Peroxisomes are highly dynamic organelles. This work shows that the plant-specific dynamin-related protein DRP5B, which was previously identified as a chloroplast division protein, is also involved in peroxisome division. DRP5B interacts with several other components of the peroxisome division machinery, suggesting that it acts cooperatively with these factors.
Abstract
Peroxisomes are highly dynamic organelles involved in various metabolic pathways. The division of peroxisomes is regulated by factors such as the PEROXIN11 (PEX11) proteins that promote peroxisome elongation and the dynamin-related proteins (DRPs) and FISSION1 (FIS1) proteins that function together to mediate organelle fission. In Arabidopsis thaliana, DRP3A/DRP3B and FIS1A (BIGYIN)/FIS1B are two pairs of homologous proteins known to function in both peroxisomal and mitochondrial division. Here, we report that DRP5B, a DRP distantly related to the DRP3s and originally identified as a chloroplast division protein, also contributes to peroxisome division. DRP5B localizes to both peroxisomes and chloroplasts. Mutations in the DRP5B gene lead to peroxisome division defects and compromised peroxisome functions. Using coimmunoprecipitation and bimolecular fluorescence complementation assays, we further demonstrate that DRP5B can interact or form a complex with itself and with DRP3A, DRP3B, FIS1A, and most of the Arabidopsis PEX11 isoforms. Our data suggest that, in contrast with DRP3A and DRP3B, whose orthologs exist across plant, fungal, and animal kingdoms, DRP5B is a plant/algal invention to facilitate the division of their organelles (i.e., chloroplasts and peroxisomes). In addition, our results support the notion that proteins involved in the early (elongation) and late (fission) stages of peroxisome division may act cooperatively.
INTRODUCTION
Peroxisomes are ubiquitous eukaryotic organelles that participate in diverse metabolic functions. In plants, these single membrane-bound subcellular structures are involved in biochemical processes, such as photorespiration, fatty acid metabolism, hydrogen peroxide degradation, synthesis of jasmonic acid, and metabolism of indole-3-butyric acid; they are also essential to embryo viability. Peroxisomes are often found in intimate physical contact with other subcellular compartments, such as mitochondria and chloroplasts, acting in concert with these organelles in a number of metabolic pathways (Nyathi and Baker, 2006; Reumann and Weber, 2006; Kaur et al., 2009).
Peroxisomes are highly dynamic organelles that change in abundance in response to environmental, metabolic, and developmental cues, to function properly under diverse conditions (Purdue and Lazarow, 2001; Yan et al., 2005). Despite continuous debates over the evolutionary origin of peroxisomes, it is commonly believed that these organelles arose from the endoplasmic reticulum during evolution and can also form de novo from the endoplasmic reticulum in cells lacking peroxisomes, at least in yeasts (Hoepfner et al., 2005; Gabaldon et al., 2006; Schluter et al., 2006; Titorenko and Mullen, 2006). Evidence from yeasts also demonstrates that peroxisomes multiply primarily from preexisting peroxisomes through constitutive or induced division, where induced division is also called proliferation. Constitutive division and proliferation both involve peroxisome elongation (growth), constriction, and fission and form at least two peroxisomes from a single preexisting peroxisome (Yan et al., 2005; Fagarasanu et al., 2007). Previous studies have identified a number of key factors in peroxisome division/proliferation, among which PEROXIN11 (PEX11), dynamin-related proteins (DRPs), and FISSION1 (FIS1) represent three evolutionarily conserved families of proteins that mediate various stages of division and proliferation (Delille et al., 2009; Kaur et al., 2009).
PEX11 is believed to play a rate-limiting role in initiating peroxisome elongation/tubulation, the first step of peroxisome division. This conclusion is based on the fact that overexpressing PEX11 promotes peroxisomal elongation, whereas deletion or silencing of the gene(s) causes fewer and/or larger peroxisomes in the cell. Yeast Saccharomyces cerevisiae carries a single PEX11 and two homologous proteins, PEX25 and PEX27, both of which contain limited sequence similarity with PEX11 and which, despite being much larger than PEX11, perform partially overlapping functions with PEX11. Mammals have three isoforms of PEX11 (PEX11 α, -β, and -γ), with PEX11 β being essential for embryo viability (Yan et al., 2005; Fagarasanu et al., 2007; Kaur and Hu, 2009). Arabidopsis thaliana contains five PEX11 isoforms, which are categorized into three subfamilies, PEX11a, PEX11b, and PEX11c to -e, all of which are integral membrane proteins of the peroxisome and perform functions similar to those of their yeast and animal orthologs (Lingard and Trelease, 2006; Orth et al., 2007). These five Arabidopsis PEX11 homologs are partially redundant in function and display distinct expression patterns (Orth et al., 2007; Desai and Hu, 2008). The PEX11 protein (Pex11p) in S. cerevisiae is able to form homooligomers, which inhibit its function (Marshall et al., 1996). Mammalian PEX11 β self-interacts in two-hybrid and coimmunoprecipitation assays (Kobayashi et al., 2007), and all five Arabidopsis PEX11 isoforms homo- and heterodimerize in a bimolecular fluorescence complementation (BiFC) system in Arabidopsis cultured cells (Lingard et al., 2008). The biological consequences of PEX11 dimerization in plants and animals and the molecular mechanism by which PEX11 functions in any given species remain elusive.
Dynamins and DRPs are large self-assembling GTPases that participate in biological processes, such as endocytosis, intracellular vesicle trafficking, cytokinesis, and organelle division, and mediate the fusion and fission of membranes by functioning as mechanochemical enzymes or signaling GTPases (Osteryoung and Nunnari, 2003; Koch et al., 2004; Praefcke and McMahon, 2004; Hoppins et al., 2007). Mutations in the mammalian Drp1 (DLP1) and the yeast Dnm1 or Vps1 genes lead to fewer and enlarged/elongated peroxisomes that have already undergone membrane constriction, indicating that DRPs function in the final fission of these organelles. Drp1 and Dnm1p are also involved in mitochondrial division, whereas Vps1p has an additional role in vacuole morphogenesis (Yan et al., 2005; Fagarasanu et al., 2007). Arabidopsis has 16 DRPs, which are divided into six families based on protein structure and sequence similarity (Hong et al., 2003). The DRP3 family includes DRP3A and DRP3B, two proteins sharing 77% amino acid sequence identity that are both dual-localized to peroxisomes and mitochondria. Peroxisomal and mitochondrial division deficiencies are observed in drp3A and drp3B mutants, with the former displaying stronger peroxisome and plant growth phenotypes than the latter (Arimura et al., 2004; Logan et al., 2004; Mano et al., 2004; Fujimoto et al., 2009; Zhang and Hu, 2009). Consistent with the notion that dimer formation is central to the GTPase activity of DRPs (Praefcke and McMahon, 2004), DRP3A and DRP3B homo- and heterodimerize in yeast two-hybrid assays (Fujimoto et al., 2009). Although the drp3A drp3B double mutants display defects in organelle division and plant growth, the plants are not severely impaired, implying that other members of the Arabidopsis DRP superfamily may be at work in peroxisomal fission (Zhang and Hu, 2009).
Yeast and mammalian species each have a single FIS1 protein, which is anchored to the membrane of peroxisomes and the outer membrane of mitochondria by the C terminus, recruiting cytosolic DRPs to the organelle membranes through interactions via the N-terminal tetratricopeptide repeat domain (Koch et al., 2003, 2005; Kuravi et al., 2006; Kobayashi et al., 2007; Serasinghe and Yoon, 2008). Arabidopsis has two FIS1 homologs, FIS1A (i.e., BIGYIN) and FIS1B, which are 58% identical to each other at the protein level and are both dual-targeted to peroxisomes and mitochondria. The fis1 loss-of-function mutants contain fewer and enlarged peroxisomes and mitochondria, whereas ectopic expression of FIS1A or FIS1B results in increased numbers of these organelles, reinforcing the rate-limiting role for FIS1 in organelle fission (Scott et al., 2006; Zhang and Hu, 2008, 2009). The mammalian FIS1 protein self-interacts on the outer membrane of mitochondria, and this oligomerization was believed to be required for the functioning of FIS1 in mitochondrial fission (Serasinghe and Yoon, 2008). Whether FIS1A and FIS1B also form oligomers and are responsible for recruiting DRPs to peroxisomes in plants have yet to be confirmed.
Chemical cross-linking and coimmunoprecipitation studies in Chinese hamster ovary cells revealed the formation of a ternary heterocomplex consisting of PEX11 β, Drp1 (DLP1), and FIS1 on the peroxisomal membrane and interaction between FIS1 and PEX11 β (Kobayashi et al., 2007). Likewise, BiFC experiments in Arabidopsis cultured cells found all five PEX11 proteins to interact with FIS1B (Lingard et al., 2008). In addition, overexpression of PEX11 β can no longer induce peroxisome proliferation in mammalian cells in which the expression of Drp1 was silenced (Li and Gould, 2003). These data together suggest that, besides FIS1, PEX11 may also act in cooperation with DRPs on peroxisomes. Whether plant PEX11, DRP, and FIS1 proteins also form a complex on peroxisomal membranes and coordinately regulate peroxisome division is unknown.
To get a complete mechanistic view of how peroxisomes divide in plants and to correlate the dynamics of the abundance of these organelles with plant physiology, we searched for additional players in peroxisome division. Here, we report that Arabidopsis DRP5B, a plant/algal-specific DRP previously shown to be required for plastid division, plays an additional role in the division of peroxisomes and contributes to the proper functioning of peroxisomes. We also analyzed the interaction between DRP5B and members of the Arabidopsis DRP3, FIS1, and PEX11 protein families and provide evidence for possible coordination of these proteins in peroxisome division in plants.
RESULTS
DRP5B (ARC5) Is Involved in the Division of Both Peroxisomes and Chloroplasts
To identify new proteins in peroxisome division, we first focused on other members of the Arabidopsis DRP superfamily. DRP5B, also called ARC5 (for accumulation and replication of chloroplasts), is the only Arabidopsis DRP besides DRP3A and DRP3B that is known to play a direct role in organelle division (Gao et al., 2003; Miyagishima et al., 2003; Glynn et al., 2008). DRP5B forms a discontinuous ring at the division site on chloroplasts, executing the fission of these organelles (Gao et al., 2003). Interestingly, green fluorescent protein (GFP)-DRP5B and Cm Dnm2, a DRP from the primitive red algae Cyanidioschyzon merolae that participates in chloroplast division, were both reported to exist as cytosolic patches as well (Miyagishima et al., 2003; Glynn et al., 2008). These previous results led us to speculate that, in addition to being involved in chloroplast division, DRP5B may be targeted to other subcellular compartments, such as peroxisomes, exerting its function in the division of multiple types of organelles.
To investigate whether DRP5B plays a role in peroxisome division, we expressed the peroxisomal marker protein YFP-PTS1, a fusion of the yellow fluorescent protein and a C-terminal Peroxisome Targeting Signal type 1 tripeptide (PTS1, Ser-Lys-Leu), in drp5B mutants. The two drp5B null alleles used are drp5B-1 (in Landsberg erecta [Ler]), which creates a stop codon in the middle of DRP5B, and drp5B-2 (SAIL 71D_11, in Columbia-0 [Col-0]), which has a T-DNA insertion in the 8th intron (Gao et al., 2003; Miyagishima et al., 2006). We also analyzed two drp5A null alleles, drp5A-1 (SALK_065118) and drp5A-2 (SALK_062383), which have a T-DNA inserted in the 4th intron and the 7th exon, respectively (Miyagishima et al., 2008). DRP5A is the other member of the DRP5 family and shares similar domain structure with DRP5B, but it was recently shown to be involved in cytokinesis instead of chloroplast division (Miyagishima et al., 2008).
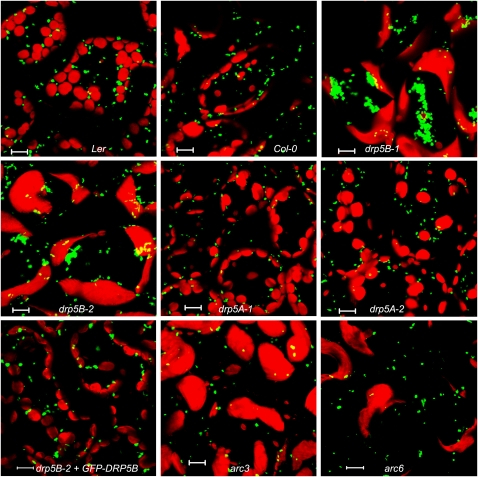
Confocal laser scanning microscopy image analysis of mesophyll cells from T3 drp5B mutants expressing YFP-PTS1 confirmed the previously described phenotypes (i.e., enlarged chloroplasts with impaired division) (Gao et al., 2003). In addition, the mutants also contain highly aggregated peroxisomes (Figure 1). These peroxisomes appear to have gone through membrane constriction but failed to complete fission and, as a result, are unable to separate from each other. Although we were unable to accurately quantify the number of peroxisomes due to their strong clustering, peroxisomes are clearly impaired in division, at least partially, in the drp5B mutants. Abnormal peroxisomal morphologies were also observed in roots and etiolated seedlings of the drp5B mutants, and these peroxisomes were more clustered and larger than those of wild-type plants (see Supplemental Figures 1A to 1F). These results together point to defects in peroxisome fission in the drp5B mutants. By contrast, neither of the DRP5A mutant alleles shows any abnormalities in peroxisome morphology (Figure 1), which largely excludes the involvement of DRP5A in peroxisome division. To further confirm that the peroxisome phenotypes in drp5B mutants were caused by the mutations in DRP5B, we expressed the peroxisome marker, DsRed2-PTS1, in the drp5B-2 mutant that carries the GFP-DRP5B gene whose expression is driven by the DRP5B native promoter (Miyagishima et al., 2008). In addition to rescuing the chloroplast division deficiency, GFP-DRP5B also largely restored the peroxisome phenotype in drp5B-2, as seen by the disappearance of most peroxisomal clusters and the reappearance of numerous spherical peroxisomes similar to those of the wild type (Figure 1). We conclude that DRP5B not only is involved in chloroplast division but also plays a role in the division of peroxisomes.
Figure 1.
Peroxisomal and Chloroplast Morphologies in Wild-Type Plants and in drp5 and arc Mutants.
Confocal images were obtained of leaf mesophyll cells from 4-week-old plants expressing the YFP-PTS1 (in Ler, Col-0, drp5B-1, drp5B-2, drp5A-1, and arc3) or DsRed2-PTS1 (in drp5B expressing GFP-DRP5B and arc6) peroxisome marker protein. Green signals come from YFP or DsRed2 and red signals are emitted from the chlorophyll. Bars = 10 μm.
The close functional association between plastids and peroxisomes (e.g., in photorespiration) prompted us to evaluate peroxisome morphology in two chloroplast division mutants, arc3 and arc6 (Vitha et al., 2003; Glynn et al., 2008), to rule out the possibility that the peroxisomal division defect in drp5B is caused indirectly by the abnormal division of chloroplasts. Chloroplast division is orchestrated by multiple molecular machineries composed of a number of proteins, among which ARC3 is localized in the stroma and required for the correct positioning of the division rings, and ARC6 spans the inner envelope and is responsible for correct positioning of the stromal ring Ftz and recruitment of DRP5B to the chloroplast surface through the outer-envelope proteins PDV1 and PDV2 (Yang et al., 2008; Okazaki et al., 2009). To visualize peroxisomes, YFP-PTS1 was introduced into arc3 and DsRed2-PTS1 was transformed into arc6. Despite having dramatically enlarged chloroplasts with impaired division, T2 plants of arc3 and arc6 expressing YFP-PTS1 and DsRed2-PTS1, respectively, show no obvious changes in peroxisome morphology and abundance (Figure 1). These data demonstrate that the peroxisome division deficiency in drp5B mutants is not a side effect of chloroplast morphology and number changes; furthermore, DRP5B is likely the only protein shared by chloroplast and peroxisome division.
DRP5B Is Dual Localized
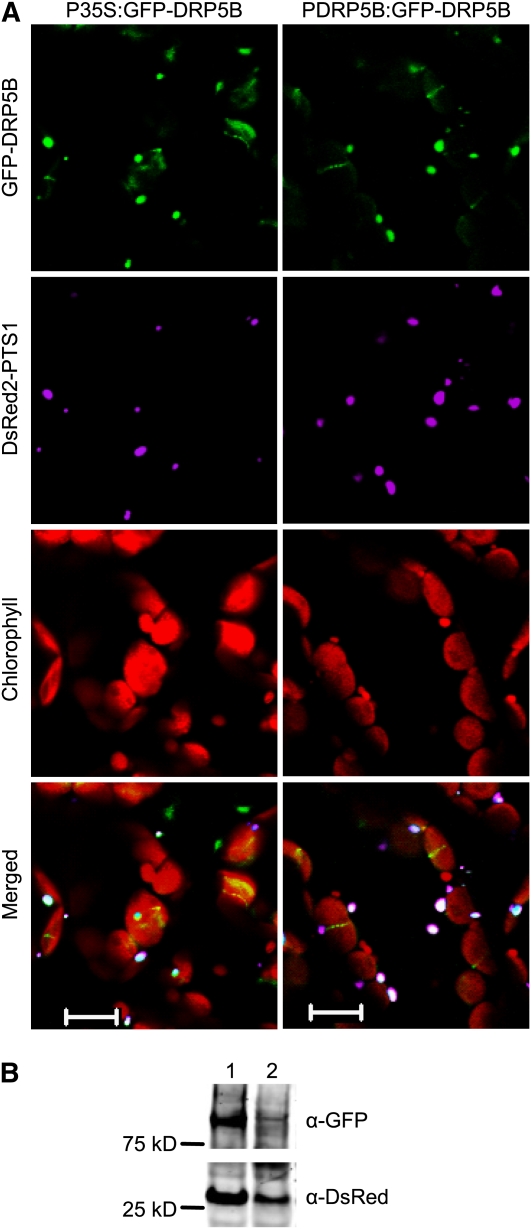
Given that DRP5B had not been demonstrated previously to exert a function in peroxisome division, we reevaluated the subcellular localization of this protein by coexpressing DsRed2-PTS1 and GFP-DRP5B in wild-type Arabidopsis. The GFP-DRP5B gene was under the control of the cauliflower mosaic virus 35S promoter (P35S:GFP-DRP5B) or the DRP5B native promoter (PDRP5B:GFP-DRP5B). T3 plants containing both DsRed2-PTS1 and GFP-DRP5B were examined using confocal microscopy. In both types of transgenic plants, GFP fluorescent signals were detected not only as a discontinuous ring structure at the chloroplast division sites, but also on peroxisomes tagged by DsRed2-PTS1, showing that DRP5B is targeted to both chloroplasts and peroxisomes (Figure 2A). Consistent with data shown by Glynn et al. (2008), weak localization of GFP-DRP5B throughout chloroplasts was also observed (Figure 2A). The punctate structures labeled by GFP-DRP5B were not found to colocalize with mitochondrial markers. Immunoblot analysis showed that the GFP-DRP5B and DsRed2-PTS1 proteins are indeed expressed in P35S:GFP-DRP5B and PDRP5B:GFP-DRP5B lines, with higher GFP-DRP5B expression detected in the former (Figure 2B). Even though GFP-DRP5B is functional in complementing the drp5B mutant phenotypes (Gao et al., 2003; this study), no apparent differences in peroxisome appearance or abundance were found between P35S:GFP-DRP5B and PDRP5B:GFP-DRP5B lines (Figure 2A). This result is in line with previous findings that overexpressing DRP3A or DRP3B does not affect peroxisome size and number (Mano et al., 2004; Zhang and Hu, 2009), providing evidence that DRP proteins by themselves are insufficient to induce organelle division.
Figure 2.
Dual Localization of GFP-DRP5B.
(A) Confocal images taken of mesophyll cells of 4-week-old plants coexpressing the P35S:DsRed2-PTS1 and P35S:GFP-DRP5B (or PDRP5B:GFP-DRP5B) transgenes. Bars = 10 μm.
(B) Immunoblot analysis detecting the GFP-DRP5B and DsRed2-PTS1 proteins from plants in (A), using α-GFP and α-DsRed antibodies, respectively. Lanes 1 and 2 contain proteins from plants expressing P35S:GFP-DRP5B and PDRP5B:GFP-DRP5B, respectively.
Since ARC6 plays a significant role in recruiting DRP5B to chloroplasts (Vitha et al., 2003; Glynn et al., 2008), we also assessed the subcellular localization of GFP-DRP5B in the arc6 mutant. DsRed2-PTS1 was expressed in arc6 plants carrying the PDRP5B:GFP-DRP5B transgene (Glynn et al., 2008), and T3 progenies containing both transgenes were examined by confocal microscopy. In arc6, although GFP-DRP5B is not targeted to ring structures on chloroplasts, similar to what was found by Vitha et al. (2003) and Glynn et al. (2008), its peroxisomal localization is mostly unaffected (see Supplemental Figure 1G online). This result reinforces our conclusion (see previous section) that the role for DRP5B in peroxisome division is largely independent of its function in chloroplast division.
DRP5B Contributes to Peroxisome Functions
To elucidate the impact of DRP5B on plant growth and development, we first investigated the expression profiles of this gene, using data collected from Arabidopsis microarray databases and the Genevestigator tool (https://www.genevestigator.ethz.ch/). DRP5B is ubiquitously expressed in all tissues and throughout development, with high expression levels in green tissues, such as cotyledons and cauline and rosette leaves (see Supplemental Figure 2A online). In addition, expression of DRP5B starts out at a relatively low level during seed germination, increases significantly as the plants develop leaves, reaches its peak during bolting, and declines after plants enter the reproductive phase (see Supplemental Figure 2B online).
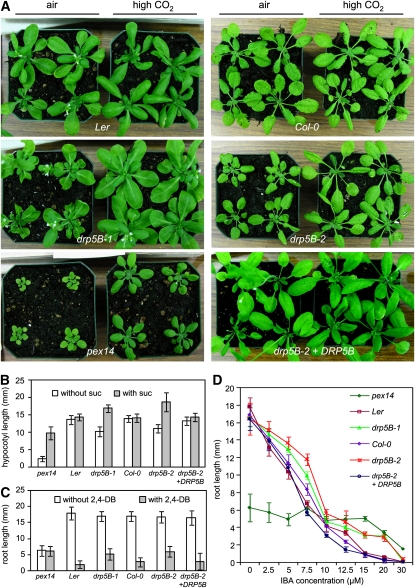
The expression pattern for DRP5B suggests that the protein it encodes may play a prominent role in green tissues, where photorespiration (the primary function for leaf peroxisomes) takes place. Photorespiration is coordinated by chloroplasts, peroxisomes, and mitochondria. It takes in O2 and releases CO2 in the light, salvaging and recycling phosphoglycolate back to the chloroplast. Because this pathway is not required under high CO2 conditions, photorespiration mutants display much stronger growth phenotypes in normal air conditions than in an environment with elevated CO2 (Reumann and Weber, 2006; Kaur et al., 2009). The pex14 null mutant, which contains a T-DNA insertion in the peroxisome biogenesis factor PEROXIN14, serves as a positive control in this study (Figure 3) as in many of our previous studies (Fan et al., 2005; Orth et al., 2007; Zhang and Hu, 2009). After growing in ambient air for 3 to 4 weeks, drp5B mutants start to show retarded growth compared with wild-type plants, and this phenotype can be rescued by growing the mutants in elevated (3000 ppm) CO2. By contrast, wild-type plants, drp5B mutants expressing GFP-DRP5B, and even other arc mutants, such as arc3, have similar plant sizes irrespective of the CO2 level in the growth environment (Figure 3A; see Supplemental Figure 2C online). These data demonstrate that DRP5B impacts photorespiration, possibly owing to its function in the division of both peroxisomes and chloroplasts, two major participants of the glycolate recycling pathway in photorespiration.
Figure 3.
The Role of DRP5B in Plant Growth and Peroxisome Activities.
(A) Comparison of 4-week-old plants grown in ambient air and under 3000 ppm CO2.
(B) Sucrose dependence assay. Hypocotyl lengths of seedlings grown for 5 d in the dark on half-strength Murashige and Skoog (MS) media with or without the supplement of 1% sucrose (w/v) are shown.
(C) and (D) Effect of 2,4-DB (C) and IBA (D) on primary root elongation. Plants were grown for 5 d in the light on half-strength LS media supplemented with 0.8 μ M 2,4-DB (C) or various concentrations of IBA (D).
For (B) to (D), n = 60 and P < 0.05 for each pairwise t test between the mutant (or complemented mutant) and its corresponding wild-type parent. Error bars are standard deviations.
We later determined the effect that DRP5B imparts on fatty acid β-oxidation, a major peroxisome function required for the conversion of triacylglycerol to sucrose to fuel postgerminative seedling establishment (reviewed in Penfield et al., 2006; Kaur et al., 2009). First, drp5B mutant seeds were germinated in media with or without sucrose. In the absence of sucrose, hypocotyls of dark-grown drp5B-1 and drp5B-2 are shorter than those of the wild type and the complemented drp5B-2 plants; this phenotype is largely rescued by application of sucrose to the media (Figure 3B). These data suggest that drp5B mutants are partially deficient in storage oil mobilization during hypocotyl elongation in germinating seedlings. Second, we measured the response of drp5B mutants to indole 3-butyric acid (IBA) and the synthetic auxin 2,4-dichlorophenoxybutyric acid (2,4-DB). IBA and 2,4-DB are protoauxins that can be metabolized to the bioactive auxins IAA and 2,4-D, respectively, through peroxisomal β-oxidation. Mutants deficient in β-oxidation would show resistance to the inhibitory effect of these compounds on primary root elongation (Hayashi et al., 1998; Zolman et al., 2000). Weak but statistically significant resistance to both 2,4-DB and IBA was shown in drp5B mutant seedlings, compared with the wild type and the rescued drp5B-2 mutant plants (Figures 3C and 3D), suggesting that abnormalities in peroxisome division in the drp5B mutants ultimately result in deficiencies in peroxisomal functions, such as β-oxidation during seedling establishment. Quantitative experiments described in this section were repeated three times with similar results; data from the third repeat are shown in Figures 3B to 3D. After performing pairwise t tests between measurements for each genotype and the respective wild-type background (Col-0 or Ler) it was generated from, it was concluded that the differences in hypocotyl and root measurements are statistically significant (see Figure 3 legend).
Coimmunoprecipitation and BiFC Assays Reveal Interaction/Protein Complex Formation between DRP5B and DRP3, FIS1, and PEX11 Proteins
Having established a role for DRP5B in peroxisome division, we turned our attention to the ability of this protein to interact with itself and with DRP3A and DRP3B, as DRP3A and DRP3B had been shown to homo- and heterodimerize in yeast two-hybrid systems (Fujimoto et al., 2009). In addition, given the reported interaction between FIS1, DRP, and PEX11 in mammalian cells (Kobayashi et al., 2007), we were also interested in testing whether DRP5B and members of the FIS1 and PEX11 families in Arabidopsis form complexes. Interaction between proteins involved in early (i.e., elongation) and late (i.e., fission) stages of peroxisome division may indicate that these distinct machineries are coordinated in function.
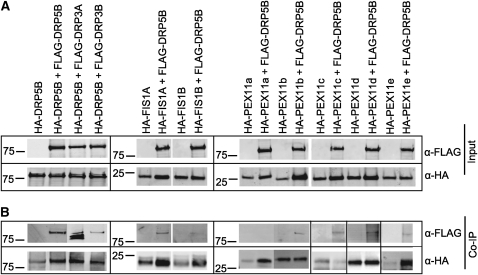
To test for protein–protein interaction, we first employed coimmunoprecipitation (co-IP). We used the binary vectors pEARLEY201 and pEARLEY202 (see Methods and Supplemental Table 1 online) to construct 35S-driven gene fusions, in which HA and FLAG tags were cloned, respectively, to the N terminus of the inquest proteins, and introduced the construct pairs into Nicotiana tabacum leaves via Agrobacterium tumefaciens infiltration. Forty-eight hours after inoculation, proteins extracted from the infiltrated leaves expressing each of the HA- and FLAG-tagged protein pairs were subjected to immunoblot analysis to ensure that the fusion proteins were being expressed (Figure 4A). Subsequently, total protein extracts were incubated with agarose beads conjugated with α-HA, and proteins pulled down by α-HA were subjected to immunodetection using α-FLAG and α-HA antibodies. Detection of the two HA- and FLAG-fusion proteins in the same co-IP would suggest that the two proteins are in the same complex.
Figure 4.
Co-IP Assays to Test for Physical Association between DRP5B and Other Known Peroxisome Division Proteins.
(A) Immunoblot analysis of proteins extracted from tobacco leaves expressing HA- and FLAG-fusion proteins.
(B) Immunoblot analysis of proteins bound to anti-HA beads.
Sizes of the molecular markers (in kD) are shown to the left of the blots. Different gels are separated by boxes.
The co-IP results showed putative complex formation between DRP5B proteins and between DRP5B and DRP3A/DRP3B, FIS1A, and PEX11b to -e; no (or minimal) association was found between DRP5B and FIS1B and between DRP5B and PEX11a (Figure 4B). These data together indicate that DRP5B is capable of interacting with itself and heterodimerizing with DRP3A and DRP3B and that it can form complexes with FIS1A and four of the five Arabidopsis PEX11 proteins in vivo.
To confirm results obtained from co-IP, we also used BiFC to test for protein–protein interaction. BiFC is an in vivo assay that not only determines whether proteins interact or reside in close proximity, but also detects cellular locations for such interactions. N- and C-terminal fragments of YFP (YN and YC) were fused to the N terminus of DRP5B, DRP3A, DRP3B, FIS1A, FIS1B, and PEX11a to -e genes to generate YN-gene and YC-gene fusion constructs using binary vectors derived from pFGC5941 (see Methods; see Supplemental Table 1 online). For subsequent evaluation of protein expression, an HA tag was added to the N terminus of YN and a 6XHis tag was fused to the N terminus of YC. All fusion genes were driven by the 35S promoter. Each YN- and YC-fusion pair, along with the peroxisomal marker cyan fluorescent protein (CFP)-PTS1, was transiently coexpressed in N. tabacum leaves using A. tumefaciens–mediated transformation. Epidermal or mesophyll cells of the inoculated tissues were analyzed by confocal microscopy after 48 h. To ensure that the proteins were expressed, we subjected proteins extracted from the inoculated tissues to immunoblot analysis, using α-HA, α-His, and α-GFP antibodies to detect the YN-, YC-, and CFP-PTS1 fusion proteins, respectively (see Supplemental Figure 3 online).
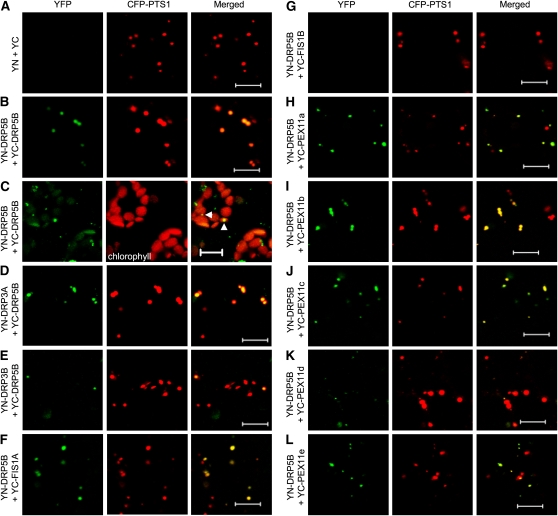
Leaf tissues infiltrated with the control vectors YC and YN showed no YFP signals (Figure 5A), whereas YN-DRP5B and YC-DRP5B, when combined, conferred YFP fluorescence on CFP-PTS1–tagged peroxisomes (Figure 5B) as well as on chloroplasts marked by chlorophyll autofluorescence (Figure 5C). Intriguingly, we were unable to detect YFP fluorescence as ring structures on chloroplasts. In addition, DRP5B interacts with DRP3A and DRP3B on peroxisomes (Figures 5D and 5E). Consistent with results from co-IP, DRP5B interacts with FIS1A, but not FIS1B, on peroxisomes (Figures 5F and 5G; see Supplemental Figure 4A online). Lingard et al. (2008) used BiFC to show in Arabidopsis cell cultures that all five Arabidopsis PEX11 proteins form homo- and heterodimers and that they each interact with FIS1B but not FIS1A or DRP3A. In our BiFC system, we were able to reproduce the positive interaction results for PEX11 proteins and between FIS1B and each of the PEX11 proteins. As shown in Figures 5H to 5L, we detected peroxisome-localized association between DRP5B and each of the five PEX11 isoforms. These data largely corroborate those obtained by co-IP (Figure 4B). One exception is the interaction between DRP5B and PEX11a. Whereas FLAG-DRP5B was not pulled down by HA-PEX11a (Figure 4B), these two proteins show strong BiFC when combined (Figure 5H).
Figure 5.
Protein–Protein Interactions Involving DRP5B as Detected by BiFC.
Confocal images were taken from N. tabacum leaf epidermal cells expressing the YN- and YC-fusions along with CFP-PTS1 ([A], [B], and [D] to [L]) or mesophyll cells expressing YN- and YC-fusion proteins only (C). YFP fluorescence (green) is an indication of BiFC, and red signals come from CFP-PTS1 or chloroplasts that emit autofluorescence. The arrowheads in (C) indicate strong YFP signals at the putative division sites of chloroplasts. Bars = 10 μ m.
DISCUSSION
DRP5B Plays a Dual Role in Organelle Division
DRP5B was originally identified for its function in chloroplast division (Gao et al., 2003). Here, we provide several lines of evidence that this protein has an additional role in the division of peroxisomes and is involved in maintaining proper peroxisomal activities. First, GFP-DRP5B localizes to chloroplast division rings as well as spherical structures labeled by the CFP-PTS1 peroxisomal marker protein. Second, besides the previously reported phenotypes, such as enlarged and dumbbell-shaped chloroplasts, drp5B mutants also exhibit enlarged peroxisomes and peroxisomes that have undergone membrane constriction but failed to complete fission and therefore are unable to separate from each other. Third, peroxisomal functions, such as photorespiration and fatty acid β-oxidation, are compromised in the drp5B mutants. Finally, deficiencies in peroxisomal morphology and function in the drp5B mutants can be rescued by expression of the wild-type DRP5B protein. In summary, DRP5B has joined DRP3A and DRP3B as plant DRPs recognized as being involved in peroxisome division. A recent comprehensive phylogenetic analysis of dynamin proteins showed that DRP3A/DRP3B and the yeast and mammalian DRP proteins known to play a role in peroxisome division (i.e., Dnm1p, Vps1p, and Drp1) form a subclade distant from the DRP5 proteins. In addition, DRP5B and its putative orthologs, which are identified only from plant and algal genomes, are derived from DRPs involved in cytokinesis, a function that is maintained by DRP5A (Miyagishima et al., 2008). Thus, in contrast with DRP3A and DRP3B, DRP5B is a plant/algal-specific dynamin in the peroxisome division machinery. Finally, the list of DRPs associated with peroxisome division in plants may not be complete, as there are over a dozen Arabidopsis DRPs (Hong et al., 2003), most of which have not been characterized with respect to their relevance to peroxisomes.
The discovery that DRP5B also participates in peroxisome fission is somewhat unexpected. DRP3A and DRP3B are highly identical in sequence and both contain the GTPase, middle, and GTPase effector domains (Hong et al., 2003). As a result, these two proteins are interchangeable in mitochondrial division and partially redundant in the division of peroxisomes (Fujimoto et al., 2009; Zhang and Hu, 2009). However, DRP5B shares little sequence similarity with the DRP3s and contains an additional pleckstrin homology domain, which may be capable of binding to membrane phospholipids (Hong et al., 2003). DRP5B and DRP3A/3B also differ in their peroxisome localization patterns. When fused to YFP or GFP, DRP3A and DRP3B were shown to be often in juxtaposition to peroxisomes (Mano et al., 2004; Fujimoto et al., 2009; Zhang and Hu, 2009), whereas P35S:GFP-DRP5B or PDRP5B:GFP-DRP5B is evenly distributed along peroxisomes (Figure 2A). Finally, peroxisomes in drp3A mutants frequently contain long membraneous tails, named peroxules, a phenotype that is not shown in drp5B mutants. These data collectively point toward the possibility that the role for DRP5B in peroxisome division is to some extent distinct from that of DRP3A and DRP3B. To test this hypothesis, it will be crucial to determine whether DRP5B can substitute for DRP3 in peroxisome division.
Another example of a single DRP participating in diverse functions comes from the Arabidopsis DRP1 family. This family is generally believed to be involved in cytokinesis and cell expansion (Konopka and Bednarek, 2008); however, DRP1C and DRP1E were also reported to act in mitochondrial morphogenesis (Jin et al., 2003). In nonplant systems, the yeast Vps1p and Dnm1p and the mammalian DLP1 (Drp1) proteins are DRPs involved in the division/vesiculation of more than one type of organelle (Wilsbach and Payne, 1993; Hoepfner et al., 2001; Koch et al., 2003; Li and Gould, 2003; Koch et al., 2004; Kuravi et al., 2006; Schrader, 2006). These results together suggest that a given DRP, which normally lacks intrinsic organelle targeting signals, can be recruited to different types of subcellular structures to facilitate membrane fission. However, recruitment of DRPs also seems to be partially specific. For example, DRP3A, DRP3B, and DRP5B do not participate in the fission of membrane structures other than peroxisomes, mitochondria, and chloroplasts, whereas DRP5A, despite being structurally similar to DRP5B, functions in cytokinesis instead of chloroplast division (Mano et al., 2004; Miyagishima et al., 2008; Fujimoto et al., 2009; Zhang and Hu, 2009).
Photorespiration, fatty acid metabolism, and jasmonic acid biosynthesis are among the metabolic pathways coordinated by peroxisomes and other organelles, including chloroplasts and mitochondria (Reumann and Weber, 2006; Kaur et al., 2009). The efficiency of these metabolic processes is thought, at least in part, to rely on the intimate physical association and functional cooperation between the organelles involved. In Arabidopsis, DRP3 and its putative anchor, FIS1, are shared by the division machineries of peroxisomes and mitochondria, and DRP5B is shared by peroxisomes and chloroplasts. The use of shared fission components could be a mechanism to render coordinated division among the metabolically linked subcellular compartments. It will be interesting to determine whether such coordinated division truly takes places in plants, and if so, whether it has biological significance.
Coordination between the Fission Proteins (DRP and FIS1) and between the Fission Machinery and the Elongation/Tubulation Factors (PEX11)
In this study, we used two independent approaches, co-IP and BiFC, to demonstrate that DRP5B can be physically associated with members of the DRP3, FIS1, and PEX11 protein families. Results from these two approaches largely agree with each other, with the exception of interaction between PEX11a and DRP5B. Furthermore, additional co-IP assays also confirmed the interaction between DRP3A/DRP3B and some members of the DRP, FIS1, and PEX11 families in planta (see Supplemental Figure 5 online). Lastly, using BiFC, we were able to show that FIS1A and FIS1B each form homodimers on peroxisomes (see Supplemental Figure 4B online), consistent with findings from studies of the mammalian FIS1 ortholog (Serasinghe and Yoon, 2008). However, FIS1A and FIS1B do not seem to heterodimerize on the peroxisome in our BiFC system (see Supplemental Figure 4C online). These data collectively are in line with the current knowledge about the interplay between these proteins in mammalian cells, where the DRP proteins homooligomerize, FIS1 helps to recruit DRP proteins, and DRP, FIS1, and PEX11 may form a ternary complex (Thoms and Erdmann, 2005; Yan et al., 2005; Delille et al., 2009). It remains to be determined whether DRP3A, DRP3B, and DRP5B assemble together into polymers and mediate peroxisome division in a concerted manner, whether FIS1 homooligomerization is required for its proper function, and whether PEX11, a protein primarily responsible for peroxisome elongation, is also directly involved in the FIS1-dependent recruitment of DRPs to peroxisomes.
Multiple factors, such as protein expression levels, rates of protein folding, and protein stability, may contribute to variations in the detection of protein–protein interaction by BiFC, leading to false positive/negative results (Lalonde et al., 2008). Despite the fact that our negative control and the DRP5B-FIS1B combination did not give rise to any YFP complementation signals in our BiFC experiments, overexpressing proteins in a small organelle-like peroxisome (0.1 to 1 μ m) may potentially cause false positive protein interactions. As such, follow-up studies are needed to authenticate the interaction between PEX11a and DRP5B. Transgenic Arabidopsis plants, cultured Arabidopsis cells, or tobacco epidermal cells overexpressing individual FIS1 or PEX11 isoforms exhibit marked increases in peroxisome numbers or dramatic elongation of the organelles (Lingard and Trelease, 2006; Orth et al., 2007; Zhang and Hu, 2008, 2009). However, in our BiFC assays, YFP complementation is more often detected on peroxisomes that are not elongated and in cells where dramatic increases in peroxisome abundance are not observed, hinting at a possible limitation for our BiFC system.
The diversification of the DRP, FIS1, and PEX11 families in plants may have led to the specific recruitment of the DRP proteins by distinct anchor proteins/protein complexes on the various types of organelles. For instance, DRP5B interacts with FIS1A, but not FIS1B, on peroxisomes (this study), whereas its recruitment to chloroplasts is obviously dependent on a group of chloroplast envelope proteins (e.g., ARC6, PDV1, and PDV2) (Gao et al., 2003; Miyagishima et al., 2006; Glynn et al., 2008). ELM1 (for Elongated Mitochondria1), a plant-specific protein that exclusively targets to the outer membrane of mitochondria, interacts with both DRP3A and DRP3B, serving as a mitochondrial anchor for (at least) DRP3A (Arimura et al., 2008). It is also possible that FIS1 and ELM1 are part of the same mitochondrial membrane complex responsible for recruiting DRP proteins. Lastly, yeast mitochondrial and peroxisomal divisions both require Mdv1p (or Caf4p), a cytosolic linker that interacts with both DRP and FIS1 proteins on these organelles (reviewed in Delille et al., 2009). Although the orthologs for Mdv1p/Caf4p were not identified in mammals, we cannot exclude the possibility that their functional analogs exist in plants.
METHODS
Plant Materials, Growth Conditions, and Transformation
Arabidopsis thaliana plants were germinated under 16-h-light (60 μ E m−2 s−1)/8-h-dark conditions on 0.6% (w/v) agar plates with half-strength MS supplemented with 1% (w/v) sucrose. After 2 weeks, seedlings were transplanted into soil and grown under a photosynthetic photon flux density of 70 to 80 μ mol m−2 s−1 at 21°C with a 14-h-light/10-h-dark period. Tobacco plants were grown under 30 to 40 μ mol m−2 s−1 of light intensity at 24°C, with a photoperiod of 14 h light/10 h dark.
CFP-PTS1, YFP-PTS1 (Fan et al., 2005; Orth et al., 2007; Zhang and Hu, 2008, 2009), and DsRed2-PTS1 were used as markers to visualize peroxisomes. To make DsRed2-PTS1, a DsRed2-Ser-Lys-Leu fragment was amplified by PCR from the vector pDsRed2-Peroxi (BD Biosciences) and cloned into a vector derived from pPZP222 (Hajdukiewicz et al., 1994) and carrying the 35S promoter. To determine subcellular localization of DRP5B, Arabidopsis plants expressing the GFP-DRP5B transgene (driven by cauliflower mosaic virus 35S or the DRP5B native promoter; provided by the Osteryoung Lab, Michigan State University) were transformed with P35S:DsRed2-PTS1. To visualize peroxisomes in various mutant backgrounds, YFP-PTS1 or DsRed2-PTS1 was expressed in drp5B-1, drp5B-2 (SAIL 71D_11), drp5A-1 (SALK_065118), drp5A-2 (SALK_062383), arc3, and arc6 (gifts from the Osteryoung Lab). The Agrobacterium tumefaciens strain C58C1 was used for all plant transformations, and selection of transgenic plants was performed as described previously (Zhang and Hu, 2009). In addition, Basta (10 μ g/mL) and gentimycin (50 μ g/mL) were used to select for plants expressing GFP-DRP5B and DsRed2-PTS1, respectively.
Confocal Laser Scanning Microscopy and Image Analysis
For colocalization and mutant analyses, rosette leaves of 4-week-old Arabidopsis plants were analyzed using a confocal laser scanning microscope (Zeiss Meta 510) to capture images of fluorescent proteins. Confocal microscopy observation was performed as previously described (Zhang and Hu, 2009). We used 458-, 488-, 514-, 543-, and 633-nm lasers for excitation of CFP, GFP, YFP, DsRed, and chlorophyll, respectively. For emission, we used 465- to 510-nm band-pass (CFP), 505- to 530-nm band-pass (GFP), 520- to 555-nm band-pass (YFP), 560- to 615-nm band-pass (DsRed2), and 650-nm long-pass (chlorophyll) filters. All images were obtained from optical sections of 6 μ m in depth.
Sugar Dependence and 2,4-DB/IBA Response Assays
For sugar dependence analysis, seeds were placed on half-strength MS agar plates supplemented with or without 1% (w/v) sucrose, stratified at 4°C for 2 d in the dark, and exposed to 24 h of light to induce germination before being placed in dark conditions. After 5 d of seedling growth in the dark, hypocotyl lengths were measured using ImageJ. To study the response to 2,4-DB and IBA, 2,4-DB (0.8 μ M) or IBA (final concentration 0, 2.5, 5, 7.5, 10, 12.5, 15, 20, and 30 μ M) was added to half-strength LS agar media supplemented with 0.5% sucrose. After 2 d of stratification, seeds were kept under low-intensity light for 5 d. Hypocotyls (for sugar dependence assay) and roots (for IBA and 2,4-DB responses) were scanned using an EPSON scanner and measured using ImageJ (http://rsb.info.nih.gov/ij/). For all statistic analyses, n = 60 and P < 0.05.
Immunoblot Analysis
Total protein was extracted from leaf discs of 4-week-old Arabidopsis plants or tobacco (Nicotiana tabacum) leaves. Homogenized leaf tissue was kept in 1 × SDS-PAGE sample buffer, boiled for 5 min, and centrifuged for 5 min. The supernatant was run on SDS-PAGE gels and transferred to Immobilon-P membrane for blotting (Millipore). Primary antibodies used to detect proteins include a rabbit polyclonal GFP antibody for CFP and GFP (Santa Cruz Biotechnology), a mouse monoclonal His antibody for the 6XHis tag (Millipore Antibodies), a rabbit monoclonal HA antibody for HA tag (Cell Signaling Technology), and a rabbit monoclonal FLAG antibody for FLAG tag (Cell Signaling). As secondary antibody, we used goat anti-rabbit IgG (for α-GFP, α-HA, and α-FLAG) or goat anti-mouse IgG (for α-His) from LI-COR Biosciences.
Co-IP
The full-length coding sequence of the tested proteins was cloned into binary vectors pEarleyGate201 or pEarleyGate202 (CD3-687 and CD3-688 from ABRC) to generate HA protein (HA fused to the N terminus of each protein) and FLAG protein (FLAG fused to the N terminus of each protein). The FLAG epitope sequence used is DYKDDDDK, and the HA epitope is YPYDVPDYA. All constructs used for co-IP in this study are listed in Supplemental Table 1 online. Agrobacteria containing each HA and FLAG protein pair were coinfiltrated into leaves of 4-week-old N. tabacum (cv Petit Havana) plants grown at 25°C (Goodin et al., 2002). After 48 h, leaf discs were collected and homogenized in lysis buffer (Nomura et al., 2006). The lysis buffer contains 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, pH 7.5, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 0.5 mM DTT, and plant protease inhibitor cocktail. The samples were then centrifuged at 20,000g for 15 min at 4°C to remove insoluble debris. The supernatant was dialyzed against dialysis buffer (10 mM Tris-HCl, pH 7.5, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT, and 0.3M KCl). The supernatant was incubated with anti-HA agarose beads (Sigma-Aldrich) overnight at 4°C, and the mixture was centrifuged at 500g for 1 min to collect agarose beads, which were then washed three times with lysis buffer and resuspended in 1 × SDS-PAGE sample buffer for immunoblot analysis. Proteins were separated on SDS-PAGE gels and transferred to Immobilon-P membrane, followed by immunodetection by α-FLAG and α-HA antibodies.
BiFC Assays
The full-length coding sequence of DRP5B, DRP3A, DRP3B, PEX11a, PEX11b, PEX11c, PEX11d, PEX11e, FIS1A, and FIS1B was individually cloned into the binary vector pFGC5941 (stock CD3-447 from ABRC, with the ChsA intron removed) to generate YN-protein (N-terminal fragment of YFP fused to the N terminus of each protein) and YC-protein (C-terminal fragment of YFP fused to the N terminus of each protein), as described previously (Bracha-Drori et al., 2004). An HA tag was added to the N terminus of YN-protein, and a 6XHis tag was added to the N terminus of YC-protein. The HA epitope sequence used in this study is YPYDVPDYA; the 6XHis sequence is KKKKKK. All BiFC constructs for this study are listed in Supplemental Table 1 online. Mixtures of Agrobacteria (strain C58C1) containing each protein pair along with the peroxisomal marker CFP-PTS1 were coinfiltrated into leaves of 4-week-old N. tabacum (cv Petit Havana) plants grown at 25°C (Goodin et al., 2002), resulting in coexpression of these proteins in the same infiltrated area. Imaging analysis of leaf epidermal or mesophyll cells in the infiltrated area was performed by confocal laser scanning microscopy as described above. Immunoblot analysis was also conducted on the infiltrated tissue to confirm coexpression of the proteins.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: PEX11a (At1g47750), PEX11b (At3g47430), PEX11c (At1g01820), PEX11d (At2g45740), PEX11e (At3g61070), FIS1A (At3g57090), FIS1B (At5g12390), DRP3A (At4g33650), DRP3B (At2g14120), and DRP5B (At3g19720).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Confocal Images Showing Peroxisomal Phenotypes in drp5B and arc6 Mutants.
Supplemental Figure 2. Expression Levels of the DRP5B Gene in Various Tissues and Growth Phenotypes of Plants in Ambient Air or Elevated CO2.
Supplemental Figure 3. Immunoblot Analysis of Proteins Extracted from Tissues Used for BiFC Assays.
Supplemental Figure 4. Additional BiFC Assays Testing the Protein–Protein Interaction between DRP5B and FIS1 Proteins and the Homodimerization and Heterodimerization of FIS1 Proteins.
Supplemental Figure 5. Additional Co-IP Assays to Test the Interactions Involving DRP3, FIS1, and PEX11 Proteins.
Supplemental Table 1. Binary Constructs Used for Co-IP and BiFC in This Study.
Acknowledgments
We thank Katherine Osteryoung and her lab members Deena Kadirjan-Kalbach and Jonathan Glynn for providing seeds of drp5B (arc5), drp5A, arc3, and arc6 mutants and the GFP-DRP5B lines, Jilian Fan for making the DsRed2-PTS1 construct, the ABRC for vectors for the BiFC and co-IP assays, Katherine Osteryoung for comments on the manuscript, and Karen Cline for editorial help. This work was supported by the National Science Foundation (MCB 0618335) and by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021) to J.H.
References
- Arimura S., Aida G.P., Fujimoto M., Nakazono M., Tsutsumi N. (2004). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45: 236–242 [DOI] [PubMed] [Google Scholar]
- Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W., Tsutsumi N. (2008). Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell 20: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Delille H.K., Alves R., Schrader M. (2009). Biogenesis of peroxisomes and mitochondria: Linked by division. Histochem. Cell Biol. 131: 441–446 [DOI] [PubMed] [Google Scholar]
- Desai M., Hu J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A., Fagarasanu M., Rachubinski R.A. (2007). Maintaining peroxisome populations: A story of division and inheritance. Annu. Rev. Cell Dev. Biol. 23: 321–344 [DOI] [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M., Arimura S., Mano S., Kondo M., Saito C., Ueda T., Nakazono M., Nakano A., Nishimura M., Tsutsumi N. (2009). Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 58: 388–400 [DOI] [PubMed] [Google Scholar]
- Gabaldon T., Snel B., van Zimmeren F., Hemrika W., Tabak H., Huynen M.A. (2006). Origin and evolution of the peroxisomal proteome. Biol. Direct 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Kadirjan-Kalbach D., Froehlich J.E., Osteryoung K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Froehlich J.E., Osteryoung K.W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodin M.M., Dietzgen R.G., Schichnes D., Ruzin S., Jackson A.O. (2002). pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schildknegt D., Braakman I., Philippsen P., Tabak H.F. (2005). Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122: 85–95 [DOI] [PubMed] [Google Scholar]
- Hoepfner D., van den Berg M., Philippsen P., Tabak H.F., Hettema E.H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155: 979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Bednarek S.Y., Blumwald E., Hwang I., Jurgens G., Menzel D., Osteryoung K.W., Raikhel N.V., Shinozaki K., Tsutsumi N., Verma D.P. (2003). A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol. 53: 261–265 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. (2007). The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76: 751–780 [DOI] [PubMed] [Google Scholar]
- Jin J.B., Bae H., Kim S.J., Jin Y.H., Goh C.H., Kim D.H., Lee Y.J., Tse Y.C., Jiang L., Hwang I. (2003). The Arabidopsis dynamin-like proteins ADL1C and ADL1E play a critical role in mitochondrial morphogenesis. Plant Cell 15: 2357–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Hu J. (2009). Dynamics of peroxisome abundance: A tale of division and proliferation. Curr. Opin. Plant Biol. 12: 781–788 [DOI] [PubMed] [Google Scholar]
- Kaur N., Reumann S., Hu J. (2009). Peroxisome biogenesis and function. In The Arabidopsis Book. C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Tanaka A., Fujiki Y. (2007). Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313: 1675–1686 [DOI] [PubMed] [Google Scholar]
- Koch A., Schneider G., Luers G.H., Schrader M. (2004). Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117: 3995–4006 [DOI] [PubMed] [Google Scholar]
- Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M.A., Schrader M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278: 8597–8605 [DOI] [PubMed] [Google Scholar]
- Koch A., Yoon Y., Bonekamp N.A., McNiven M.A., Schrader M. (2005). A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 16: 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 147: 1590–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi K., Nagotu S., Krikken A.M., Sjollema K., Deckers M., Erdmann R., Veenhuis M., van der Klei I.J. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119: 3994–4001 [DOI] [PubMed] [Google Scholar]
- Lalonde S., Ehrhardt D.W., Loque D., Chen J., Rhee S.Y., Frommer W.B. (2008). Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 53: 610–635 [DOI] [PubMed] [Google Scholar]
- Li X., Gould S.J. (2003). The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278: 17012–17020 [DOI] [PubMed] [Google Scholar]
- Lingard M.J., Gidda S.K., Bingham S., Rothstein S.J., Mullen R.T., Trelease R.N. (2008). Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard M.J., Trelease R.N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell Sci. 119: 1961–1972 [DOI] [PubMed] [Google Scholar]
- Logan D.C., Scott I., Tobin A.K. (2004). ADL2a, like ADL2b, is involved in the control of higher plant mitochondrial morphology. J. Exp. Bot. 55: 783–785 [DOI] [PubMed] [Google Scholar]
- Mano S., Nakamori C., Kondo M., Hayashi M., Nishimura M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38: 487–498 [DOI] [PubMed] [Google Scholar]
- Marshall P.A., Dyer J.M., Quick M.E., Goodman J.M. (1996). Redox-sensitive homodimerization of Pex11p: A proposed mechanism to regulate peroxisomal division. J. Cell Biol. 135: 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Kuwayama H., Urushihara H., Nakanishi H. (2008). Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. USA 105: 15202–15207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Nishida K., Mori T., Matsuzaki M., Higashiyama T., Kuroiwa H., Kuroiwa T. (2003). A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15: 655–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., DebRoy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Nyathi Y., Baker A. (2006). Plant peroxisomes as a source of signalling molecules. Biochim. Biophys. Acta 1763: 1478–1495 [DOI] [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Suzuki K., Mori T., Ichikawa T., Matsui M., Nakanishi H., Miyagishima S.Y. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth T., Reumann S., Zhang X., Fan J., Wenzel D., Quan S., Hu J. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Pinfield-Wells H.M., Graham I.A. (2006). Storage reserve mobilization and seedling establishment in Arabidopsis In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Nunnari J. (2003). The division of endosymbiotic organelles. Science 302: 1698–1704 [DOI] [PubMed] [Google Scholar]
- Praefcke G.J., McMahon H.T. (2004). The dynamin superfamily: universal membrane tubulation and fission molecules?. Nat. Rev. Mol. Cell Biol. 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Purdue P.E., Lazarow P.B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17: 701–752 [DOI] [PubMed] [Google Scholar]
- Reumann S., Weber A.P. (2006). Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled–others remain. Biochim. Biophys. Acta 1763: 1496–1510 [DOI] [PubMed] [Google Scholar]
- Schluter A., Fourcade S., Ripp R., Mandel J.L., Poch O., Pujol A. (2006). The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol. Biol. Evol. 23: 838–845 [DOI] [PubMed] [Google Scholar]
- Schrader M. (2006). Shared components of mitochondrial and peroxisomal division. Biochim. Biophys. Acta 1763: 531–541 [DOI] [PubMed] [Google Scholar]
- Scott I., Tobin A.K., Logan D.C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57: 1275–1280 [DOI] [PubMed] [Google Scholar]
- Serasinghe M.N., Yoon Y. (2008). The mitochondrial outer membrane protein hFis1 regulates mitochondrial morphology and fission through self-interaction. Exp. Cell Res. 314: 3494–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S., Erdmann R. (2005). Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272: 5169–5181 [DOI] [PubMed] [Google Scholar]
- Titorenko V.I., Mullen R.T. (2006). Peroxisome biogenesis: The peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 174: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach K., Payne G.S. (1993). Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 12: 3049–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Rayapuram N., Subramani S. (2005). The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 17: 376–383 [DOI] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2008). FISSION1A and FISSION1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol. Plant 1: 1036–1047 [DOI] [PubMed] [Google Scholar]
- Zhang X., Hu J. (2009). Two small protein families, DYNAMIN-RELATED PROTEIN3 and FISSION1, are required for peroxisome fission in Arabidopsis. Plant J. 57: 146–159 [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]