This work presents the crystal structure of mint heteromeric prenyltransferase, which is responsible for the biosynthesis of geranyl pyrophosphate, a precursor of the monoterpene menthol. By combining biochemical and genetic complementation approaches, the authors show that the molecular mechanism regulating specific product formation is mediated by protein–protein interactions.
Abstract
Terpenes (isoprenoids), derived from isoprenyl pyrophosphates, are versatile natural compounds that act as metabolism mediators, plant volatiles, and ecological communicators. Divergent evolution of homomeric prenyltransferases (PTSs) has allowed PTSs to optimize their active-site pockets to achieve catalytic fidelity and diversity. Little is known about heteromeric PTSs, particularly the mechanisms regulating formation of specific products. Here, we report the crystal structure of the (LSU · SSU)2-type (LSU/SSU = large/small subunit) heterotetrameric geranyl pyrophosphate synthase (GPPS) from mint (Mentha piperita). The LSU and SSU of mint GPPS are responsible for catalysis and regulation, respectively, and this SSU lacks the essential catalytic amino acid residues found in LSU and other PTSs. Whereas no activity was detected for individually expressed LSU or SSU, the intact (LSU · SSU)2 tetramer produced not only C10-GPP at the beginning of the reaction but also C20-GGPP (geranylgeranyl pyrophosphate) at longer reaction times. The activity for synthesizing C10-GPP and C20-GGPP, but not C15-farnesyl pyrophosphate, reflects a conserved active-site structure of the LSU and the closely related mustard (Sinapis alba) homodimeric GGPPS. Furthermore, using a genetic complementation system, we showed that no C20-GGPP is produced by the mint GPPS in vivo. Presumably through protein–protein interactions, the SSU remodels the active-site cavity of LSU for synthesizing C10-GPP, the precursor of volatile C10-monoterpenes.
INTRODUCTION
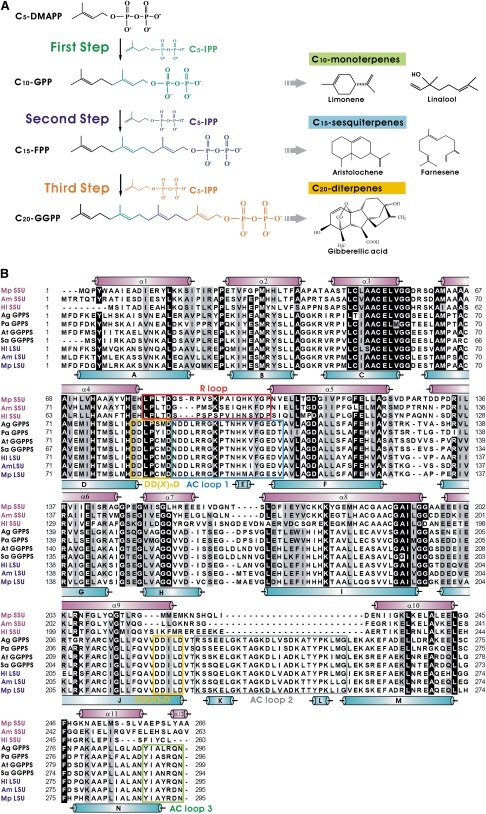
Linear prenyl pyrophosphates (LPPs) are the precursors for the more than 55,000 terpenes (isoprenoids) that have been identified in various organisms. Many are essential for important biological processes, such as protein prenylation (Ras, Rab, and Rho), proper functioning of the electron transport chain (quinine and heme a), glycoprotein biosynthesis (dolichol), and the metabolism of growth hormones (gibberellin, cytokinin, and sterol) (Liang et al., 2002; McTaggart, 2006; Gershenzon and Dudareva, 2007; Christianson, 2008; Kirby and Keasling, 2009). Some terpenes have considerable commercial interest as medicines (taxol for anticancer and artemisinin for antimalaria), flavors and fragrances (menthol and linalool), and nutritional supplements (carotenoid and vitamin A) (Pichersky et al., 2006; Gershenzon and Dudareva, 2007; Kirby and Keasling, 2009).
In plants, terpenes are derived from the universal five-carbon (C5) precursor isopentenyl pyrophosphate (C5-IPP) (Pichersky et al., 2006). Starting with C5-dimethylallyl pyrophosphate (C5-DMAPP), head-to-tail condensation reactions with one to three molecules of C5-IPP can generate C10-geranyl pyrophosphate (C10-GPP), C15-farnesyl pyrophosphate (C15-FPP), and C20-geranylgeranyl pyrophosphate (C20-GGPP). These products are generated by reactions involving the corresponding enzymes: C10-GPP synthase (GPPS), C15-FPP synthase (FPPS), and C20-GGPP synthase (GGPPS) (Figure 1A). These LPPs are the key intermediates for biosynthesis of diverse terpenes (Pichersky et al., 2006; Kirby and Keasling, 2009). Because the lengths of LPPs determine their distinct physiological roles, the production of LPPs is precisely regulated by their respective prenyltransferases (PTSs), groups of highly conserved enzymes in the cells (Kellogg and Poulter, 1997; Ogura and Koyama, 1998; Wang and Ohnuma, 1999; Liang et al., 2002; Szkopinska and Plochocka, 2005). PTSs can be further classified into cis and trans types on the basis of the type of double bond formed during C5-IPP condensation (Liang et al., 2002). Here, we focus on the trans-type PTSs.
Figure 1.
Catalytic Reactions and Multiple Sequence Alignment of Plant PTSs.
(A) Schematic diagram of catalytic reactions of PTSs.
(B) SSUs from Mp GPPS (Mp SSU), Am GPPS (Am SSU), and Hl GPPS (Hl SSU), Abies grandis GPPS (Ag GPPS), P. abies GPPS (Pa GPPS), Arabidopsis thaliana GGPPS (At GGPPS), S. alba GGPPS (Sa GGPPS), and LSUs from Hl GPPS (Hl LSU), Am GPPS (Am LSU), and Mp GPPS (Mp LSU) are included in the alignment. Regions in the SSU and LSU of Mp GPPS corresponding to their respective structural α-helices are denoted by purple and cyan cylinders. Identical and similar amino acid residues are shaded in black and gray, respectively. The conserved functional motifs, DD(X)nD, are denoted by yellow boxes. The R loop of SSU is boxed in red. The AC loops 1, 2, and 3 of LSU are boxed in blue, gray, and green, respectively. All sequences presented here have the N-terminal signal peptides omitted.
The function and structure of homomeric PTSs, such as FPPS and GGPPS, have been well studied (Ohnuma et al., 1996; Tarshis et al., 1996; Hemmi et al., 2003; Hosfield et al., 2004; Chang et al., 2006; Gabelli et al., 2006; Kavanagh et al., 2006a, 2006b; Kloer et al., 2006; Rondeau et al., 2006; Guo et al., 2007; Noike et al., 2008). The sequences of different enzymes generally contain <30% conserved amino acids (see Supplemental Figure 1 online). The three-dimensional structure contains a conserved α-helical bundle that surrounds a cavity that is located adjacent to the active site containing two conserved DD(X)nD motifs (D indicates Asp, X indicates any residue; n = 2 or 4) facing each other on opposite helices over the central cavity. These motifs are important for substrate and cofactor binding. Previous studies suggest the existence of a molecular ruler mechanism whereby the contour (shape and size) of the catalytic cavity (tunnel) of PTSs determines the product specificity and substrate selectivity (Ohnuma et al., 1996; Tarshis et al., 1996; Hemmi et al., 2003; Guo et al., 2004a, 2004b; Sun et al., 2005; Chang et al., 2006; Kavanagh et al., 2006a; Kloer et al., 2006; Thulasiram et al., 2007; Noike et al., 2008).
By contrast, heteromeric PTSs have so far only been identified in a few prokaryotes and eukaryotes, such as Bacillus subtilis, Homo sapiens, Humulus lupulus, Antrirrhinum majus, and Mentha piperita (Fujii et al., 1982, 1983; Koike-Takeshita et al., 1995; Zhang et al., 1997; Burke et al., 1999; Burke and Croteau, 2002; Saiki et al., 2003, 2005; Tholl et al., 2004; Ye et al., 2007; Wang and Dixon, 2009) (see Supplemental Figure 1 online). They are composed of two different types of subunits: one is significantly homologous (50% identity) to homomeric PTSs, while the other is less homologous (15% identity), lacks the DD(X)nD motif, and is recognized as a noncatalytic subunit (see Supplemental Figures 2 and 3 online). The noncatalytic subunit is not only required for enzymatic activities of the heteromeric PTSs but also serves to modify catalytic fidelity or to promote catalytic activity in plants.
M. piperita GPPS (Mp GPPS), involved in the biosynthesis of essential oil (menthol) in mint glandular trichomes, is such a two-component heteromeric PTS consisting of a large and a small subunit, denoted LSU and SSU, respectively. LSU is a PTS-like protein with ∼75% sequence identity to plant GGPPS, and presumably its structure is highly similar to that of Sinapis alba GGPPS (Sa GGPPS) (Kloer et al., 2006). SSU is less similar in sequence to other PTS proteins (Figure 1B). Both LSU and SSU are catalytically inactive by themselves (Burke et al., 1999, 2004; Burke and Croteau, 2002; Croteau et al., 2005). Previous studies have shown that Mp GPPS synthesizes C10-GPP, which is the common precursor of C10-monoterpenes, many of which are plant volatiles involved in important biological activities, such as producing floral or fruit scents that attract pollinators and predators of herbivores, emitting signals that ward off pathogens, and acting as mediators of interplant communication (Kessler and Baldwin, 2001; Runyon et al., 2006; Gershenzon and Dudareva, 2007).
Thus far, no structure of a heteromeric PTS is available (see Supplemental Figure 1 online). In an effort to elucidate the molecular mechanism of intersubunit interaction in heteromeric PTSs, we determined the crystal structure of a new (LSU · SSU)2-type heterotetrameric Mp GPPS, in a pseudomature form with the plastid targeting presequence removed. The structure reveals that LSU serves as a catalytic unit, while SSU acts as a regulatory unit. Further kinetic studies and in vivo assay showed that Mp GPPS synthesizes C10-GPP as the major product. No C20-GGPP was produced in vivo, even though LSU possesses a sufficiently large cavity for the accommodation of C20-GGPP. These results provide a significant insight into intersubunit regulation in heteromeric PTS.
RESULTS
Crystal Structure of the Heterotetrameric Mp GPPS
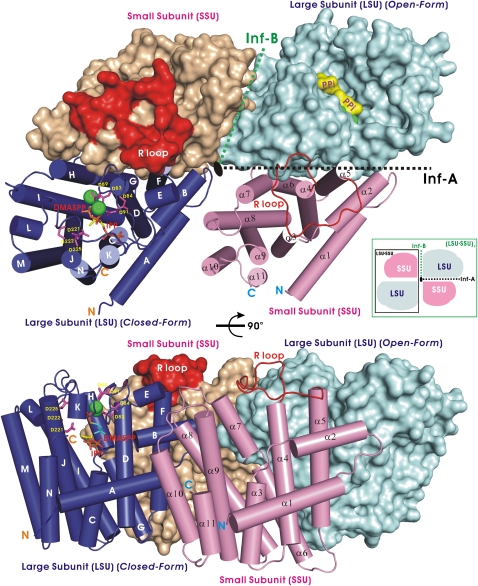
X-ray crystallographic analysis of the Mp GPPS revealed a novel (LSU · SSU)2 architecture composed of two LSU · SSU heterodimers (Figure 2). The LSU and SSU in each LSU · SSU heterodimer are related by a pseudodyad axis, and two LSU · SSU dimers form a tetramer (LSU · SSU)2 about a third dyad, all of which are parallel to one another. This arrangement differs from that of most other tetrameric assemblies, including those with tetrahedral 222-symmetry (hemoglobin) or square fourfold symmetry (neuraminidase) (Russell et al., 2006). Similar structural assembly has been reported for photosystem II, which contains terpenes (Guskov et al., 2009). Crystallographic symmetry elements are unable to relate the protein subunits in any other way due to the orthorhombic space group of P212121 containing only screw axes. When analyzed by gel filtration, a stable heterotetramer (123 kD) of Mp GPPS formed in solution independent of protein concentration (see Supplemental Figure 4 online). Furthermore, previous studies using immunocytochemical localization have demonstrated that LSU and SSU of Mp GPPS coexist within the leucoplasts of the mint glandular trichomes (Turner and Croteau, 2004).
Figure 2.
Architecture of the (LSU · SSU)2-Type Heterotetrameric Mp GPPS.
One LSU subunit is shown as a filled surface model in cyan, and the other LSU subunit is presented as cylinders in blue. Their associated SSU subunits are shown as magenta cylinders and a wheat-colored surface model, respectively. The conserved DD(X)nD motifs are shown as sticks in magenta and the Mg2+ ions as green balls in the LSU. The bound C5-DMASPP (a substrate analog) and C5-IPP are also shown as sticks. In the surface models, the two pyrophosphate groups (PPi) are highlighted in yellow, and the R loop of SSU is colored in red. The extensive interface A (inf-A) associates LSU and SSU into an LSU · SSU dimer. Interface B (inf-B) mediates interactions between two LSU · SSU dimers in an (LSU · SSU)2 tetramer. The bottom part of the figure shows the model rotated 90°.
Our heterotetrameric crystal structures also contain bound ligands, including (1) Mg2+ ions (denoted Mp GPPS-Mg2+), (2) IPP (Mp GPPS-IPP), (3) the nonhydrolyzable DMAPP analog dimethylallyl thiopyrophosphate (DMASPP), IPP, and Mg2+ ions (Mp GPPS-Mg2+/IPP/DMASPP), and (4) GPP and Mg2+ ions (Mp GPPS-Mg2+/GPP; see Supplemental Table 1 and Supplemental Figure 5 online). Taken together, the crystallographically refined models encompass both full-length LSU (residues 1 to 295) and SSU (residues 1 to 266). Excluding regions of ligand-induced conformational change, all structures are virtually identical with an average root mean square deviation of 0.63 Å for 1059 Cαatoms (see Supplemental Figure 6 online). The LSU and SSU in an LSU · SSU dimer are associated by helices B, F, G, and H of LSU and helices α-2, α-5, α-6, and α-7 of SSU, forming interface A (1980 Å2) (Figure 2). Between two LSU · SSU dimers in an (LSU · SSU)2 tetramer, helices A and B, and helices α-7, α-8, and α-10 form interface B (380 Å2) (Figure 2). Interface A is similar to the subunit interface of most homodimeric PTSs but is larger in area, particularly compared with the 1570 Å2 interface of Sa GGPPS. When LSU or SSU was expressed alone in Escherichia coli, each purified protein did not form a dimer, as analyzed using gel filtration chromatography. These results indicate that the molecular surface for dimer formation is quite different in Mp GPPS and Sa GGPPS.
In fact, interface A has more hydrogen bonds and salt bridges, which contribute to a tighter interaction, relative to the other homodimeric PTSs (see Supplemental Table 5 online). When analyzing residues in the interface area of mint LSU and that between the two monomers in Sa GGPPS, we found three key differences (Ser-135, Ala-144, Lys-155 of Sa GGPPS versus Ala-126, Glu-134, and Val-145 of LSU, respectively) that may account for the LSU being unable to form a homodimer. In Sa GGPPS, the side chain of Lys-155 forms a direct salt bridge across the dimer interface to Glu-131 of the other subunit, and it also interacts indirectly with the polar residue of Ser-135, mediated by interfacial water molecules. Interestingly, Lys-155 is a conserved residue, also found in other plant GGPPSs and other LSUs possessing GGPPS activity (Tholl et al., 2004; Wang and Dixon, 2009) (Figure 1B). The LSU of Mp GPPS cannot accommodate those interactions because the Lys-155 and Ser-135 of Sa GGPPS are replaced by Val-145 and Ala-126 of LSU, respectively, eliminating the salt bridge interaction. In addition, Ala-144 of Sa GGPPS is replaced by Glu-134 of LSU. If two LSUs are modeled into a dimer such as that of Sa GGPPS, the two twofold related opposing Glu-134 side chains will result in negatively charged repulsion. In yeast GGPPS, mutating only one interface residue of Met-111 into Glu resulted in the disruption of dimer formation, and mutating Leu-8 and Ile-9 in the N-terminal helix, which is also involved in dimer formation, into Gly had a similar disruptive effect (Lo et al., 2009). Consequently, the LSU monomer prefers to associate with SSU rather than to form a homodimer. The monomeric LSU might not have a stable structure to afford an enzyme function.
Catalytic Activities of Mp GPPS
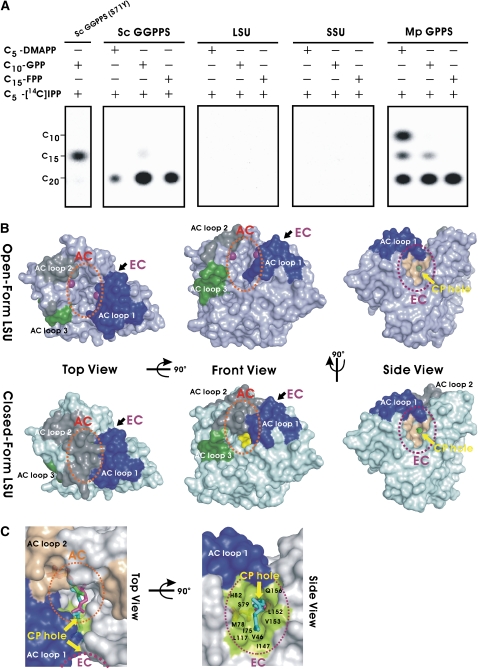
Neither LSU nor SSU alone showed detectable PTS activity despite the marked sequence similarity of LSU to those of plant GGPPSs (Burke et al., 1999; Burke and Croteau, 2002; Burke et al., 2004) (Figure 3A). These results are consistent with those using other heteromeric PTSs found in human and yeast DPPS, human and mouse SPPS, and bacterial HPPS and HEPPPS (Fujii et al., 1982, 1983; Koike-Takeshita et al., 1995; Zhang et al., 1997; Saiki et al., 2003, 2005) (see Supplemental Figure 1 online). By contrast, LSU of A. majus GPPS (Am GPPS) and H. lupulus GPPS (Hl GPPS) are active under in vitro assay conditions (Tholl et al., 2004; Wang and Dixon, 2009).
Figure 3.
In Vitro Product Analysis of Mp GPPS and Conformations of the LSU.
(A) Functional assays (thin layer chromatography) of product synthesis of individual LSU and SSU subunits and complexes of Mp GPPS with three allylic substrates (left column) and C5-[14C] IPP. The products of wild-type Sc GGPPS and a mutant (S71Y), synthesizing C20-GGPP and C15-FPP, were used as markers (Chang et al., 2006).
(B) Surface representations of the open-form (Mp GPPS-Mg2+) and the closed-form [Mp GPPS-Mg2+/IPP/DMASPP (I)] of LSU. The C5-DMASPP (green) and C5-IPP (yellow) ligands are shown on the surface models and the Mg2+ ions as purple balls. AC loops 1, 2, and 3 are highlighted in blue, gray, and green, respectively. Yellow arrows indicate the CP hole for product elongation beyond C10-GPP from the AC (orange dotted circle) into the EC (purple dotted circle).
(C) Superposition of the structures of Sa GGPPS and Mp GPPS. It is likely that C20-GGPP (cyan, from Sa GGPPS) extends through the CP hole into the hydrophobic EC of LSU (green surface). C5-IPP (green, from Mp GPPS-IPP) and C10-GPP (magenta, from Mp GPPS-Mg2+/GPP) are represented as sticks.
When SSU and LSU are coexpressed in E. coli, the purified heterotetrameric Mp GPPS generates not only the anticipated C10-GPP, but also the unexpected C20-GGPP in assays using ample amounts of C5-[14C] IPP and C5-DMAPP as substrates (Figure 3A). To further investigate the substrate specificity of Mp GPPS, the enzyme was incubated with other allylic substrates of C10-GPP, C15-FPP, and C20-GGPP in the presence of C5-[14C] IPP (Figure 3A). In addition, the well-studied homodimeric Saccharomyces cerevisiae GGPPS (Sc GGPPS) served as a positive control to verify the in vitro assay condition (i.e., whether its specific product of C20-GGPP is generated) (Hemmi et al., 2003; Chang et al., 2006; Lo et al., 2009). These results indicate that Mp GPPS can also recognize C10-GPP and C15-FPP as allylic substrates and can react with C5-[14C] IPP to produce C20-GGPP as the final product. The two major products were also unambiguously detected in the time-course experiment: C10-GPP when C5-DMAPP was incubated with C5-[14C] IPP and C20-GGPP when C10-GPP with C5-[14C] IPP or C15-FPP and C5-[14C] IPP were used (see Supplemental Figure 7 online). By contrast, C15-FPP only occurred as an intermediate in small amounts.
Intriguingly, although most homomeric and heteromeric PTSs have high fidelity in their catalyzed reactions, a few homomeric PTSs (found in Menthanobacterium thermoautotrophicum, Toxoplasma godii, Myzus persicae, Zea mays cv B73, Picea abies, and heteromeric Hl GPPS) are putatively bifunctional PTSs, while Cryptosporidium parvum PTS is a nonspecific polyprenyl pyrophosphate synthase (Chen and Poulter, 1993; Cervantes-Cervantes et al., 2006; Ling et al., 2007; Artz et al., 2008; Schmidt and Gershenzon, 2008; Vandermoten et al., 2008; Schmidt et al., 2009; Wang and Dixon, 2009). In addition, our kinetic measurements also reveal that Mp GPPS can use C5-DMAPP, C10-GPP, and C15-FPP as substrates and, consistent with our in vitro assays, have a higher affinity for C15-FPP (see Supplemental Table 2 online). The C5-DMAPP kcat value is significantly higher than C10-GPP and C15-FPP by ∼1000-fold and 70-fold, respectively. In sum, C5-DMAPP is the best suitable allylic substrate for Mp GPPS relative to C10-GPP and C15-FPP, as judged by the value of kcat/Km.
The Active Site of Mp GPPS
To investigate the role that LSU plays in Mp GPPS activity, we constructed the [LSU(D83A/D84A/D89A) · SSU]2 mutant in which the three conserved Asp residues of the DD(X)nD motif, located on loop DE of LSU and important in substrate binding and catalysis, were substituted with Ala. The heterotetrameric [LSU(D83A/D84A/D89A) · SSU]2 mutant was completely inactive (see Supplemental Figure 8 online). These results suggest that the enzymatic activity of Mp GPPS is contributed by LSU but not by SSU. By comparing Mp GPPS and homodimeric PTS structures, we conclude that SSU stabilizes LSU by adopting the position of an identical subunit and creating the proper architecture of a functional catalytic site.
In this regard, three notable regions surrounding the active site cavity (AC) of LSU, denoted as AC loops 1, 2, and 3, were identified by comparing the superposed structures (Figure 3B; see Supplemental Figure 6 online). AC loop 1 between helices D and F contains several critical residues, Asp-83, Asp-84, Asp-89, Asp-91, Arg-94, and Arg-95, that interact with both allylic and homoallylic substrates (see Supplemental Figure 9 online). In Sa GGPPS and Sulfolobus solfataricus HPPS (Ss HPPS) (Sun et al., 2005; Kloer et al., 2006), two homodimeric PTSs having the highest DALI scores, (highest structural similarities) to Mp GPPS, and the regions corresponding to AC loop 1 showed dramatic ligand binding-induced conformational changes necessary for substrate entry or product release (Holm and Sander, 1993) (see Supplemental Figure 10 online). The conformational switch for converting the unliganded (open) to the ligand-bound (closed) structures is located mainly around helices J–N of LSU, including AC loops 2 and 3, which is similar to other homomeric PTSs (Hosfield et al., 2004; Sun et al., 2005; Kloer et al., 2006; Rondeau et al., 2006). AC loop 2 functions as a gate for allylic substrate entry, and AC loop 3 is involved in homoallylic substrate binding. Moreover, it was observed that two conserved DD(X)nD motifs, which are located far apart from each other in the open form, would come closer together by ligand-induced interactions (Figure 3B; see Supplemental Figure 9 online). In the catalytic site, obvious conformational changes also cause a shift in the side chain orientation of Lys-44, Arg-95, and Lys-235 for interaction with substrates (see Supplemental Figure 9 online).
By contrast, the SSU of Mp GPPS does not contain either of the catalytically important DD(X)nD motifs. The regions in SSU corresponding to AC loops 1, 2, and 3 of LSU are not very conserved in terms of their length and sequence, even among the SSU of different plant GPPS (Figure 1B). Nevertheless, the loop connecting helices α-4 and α-5 of SSU, denoted R loop here, adopts a similar disposition as does its equivalent AC loop 1 of LSU (Figure 2; see Supplemental Figure 11 online). AC loop 2 is completely absent in SSU, and the C-terminal segment shows a conformation quite different from that of the AC loop 3 in LSU. The R loop of SSU may restrict movement of AC loop 1 of LSU, in much the same way as do the two AC loops 1 interact with each other in the homodimeric PTSs (Sun et al., 2005; Kloer et al., 2006). Further studies are needed to elucidate the precise function of this loop.
Roles of the Three AC Loops
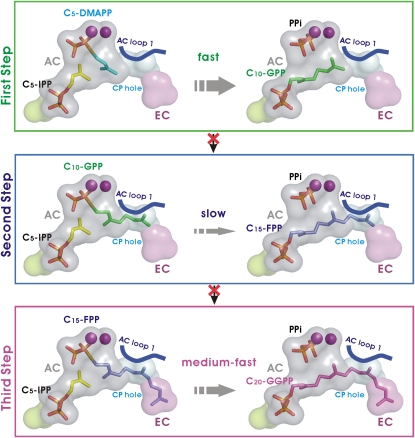
The crystal structure of Mp GPPS shows that the active-site cavity itself is limited in size, being just large enough for C10-GPP (Figures 3B and 3C). However, adjacent to it is a second elongation cavity (EC) that can accommodate longer prenyl products (i.e., C15-FPP and C20-GGPP; Figure 3C) and is connected to the AC through a cavity penetration (CP) hole. Overall, this arrangement resembles a flexible hourglass. This two-chamber architecture (Figures 3B and 3C) is probably a result of catalytic site remodeling of LSU by SSU. The strong interactions between LSU and SSU restrict the enzyme's specificity to the production of C10-GPP. By contrast, the homodimeric Sa GGPPS has an unrestricted single chamber in each subunit, allowing the synthesis of C20-GGPP. The hourglass-like cavity helps explain the reaction kinetics and product specificity (Figure 3B; see Supplemental Table 2 online). When C20-GGPP is synthesized from C5-DMAPP and C5-IPP, the second step (generating C15-FPP) is much slower than the first and third steps (generating C10-GPP and C20-GGPP, respectively; see Figure 1A), allowing time for the open-close movements of AC loop 1 and the release of the first product, C10-GPP. Once C15-FPP is produced, its hydrocarbon moiety immediately penetrates the CP hole, and the reaction proceeds to form C20-GGPP (Figure 3C).
This hypothesis predicts a bottleneck in the C15-FPP production (C10-GPP with C5-IPP) in the second step, which is consistent with our kinetic data in that the value of kcat/Km in the first step is higher than that of the second step by ∼1000-fold (see Supplemental Table 2 online). Our results also suggest that catalytic reaction may be diversified via subunit interactions. This differs from previous studies of homomeric PTSs in which the product chain length was observed to be increased by minimal changes of critical residues to reduce their steric effect and expand the catalytic cavity (Guo et al., 2004a, 2004b; Sun et al., 2005; Chang et al., 2006).
Presumably, the remodeling of a GGPPS-like single-chamber active site for C20-GGPP into a two-chamber, hourglass-like architecture, which prefers C10-GPP over C20-GGPP, can be a result of amino acid substitutions in the vicinity of the active site. However, structural comparison did not reveal any remarkably different residue in the active-site region (see Supplemental Figure 12 online). In fact, all different residues between Sa GGPPS and the LSU of Mp GPPS are found either on the protein surface or at the subunit interface, and all active-site residues are conserved between these two highly homologous proteins (Figure 1B). Further from the active site, near the LSU-SSU interface, occur three residues that are different from those in Sa GGPPS, namely, Cys-161, Val-160, and Ser-107. The former two correspond to Ser-171 and Ile-170 in Sa GGPPS, and they are not likely to change the chamber structure. The smaller Val-160 could make the cavity even larger. Although Ser-107 of LSU, near the C terminus of AC loop 1, makes a hydrogen bond with Arg-89 in the R loop of SSU, the significance of this interaction is uncertain. Consequently, the two-chamber formation of active site in Mp GPPS should be a result of intersubunit interactions.
Based on a structural comparison between the open and closed forms of LSU, we found that AC loop 2 is a highly mobile region, relative to AC loops 1 and 3 (Figure 3B; see Supplemental Figure 13 online). In addition, AC loop 2 has a similar conformation in Mp GPPS-Mg2+ and Mp GPPS-IPP and acts like a lid in the structure of Mp GPPS-Mg2+/IPP/DMASPP (see Supplemental Figure 13 online). This result indicates that the extensive conformational change on AC loop 2 is induced by allylic substrate rather than by the homoallylic substrate and that AC loop 2 serves to shield the allylic substrate from solvent during the catalytic reaction (see Supplemental Figure 13 online).
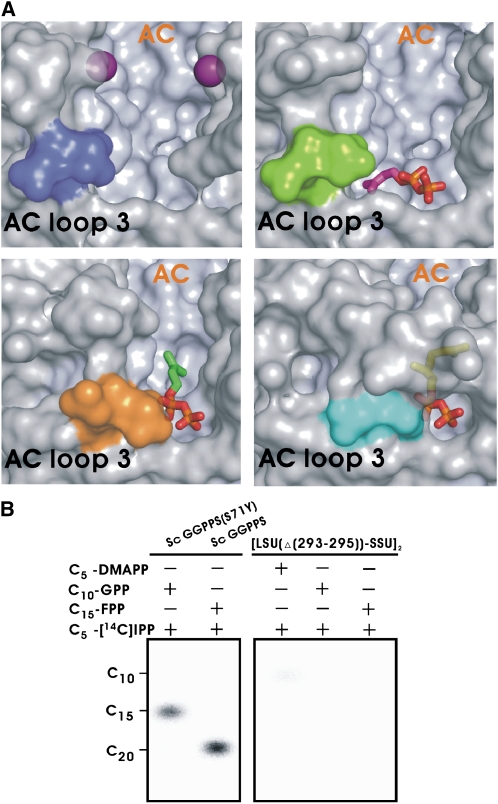
Two physically distinct ligand binding sites, for allylic substrate (C5-DMAPP) with Mg2+, C10-GPP, and homoallylic substrate (C5-IPP), and one novel misoriented binding site of C5-IPP are represented in our ligand-bound structures (see Supplemental Figures 5 and 14 online). In a ternary complex, two orientations of the bound C5-IPP in the individual binding pocket are seen, illustrating how the IPP molecule enters in the proper orientation crucial for reacting with the allylic substrate (Figure 4A; see Supplemental Figure 14 online). AC loop 3, especially the three conserved residues (Arg-293, Asp-294, and Asn-295) at the C terminus of LSU, seems to regulate the binding of C5-IPP in the correct position for the ensuing catalytic reaction (Figure 4A). Consistently, the C-terminal deletion mutant of [LSU(Δ(293-295)) · SSU]2 is essentially inactive; only a minute amount of C10-GPP can be detected when C5-DMAPP reacts with C5-IPP (Figure 4B), attesting to the importance of AC loop 3 in catalysis.
Figure 4.
Function of AC Loop 3.
(A) C-terminal conformational changes of LSU upon C5-IPP binding. The three conserved terminal residues (RDN) are shown as colored molecular surfaces (blue for Mp GPPS-Mg2+, green for Mp GPPS-Mg2+/IPP/DMASPP [II], orange for Mp GPPS-IPP, and cyan for Mp GPPS-Mg2+/GPP). The Mg2+ ions are shown as purple balls, and the C5-IPP and C10-GPP ligands are shown as sticks.
(B) In vitro analysis (thin layer chromatography) of products of [LSU(Δ(293-295)) · SSU]2, using C20-GGPP and C15-FPP synthesized by Sc GGPPS and the mutant S71Y as markers.
In Vivo Function of Mp GPPS
Because the LSU has EC to accommodate C20-GGPP, and two major products (C10-GPP and C20-GGPP) can be detected by in vitro assay, it is possible that the heterotetrameric Mp GPPS may be a bifunctional enzyme, similar to the heteromeric Hl GPPS (Wang and Dixon, 2009). However, there has not been any biological evidence that C20-GGPP is observed as a product of Mp GPPS in nature. Instead, previous studies have shown that the transcription level of Mp GPPS correlates closely with C10-monoterpene biosynthesis in mint (McConkey et al., 2000; Croteau et al., 2005).
In order to investigate whether Mp GPPS possesses GGPPS activity in nature, we used a genetic complementation method by substituting crtE-encoded GGPPS in the crt gene cluster of Pantoea ananatis (formerly Erwinia uredovora), which is used for the biosynthesis of carotenoid (a prominent yellow pigment) (Misawa et al., 1990; Zhu et al., 1997; Kainou et al., 1999; Engprasert et al., 2004; Ye et al., 2007). Since E. coli does not possess an intrinsic gene encoding GGPPS, E. coli cells harboring pACCAR25 ΔcrtE, which carries crtB (phytoene desaturase), crtI (phytoene desaturase), crtY (lycopene cyclase), crtZ (β-carotene hydroxylase), and crtX (zeaxanthin β-glucosidase), cannot, without the presence of the crtE gene, accumulate carotenoid unless C20-GGPP is generated (see Supplemental Figure 15A online). The transformants carrying the pACCAR25 ΔcrtE and human or yeast GGPPS gene (positive control) are expected to show a notable yellow color, indicating the function of crtE has been substituted, whereas the empty vectors and the constructs expressing GPPS from orchid (Phalaenopsis bellina), FPPS from E. coli, or Sc GGPPS (S71Y) mutant are used as negative controls (Kainou et al., 1999; Hosfield et al., 2004; Chang et al., 2006; Kavanagh et al., 2006a; Hsiao et al., 2008) (see Supplemental Figure 15B online). LSU alone again had no GGPPS activity by such in vivo assay methods, despite the LSU sequence being ∼70% identical to those of other plant GGPPS.
Interestingly, neither cotransformation of LSU and SSU nor transformation of a duet vector with LSU and SSU, namely, LSU-SSU/pET-32, in E. coli carrying pACCAR25 ΔcrtE, resulted in detectable yellow pigmentation when measured for the optical absorption of the extracted pigments from the transformants (see Supplemental Figures 15B and 15C online). We then considered whether Mp GPPS is unable to produce the yellow pigment of carotenoid due to the limited substrate pool of C15-FPP in E. coli cytosol. According to our kinetic results (Figure 1A; see Supplemental Table 2 online), the second condensation step of Mp GPPS from C10-GPP to C15-FPP is a bottleneck. E. coli harboring pACCAR25 ΔcrtE coexpressing either Mp GPPS plus Ec FPPS, or Mp GPPS plus Sc GGPPS (S71Y), was employed to supply sufficient C15-FPP for Mp GPPS. However, this construct still failed to produce C20-GGPP (see Supplemental Figures 15B and 15C online). Taken together, these data suggest that Mp GPPS does not preserve the function of GGPPS in vivo even though its LSU is very similar to Sa GGPPS in terms of amino acid sequence and the three-dimensional structure (Kloer et al., 2006) (see Supplemental Figure 12 online).
DISCUSSION
Our structural and mutagenetic studies offer a new insight into how the product specificity and fidelity of the (LSU.SSU)2-type Mp GPPS is determined via intersubunit regulation through a novel molecular mechanism. In sum, the SSU limits the ability to conduce catalytic reaction beyond the C10-GPP by restricting the connection between AC and EC. Based on structure and sequence comparisons with plant GGPPSs, the two-chamber architecture of LSU in the Mp GPPS structure provides additional cavities (CP hole and EC) for accommodating the longer product of C20-GGPP (Figure 5; see Supplemental Figure 12 online). Our proposed two-chamber mechanism in which the product chain length is regulated by intersubunit interaction is distinct from the previous single-chamber molecular ruler mechanism for well-studied homomeric PTSs, which use bulky residues to serve as flooring at the bottom of each enzyme active site to block further product chain elongation (Ohnuma et al., 1996; Tarshis et al., 1996; Hemmi et al., 2003; Guo et al., 2004a, 2004b; Sun et al., 2005; Chang et al., 2006; Noike et al., 2008). When the elongation barrier is removed in the active site of these enzymes by substituting the bulky residue with a smaller one, longer chain length products are generated.
Figure 5.
Schematic Model of the Two-Chamber Architecture for Product Regulation.
The cavities for C5-IPP (misoriented), AC, CP hole, and EC are presented in green, gray, cyan, and purple, respectively. The homoallylic substrate of C5-IPP, the allylic substrates of C5-DMAPP, C10-GPP, C15-FPP, and C20-GGPP, and PPi are shown as sticks. The Mg2+ ions are presented as purple balls. The AC loop 1 of LSU is depicted as a blue line. The size of the gray arrow indicates the level of catalytic efficiency in the three individual steps of the catalytic reactions (see Figure 1A). The red crosses indicate that the catalytic reactions beyond the first step in vivo are blocked via intersubunit interactions.
In addition, another group of enzymes called terpene synthases, which share a similar α-helical fold with homomeric PTSs, cyclize the allylic substrates to produce a variety of terpene hydrocarbon scaffolds. Accumulated studies have demonstrated that the contours of the active site of terpene synthases can be changed to generate more diversified products by replacing the critical residues around their individual active-site cavities (Yoshikuni et al., 2006; Thulasiram et al., 2007; Xu et al., 2007; O'Maille et al., 2008). In parallel, our studies of heteromeric Mp GPPS reveal that the shape and size of the active-site cavity can be molded by intersubunit interactions (Figure 5). This new mechanism may shed light on the previous mysteries of heteromeric PTSs regarding their molecular basis for product chain length determination.
As the products generated through PTSs serve as critical precursors for several physiological processes, such as protein prenylation, PTSs need to be regulated precisely (Liang et al., 2002; Szkopinska and Plochocka, 2005; McTaggart, 2006). Heteromeric PTSs possess two-component systems, comprised of a noncatalytic and a PTS-like subunit, though the question concerning the role played by the noncatalytic subunit in vivo remains. In plants, biosynthesis of terpenes is compartmentalized in such a way that C10-GPP and C20-GGPP are generated via the plastidic methylerythritol phosphate pathway, whereas C15-FPP and its derivatives (C15-sesqui- and C30-triterpenes) are produced through the cytosolic mevalonic acid pathway (Croteau et al., 2005; Pichersky et al., 2006; Kirby and Keasling, 2009). The LSU and SSU of Mp GPPS have also been shown to occur within the leucoplast (a nonpigmented chloroplast) of the glandular trichome, which is the specialized mint tissue responsible for the production and accumulation of essential oil, a derivative of C10-GPP (Turner and Croteau, 2004). Consequently, the unique SSU of Mp GPPS has evolved to interact with LSU to produce C10-GPP from C5-DMAPP and C5-IPP, which are generated by the plastidic methylerythritol phosphate pathway. In addition, as shown in previous studies of Am GPPS and Hl GPPS, expression levels of their SSU mRNA and protein display a tissue specificity, particularly in flowers and glandular trichomes, where C10-monoterpenes are synthesized. However, their LSUs constitutively express in vegetable and C10-monoterpene storage/emission organs (Tholl et al., 2004; Wang and Dixon, 2009). In summary, the expression of SSU, but not LSU, is closely correlated with the C10-monoterpene biosynthesis. It is therefore intriguing to consider that SSUs have evolved to ensure their catalytic specificity and fidelity in a particular specialized tissue by intersubunit regulation. In addition, these findings also encourage investigation of other heteromeric PTSs in vivo.
Finally, the role played by LSU of Mp GPPS in the production of C20-GGPP in vivo remains to be clarified because no GGPPS activity was detected in LSU of Mp GPPS, in contrast with LSU of Am GPPS and Hl GPPS, which were found to be functional GGPPSs by in vitro assay (Tholl et al., 2004; Wang and Dixon, 2009). Whether these two enzymes are active in vivo could be tested by methods such as the genetic complementation assay used here. Judging from the accumulated carotenoid in our in vivo assay, heteromeric Mp GPPS and LSU alone in E. coli cannot produce detectable amounts of C20-GGPP, which is important in protein geranylgeranylation (Rac, Rap, and Rho) and the biosynthesis pathway of carotenoid and gibberellin in plant plastid (McTaggart, 2006; Pichersky et al., 2006; Gershenzon and Dudareva, 2007). No GGPPS has been identified in the ESTs from the mint glandular trichome (Croteau et al., 2005), but GPPS and GGPPS are generally considered to coexist in plastids (Croteau et al., 2005; Pichersky et al., 2006; Kirby and Keasling, 2009). Consequently, it is worthwhile to investigate how C20-GGPP is manufactured in the leucoplast of mint glandular trichome, if not by the quasibifunctional Mp GPPS.
Taken together, our findings provide a new insight into how the catalytic reactions of PTSs are regulated via intersubunit interactions. Remarkably, only a few heteromeric PTSs have been discovered in prokaryotes and eukaryotes in previous studies, suggesting that the candidate genes encoding the noncatalytic subunit of heteromeric PTSs might have been neglected (see Supplemental Figure 1 online). A renewed search of the possible genes of heteromeric PTSs is warranted.
METHODS
Cloning and Mutagenesis
The truncated versions of Mentha piperita SSU (residues 1 to 266) and LSU (residues 1 to 295) without their plastid targeting sequences in pET-37b (Novagen) with C-terminal His-tag and pET-32a (Novagen) were denoted SSU/pET-37 and LSU/pET-32, respectively, as previously described (Burke and Croteau, 2002). For crystallization, site-directed mutagenesis was performed to yield SSU/pET-37 D2 by deleting an additional seven residues between the C terminus of SSU and His-tag. Other mutants were also prepared by site-directed mutagenesis. The truncated version of LSU was subcloned into pBAD-TOPO (Invitrogen) to create LSU/pBAD. The PCR products of human GGPPS and Escherichia coli FPPS were cloned into pET-46 Ek/LIC and pET-30 Xa/LIC (Novagen), respectively. For a duet vector, the DNA fragment amplified by PCR using primers Sal ISSUF and Not ISSUR was cloned into the Sal I/Not I site of LSU/pET-32, namely, LSU-SSU/pET-32. The primers are shown in Supplemental Tables 3 and 4 online.
Protein Expression and Purification
LSU/pET-32 and SSU/pET-37 D2 were cotransformed to E. coli BL21 (DE3) and induced with 0.5 mM isopropyl β-thiogalactopyranoside (IPTG) at 10°C for 60 h. Cells were collected, resuspended in extraction buffer [50 mM Tris, pH 8.5, 40 mM imidazole, 0.75 M NaCl, 25% (w/v) glycerol, 0.2 M sorbitol, 10 mM MgCl2, 2 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP), and 2 ng mL−1 of benzonase (Novagen) with added Protease Inhibitor Cocktail (Roche)], and lysed by Cell Disruption Solutions (Constant Systems). After ultracentrifugation at 205,000g (Beckman Ti45) for 60 min at 4°C, protein complexes were purified by a Ni column (GE Healthcare) and further isolated (LSU·SSU)2 by gel filtration (Superdex 200; GE Healthcare) in GF buffer (25 mM Tirs, pH 8.5, 0.2 M NaCl, 5% [w/v] glycerol, and 2 mM TCEP). E. coli BL21 (DE3) cells were transformed with SSU/pET-37 D2 to express SSU at 10°C using 0.2 mM IPTG. LSU/pBAD was transformed into E. coli BL21 (DE3) cells and grown in medium (M9 salts, 2% casamino acids, 0.2% glucose, 1 mM MgCl2) to express LSU by the addition of l-arabinose to 0.002% at 10°C. Purification of Mp GPPS mutants and individual SSU and LSU followed a similar procedure to that described above. LSU and SSU were identified by liquid chromatography–nanoelectrospray ionization-tandem mass spectrometry search using the program MASCOT (Perkins et al., 1999). The (LSU·SSU)2 was further determined on a Superdex 200 10/300 GL High Performance column (GE Healthcare) in GF buffer by comparing it with those standards (see Supplemental Figure 5 online).
Crystallographic Analyses
Crystallization was performed by the hanging-drop vapor diffusion method (Hampton Research) at 20°C for 3 to 4 weeks. The Mp GPPS-Mg2+ crystals were obtained by mixing protein solution (4 mg mL−1) with an equal volume of reservoir solution (100 mM Bis-Tris, pH 6.5, 200 mM CH3COONH4, 13 to 19% [w/v] PEG 3350, and 2 to 5% [w/v] PEG 1000) and soaked in the reservoir solution containing 2.5 mM MgCl2 and 15% (v/v) ethylene glycol as a cryoprotectant for 12 h at 20°C prior to data collection. For preparing Mp GPPS-Mg2+/IPP/DMASPP or Mp GPPS-Mg2+/GPP crystals, protein solutions in the presence of 2.5 mM ligands (MgCl2, C5-IPP, and C5-DMASPP or MgCl2 and C10-GPP) were used, and the crystallization conditions were basically the same as described above. The Mp GPPS-Mg2+/IPP/DMASPP or Mp GPPS-Mg2+/GPP crystals were then soaked for 3 s in the reservoir solution containing 2.5 mM of the appropriate ligands and 15% (v/v) ethylene glycol before flash-cooling in liquid nitrogen. For the Mp GPPS-IPP complex, the crystals of Mp GPPS-Mg2+/IPP/DMASPP were soaked in the reservoir solution containing 15% (v/v) ethylene glycol (but not MgCl2) for 15 h at 20°C, during which the bound C5-DMASPP and Mg2+ ions were released from the active site with a concomitant conformational change of the AC loop 2. Diffraction data were collected at beamlines BL13B1 and BL13C1 of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan), Taiwan Contract BL12B2 station at SPring-8 (Hyogo, Japan), as well as BL 5A at the Photon Factory (Tsukuba, Japan). Data were processed using HKL2000 (Otwinowski and Minor, 1997). Five percent randomly selected reflections were set aside for caluculating Rfree (Brunger, 1993). The homodimeric Sa GGPPS (PDB: 2J1O) was used as search model for molecular replacement to determine the initial phases by crystallography and NMR system (Brunger et al., 1998). The (LSU·SSU)2 model was obtained by searching for a second dimer after the first dimer was located. Density modification with twofold noncrystallography symmetry averaging improved the model phases and allowed manual rebuilding with program O and XtalView (Jones et al., 1991; Cowtan and Main, 1996; McRee, 1999). The position and conformation of ligand molecules were modeled by HIC-Up and validated by Fo-Fc map (Kleywegt, 2007). The structure models were determined by repeated cycles of minimization, annealing, and B factor refinement with crystallography and NMR system, and their stereochemical quality was checked by PROCHECK (Laskowski et al., 1993). Diffraction data and refinement statistics are summarized in Supplemental Table 1 online. Molecular graphics were generated with PyMOL (Delano, 2002).
In Vitro Enzymatic Analyses and Kinetic Parameters
In vitro assays were performed in reaction solution (100 mM HEPES, pH 7.5, 0.1% Triton X-100, 5 mM MgCl2, and 50 mM KCl) with 1 μ M enzyme and incubated for 6 h at 25°C except for the time course experiment. The in vitro assays and kinetic measurements followed our published procedures (Kuo and Liang, 2002; Guo et al., 2004a, 2004b; Sun et al., 2005; Chang et al., 2006; Lo et al., 2009). The following substrate mixtures (50 μ M C5-DMAPP with 200 μ M C5-[14C] IPP, 50 μ M C10-GPP with 150 μ M C5-[14C] IPP, 50 μ M C15-FPP with 100 μ M C5-[14C] IPP, and 50 μ M C20-GGPP with 50 μ M C5-[14C] IPP) were used in assays. The products were identified by thin layer chromatography on silica gel 60 RP-18 F254S (Merck) using acetone:water (18:2) as the mobile phase. For Km and kcat measurements of allylic substrates, 0.05 μ M purified Mp GPPS was added in reaction solution with 0.5 to 600 μ M C5-DMAPP, 0.5 to 500 μ M C10-GPP, or 0.5 to 400 μ M C15-FPP in the saturating concentration of C5-[14C] IPP (250 μ M). The C5-IPP Km values for C5-DMAPP (750 μ M), C10-GPP (500 μ M), and C15-FPP (170 μ M) were determined at C5-[14C] IPP concentrations from 0.25 to 300 μ M. Data were analyzed by nonlinear regression of Michaelis-Menten equation to obtain Km and kcat values. Substrates were purchased from Sigma-Aldrich, Echelon Biosciences, and Amersham Pharmacia Biotech.
In Vivo Genetic Complementation Assay
The pACCAR25 ΔcrtE, including the crt gene cluster with the exception of the deleted crtE encoding GGPPS, were developed for identification of GGPPS activity (Misawa et al., 1990; Zhu et al., 1997; Kainou et al., 1999; Engprasert et al., 2004; Ye et al., 2007). The empty vectors of pET-16 and pET-28, and following constructs of Hs GGPPS/pET-46 (human GGPPS), Sc GGPPS/pET-32 (yeast GGPPS), Pb GPPS/pET-15 (orchid GPPS), Ec FPPS/pET-30 (E. coli FPPS), and Sc GGPPS(S71Y)/pET-32 (yeast GGPPS mutant), were used here. The constructs were cotransformed into E. coli BL21 (DE3) carrying pACCAR25 ΔcrtE supplying antibiotics used for selection [chloramphenicol for pACCAR25 ΔcrtE; ampicillin for pET-16, Hs GGPPS/pET-46, Sc GGPPS/pET-32, Pb GPPS/pET-15, Sc GGPPS(S71Y)/pET-32, LSU-SSU/pET-32, or LSU/pET-32; kanamycin for pET-28, Ec FPPS/pET-30, or SSU/pET-37 D2], and the cultured cells were induced with 0.5 mM IPTG for 72 h at 20°C in Luria-Bertani medium until OD600 reached 0.8. For quantification of carotenoid, the same wet weight pellets were harvested by centrifugation and dissolved in 90% (v/v) acetone to extract yellow pigment. The concentration of carotenoid was measured by absorption at a wavelength of 450 nm (Perkin-Elmer Lambda Bio40).
Accession Numbers
Coordinates and structure factors of Mp GPPS-Mg2+, Mp GPPS-IPP, Mp GPPS-Mg2+/IPP/DMASPP (I), Mp GPPS-Mg2+/IPP/DMASPP (II), and Mp GPPS-Mg2+/GPP have been deposited in the Protein Data Bank (http://www.rcsb.org) with the accession codes 3KRA, 3KRC, 3KRF, 3KRO, and 3KRP, respectively. Accession codes for other PTSs, when available, are shown in Supplemental Figure 1 online. Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: SSU of Mp GPPS (Mp SSU), ABW86880; SSU of Am GPPS (Am SSU), AAS82859; SSU of Hl GPPS (Hl SSU), ACQ90681; Abies grandis GPPS (Ag GPPS), AAN01133; Picea abies GPPS (Pa GPPS), ACA21458; Arabidopsis thaliana GGPPS (At GGPPS), NP_195399; Sinapis alba GGPPS (Sa GGPPS), CAA67330; LSU of Hl GPPS (Hl LSU), ACQ90682; LSU of Am GPPS (Am LSU), AAS82860; Mp GPPS (Mp LSU), ABW86879; human GGPPS, NM_004837; yeast GGPPS, SCU31632; E. coli FPPS, NZ_AAMK02000019; and orchid GPPS, EU023907.
Author Contributions
T.-H.C., T.-P.K., and A.H.-J.W. designed research and wrote the manuscript. T.-H.C., and F.-L.H. performed protein preparation. T.-H.C. and F.-L.H. performed enzymatic assays, crystallographic analysis, and in vivo assays. T.-H.C. and T.-P.K. solved and refined the structures. K.-H.T., P.-H.L., and T.-H.C. measured the kinetic parameters. T.-H.C., T.-P.K., P.-H.L., and A.H.-J.W. analyzed data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Classification of Prenyltransferases (trans-Type Only).
Supplemental Figure 2. Amino Acid Sequence Alignment of Heteromeric PTS Subunits That Lack the DD(X)nD Motif.
Supplemental Figure 3. Amino Acid Sequence Alignment of Heteromeric PTS Subunits That Contain the DD(X)nD Motif.
Supplemental Figure 4. Analytic Gel Filtration Assays of Heterotetrameric Mp GPPS.
Supplemental Figure 5. Electron Density Maps for the Ligands.
Supplemental Figure 6. Superpositions of Mp GPPS in Complex with a Variety of Ligands.
Supplemental Figure 7. Time-Course Assays of Substrate and Product Specificities of Mp GPPS.
Supplemental Figure 8. Products Synthesized by the [LSU(D83A/D84A/D89A)·SSU]2 Mutant.
Supplemental Figure 9. Active Sites of Mp GPPS for Ligand Binding.
Supplemental Figure 10. AC Loop 1 Conformational Changes of PTSs.
Supplemental Figure 11. Comparison of SSU and Sa GGPPS
Supplemental Figure 12. Superpositions of LSU of Mp GPPS and Sa GGPPS.
Supplemental Figure 13. Conformational Change of AC Loop 2 Involved in the Allylic Substrate Binding Step.
Supplemental Figure 14. Ligand Binding Locations.
Supplemental Figure 15. In Vivo Activity of GGPPS.
Supplemental Figure 16. Ligand-Induced Conformational Change of Mp GPPS.
Supplemental Table 1. Data Collection and Refinement Statistics.
Supplemental Table 2. Kinetic Parameters of (LSU·SSU)2-Type Heterotetrameric Mp GPPS.
Supplemental Table 3. The Mutagenic Primers Used to Construct Recombinant Mp GPPS.
Supplemental Table 4. The Constructs Used for in Vitro and in Vivo Assays.
Supplemental Table 5. Parameters of Protein Interface Area.
Acknowledgments
We thank Rodney B. Croteau (Washington State University) for providing cDNA constructs of Mp GPPS, Makoto Kawamukai (Shimane University, Japan) for the gift of pACCAR25 ΔcrtE plasmid, Yu-Yun Hsiao (National Cheng Kung University, Taiwan) for supplying the plasmid of Pb GPPS/pET-15, Rey-Ting Guo (Academia Sinica, Taiwan) for helping with the protein expression, and Kai-Fa Huang (Academia Sinica, Taiwan) for assistance in x-ray data collection. The Protein Crystallography Facility of National Synchrotron Radiation Research Center is supported by the National Research Program for Genomic Medicine. Mass spectrometry analyses were performed by the Core Facilities for Proteomics Research located at the Institute of Biological Chemistry, Academia Sinica. This work was supported by Academia Sinica and Core Facility for Protein Production and X-Ray Structural Analysis (NSC97-3112-B-001-035-B4) to A.H.-J.W.
References
- Artz J.D., Dunford J.E., Arrowood M.J., Dong A., Chruszcz M., Kavanagh K.L., Minor W., Russell R.G., Ebetino F.H., Oppermann U., Hui R. (2008). Targeting a uniquely nonspecific prenyl synthase with bisphosphonates to combat cryptosporidiosis. Chem. Biol. 15: 1296–1306 [DOI] [PubMed] [Google Scholar]
- Brunger A.T. (1993). Assessment of phase accuracy by cross validation: The free R value. Methods and applications. Acta Crystallogr. D Biol. Crystallogr. 49: 24–36 [DOI] [PubMed] [Google Scholar]
- Brunger A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burke C., Croteau R. (2002). Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J. Biol. Chem. 277: 3141–3149 [DOI] [PubMed] [Google Scholar]
- Burke C., Klettke K., Croteau R. (2004). Heteromeric geranyl diphosphate synthase from mint: construction of a functional fusion protein and inhibition by bisphosphonate substrate analogs. Arch. Biochem. Biophys. 422: 52–60 [DOI] [PubMed] [Google Scholar]
- Burke C.C., Wildung M.R., Croteau R. (1999). Geranyl diphosphate synthase: Cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc. Natl. Acad. Sci. USA 96: 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Cervantes M., Gallagher C.E., Zhu C., Wurtzel E.T. (2006). Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol. 141: 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.H., Guo R.T., Ko T.P., Wang A.H., Liang P.H. (2006). Crystal structure of type-III geranylgeranyl pyrophosphate synthase from Saccharomyces cerevisiae and the mechanism of product chain length determination. J. Biol. Chem. 281: 14991–15000 [DOI] [PubMed] [Google Scholar]
- Chen A., Poulter C.D. (1993). Purification and characterization of farnesyl diphosphate/geranylgeranyl diphosphate synthase. A thermostable bifunctional enzyme from Methanobacterium thermoautotrophicum. J. Biol. Chem. 268: 11002–11007 [PubMed] [Google Scholar]
- Christianson D.W. (2008). Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 12: 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K.D., Main P. (1996). Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D Biol. Crystallogr. 52: 43–48 [DOI] [PubMed] [Google Scholar]
- Croteau R.B., Davis E.M., Ringer K.L., Wildung M.R. (2005). (-)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 92: 562–577 [DOI] [PubMed] [Google Scholar]
- Delano W.L. (2002). The PyMOL Molecular Graphic System. (http://www.pymol.org/: DeLano Scientific) [Google Scholar]
- Engprasert S., Taura F., Kawamukai M., Shoyama Y. (2004). Molecular cloning and functional expression of geranylgeranyl pyrophosphate synthase from Coleus forskohlii Briq. BMC Plant Biol. 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Koyama T., Ogura K. (1982). Hexaprenyl pyrophosphate synthetase from Micrococcus luteus B-P 26. Separation of two essential components. J. Biol. Chem. 257: 14610–14612 [PubMed] [Google Scholar]
- Fujii H., Koyama T., Ogura K. (1983). Essential protein factors for polyprenyl pyrophosphate synthetases. Separation of heptaprenyl pyrophosphate synthetase into two components. FEBS Lett. 161: 257–260 [DOI] [PubMed] [Google Scholar]
- Gabelli S.B., McLellan J.S., Montalvetti A., Oldfield E., Docampo R., Amzel L.M. (2006). Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design. Proteins 62: 80–88 [DOI] [PubMed] [Google Scholar]
- Gershenzon J., Dudareva N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Guo R.T., et al. (2007). Bisphosphonates target multiple sites in both cis- and trans-prenyltransferases. Proc. Natl. Acad. Sci. USA 104: 10022–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R.T., Kuo C.J., Chou C.C., Ko T.P., Shr H.L., Liang P.H., Wang A.H. (2004b). Crystal structure of octaprenyl pyrophosphate synthase from hyperthermophilic Thermotoga maritima and mechanism of product chain length determination. J. Biol. Chem. 279: 4903–4912 [DOI] [PubMed] [Google Scholar]
- Guo R.T., Kuo C.J., Ko T.P., Chou C.C., Liang P.H., Wang A.H. (2004a). A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products. Biochemistry 43: 7678–7686 [DOI] [PubMed] [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Hemmi H., Noike M., Nakayama T., Nishino T. (2003). An alternative mechanism of product chain-length determination in type III geranylgeranyl diphosphate synthase. Eur. J. Biochem. 270: 2186–2194 [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. (1993). Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Hosfield D.J., Zhang Y., Dougan D.R., Broun A., Tari L.W., Swanson R.V., Finn J. (2004). Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis. J. Biol. Chem. 279: 8526–8529 [DOI] [PubMed] [Google Scholar]
- Hsiao Y.Y., Jeng M.F., Tsai W.C., Chuang Y.C., Li C.Y., Wu T.S., Kuoh C.S., Chen W.H., Chen H.H. (2008). A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)2–4D motif. Plant J. 55: 719–733 [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. (1991). Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kainou T., Kawamura K., Tanaka K., Matsuda H., Kawamukai M. (1999). Identification of the GGPS1 genes encoding geranylgeranyl diphosphate synthases from mouse and human. Biochim. Biophys. Acta 1437: 333–340 [DOI] [PubMed] [Google Scholar]
- Kavanagh K.L., Dunford J.E., Bunkoczi G., Russell R.G., Oppermann U. (2006a). The crystal structure of human geranylgeranyl pyrophosphate synthase reveals a novel hexameric arrangement and inhibitory product binding. J. Biol. Chem. 281: 22004–22012 [DOI] [PubMed] [Google Scholar]
- Kavanagh K.L., Guo K., Dunford J.E., Wu X., Knapp S., Ebetino F.H., Rogers M.J., Russell R.G., Oppermann U. (2006b). The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc. Natl. Acad. Sci. USA 103: 7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg B.A., Poulter C.D. (1997). Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1: 570–578 [DOI] [PubMed] [Google Scholar]
- Kessler A., Baldwin I.T. (2001). Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kirby J., Keasling J.D. (2009). Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu. Rev. Plant Biol. 60: 335–355 [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. (2007). Crystallographic refinement of ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 63: 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloer D.P., Welsch R., Beyer P., Schulz G.E. (2006). Structure and reaction geometry of geranylgeranyl diphosphate synthase from Sinapis alba. Biochemistry 45: 15197–15204 [DOI] [PubMed] [Google Scholar]
- Koike-Takeshita A., Koyama T., Obata S., Ogura K. (1995). Molecular cloning and nucleotide sequences of the genes for two essential proteins constituting a novel enzyme system for heptaprenyl diphosphate synthesis. J. Biol. Chem. 270: 18396–18400 [DOI] [PubMed] [Google Scholar]
- Kuo T.H., Liang P.H. (2002). Reaction kinetic pathway of the recombinant octaprenyl pyrophosphate synthase from Thermotoga maritima: How is it different from that of the mesophilic enzyme. Biochim. Biophys. Acta 1599: 125–133 [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M.J. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr 26: 283–291 [Google Scholar]
- Liang P.H., Ko T.P., Wang A.H. (2002). Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269: 3339–3354 [DOI] [PubMed] [Google Scholar]
- Ling Y., Li Z.H., Miranda K., Oldfield E., Moreno S.N. (2007). The farnesyl-diphosphate/geranylgeranyl-diphosphate synthase of Toxoplasma gondii is a bifunctional enzyme and a molecular target of bisphosphonates. J. Biol. Chem. 282: 30804–30816 [DOI] [PubMed] [Google Scholar]
- Lo C.H., Chang Y.H., Wright J.D., Chen S.H., Kan D., Lim C., Liang P.H. (2009). Combined experimental and theoretical study of long-range interactions modulating dimerization and activity of yeast geranylgeranyl diphosphate synthase. J. Am. Chem. Soc. 131: 4051–4062 [DOI] [PubMed] [Google Scholar]
- McConkey M.E., Gershenzon J., Croteau R.B. (2000). Developmental regulation of monoterpene biosynthesis in the glandular trichomes of peppermint. Plant Physiol. 122: 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee D.E. (1999). XtalView/Xfit-A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156–165 [DOI] [PubMed] [Google Scholar]
- McTaggart S.J. (2006). Isoprenylated proteins. Cell. Mol. Life Sci. 63: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N., Nakagawa M., Kobayashi K., Yamano S., Izawa Y., Nakamura K., Harashima K. (1990). Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172: 6704–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noike M., Katagiri T., Nakayama T., Koyama T., Nishino T., Hemmi H. (2008). The product chain length determination mechanism of type II geranylgeranyl diphosphate synthase requires subunit interaction. FEBS J. 275: 3921–3933 [DOI] [PubMed] [Google Scholar]
- Ogura K., Koyama T. (1998). Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 98: 1263–1276 [DOI] [PubMed] [Google Scholar]
- Ohnuma S., Hirooka K., Hemmi H., Ishida C., Ohto C., Nishino T. (1996). Conversion of product specificity of archaebacterial geranylgeranyl-diphosphate synthase. Identification of essential amino acid residues for chain length determination of prenyltransferase reaction. J. Biol. Chem. 271: 18831–18837 [DOI] [PubMed] [Google Scholar]
- O'Maille P.E., Malone A., Dellas N., Andes Hess B., Jr., Smentek L., Sheehan I., Greenhagen B.T., Chappell J., Manning G., Noel J.P. (2008). Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Pichersky E., Noel J.P., Dudareva N. (2006). Biosynthesis of plant volatiles: nature's diversity and ingenuity. Science 311: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondeau J.M., Bitsch F., Bourgier E., Geiser M., Hemmig R., Kroemer M., Lehmann S., Ramage P., Rieffel S., Strauss A., Green J.R., Jahnke W. (2006). Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs. ChemMedChem 1: 267–273 [DOI] [PubMed] [Google Scholar]
- Runyon J.B., Mescher M.C., De Moraes C.M. (2006). Volatile chemical cues guide host location and host selection by parasitic plants. Science 313: 1964–1967 [DOI] [PubMed] [Google Scholar]
- Russell R.J., Haire L.F., Stevens D.J., Collins P.J., Lin Y.P., Blackburn G.M., Hay A.J., Gamblin S.J., Skehel J.J. (2006). The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443: 45–49 [DOI] [PubMed] [Google Scholar]
- Saiki R., Nagata A., Kainou T., Matsuda H., Kawamukai M. (2005). Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans. FEBS J. 272: 5606–5622 [DOI] [PubMed] [Google Scholar]
- Saiki R., Nagata A., Uchida N., Kainou T., Matsuda H., Kawamukai M. (2003). Fission yeast decaprenyl diphosphate synthase consists of Dps1 and the newly characterized Dlp1 protein in a novel heterotetrameric structure. Eur. J. Biochem. 270: 4113–4121 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Gershenzon J. (2008). Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies). Phytochemistry 69: 49–57 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Wächtler B., Temp U., Krekling T., Séguin A., Gershenzon J. (2009). A bifunctional geranyl and geranylgeranyl diphosphate synthase is involved in terpene oleoresin formation in Norway spruce (Picea abies). Plant Physiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H.Y., Ko T.P., Kuo C.J., Guo R.T., Chou C.C., Liang P.H., Wang A.H. (2005). Homodimeric hexaprenyl pyrophosphate synthase from the thermoacidophilic crenarchaeon Sulfolobus solfataricus displays asymmetric subunit structures. J. Bacteriol. 187: 8137–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkopinska A., Plochocka D. (2005). Farnesyl diphosphate synthase; regulation of product specificity. Acta Biochim. Pol. 52: 45–55 [PubMed] [Google Scholar]
- Tarshis L.C., Proteau P.J., Kellogg B.A., Sacchettini J.C., Poulter C.D. (1996). Regulation of product chain length by isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. USA 93: 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., Kish C.M., Orlova I., Sherman D., Gershenzon J., Pichersky E., Dudareva N. (2004). Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16: 977–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiram H.V., Erickson H.K., Poulter C.D. (2007). Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science 316: 73–76 [DOI] [PubMed] [Google Scholar]
- Turner G.W., Croteau R. (2004). Organization of monoterpene biosynthesis in Mentha. Immunocytochemical localizations of geranyl diphosphate synthase, limonene-6-hydroxylase, isopiperitenol dehydrogenase, and pulegone reductase. Plant Physiol. 136: 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermoten S., Charloteaux B., Santini S., Sen S.E., Beliveau C., Vandenbol M., Francis F., Brasseur R., Cusson M., Haubruge E. (2008). Characterization of a novel aphid prenyltransferase displaying dual geranyl/farnesyl diphosphate synthase activity. FEBS Lett. 582: 1928–1934 [DOI] [PubMed] [Google Scholar]
- Wang G., Dixon R.A. (2009). Heterodimeric geranyl(geranyl)diphosphate synthase from hop (Humulus lupulus) and the evolution of monoterpene biosynthesis. Proc. Natl. Acad. Sci. USA 106: 9914–9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Ohnuma S. (1999). Chain-length determination mechanism of isoprenyl diphosphate synthases and implications for molecular evolution. Trends Biochem. Sci. 24: 445–451 [DOI] [PubMed] [Google Scholar]
- Xu M., Wilderman P.R., Peters R.J. (2007). Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. USA 104: 7397–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Fujii M., Hirata A., Kawamukai M., Shimoda C., Nakamura T. (2007). Geranylgeranyl diphosphate synthase in fission yeast is a heteromer of farnesyl diphosphate synthase (FPS), Fps1, and an FPS-like protein, Spo9, essential for sporulation. Mol. Biol. Cell 18: 3568–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikuni Y., Ferrin T.E., Keasling J.D. (2006). Designed divergent evolution of enzyme function. Nature 440: 1078–1082 [DOI] [PubMed] [Google Scholar]
- Zhang Y.W., Koyama T., Ogura K. (1997). Two cistrons of the gerC operon of Bacillus subtilis encode the two subunits of heptaprenyl diphosphate synthase. J. Bacteriol. 179: 1417–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.F., Suzuki K., Okada K., Tanaka K., Nakagawa T., Kawamukai M., Matsuda K. (1997). Cloning and functional expression of a novel geranylgeranyl pyrophosphate synthase gene from Arabidopsis thaliana in Escherichia coli. Plant Cell Physiol. 38: 357–361 [DOI] [PubMed] [Google Scholar]