filamentous fungi: Establishment, maintenance and new axes. Curr. Opin. Microbiol. 5: 580–585 [DOI] [PubMed] [Google Scholar]
- Ogino K., Takeda T., Matsui E., Iiyama H., Taniyama C., Arai K., Masai H. (2001). Bipartite binding of a kinase activator activates Cdc7-related kinase essential for S phase. J. Biol. Chem. 276: 31376–31387 [DOI] [PubMed] [Google Scholar]
- Osmani A.H., McGuire S.L., Osmani S.A. (1991). Parallel activation of the nimA and p34cdc2 cell cycle-regulated protein-kinases is required to initiate mitosis in Aspergillus nidulans. Cell 67: 283–291 [DOI] [PubMed] [Google Scholar]
- Osmani S.A., Engle D.B., Doonan J.H., Morris N.R. (1988). Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell 52: 241–251 [DOI] [PubMed] [Google Scholar]
- Park G., Xue C., Zhao X., Kim Y., Orbach M., Xu J.-R. (2006). Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell 18: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., King R.W., Hoog C., Kirschner M.W. (1996). Identification of BIME as a subunit of the anaphase-promoting-complex. Science 274: 1199–1201 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N.G., Palmer A.E., Tsien R.Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572 [DOI] [PubMed] [Google Scholar]
- Steinberg G., Perez-Martin J. (2008). Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 18: 61–67 [DOI] [PubMed] [Google Scholar]
- Talbot N.J., Ebbole D.J., Hamer J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Bornens M. (2006). Cell shape and cell division. Curr. Opin. Cell Biol. 18: 648–657 [DOI] [PubMed] [Google Scholar]
- Thines E., Weber R.W.S., Talbot N.J. (2000). MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12: 1703–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S.L., Talbot N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39: 385–417 [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C., Barooah M., Egan M., Wakely G., Talbot N.J. (2006). Cell cycle-regulated autophagic cell death is necessary for plant infection by the rice blast fungus. Science 312: 580–583 [DOI] [PubMed] [Google Scholar]
- Wang Z., Thornton C.R., Kershaw M.J., Debao L., Talbot N.J. (2003). The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol. Microbiol. 47: 1601–1612 [DOI] [PubMed] [Google Scholar]
- Whiteway M., Bachewich C. (2007). Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61: 529–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.A., Talbot N.J. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195 [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Urban M., Sweigard J.A., Hamer J.E. (1997). The CPKA gene of Magnaporthe grisea is essential for appressorial penetration. Mol. Plant Microbe Interact. 10: 187–194 [Google Scholar]
- Zhao X., Kim Y., Park G., Xu J.R. (2005). A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17: 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.