This work provides a description of the transcriptional and epigenetic state of the female gametes and the early embryo in Arabidopsis. It shows that while the endosperm is transcriptionally active, the embryo is relatively quiescent, a difference that is established prefertilization in the female gametes, maintained postfertilization, and which may play a role in the reprogramming of the young plant embryo.
Abstract
Whether deposited maternal products are important during early seed development in flowering plants remains controversial. Here, we show that RNA interference–mediated downregulation of transcription is deleterious to endosperm development but does not block zygotic divisions. Furthermore, we show that RNA POLYMERASE II is less active in the embryo than in the endosperm. This dimorphic pattern is established late during female gametogenesis and is inherited by the two products of fertilization. This juxtaposition of distinct transcriptional activities correlates with differential patterns of histone H3 lysine 9 dimethylation, LIKE HETEROCHROMATIN PROTEIN1 localization, and Histone H2B turnover in the egg cell versus the central cell. Thus, distinct epigenetic and transcriptional patterns in the embryo and endosperm are already established in their gametic progenitors. We further demonstrate that the non-CG DNA methyltransferase CHROMOMETHYLASE3 (CMT3) and DEMETER-LIKE DNA glycosylases are required for the correct distribution of H3K9 dimethylation in the egg and central cells, respectively, and that plants defective for CMT3 activity show abnormal embryo development. Our results provide evidence that cell-specific mechanisms lead to the differentiation of epigenetically distinct female gametes in Arabidopsis thaliana. They also suggest that the establishment of a quiescent state in the zygote may play a role in the reprogramming of the young plant embryo.
INTRODUCTION
In animals, the early phase of embryogenesis relies primarily on maternal mRNAs and proteins stored in the oocyte prior to fertilization, with the young embryo being transcriptionally quiescent (Newman-Smith and Rothman, 1998; Bultman et al., 2006; De Renzis et al., 2007). This quiescence is critical for the acquisition of totipotency and the differentiation of germ cells (reviewed in Seydoux and Braun, 2006). The maternal-to-zygotic transition defines the period when this maternal influence ends, and the development of the embryo requires de novo activities produced by the zygotic genome. It occurs one to several cell division cycles following fertilization and entails the degradation of the maternal transcript population, the turnover of maternally stored proteins, and the initiation of zygotic transcription (zygotic genome activation; reviewed in Schier, 2007; Baroux et al., 2008; Tadros and Lipshitz, 2009).
The extent of maternal influence on early seed development in angiosperms is less clear. Unlike animals, plants do not set aside germ cells early in development. Instead, the multicellular gametophytes differentiate from meiotic products in the adult plant. In the female reproductive organs (ovules), one such cell undergoes meiosis, producing four megaspores, only one of which survives. In the majority of angiosperms, the surviving megaspore goes through three rounds of nuclear division without cytokinesis, forming a syncytium containing eight genetically identical haploid nuclei. Subsequent cellularization and differentiation results in a seven-celled embryo sac (female gametophyte) comprising two gametes, the haploid egg, and the homo-diploid central cells, as well as five accessory cells (two synergids and three antipodal cells). Similarly, the male gametophyte (pollen) is produced in the male reproductive organs (anthers) and comprises one accessory (vegetative) cell and two sperm cells derived upon mitosis from a single haploid microspore. Thus, in most plant species, each pair of gametes is genetically identical. Fertilization of the egg cell by one of the sperm cells gives rise to the zygote, while fertilization of the central cell by the second sperm cell produces the endosperm, an embryo-supporting tissue. Embryo and endosperm develop in a coordinated manner together and enclosed in the sporophytic, maternal integuments, forming the seed.
Genetic analyses in Arabidopsis thaliana indicate that early seed development relies, at least in part, on maternal factors (Grossniklaus et al., 1998; Kinoshita et al., 1999; Luo et al., 2000; Moore, 2002; Guitton and Berger, 2005; Pagnussat et al., 2005; Ngo et al., 2007). Expression analyses from Arabidopsis and maize (Zea mays) have shown that transcripts detected during early seed development, up to 3 d after pollination (DAP), are predominantly maternal (Vielle-Calzada et al., 2000; Grimanelli et al., 2005). This has been observed in both the embryo and the endosperm and raised the suggestion that early seed development might depend strongly on maternal transcripts. However, the origin of these maternal transcripts is unclear: since most expression studies only measure steady state mRNA levels, it is usually not possible to determine whether a maternal transcript detected in the embryo is gametophytic (deposited prior to fertilization), zygotic (produced de novo after fertilization), or both. In fact, several paternal alleles (endogenous and transgenic) are expressed soon after fertilization (Weijers et al., 2001; Scholten et al., 2002; Köhler et al., 2005; Meyer and Scholten, 2007; Bayer et al., 2009). In addition, many early embryo lethal phenotypes segregate as classical zygotic mutations, suggesting that both maternal and paternal alleles are active (Tzafrir et al., 2004). Thus, it is still unknown whether maternal transcripts are sufficient to sustain early seed growth. As very little is known about zygotic genome activation and the maternal-to-zygotic transition in plants, important questions remain unresolved. In particular, we were interested to determine the extent to which maternally deposited information regulates early development of the fertilization products and whether a phase of transcriptional quiescence follows fertilization.
In this study, we show that the zygote remains relatively quiescent and that the embryo can undergo several divisions in the absence of de novo transcription, thus relying on deposited maternal products. By contrast, the endosperm strictly requires de novo transcription starting at fertilization. We further link these differences in transcriptional activity to global chromatin states inherited from the female gametes. Finally, we show by analyzing DNA methylation mutants that different mechanisms are involved in establishing gamete-specific epigenetic patterns in the egg and central cells.
RESULTS
Embryo and Endosperm Have Different Transcriptional Requirements
One strategy to determine the maternal-to-zygotic transition is to block RNA POLYMERASE II (POLII) activity to prevent de novo transcription after fertilization and observe how far the fertilization products develop using the cytoplasmically inherited products from the gametes. In animals, both toxicological experiments and RNA interference (RNAi) strategies have been used to inhibit POLII activity (Powell-Coffman et al., 1996; reviewed in Baroux et al., 2008; Tadros and Lipshitz, 2009). Because toxicological approaches are not easily applicable to Arabidopsis seeds, we chose to downregulate the Arabidopsis gene encoding the main subunit of POLII gene by RNAi.
First, we checked that RNAi against POLII efficiently blocks transcription by expressing the RNAi construct in the haploid functional megaspore, the product of female meiosis in plants that gives rise to the female gametophyte. The functional megaspore is an appropriate control for such experiments because its development requires de novo transcription of developmentally regulated genes. For example, AGP18, an arabinogalactan protein, is necessary for the early development of the female gametophyte and is specifically expressed in the differentiating megaspore (Acosta-Garcia and Vielle-Calzada, 2004). Heterozygous loss-of-function mutants for the AGP18 locus segregate as gametophytic lethal, with half of the gametophytes aborted in the ovules. This shows that female gametophyte development requires de novo transcription of AGP18 at the functional megaspore stage and that possible carryover of wild-type transcripts from the heterozygous meiocytes is not sufficient to complement for lack of AGP18 activity in the gametophyte. Moreover, it has been shown that female gametophytes carrying defective alleles for POLII arrest early in development due to the failure of female megaspores to complete the three rounds of mitosis required for the development of mature gametophytes (Onodera et al., 2008). Using the pFM1 promoter (Huanca-Mamani et al., 2005), which is specifically expressed in the functional megaspore, we saw that transgenic lines with an RNAi construct targeting POLII resulted in immediate developmental arrest at the functional megaspore stage (see Supplemental Figure 1A and Supplemental Table 1 online), thus showing that RNAi against POLII is an efficient way to avert transcription. Another question, however, was whether the RNAi machinery is functional in the seed at early stages. To answer this question, we induced RNAi using the promoter that encodes N-ACETYLGLUCOSAMINIDASE (pNG) (Ronceret et al., 2008a) against the NG gene itself. Reporter gene analysis and mRNA in situ experiments have shown that activity of the pNG promoter is detected in the central cell and the egg apparatus (egg cell and synergids) only in mature unfertilized embryo sacs, immediately prior to fertilization, and only maternally in the early embryo and endosperm (Ronceret et al., 2008a) (see Supplemental Figure 1B online). We looked at 10 independent RNAi lines, four of which phenocopied the loss-of-function phenotype of a T-DNA–induced ng mutant (normal endosperm growth but embryo arrest at the one- or two-cell stage; Ronceret et al., 2008a) (see Supplemental Figure 1C and Supplemental Table 1 online). This shows that the RNAi machinery is functional early during seed development and unambiguously at the one-cell stage.
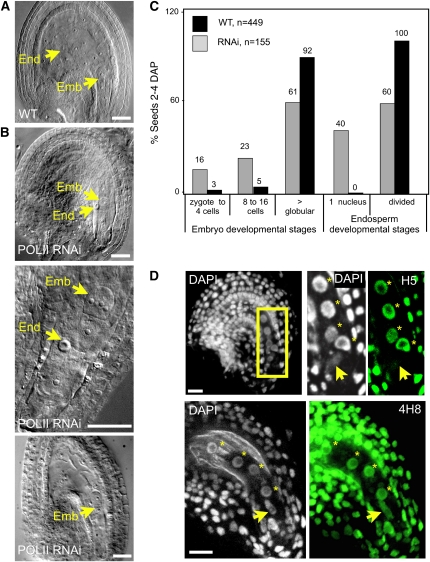
We then expressed our RNAi construct against POLII under the pNG promoter and followed embryo and endosperm divisions in response to POLII downregulation. Twenty-two independent transgenic lines were generated, seven of which showed reduced fertility (see Supplemental Table 1 online). In five of these lines, the embryo developed until the preglobular stage (16 to 32 cells), while the primary endosperm nucleus arrested as a single, enlarged nucleus (Figures 1A to 1C). To verify the relation between POLII downregulation and the resulting phenotype, we used an antibody (H5) directed against the active isoform of the main subunit of POLII. The H5 antibody specifically recognizes the heptamer of the POLII C-terminal domain when phosphorylated on Ser-2, which is a hallmark of POLII engaged in transcript elongation (Palancade and Bensaude, 2003). In these RNAi lines, abnormal seeds showed no detectable active POLII as determined by indirect immunolocalization (see Supplemental Figure 1D and Supplemental Table 1 online), strongly suggesting that the arrest was a phenotypic response to the downregulation of POLII. To further demonstrate that the phenotype was the result of RNAi induction, we introduced the pNG-POLII transgene leading to arrest of endosperm development in a genetic background deficient for the RDR2 RNA-dependent RNA polymerase, which is involved in the biogenesis of small interfering RNAs (Xie et al., 2004). In the selfed progeny of pNG-POLII × rdr2-1/rdr2-1 crosses, the frequency of aborted seeds was 50% less that in the pNG-POLII RNAi lines (see Supplemental Table 1 online). This indicates that rdr2 suppressed the pNG-POLII RNAi phenotype, producing wild-type female gametophytes. In the two remaining lines, the embryo arrested similarly, but the endosperm underwent a few divisions, albeit much slower than the wild type (Figures 1A and 1B). This was particularly evident at the one-cell embryo stage (Figures 1A and 1B), where the number of endosperm nuclei was less than half of the wild-type count. We interpreted these phenotypes as resulting from partial downregulation of transcriptional activity.
Figure 1.
Transcriptional Requirements in Embryo and Endosperm.
(A) Whole-mount clearing of a wild-type seed at the one-cell embryo stage. Emb, embryo; End, endosperm.
(B) Whole-mount clearing of developmental abnormalities induced by downregulating POLII in pNG-POLII RNAi lines at the one-cell (top) or 4/8-cell (center) embryo stages with arrested central cell development and seed at 2 DAP with a slow-growing endosperm (bottom).
(C) Distribution of embryo and endosperm developmental stages in RNAi lines with arrested central cell development and wild-type seeds 2 to 4 DAP.
(D) Whole-mount seeds counterstained with DAPI (marking chromatin) after immunostaining of the active form of RNA POLII (H5 antibody) or RNA POLII independently of its transcriptional engagement (4H8 antibody), as indicated. Projections of consecutive sections are shown. The seeds are at the zygotic stage, and the endosperm has undergone two cleavages. The box corresponds to the close-up. The arrow indicates the zygote nucleus; endosperm nuclei are indicated by stars.
Bars = 10 μm.
[See online article for color version of this figure.]
Our results indicate that levels of POLII downregulation achieved under the pNG promoter are sufficient to block or severely impair endosperm development but do not affect the development of the early embryo. This suggests that both tissues have different transcriptional requirements during early development: the endosperm is dependent on de novo transcription, while maternally inherited products appear to be sufficient to sustain embryo growth until the preglobular stage.
While these results indicate a substantial maternal influence in the embryo, they do not rule out de novo, eventually redundant, zygotic transcription. We wanted to check whether these different transcriptional requirements correlated with differences in global POLII activity between embryo and endosperm. Using the H5 antibody and indirect immunofluorescence on whole-mount Arabidopsis seeds, we observed a clear signal in the endosperm nuclei, but no or little signal in the zygote (Figure 1D). Using another antibody that reacts with the same epitope but irrespective of phosphorylation status, and thus POLII activity (4H8; see Methods), we observed that both fertilization products contained detectable amounts of POLII (Figure 1D). These results collectively confirm two distinct transcriptional patterns in the fertilization products: sustained transcription in the endosperm and relative quiescence in the zygote, where transcriptional activity remains below our detection threshold.
Transcriptional Activity in the Seed Correlates with Global Differences in Dimethylation of Lysine 9 on Histone H3
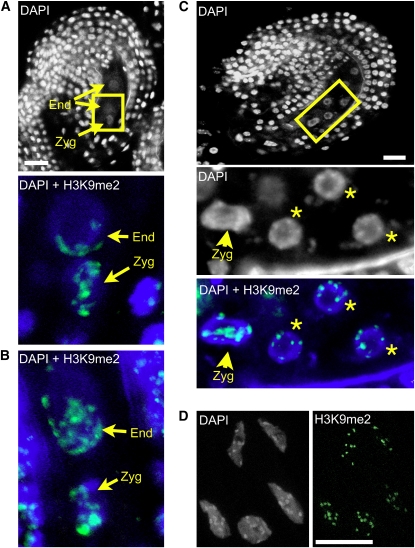
In plants as in animals, the combination of histone tail modifications and DNA methylation patterns determines the structure of chromatin and its transcriptional competence (reviewed in Fuchs et al., 2006; Vaillant and Paszkowski, 2007). DNA hypermethylation and dimethylation of lysine 9 on histone H3 (H3K9me2) are linked to a transcriptionally repressive heterochromatic state; hypomethylation of DNA and H3K9, by contrast, are usually linked to a permissive euchromatic state. To determine whether the different patterns of transcriptional activity observed in embryo versus endosperm are linked to differences in chromatin states, we analyzed the patterns of H3K9me2 in early seeds. The antibody against H3K9me2 has been used extensively for chromatin analyses in plant (for example, Tariq et al., 2003; Mathieu et al., 2007; Zhang et al., 2008) and animal systems (Barski et al., 2007) and was shown to be highly specific. We confirmed its specificity in the context of whole-mount immunolocalizations, as we did not detect any signal in ovules of a mutant line deficient for KRYPTONITE, the main H3K9 methyltransferase in Arabidopsis (see Supplemental Figure 2A online). In wild-type seeds, just after fertilization, signals were detected in the maternal sporophytic cells and both fertilization products. Interestingly, the distribution pattern greatly differed between the endosperm and embryo. In the endosperm, nuclei before the third cleavage showed a depletion of H3K9me2 signals, and dispersed signals were observed at one pole of the nucleus, facing the micropyle (Figures 2A and 2B; n = 50 ovules). By contrast, the zygote was enriched in H3K9me2 signals with strong and well-defined foci (Figures 2A to 2C). The polarization observed in the primary endosperm nuclei is reminiscent of the nuclear distribution of a paternally transmitted centromeric H3 fused to green fluorescent protein (HTR12-GFP; Ingouff et al., 2007), suggesting that the H3K9me2 distribution pattern might reflect polarization of paternally inherited chromatin in endosperm nuclei. This polarization was only transient, and after the third cleavage, distinct foci were visible and colocalized with 4',6-diamidino-2-phenylindole (DAPI)-stained chromocenters in all endosperm nuclei, similar to the pattern seen in somatic nuclei, but distinct from the zygotic pattern at that stage (Figures 2C and 2D; n = 50 ovules). Thus, the endosperm and zygotic lineages are highly dimorphic for their patterns of H3K9me2.
Figure 2.
Patterns of H3K9me2 in the Early Seed.
(A) Wild-type (Columbia-0 ecotype) early seed following the first division of the primary endosperm nucleus; strong and well-defined H3K9me2 foci are visible in the zygote, while dispersed foci distributed in a polar fashion are detected in endosperm nuclei. Top: DAPI staining (white) of the whole early seed. Bottom: close-up (indicated by the yellow rectangle) showing overlay of DAPI (blue) and H3K9me2 (green). Zyg, zygote; End, endosperm.
(B) Additional example, early seed with primary endosperm nucleus. DAPI, blue; H3K9me2, green.
(C) Seed after three rounds of divisions in the endosperm showing well-defined chromocentric H3K9me2 foci in the endosperm, while the distribution in the zygote remains similar to the previous stage (A). DAPI, white; overlay of DAPI, blue; H3K9me2 signals, green. Endosperm nuclei are indicated by stars.
(D) Somatic cells from ovule integuments showing well-defined H3K9me2 foci corresponding to the heterochromatic chromocenters. DAPI, white, left; H3K9me2 signals, green, right.
All images are projections of consecutive optical sections. Bars = 10 μm.
The Transcriptional and Chromatin States in Embryo and Endosperm Are Inherited from Epigenetically Dimorphic Female Gametes
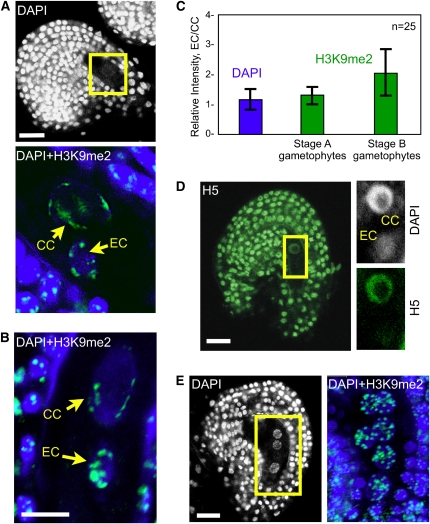
To determine whether the dimorphisms observed in the seed are established in the female gametes before fertilization, or de novo after fertilization, we analyzed the distribution of H3K9me2 and active POLII (H5) in the mature female gametophyte. In the egg nucleus, H3K9me2 signals showed a distribution pattern comparable to the zygote with well-defined and strongly staining foci (Figures 3A and 3B). By contrast to the neighboring egg cell, the central cell showed a more dispersed staining pattern with elongated or less-defined signals located at the periphery of the nucleus (Figures 3A and 3B). These H3K9me2 signals did not colocalize with recognizable DAPI-stained chromocenters, possibly indicating decondensed pericentromeric regions (see Supplemental Figure 2B online). Furthermore, H3K9me2 signal intensity in the central cell differed between the gametophytes staged immediately after polar nuclei fusion and those at complete maturity (as defined by the orientation and shape of the nuclei; Faure et al., 2002). In the former, immunostaining intensity was quantitatively comparable to that of the egg nucleus (Figures 3A and 3C). By contrast, in the latter, fully mature embryo sacs, H3K9me2 signals were quantitatively less intense in the central cell than in the egg cell (Figures 3B and 3C). This decrease in fluorescence intensity was generally concomitant with a polarization of the signals in the nuclear side facing the egg nucleus (Figure 3B; n = 50 ovules), as observed in the endosperm nuclei immediately following fertilization.
Figure 3.
POLII Activity and H3K9 Dimethylation in the Ovule.
(A) Ovule following polar nuclei fusion. Top: DAPI staining of the whole ovule. Bottom: Close-up showing overlay of DAPI (blue) and H3K9me2 (green) and projection of consecutive optical sections. EC, egg cell; CC, central cell.
(B) Ovule at maturity prior to fertilization, overlay of DAPI (blue) and H3K9me2 (green), and projection of consecutive optical sections.
(C) Quantification of the relative H3K9me2 fluorescence intensity (green bars) between the egg cell and central cell (EC/CC) at two developmental stages (25 each) as shown in (A) and (B), respectively. The relative intensity is expressed as the ratio EC/CC of the mean intensity per pixel. Quantification of DAPI signals (blue bar) controls the accuracy of the measurements: despite differences between the egg and central cells in nuclear size, ploidy level and DNA compaction, the DAPI signal EC/CC ratio is equal to 1. The error bars represent sd.
(D) Mature ovule prior to fertilization stained with DAPI (white) and H5 (green) showing different levels of active POLII in the egg and central cell nuclei; projection of consecutive optical sections.
(E) Immature ovule prior to gametophyte cellularization showing comparable H3K9me2 distribution in all gametophytic nuclei; single optical section of the whole ovule stained with DAPI (left) reveals three gametophyte nuclei. Overlay of DAPI (blue) and H3K9me2 (green) signals (right) following projection of consecutive optical sections shows six out of the eight gametophytic nuclei.
Bars = 10 μm.
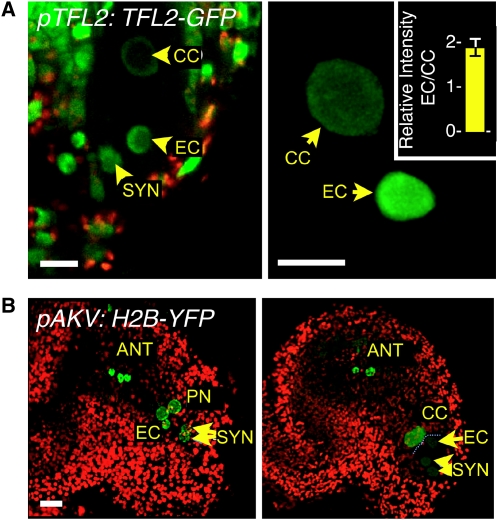
Although H3K9me2 is a genome-wide repressive mark (Johnson et al., 2007; Turck et al., 2007), it is cytologically detected in Arabidopsis nuclei essentially in heterochromatic chromocenters (Fuchs et al., 2006). To investigate another repressive mark associated with euchromatic genes, we analyzed the distribution of LIKE HETEROCHROMATIN PROTEIN1/TERMINAL FLOWER2 (LHP1/TFL2) fused in translation to GFP (pTFL2:TFL2-GFP; Nakahigashi et al., 2005). This reporter protein was previously found to be distributed throughout the euchromatic compartment (Libault et al., 2005; Nakahigashi et al., 2005), consistent with its association with silenced loci enriched in H3K27me3 (Turck et al., 2007; Zhang et al., 2007; Exner et al., 2009). Confocal imaging of mature female gametophytes showed that GFP signals in the central cell nucleus were much lower than in the egg nucleus (Figure 4A). This was confirmed following quantification of the fluorescence intensity in entire nuclei. The intensity ratio of egg cell to central cell was 2.04 (± 0.20, n = 20 ovules) after correcting for the difference in DNA content between the haploid egg cell and the homodiploid central cell (Figure 4A). This clearly indicates a relative enrichment in repressive chromatin in the egg cell, corresponding to a quantitatively more silenced state of euchromatic domains.
Figure 4.
TFL2 and Histone H2B Distribution in the Ovule.
(A) and (B) GFP or YFP signals are displayed as green, and chlorophyll autofluorescence is displayed in red. ANT, antipodal cells; EC, egg cell; PN, polar nuclei; CC, central cell; SYN, synergids. Bars = 10 μm.
(A) Mature ovule prior to fertilization expressing a pTFL2:TFL2-GFP transgene monitoring LHP1 distribution. Left: projection of consecutive optical sections showing the nuclei of the embryo sac and the ovule integuments. Right: Close-up of a three-dimensional reconstruction of central cell and egg cell nuclei, as used for quantification of fluorescence intensity signals (see Methods) shown in the inset (n = 20 ovules). The error bars represent sd.
(B) Immature (left) and mature (right) ovules after cellularization of the female gametophyte; projection of consecutive optical sections of a transgenic line expressing a pAKV:H2B-YFP transgene monitoring H2B dynamics.
Consistent with these observations, immunolocalization of the active form of POLII using the H5 antibody gave a strong signal in the central cell nucleus, while the signal was barely detectable in the egg nucleus (Figure 3D). This is similar to what we observed in the primary endosperm nucleus and the zygote. Using the 4H8 antibody that reacts with the same epitope irrespective of its phosphorylation status, we observed that both cell types contained detectable amounts of POLII (see Supplemental Figure 3 online). Taken together, these results indicate that the distinct epigenetic and transcriptional patterns observed in embryo and endosperm are already established in their gametic progenitors, the egg and central cells, respectively.
The Epigenetic Dimorphism between Egg and Central Cell Is Established Late during Female Gametogenesis
The previous observations were made on cellularized female gametophytes, and the question arose whether the observed epigenetic dimorphism in the female gametes was established in the female gametophyte prior to or after cellularization. Thus, we analyzed H3K9me2 distribution patterns in syncytial female gametophytes at a stage in which the three mitoses have been completed, but individual cells have not yet formed. All eight nuclei showed similar H3K9me2 signals, distributed in surprisingly numerous (15 to 20), well-defined foci (Figure 3E, n = 50 ovules). This pattern differed from that of both the egg and central cells. Similarly, the progenitor nuclei of the central cell and egg cell did not show quantitative differences from each other in LHP1-GFP intensity following the third mitosis (see Supplemental Figure 4 online). This indicates that cell-specific chromatin patterns in the mature gametophyte are established after cellularization. Possibly, such chromatin remodeling events encompass additional chromatin modifications or constituents as suggested by the dynamics of a yellow fluorescent protein (YFP)-tagged histone H2B in the gametes. When expressed under a gametophyte-specific promoter, pAKV (Rotman et al., 2005), H2B-YFP was incorporated into all eight nuclei of the gametophyte prior to cellularization but was selectively depleted in the nuclei of the egg apparatus (egg cell and synergids) in mature gametophytes (Figure 4B). While indirect, this finding nevertheless suggests active and differential core histone remodeling during the last stage of gamete differentiation. The timing of H2B-YFP depletion is strikingly similar to the changes observed for H3K9me2 (Figure 3E; see Supplemental Figure 2B online), both taking place during or after the fusion of the two polar nuclei. Taken together, these data indicate that the differentiation of the female gametes is accompanied by the establishment of a global epigenetic dimorphism between these two cell types, which takes place after cellularization.
Specific Patterns of H3K9me2 in the Egg Cell and Central Cell Require CMT3 and DEMETER-LIKE Activity, Respectively
To further understand the mechanisms involved in establishing these dimorphic epigenetic patterns, particularly in the egg cell, we screened Arabidopsis loss-of-function mutants affecting silencing pathways (hereafter and Supplemental Table 2 online). We analyzed mutations in (1) MET1, DRM1, DRM2, and CMT3 DNA methyltransferases (Vaillant and Paszkowski, 2007); (2) NRPD1a and NRPD1b, two subunits of RNA POLYMERASE IV and V involved in RNA-directed DNA methylation (Vaillant and Paszkowski, 2007); (3) DEMETER (DME), a DNA glycosylase conferring demethylation in the central cell and endosperm (Choi et al., 2002; Gehring et al., 2006, 2009; Hsieh et al., 2009); and (4) DEMETER-LIKE (DML) DNA glycosylases in a triple dml1 dml2 dml3 mutant (with ros1-3, dml2-1, and dml3-1 alleles; Penterman et al., 2007). For all mutant lines, we performed immunolocalization experiments to detect H3K9me2 in cellularized gametophytes after fusion of the polar nuclei, the earliest stage showing a dimorphism between egg and central cell nuclei, and looked for potential alterations of this dimorphism.
Both drm1 drm2 and dme embryo sacs showed patterns of H3K9me2 similar to the wild type (data not shown). By contrast, met1, nrpd1a nrpd1b, cmt3, and dml1 dml2 dml3 mutants affected H3K9me2 distribution in the embryo sac. But while met1 and nrpd1a nrpd1b affected indiscriminately H3K9me2 in both female gametes (see Supplemental Figure 4 online and below), cmt3-7 and dml1 dml2 dml3 specifically affected the egg and central cells, respectively (Figure 5 and below). In met1 ovules (see Supplemental Figures 5A and 5B online), all cell types in the ovule showed reduced and dispersed signals, very similar to what has been previously described on somatic cells (Tariq et al., 2003; Mathieu et al., 2007). Disruption of H3K9me2 patterns affected similarly both gametes, without apparent specificity; neither the central cell nor the egg cell showed wild-type patterns. Similarly, in the double nrpd1a nrpd1b mutant (see Supplemental Figures 5A and 5C online), we observed a global effect on H3K9me2, with numerous and dispersed signals in all cell types of the ovule, similar to the previously described response to POLIV inactivation in somatic cells (Onodera et al., 2005). The signals were distinct from wild-type patterns in both the egg cell and the central cell.
Figure 5.
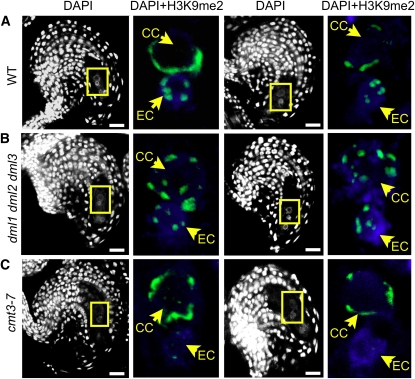
Effect of CMT3 and DML Loss of Function on H3K9me2 in the Ovule.
(A) to (C) Ovules after fusion of the polar nuclei, as in Figure 3A, counterstained with DAPI (white). Left: Single optical sections; right: close-ups showing DAPI (blue) and H3K9me2 (green) overlays; projections of consecutive optical sections. Patterns shown below were observed in at least 30 ovules of each wild-type or mutant line. Two examples for each line are shown. EC, egg cell; CC, central cell. Bars = 10 μm.
(A) Wild-type ovule.
(B) dml1 dml2 dml3 mutant ovule showing an egg cell-like pattern of H3K9me2 in the central cell nucleus.
(C) cmt3 mutant ovule showing altered H3K9me2 signal specifically in the egg cell nucleus.
In the triple dml1 dml2 dml3 mutant, H3K9me2 distribution in the egg cell appeared unaffected. By contrast, the specific pattern observed in the central cell nuclei of wild-type plants was lost. H3K9me2 signals in the central cell of mutant plants showed an egg cell-like pattern, except for the number of foci (10 instead of 5), consistent with the homodiploid nature of the central cell nucleus (Figure 5B; n > 30 ovules). This indicates that DML enzymes are required for H3K9me2 distribution patterns in the central cell nucleus specifically, likely indirectly via its effect on cytosine methylation. To determine which DML enzyme was responsible for the phenotype, we analyzed H3K9me2 signals in the single mutants but could not phenocopy the triple mutant in any of them, indicating functional redundancy of the three enzymes.
Conversely, the cmt3 mutation had no impact on the central cell, but H3K9me2 signals became undetectable in the egg cell nucleus (Figure 5C; n > 50 ovules). Before cellularization, however, H3K9me2 patterns were similar in cmt3 and wild-type ovules (see Supplemental Figure 6 online). These results indicate that CMT3 is required specifically to establish or maintain H3K9me2 in the egg cell nucleus following cellularization.
CMT3-Deficient Embryos Display Transiently Abnormal Early Development
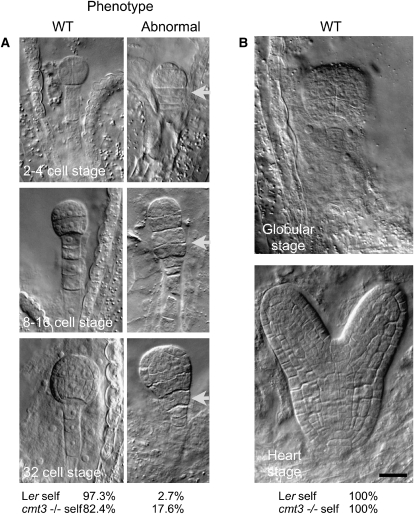
In animals, the period of transcriptional quiescence preceding the maternal-to-zygotic transition is essential to genome reprogramming in the embryo. We asked whether the loss of the repressive chromatin mark H3K9me2 in cmt3 eggs affected embryogenesis. As reported in numerous papers, cmt3 mutants are fully fertile. Yet, previous reports mentioned that up to 3% abnormal embryo development is observed in cmt3 mutants when including embryo stages from 0 to 6 DAP, which corresponds to the torpedo stage in Arabidopsis (Xiao et al., 2006). We discriminated in our analyses between early (preglobular) and later stages of development (until the heart stage). Interestingly, we noticed that later-stage embryos (globular to heart) were indistinguishable from wild-type embryos. By contrast, a clear developmental phenotype was evident at earlier stages (Figure 6; see Supplemental Table 3 online). Reciprocal crosses to a wild-type plant (Landsberg erecta [Ler]) further showed that the phenotype was linked to the transmission of a defective maternal allele (see Supplemental Table 3 online). The earliest cmt3 phenotype was unambiguously observed as early as the two-cell embryo stage (Figure 6) with abnormal cell divisions evident in the uppermost cell of the suspensor. Later, at the 8- to 16-cell and 32-cell embryo stages, there was no clear demarcation between embryo and suspensor because of excess longitudinal cell divisions in the suspensor. Abnormalities in planes of cell division were also observed in the embryo proper. These observations indicate that maternal CMT3 affects the timing and/or spatial orientation of embryonic divisions. These early phenotypes are strikingly reminiscent of other mutants affecting pattern formation in Arabidopsis, including mutants in DNA methyltransferase MET1 (Xiao et al., 2006) and suggest that cmt3-defective plants similarly uncouple cell division from patterning.
Figure 6.
Phenotypic Characterization of cmt3-Defective Plants.
Whole-mount clearing of wild-type (Ler) and homozygous mutant (cmt3−/−) seeds obtained from self-pollinated plants. All images are at the same magnification.
(A) cmt3 embryos display a significant proportion of abnormal divisions (P < 0.01, t test) observed from the two- to four-cell to the 32-cell stages, prominently at the transition zone between the suspensor and the embryo (arrow). The frequency of wild-type and defective embryos is indicated as a percentage of total seed counts at the bottom (Ler self, n = 451, and cmt3−/− self, n = 216).
(B) At the globular stage, cmt3 embryos recover a wild-type phenotype, similar to Ler. Bar = 10 μm.
DISCUSSION
The extent and role of embryonic transcription during early seed development remained unclear until now, largely due to the difficulty of distinguishing cytoplasmically stored transcripts (from gametic transcription) from transcripts produced de novo in the fertilization products. mRNA expression analyses reveal similarly deposited transcripts, de novo activity, and a combination of both (Tadros and Lipshitz, 2009). Here, we investigated the transcriptional competence of the fertilization products by analyzing the presence of the active form of POLII and the presence of histone modifications associated with the transcriptional competence of chromatin. We found that the embryo is relatively quiescent, while the endosperm is transcriptionally active. This differentiation was established prefertilization and maintained in the zygote and the endosperm postfertilization.
Consistently, the downregulation of transcription in the fertilization products was deleterious to endosperm development but did not block zygotic divisions. This indicates that, specifically in the embryo, deposited products were sufficient to compensate for the diminished de novo transcription and, thus, that the zygote contains enough maternal products to sustain the first cell divisions. How far these deposited products might sustain embryo development is unclear. The stage of developmental arrest in these experiments may indicate the stage at which the embryo functionally requires de novo zygotic transcription, a central event of the maternal-to-zygotic transition (Baroux et al., 2008; Tadros and Lipshitz, 2009). The variability of developmental arrest between siblings of the POLII-RNAi lines may denote an unequal amount of inherited maternal products. Alternatively, some embryos may benefit from a partial recovery of POLII or a precocious maternal-to-zygotic transition. Testing this hypothesis would require analyzing defective embryos in real time. Technically, however, we cannot discriminate at early stages embryos with wild-type development from defective embryos prior to developmental arrest, and the scoring of the RNAi-induced phenotype is thus performed ex-post (i.e., after developmental arrest). Assuming early POLII recovery, we would expect those partially rescued embryos to develop until the globular stage as do glauce mutant embryos in the absence of endosperm (Ngo et al., 2007). Since the arrest is earlier in our POLII-RNAi lines (preglobular), we favor the first hypothesis that developmental arrest marks the maternal-to-zygotic transition. It also remains an open question whether the activity of pNG is equivalent in both the egg cell and central cell. Both β-glucuronidase staining (see Supplemental Figure 1B online) and mRNA in situ experiments (Ronceret et al., 2008a) indicate that the promoter is expressed in both cell types, but neither method provides accurate measurements of relative activity. Importantly, however, pNG-NG RNAi transgenic lines phenocopied a loss-of-function NG allele, strongly suggesting sufficient promoter activity in the embryo. Furthermore, a pNG-POLII RNAi is likely not cell autonomous within the gametophyte; thus, expression in both cell types likely has cross-effects on both fertilization products.
Collectively, these experiments support previous data (Vielle-Calzada et al., 2000; Moore, 2002; Grimanelli et al., 2005; Guitton and Berger, 2005; Pagnussat et al., 2005; Ngo et al., 2007), suggesting that the early development of the embryo is under significant maternal regulation. This does not rule out the possibility that in plants, as in many animal systems, a number of developmentally critical genes might be expressed early. Such genes have indeed been identified (Ronceret et al., 2005, 2008b; Bayer et al., 2009), but whether they require de novo transcription, or are just translated after fertilization, is yet unclear.
Experimental evidence indicates large epigenetic differences between the two fertilization products. It has recently been shown that virtually the entire endosperm genome is demethylated at 8 DAP, in sharp contrast with the embryo genome (Gehring et al., 2009; Hsieh et al., 2009). Moreover, the paternal chromatin undergoes distinct remodeling in the zygote and the endosperm following fertilization (Ingouff et al., 2007), and imprinting (i.e., parent-of-origin-dependent, monoallelic expression) is more prevalent in the endosperm than in the embryo. With the exception of a limited number of imprinted loci, which are differentially methylated between the egg and central cells (Gutierrez-Marcos et al., 2006), or specifically demethylated in the central cell by DME (Choi et al., 2002; Gehring et al., 2006; Jullien et al., 2008), little is known about the epigenetic state of the two female gametes. It is thus unclear whether global epigenetic differentiation mechanisms exist between the two cell types and, consequently, whether global epigenetic differences in the seed are established de novo postfertilization or whether differentiation is established in the female gametes prefertilization. By monitoring in situ in the ovule the dynamics of chromatin states in the female gametes, we show that the egg cell and central cell chromatin undergo extensive reprogramming, resulting in an epigenetic dimorphism between the two female gametes. These modifications are established at late stages of gametophyte maturation (after the establishment of the different cell types), affect both euchromatic and heterochromatic regions, and are inherited in the zygote upon fertilization (Figures 2 to 4). This suggests that global and likely active mechanisms of reprogramming occur in the female gametes and that the distinct chromatin and transcriptional states observed in the embryo and endosperm are being established in their gametic progenitor cells.
Importantly, based on the nuclear distribution of the repressive mark H3K9me2, we found specific roles for CMT3 in regulating this mark in egg chromatin and for DML activity in modulating its distribution pattern in the central cell chromatin. Thus, different mechanisms are acting in the two female gametes. Both of them likely act via DNA methylation, resulting in a hypomethylated and transcriptionally active central cell and a hypermethylated and quiescent egg cell. These observations are in line with recent findings (Gehring et al., 2009; Hsieh et al., 2009), demonstrating at the torpedo stage a genome-wide DNA demethylation of the maternal genome in the endosperm. Endosperm hypomethylation might play a role in reinforcing transposon regulation in the embryo, similar to what has been described in the male gametophytes, where transcriptional activity in the vegetative nucleus is involved in protecting the sperm cells from transposable elements (Slotkin et al., 2009). We showed here that the dimorphism in silencing states between the egg cell/zygote and central cell/endosperm affects euchromatic in addition to heterochromatic domains, and it will be extremely interesting to determine whether interactions between the two female gametes play a role in regulating genic transcription in the egg cell and the young embryo. Additional examination of DNA methylation patterns in the gametes at the sequence level, while technically challenging, would help to further define this role.
Interestingly, we could not detect any changes in H3K9me2 distribution in dme-1S mutant gametophytes, while another allele, dme-2, showed genome-wide alteration of DNA methylation levels in the endosperm at later stages (Gehring et al., 2009; Hsieh et al., 2009). Predicting the effect of dme loss of function on H3K9me2 is not trivial. In a cmt3 mutant, non-CG sites are predicted to be hypomethylated, and we observed a strong reduction of H3K9me2 in the egg cell. Because dme mutants also show non-CG hypomethylation in the endosperm, we expected, but did not observe, a reduction of H3K9me2 in the central cell. We cannot exclude, however, differences between the mutant alleles used in the studies or differences not detected by the H3K9me2 antibody, which monitors this modification essentially at cytogenetically defined heterochromatin.
It has been shown previously (Xiao et al., 2006) that normal DNA methylation patterns are required either directly or indirectly for generating and maintaining auxin gradients during embryogenesis, a key aspect of embryonic pattern formation. Interestingly, we showed here that 27.5% of embryos derived from a cmt3 mutant egg show altered patterning, possibly as a consequence of a partial breakdown of methylation-dependent silencing in the zygote. Alternatively, however, the lack of maternal CMT3 may act on the zygotic (maternal and/or paternal) genome to deregulate patterning genes. These cmt3-induced alterations are visible only transiently until the preglobular stage, and the mutant plants are fully fertile, indicating that they are fully compensated later during development. This illustrates the remarkable robustness, or canalization (Waddington, 1942), of plant embryo development in response to epigenetic perturbations, as already observed for other mutants (Saze et al., 2003; Mathieu et al., 2007).
In conclusion, we showed here that, as in animals, the plant zygote is transcriptionally relatively quiescent. We further showed that maternally stored products can sustain the first few cell divisions, thereby providing evidence for evolutionary convergence between plants and animals. In animals, this period of quiescence following fertilization is important for protecting the zygote as it undergoes reprogramming (Seydoux and Braun, 2006), including probably from transposable elements (O'Donnell and Boeke, 2007; Brennecke et al., 2008). In plants, this period of transcriptional quiescence, possibly resulting from endosperm–embryo interactions, may also offer the opportunity for extensive genome reprogramming postfertilization.
METHODS
Plant Materials
Arabidopsis thaliana accessions Columbia-0 and Ler were used as wild-type controls. Mutant lines are listed and referenced in Supplemental Table 2 online. The pTFL2:TFL2-GFP and pAKV:H2B-YFP lines were generously provided by K. Goto and W.C. Yang, respectively (Nakahigashi et al., 2005; Rotman et al., 2005).
Antibodies
All antibodies were obtained from ABCAM. Transcriptional activity was followed using an antibody (H5; ab24758) raised against POLII that specifically targets the C-terminal domain of the main subunit of POLII when phosphorylated on Ser-2. As shown previously, the C-terminal domain phosphorylation pattern is modified as POLII engages in transcript elongation (Palancade and Bensaude, 2003). The H5 antibody has thus been proposed as a landmark of transcriptional activity. The 4H8 antibody (ab5408) targets the same POLII domain irrespective of phosphorylation status and was used to visualize POLII presence, irrespective of transcriptional activity. Chromatin analysis was performed using an antibody against H3K9me2 (ab1220). We also attempted immunolocalization of antibodies against 5-methyl cytosines (mC), but without success. A peculiarity of mC antibodies detection is a step of DNA denaturation, which we believe is difficult to achieve consistently for gametic cells, which are deeply embedded in the ovule tissues.
Cytology
Pistils and siliques at various developmental stages were fixed overnight at 4°C in 4% paraformaldehyde: 1 × PBS:2% Triton fixative, washed three times in 1 × PBS, and dissected to isolate the ovules and early seeds. The dissected ovules and seeds were embedded in acrylamide as described (Bass et al., 1997) to facilitate manipulation and maintain the three-dimensional architecture of the tissues. Samples were digested in an enzymatic solution (1% driselase, 0.5% cellulase, 1% pectolyase, and 1% BSA; all from Sigma-Aldrich) for 25 min to 1 h at 37°C, depending on the developmental stage, subsequently rinsed three times in 1 × PBS, and permeabilized for 2 h in 1 × PBS:2% Triton. They were then incubated overnight at 4°C with primary antibodies used at the following dilutions: 1:400 for H3K9me2, 1:200 for H5, and 1:400 for 4H8. The slides were washed day-long in 1 × PBS:0.2% Triton and coated overnight at 4°C with secondary antibodies (Alexa Fluor 488 conjugate; Molecular Probes) used at 1:400 dilution. After washing in 1 × PBS:0.2% Triton for a minimum of 6 h, the slides were incubated with DAPI (1 μg/ml in 1 × PBS) for 1 h, washed for 2 h in 1 × PBS, and mounted in PROLONG medium (Molecular Probes). Complete three-dimensional ovule or seed images were captured on a laser scanning confocal microscope (Leica SP2) equipped for DAPI (405 nm) and fluorescein isothiocyanate (488 nm) excitation and either × 40 or × 63 objectives. Projections of selected optical sections were generated for this report and edited using Graphic Converter (LemkeSOFT). Fifty ovules or seeds were scored for each developmental stage in wild-type plants. Minimally 25 ovules were scored for each mutant (except cmt3, n > 50, and dml1 dml2 dml3, n > 30). Quantification of intensity signals was performed using ImageJ (http://rsbweb.nih.gov/ij/). Relative fluorescence intensity between the egg and central cell in each ovule was calculated as the ratio of signal intensity per pixel in the two cell types. We used maximum intensity projections, average intensity projections, or sums of intensity measured on each individual optical section; all three methods resulted in comparable ratios. Whole-mount ovule or seed clearing and β-glucuronidase staining were performed as described (Weijers et al., 2001; Acosta-Garcia and Vielle-Calzada, 2004).
Quantification of LHP1 /TFL2-GFP and H2B-YFP Fluorescence Intensity in Whole-Mount Ovules
Unfertilized carpels were placed in a drop of 1 M glycine (pH 9.4) on a microscope slide, and the carpel walls were removed for imaging. Images were collected throughout the embryo sac (z steps of 170 to 240 nm) using a confocal laser scanning microscope (Leica SP2) with an excitation line at 488 nm and emission window at 500 to 530 nm. The intensity sum of the TFL2-GFP fluorescence signal was collected in three-dimensional reconstructions using Imaris 6.3 software (Bitplane) on manually defined, individual nuclei (contour surface tool). For each ovule, the ratio of the intensity sum in the egg cell nucleus over the intensity sum in the central cell nucleus divided by 2 was calculated. The twofold correction for the central cell nucleus accounts for its homodiploid state and reports the fluorescence per haploid genome as it is in the egg cell nucleus.
Generation of RNAi Lines
Four sets of RNAi lines were generated using the pFGC5941 RNAi vector (available from ABRC/The Arabidopsis Information Resource). The first vector (pFM1-POLII) contained a 414-bp fragment of the POLII coding region amplified by PCR and cloned in pCR-II vectors (Invitrogen) using the following primers: POLII-forward (5′-ACTCTAGAGGCGCGCCTGGGAG GACGAGAAGGTCTTATTG-3′, containing restriction sites XbaI and AscI) and POLII-reverse (5′-CGGGATCCATTTAAATGCCAAGTGCTATCACCATTTGTTG-3′, containing restriction sites BamHI and SwaI). The fragment was inserted in the pFGC5941 RNAi vector first in sense orientation after digesting with AscI and SwaI and then in antisense orientation after digestion with BamHI and XbaI, generating a pFGC5941-POLII vector. The original plasmids contained a cauliflower mosaic virus (CaMV) 35S promoter. To generate the pFM1-POLII RNAi cassette, primers pFM1-S3 (5′-GCGAATTCATACTAGCATGTATCCAC-3′, containing the restriction site EcoRI) and pFM1-AS2 (5′-CATGCCATGGTGGAACTTTATCGGTTT-3′, containing the restriction site NcoI) were used to amplify pFM1. The pFGC5941-POLII plasmid was digested with EcoRI and NcoI to excise the CaMV35S promoter, and the pFM1 promoter was inserted into the same location using the EcoRI and NcoI restriction sites. The second vector (pNG-POLII) contained the same POLII target under the promoter that encodes pNG (Ronceret et al., 2008a). pNG was a gift from M. Devic and was provided in a pGEM-T Easy vector (Promega), where it had been cloned using EcoRI and NcoI restriction sites. To generate the pNG-POLII-RNAi cassette, the pFGC5941-POLII plasmid was digested with EcoRI and NcoI to excise the CaMV35S promoter, and the pNG promoter was inserted into the same site. The third vector (pNG-NG) contained the same pNG promoter driving a hairpin transcript directed against the NG gene itself. To generate the pNG-NG RNAi cassette, primers NG-forward (5′-CTTCTAGAGGCGCGCCTCCAACTATTGATGGTCTGCTTGAC-3′, containing restriction sites XbaI and AscI) and NG-reverse (5′-CGGGATCCATTTAAATTTCAAATACCAATGGAGCCCAG-3′, containing restriction sites BamHI and SwaI) were used to PCR amplify a 267-bp fragment from the coding sequence. The amplified fragment was then inserted in both sense and antisense orientations in the pFGC5941 plasmid already containing the pNG promoter, as described above. An additional construct was used to test the effect of the pNG promoter on seed viability. The RNAi construction included the promoter and the intron, as above, but not the target sequence. We could not detect significant effects of the promoter in transgenic plants (see Supplemental Table 1 online). The flower-dipping protocol was used to produce the transgenic lines. Seed development was scored by manually dissecting siliques and counting normally developed and aborted seeds. For further analysis, siliques were collected 2 to 4 DAP and observed using whole-mount clearing. pFM1-POLII RNAi lines were analyzed in the T2 generation. pNG-POLII and pNG-NG RNAi lines, however, were analyzed as primary transformants heterozygous for the transgene; we reasoned that efficient downregulation of POLII or NG in pollen would likely affect viability and was unlikely to allow proper transmission of the transgene to the next generation and, thus, that only weak events would be transmitted.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g35800 (POLII), At5g13690 (NG), At4g11130 (RDR2), At5g13960 (KRYPTONITE), At5g15380 (DRM1), At5g14620 (DRM2), At5g14620 (DME1), At5g49160 (MET1), At1g63020 (NRPD1a), At2g40030 (NRPD1b), At2g36490 (ROS1 or DML1), At3g10010 (DML2), At4g34060 (DML3), and At1g69770 (CMT3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Control Lines Showing Effective RNA Interference in Ovules and Seeds.
Supplemental Figure 2. Dynamic of H3K9me2 during the Differentiation of the Central Cell.
Supplemental Figure 3. Detection of POLII in the Mature Ovule.
Supplemental Figure 4. TFL2 Distribution during Ovule Development.
Supplemental Figure 5. H3K9me2 Patterns in met1-3 and nrpd1a-2 nrpd1b-11 Embryo Sacs.
Supplemental Figure 6. H3K9me2 Pattern before Cellularization in cmt3-7 Mutant Plants.
Supplemental Table 1. Segregation Data in RNAi Lines.
Supplemental Table 2. Mutant Lines Used in the Study.
Supplemental Table 3. Effect of CMT3 Loss of Function on Embryogenesis.
Supplementary Material
Acknowledgments
We thank M. Devic for the pNG promoter, J.C. Carrington for the rdr2-1 line, O. Mathieu for the met1-3 line, T. Lagrange for the nrpd1 mutants, R.L. Fischer for the triple dml mutant line, K. Goto for the pTFL2:TFL2-GFP line, W.C. Yang for the pAKV:H2B-YFP line, N. Lautrédou-Audouy for help with confocal laser scanning microscopy, M. Devic and S.C. Gillmor for critical reading of the manuscript, and the reviewers for constructive comments. Work at the Institut de Recherche pour le Développement was funded by an Agence Nationale de la Recherche grant to D.G. and D.A. M.A.V. was the recipient of a graduate scholarship from CONACyT. J.P.V.-C. is an International Scholar of the Howard Hughes Medical Institute. Work in U.G.’s laboratory was supported by the University of Zurich, a grant from the Swiss National Science Foundation to U.G., and a Marie-Heim-Vögtlin fellowship to C.B.
References
- Acosta-Garcia G., Vielle-Calzada J.P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16: 2614–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C., Autran D., Gillmor C.S., Grimanelli D., Grossniklaus U. (2008). The maternal-to-zygotic transition in animals and plants. Cold Spring Harb. Symp. Quant. Biol. 73: 89–100 [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bass H.W., Marshall W.F., Sedat J.W., Agard D.A., Cande W.Z. (1997). Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J. Cell Biol. 137: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488 [DOI] [PubMed] [Google Scholar]
- Bultman S.J., Gebuhr T.C., Pan H., Svoboda P., Schultz R.M., Magnuson T. (2006). Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 20: 1744–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J. (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Gehring M., Johnson L., Hannon M., Harada J.J., Goldberg R.B., Jacobsen S.E., Fischer R.L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110: 33–42 [DOI] [PubMed] [Google Scholar]
- De Renzis S., Elemento O., Tavazoie S., Wieschaus E.F. (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Aichinger E., Shu H., Wildhaber T., Alfarano P., Caflisch A., Gruissem W., Köhler C., Hennig L. (2009). The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS One 4: e5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J.E., Rotman N., Fortuné P., Dumas C. (2002). Fertilization in Arabidopsis thaliana wild type: Developmental stages and time course. Plant J. 30: 481–488 [DOI] [PubMed] [Google Scholar]
- Fuchs J., Demidov D., Houben A., Schubert I. (2006). Chromosomal histone modification patterns - From conservation to diversity. Trends Plant Sci. 11: 199–208 [DOI] [PubMed] [Google Scholar]
- Gehring M., Bubb K.L., Henikoff S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 12: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J.H., Hsieh T.F., Penterman J., Choi Y., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DEMETER DNA glycosylase establishes Polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimanelli D., Perotti E., Ramirez J., Leblanc O. (2005). Timing of the maternal-to-zygotic transition during early seed development in maize. Plant Cell 17: 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Vielle-Calzada J.P., Hoeppner M.A., Gagliano W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Guitton A.E., Berger F. (2005). Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15: 750–754 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos J.F., Costa L.M., Dal Pra M., Scholten S., Kranz E., Perez P., Dickinson H.G. (2006). Epigenetic asymmetry of imprinted genes in plant gametes. Nat. Genet. 38: 876–878 [DOI] [PubMed] [Google Scholar]
- Hsieh T.F., Ibarra C.A., Silva P., Zemach A., Eshed-Williams L., Fischer R.L., Zilberman D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huanca-Mamani W., Garcia-Aguilar M., Leon-Martinez G., Grossniklaus U., Vielle-Calzada J.P. (2005). CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102: 17231–17236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Hamamura Y., Gourgues M., Higashiyama T., Berger F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Bostick M., Zhang X., Kraft E., Henderson I., Callis J., Jacobsen S.E. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien P.E., Mosquna A., Ingouff M., Sakata T., Ohad N., Berger F. (2008). Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 6: e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Yadegari R., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Imprinting of the MEDEA Polycomb gene in the Arabidopsis endosperm. Plant Cell 11: 1945–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Page D.R., Gagliardini V., Grossniklaus U. (2005). The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Libault M., Tessadori F., Germann S., Snijder B., Fransz P., Gaudin V. (2005). The Arabidopsis LHP1 protein is a component of euchromatin. Planta 222: 910–925 [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau P., Dennis E.S., Peacock W.J., Chaudhury A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- Meyer S., Scholten S. (2007). Equivalent parental contribution to early plant zygotic development. Curr. Biol. 17: 1686–1691 [DOI] [PubMed] [Google Scholar]
- Moore J.M. (2002). Isolation and Characterization of Gametophytic Mutants in Arabidopsis thaliana. PhD dissertation (Stony Brook, NY: State University of New York at Stony Brook; ). [Google Scholar]
- Nakahigashi K., Jasencakova Z., Schubert I., Goto K. (2005). The Arabidopsis HETEROCHROMATIN PROTEIN1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 46: 1747–1756 [DOI] [PubMed] [Google Scholar]
- Newman-Smith E.D., Rothman J.H. (1998). The maternal-to-zygotic transition in embryonic patterning of Caenorhabditis elegans. Curr. Opin. Genet. Dev. 8: 472–480 [DOI] [PubMed] [Google Scholar]
- Ngo Q.A., Moore J.M., Baskar R., Grossniklaus U., Sundaresan V. (2007). Arabidopsis GLAUCE promotes fertilization-independent endosperm development and expression of paternally inherited alleles. Development 134: 4107–4117 [DOI] [PubMed] [Google Scholar]
- O'Donnell K.A., Boeke J.D. (2007). Mighty Piwis defend the germline against genome intruders. Cell 129: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Nunes P.C., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Onodera Y., Nakagawa K., Haag J.R., Pikaard D., Mikami T., Ream T., Ito Y., Pikaard C.S. (2008). Sex-biased lethality or transmission of defective transcription machinery in Arabidopsis. Genetics 180: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Ngo Q.A., Rajani S., Mayalagu S., Johnson C.S., Capron A., Xie L.F., Ye D., Sundaresan V. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Palancade B., Bensaude O. (2003). Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270: 3859–3870 [DOI] [PubMed] [Google Scholar]
- Penterman J., Zilberman D., Huh J.H., Ballinger T., Henikoff S., Fischer R.L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Coffman J.A., Knight J., Wood W.B. (1996). Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev. Biol. 178: 472–483 [DOI] [PubMed] [Google Scholar]
- Ronceret A., Gadea-Vacas J., Guilleminot J., Devic M. (2008a). The alpha-N-acetyl-glucosaminidase gene is transcriptionally activated in male and female gametes prior to fertilization and is essential for seed development in Arabidopsis. J. Exp. Bot. 59: 3649–3659 [DOI] [PubMed] [Google Scholar]
- Ronceret A., Gadea-Vacas J., Guilleminot J., Lincker F., Delorme V., Lahmy S., Pelletier G., Chaboute M.E., Devic M. (2008b). The first zygotic division in Arabidopsis requires de novo transcription of thymidylate kinase. Plant J. 53: 776–789 [DOI] [PubMed] [Google Scholar]
- Ronceret A., Guilleminot J., Lincker F., Gadea-Vacas J., Delorme V., Bechtold N., Pelletier G., Delseny M., Chaboute M.E., Devic M. (2005). Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 44: 223–236 [DOI] [PubMed] [Google Scholar]
- Rotman N., Durbarry A., Wardle A., Yang W.C., Chaboud A., Faure J.E., Berger F., Twell D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15: 244–248 [DOI] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Schier A.F. (2007). The maternal-zygotic transition: Death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Scholten S., Lorz H., Kranz E. (2002). Paternal mRNA and protein synthesis coincides with male chromatin decondensation in maize zygotes. Plant J. 32: 221–231 [DOI] [PubMed] [Google Scholar]
- Seydoux G., Braun R.E. (2006). Pathway to totipotency: Lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H.D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Tariq M., Saze H., Probst A.V., Lichota J., Habu Y., Paszkowski J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100: 8823–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I., Pena-Muralla R., Dickerman A., Berg M., Rogers R., Hutchens S., Sweeney T.C., McElver J., Aux G., Patton D., Meinke D. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant I., Paszkowski J. (2007). Role of histone and DNA-methylation in gene regulation. Curr. Opin. Plant Biol. 10: 528–533 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Baskar R., Grossniklaus U. (2000). Delayed activation of the paternal genome during seed development. Nature 404: 91–94 [DOI] [PubMed] [Google Scholar]
- Waddington C.H. (1942). Canalization of development and the inheritance of acquired characters. Nature 150: 563–565 [DOI] [PubMed] [Google Scholar]
- Weijers D., Geldner N., Offringa R., Jurgens G. (2001). Seed development: Early paternal gene activity in Arabidopsis. Nature 414: 709–710 [DOI] [PubMed] [Google Scholar]
- Xiao W., Custard K.D., Brown R.C., Lemmon B.E., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DNA-methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B.J., Khoransanizadeh S., Gaudin V., Jacobsen S.E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys21 trimethylation. Nat. Struct. Mol. Biol. 14: 869–871 [DOI] [PubMed] [Google Scholar]
- Zhang W., Lee H.R., Koo D.H., Jiang J. (2008). Epigenetic modification of centromeric chromatin: hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.