This study examines the basis of functional divergence amongst the closely related Arabidopsis AGO4, AGO6, and AGO9 proteins. Using associated small RNAs, these proteins direct DNA epigenetic modifications in a tissue-specific manner.
Abstract
Argonaute (AGO) effectors of RNA silencing bind small RNA (sRNA) molecules and mediate mRNA cleavage, translational repression, or epigenetic DNA modification. In many organisms, these targeting mechanisms are devolved to different products of AGO multigene families. To investigate the basis of AGO functional diversification, we characterized three closely related Arabidopsis thaliana AGOs (AGO4, AGO6, and AGO9) implicated in RNA-directed DNA methylation. All three AGOs bound 5′ adenosine 24-nucleotide sRNAs, but each exhibited different preferences for sRNAs from different heterochromatin-associated loci. This difference was reduced when AGO6 and AGO9 were expressed from the AGO4 promoter, indicating that the functional diversification was partially due to differential expression of the corresponding genes. However, the AGO4-directed pattern of sRNA accumulation and DNA methylation was not fully recapitulated with AGO6 or AGO9 expressed from the AGO4 promoter. Here, we show that sRNA length and 5′ nucleotide do not account for the observed functional diversification of these AGOs. Instead, the selectivity of sRNA binding is determined by the coincident expression of the AGO and sRNA-generating loci, and epigenetic modification is influenced by interactions between the AGO protein and the different target loci. These findings highlight the importance of tissue specificity and AGO-associated proteins in influencing epigenetic modifications.
INTRODUCTION
Argonaute (AGO) and related proteins are the effectors of RNA silencing mechanisms in which mRNAs are cleaved, translation is suppressed, or epigenetic modifications are introduced at the DNA or chromatin level. The target RNA cleavage mechanism is dependent on an RNase-H–like structure and activity of the AGO proteins that is referred to as slicer. Eukaryotes have three groups of AGO proteins that are classified according to the sequence of their PAZ and PIWI domains as AGOs, PIWIs, or AGO-like. Many organisms, including Caenorhabditis elegans, have representatives of each class, whereas others, such as Schizosaccharomyces pombe, have just one AGO-related protein. A common feature of all AGO-related proteins studied until now is that they are associated with small RNA (sRNA) guides so that RNA silencing is targeted in a sequence-specific manner. However, the eukaryotic AGOs have functionally diversified such that they are capable of regulating gene expression or epigenetic modification in a differential manner. This AGO diversification could be a result of a number of factors including, but not limited to, their association with different types of sRNAs, the different biochemical activities of the AGO, or their spatial and temporal AGO expression pattern (Farazi et al., 2008).
There are many different types of sRNAs that differ in their biogenesis and molecular features (Vaucheret, 2006; Farazi et al., 2008). The three main classes of sRNAs in the model plant Arabidopsis thaliana are microRNAs (miRNAs), trans-acting siRNAs (tasiRNAs), and heterochromatin-associated RNAs (hcRNAs). miRNAs and tasiRNAs are mainly 21 nucleotides in length, while hcRNAs are mainly 24 nucleotides. miRNAs derive from double-stranded RNA hairpin precursors and regulate gene expression by mRNA cleavage or translational inhibition. tasiRNAs are capable of the same type of gene regulation but derive from double-stranded RNA produced by an RNA-dependent RNA polymerase. hcRNAs are produced by an RNA-dependent RNA polymerase and they direct asymmetric cytosine DNA methylation (Brodersen and Voinnet, 2006; Vaucheret, 2006).
Arabidopsis encodes 10 AGOs whose functional diversity has been deduced from the nature of the associated sRNAs combined with genetic analysis. For example, AGO1 binds miRNAs and has a severe mutant growth phenotype that is consistent with the known miRNA targets (Baumberger and Baulcombe, 2005; Qi et al., 2005). AGO7 predominantly binds MIR390 that is an initiator of tasiRNA production (Montgomery et al., 2008). These tasiRNAs target mRNAs encoding proteins involved in hormone responses, and AGO7 mutant plants exhibit a corresponding modification of growth (Hunter et al., 2003, 2006; Fahlgren et al., 2006). AGO4 binds repeat and heterochromatin-associated siRNAs, and its mutant phenotype is associated with loss of epigenetic modifications at many chromosomal loci (Zilberman et al., 2003; Qi et al., 2006). The functional diversity of AGO-related proteins in other organisms is also associated with differences in sRNA binding. In Drosophila melanogaster, for example, there are different AGOs predominantly associated with miRNAs (Ago1), siRNAs (Ago2), and piwi-intereacting RNAs (piRNAs; PIWI, Aub, and Ago3) (Okamura et al., 2004; Saito et al., 2006; Vagin et al., 2006; Brennecke et al., 2007).
From these findings, there is a general question as to how the different classes of sRNA are channeled into the appropriate AGO proteins and how that partition relates to functional diversification. In part, the answer relates to properties of the sRNA. Recent analyses in Arabidopsis revealed that different AGO proteins select for sRNA with a specific length and 5′ terminal nucleotide (Kim, 2008). For example, AGO1 binds 21-nucleotide sRNAs with a 5′ U, AGO2 binds 21 and 22 nucleotides with a 5′ A, while AGO4 predominantly associates with 24-nucleotide sRNAs with a 5′ A (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008). The structure of the sRNA duplex may also influence binding of different classes of sRNA as illustrated by analyses of the Drosophila AGOs. After release from the miRNA precursor, the miRNA/miRNA* duplex often contains mismatched nucleotides, which are necessary for its incorporation into Drosophila AGO1. Conversely, perfectly complementary siRNA duplexes are bound by Drosophila AGO2, but changing the duplex structure to incorporate mismatches enabled the siRNA to associate with AGO1 (Forstemann et al., 2007).
These AGO-sRNA binding differences may be affected directly by the RNA binding activity of the AGO proteins as suggested above but could also be influenced indirectly by accessory proteins that bind to AGOs or the availability of particular sRNA species in the cell type in which the AGO is expressed. The expression pattern of AGO-related proteins may also affect their functional differentiation as illustrated by the PIWI proteins that are expressed in conjunction with the germ line of many organisms and associate with the germ line–specific piRNAs (Vagin et al., 2006; Malone et al., 2009).
To further our understanding of functional differentiation in AGO proteins, we have focused on AGO4 and two of its closest paralogs in Arabidopsis: AGO6 and AGO9 (Morel et al., 2002). We refer to these proteins as the AGO4 group. AGO4, AGO6, and AGO9 mutants have different molecular phenotypes in terms of sRNA accumulation and loss of DNA methylation, suggesting that these proteins are functionally distinct (Vaucheret, 2008). AGO4 and AGO6 mutations were identified in separate mutation screens from their suppression of RNA silencing phenotypes (Zilberman et al., 2003; Zheng et al., 2007). However, AGO4/AGO6 double mutants have a more severe phenotype than either of the single mutants, and so it has been suggested that they are partially redundant (Zheng et al., 2007). Phenotypes have not been identified for AGO9. A fourth AGO, AGO8, is present in Arabidopsis and is part of the AGO4 group (Morel et al., 2002). However, expression of AGO8 is very low, and it has been suggested that it is a pseudogene (Takeda et al., 2008).
Our initial approach to understanding functional diversification of the AGO4 group involved characterization of the sRNAs associated with AGO4, AGO6, and AGO9 using high-throughput sequencing. The results indicated that these three different AGOs predominantly associate with the same general class of sRNAs: 24-nucleotide 5′ adenosine sRNAs derived from the repeat and heterochromatin-associated sRNA loci. However, at some sRNA loci, we observed differential representation of AGO-associated sRNAs. We therefore performed a series of experiments to understand the functional diversity of the AGO4 group and to differentiate between possibilities that this functional diversity is mediated by characteristics of sRNA binding, tissue specificity, or biochemical activity.
Our results indicate that this differential representation derives in part from the pattern of expression of these sRNA loci and the corresponding AGO. For example, AGO6 and AGO9, when expressed under the AGO4 promoter, bound to a population of sRNAs that was more similar to the AGO4-associated population compared with the AGO6- and AGO9-bound population observed under their native promoter. However, AGO6 and AGO9 expressed from the AGO4 regulatory sequences were unable to complement all AGO4-dependent loci, indicating that coincident expression of the AGO and sRNA is insufficient to determine the target. Other factors downstream of AGO-sRNA binding, such as proteins associated with individual complexes or unknown biochemical functions of each AGO, may also contribute to the methylation and chromatin status of an Arabidopsis sRNA locus.
RESULTS
A Comparison of AGO4-, AGO6-, and AGO9-Associated sRNAs
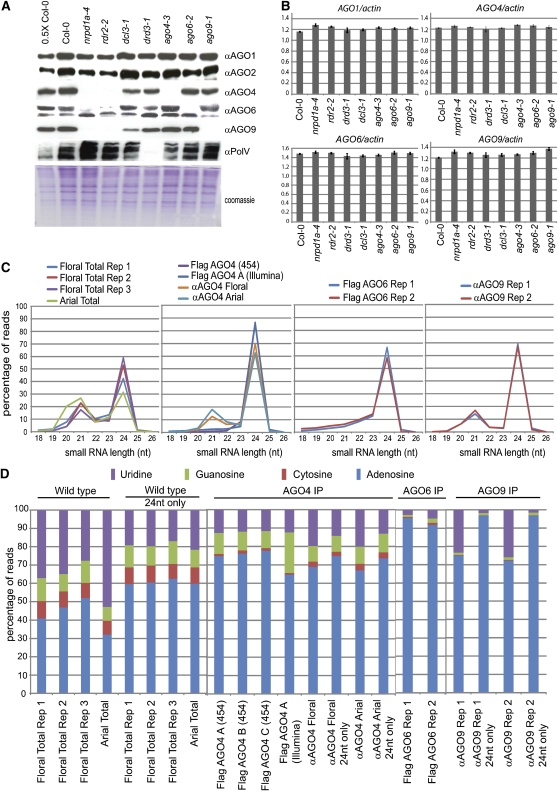
The binding of AGO4 to sRNA has been well characterized: it binds preferentially to 24-nucleotide sRNAs that are generated in a complex pathway involving a silencing-related polymerase (POLIV), an RNA-dependent polymerase (RDR2), and a Dicer (DCL3) (Qi et al., 2006; Vaucheret, 2006; Mi et al., 2008; Mosher et al., 2008). Correspondingly, in mutant plants that do not produce RDR2 or DCL3, AGO4 protein levels are lower than in wild-type plants (Li et al., 2006; Wierzbicki et al., 2009). Using immunoblotting with specific antipeptide AGO4 antibodies, we confirmed this result in plants that are mutant for the largest subunit of POLIV complex (NRPD1a) or for RDR2 (Figure 1A). These rdr2-2 and nrpd1a-4 mutants also have lower than wild-type levels of AGO6 and AGO9, whereas AGO1 and AGO2, which bind 21- and 22-nucleotide sRNAs produced independently of POLIV, RDR2, or DCL3, accumulate to near wild-type levels (Figure 1A). It is likely, therefore, that AGO6 and AGO9, like AGO4, bind sRNAs generated through the POLIV/RDR2/DCL3 pathway. The reduced levels of the AGO4 group proteins were not a result of reduced mRNA as measured by quantitative RT-PCR (Figure 1B). Presumably the AGO4 group proteins are unstable in the absence of the 24-nucleotide sRNA.
Figure 1.
The AGO4 Group Proteins Bind 24-Nucleotide sRNAs Produced by the RdDM Pathway.
(A) Immunoblots demonstrate that antipeptide antibodies to AGO4, AGO6, and AGO9 are specific, and the proteins show decrease in some RdDM mutants.
(B) RNA stability is not affected in the RNA interference mutants in which protein levels are decreased. The vertical axis represents the ratio of the average C(t) value of each AGO mRNA to ACTIN2 levels. Error bars represent an se of the ratio.
(C) AGO4, AGO6, and AGO9 preferentially bind 24-nucleotide sRNAs. Each panel contains the sRNA size profile of the total population or IP data sets. FLAG AGO4 (454) depicts the FLAG AGO4 C data set.
(D) The AGO4 group proteins have a 5′ nucleotide bias toward adenosine. Each panel denotes the percentage of sRNAs with 5′ nucleotide identities. For the total population, AGO4 IP and AGO9 IP immunopurified with the antipeptide antibody, and the 5′ nucleotide composition for the 24-nucleotide size class is indicated below the appropriate bar. Rep, replicate.
We included one other mutant, nrpe1/drd3-1, in our analysis of AGO4 group protein accumulation. This protein encodes the largest subunit of the POLV polymerase in Arabidopsis (Kanno et al., 2005; Pontier et al., 2005). AGO4 interacts in vivo and in vitro with the C-terminal domain of POLV (Li et al., 2006; El-Shami et al., 2007), but loss of POLV (nrpe1/drd3-1 mutant) has no effect on AGO4 accumulation (Figure 1A). Similarly, AGO9 accumulated at wild-type levels in an nrpe1/drd3-1 mutant, whereas AGO6 was present at reduced levels (but not absent) (Figure 1A). This result suggests that AGO6 may bind POLV itself or POLV-dependent sRNAs to a greater degree than AGO4 or AGO9 or, in a more general sense, that AGO4, AGO6, and AGO9 may bind to different subsets of POLIV pathway–generated sRNAs.
To test this possibility, we used high-throughput sequencing to characterize the sRNAs bound to AGO4 group proteins purified by immunoprecipitation (IP). AGO4 was immunopurified from floral or arial (mixed inflorescence, stem, cauline leaves, and siliques) tissue using either the epitope FLAG construct that complements the ago4-3 mutation or using an antipeptide antibody (see Supplemental Figure 1 online; Figure 4C). AGO6 was immunopurified from floral tissue using a FLAG epitope–tagged genomic construct that complements the ago6-2 mutation (see Supplemental Figure 2 online). AGO9 was immunopurified from arial tissue using an antipeptide antibody. For comparison, we also characterized the total population of sRNAs obtained from floral and arial tissue.
Figure 4.
Functional Divergence among AGO4, AGO6, and AGO9.
(A) Immunoblot analysis (αμFLAG antibody) shows a similar level of AGO4, AGO6, and AGO9 when expressed under the AGO4 promoter. AGO1 levels were used as a loading control.
(B) RNA gel blot analyses of PAGO4:FLAG AGO6 and PAGO4:FLAG AGO9 in the ago4-3 background (bkgd) show locus-specific complementation of AGO4-dependent sRNAs.
(C) Bisulfite analyses of AtSN1 and SIMPLEHAT2 indicate that asymmetric methylation complementation by AGO6 and AGO9 correlates with the observed sRNA accumulation at each locus (PAGO4:FLAG AGO9 partially complements, while PAGO4:FLAG AGO6 does not complement). For AtREP2, PAGO4:FLAG AGO9 fully complemented CpHpG and CpHpH methylation, while PAGO4:FLAG AGO6 had partial, but significant, complementation of CpHpH but not CpHpG. The different cytosine contexts are indicated above the top panel. The vertical axis represents the average percentage of methylated cytosines (per context). Error bars represent 95% confidence limits. The SIMPLEHAT2 locus contained only cytosines in a CpHpH context. Sample labels appear below the AtREP2 panel. H represents the nucleotides A, T, or C.
The total sRNA population was predominantly 21 and 24 nucleotides in length with the 24-nucleotide species being more abundant in the floral samples (Figure 1C). By contrast, the RNAs associated with AGO4, 6, or 9 were almost exclusively 24 nucleotides long (Figure 1C). A minor class of 21-nucleotide RNAs was present but only when the antipeptide antibodies were used for the immunoprecipitation: the 21-nucleotide sRNAs were absent from samples where the AGO was purified using the FLAG epitope. It is therefore likely that the 21-nucleotide RNAs were contaminants associated with the use of the antipeptide antibodies, and, for extended analysis of sRNAs associated with the AGO4 group proteins, we focused on the 24-nucleotide RNAs.
The 5′ nucleotide of the sRNA is an important determinant of sRNA-AGO association in Arabidopsis (Mi et al., 2008; Montgomery et al., 2008; Takeda et al., 2008). However, functional differentiation of the AGO4 group cannot be fully explained by this 5′ nucleotide preference because each of these proteins bound 24-nucleotide sRNAs with predominantly 5′ A residues. Of the 24-nucleotide population, this preference for A was more pronounced with AGO6 (averaged 94%) and AGO9 (averaged 97%) than with AGO4 (averaged 74%), but in all three sets of samples, the bias to A was stronger than in the 24-nucleotide RNAs in the total RNA without immunopurification (averaged 60%) (Figure 1D).
From the combined analysis of size and first nucleotide preference of the AGO-bound sRNAs, we conclude that functional differentiation of the AGO4 group is not based on these proteins binding preferentially sRNAs with a different length or 5′ nucleotide. AGO4 does bind a higher proportion of sRNAs with a 5′ G, C, and U than does AGO6 or AGO9 (P < 2.2 × 10−16) (Figure 1D). However, these 5′ non-A sRNAs are only a minor proportion of the AGO4-bound sRNAs and are not likely to have a large effect on functional differences. Other AGOs outside the AGO4 group may have a strong preference for 5′ non-A sRNAs and account for the prevalence of 5′ C residues in the total fraction of 24-nucleotide sRNAs (Figure 1D).
Genome Segments Corresponding to AGO4-, AGO6-, and AGO9-Associated sRNAs
We next assessed the possibility that the three AGO proteins bound 24-nucleotide RNAs derived from different features and locations within the Arabidopsis genome. To create a map of predicted sRNA loci, we used a circular binary segmentation algorithm (Olshen et al., 2004) on sequence reads uniquely matching to the Arabidopsis genome from total and AGO4 group IP data sets (see Supplemental Table 1 and Supplemental Methods online). This analysis partitioned the Arabidopsis genome into 31,092 segments. Of these segments, 23,327 contained sRNAs from the AGO4 IP data set, 23,563 from the AGO6 IP data sets, and 24,460 from the AGO9 IP data sets. For this analysis, we equate segments containing sRNAs above background levels (see Supplemental Methods online) as genomic loci that are sources of sRNA production.
To identify genomic features associated with sRNA accumulation, we annotated all segments by overlap with predicted protein-coding genes, methylated regions, and different types of repeats. We then asked what types of genomic features are associated with AGO4 group loci (segments with sRNA levels above background) and whether these features differ from the proportion of features present in all segments (loci and those segments without any sRNAs) (Figure 2; see Supplemental Methods online).
Figure 2.
The AGO4 Group Preferentially Associates with Repeat and Heterochromatin sRNA Loci.
The vertical axis indicates the number of sRNA loci with >50% of the locus overlapping the genomic feature indicated above the chart. Error bars represent 1 sd above and below the average number of loci that overlap a random rearrangement of the genomic features on the genome (based on 100 randomizations).
This analysis confirmed that the sRNAs associated with AGO4, AGO6, and AGO9 overlap a similar set of genomic features (Figure 2). Compared with segments generally, protein coding genes and inverted repeats were highly underrepresented. Methylated regions, all classes of transposons, tandem repeats, and miRNAs were highly overrepresented in loci associated with sRNAs that bind AGO4 group proteins (Figure 2). Overrepresentation of miRNAs in these data sets was surprising but consistent with a previous finding that AGO4 binds a subset of miRNA species (Qi et al., 2006). There was only weak overrepresentation of simple and low complexity repeats in the sRNAs bound to AGO4 group proteins (Figure 2).
However, the different AGO4 group proteins did not bind sRNAs from each locus equally. Using an empirical Bayesian method, assuming a negative binomial distribution for counts of all 24-nucleotide sRNAs matching a locus (T.J. Hardcastle,www.bioconductor.org), we found very little differential locus representation in biological replicates of AGO IP samples. By contrast, for pairwise comparisons between the AGO IP data sets (Table 1), there were many differences. On this basis, sRNA loci were most often similarly represented in the AGO4 and AGO6 data sets, and they were the most different between the AGO4 and AGO9 IP samples (Table 1).
Table 1.
Differential Representation between sRNA Data Sets
| Descriptiona | Percentage of Segments That Are Differentially Represented Loci | Percentage of Segments That Are Equivalently Represented Loci | Percentage of Segments That Are Locib |
| Replicate data sets | |||
| Floral | 4.7 | 62.9 | 67.6 |
| AGO4 IPs (454 only) | 3.3 | 34.9 | 38.2 |
| AGO4 IPs (454 versus Illumina)c | 28.3 | 28.8 | 57.1 |
| AGO6 IPs | 0.0 | 65.1 | 65.1 |
| AGO9 IPs | 0.0 | 53.7 | 53.7 |
| Comparison between tissue types | |||
| Floral versus aerial | 13.0 | 57.0 | 70.0 |
| Comparison between IP samples | |||
| AGO4 IP versus AGO6 IPd | 37.8 | 32.5 | 70.3 |
| AGO4 IP versus AGO9 IPe | 53.1 | 11.2 | 64.4 |
| AGO6 IP versus AGO9 IP | 48.6 | 17.8 | 66.4 |
| Comparison between IPs under the AGO4 promoterd | |||
| PAGO4:AGO4 IP versus PAGO4:AGO6 IP | 16.6 | 44.2 | 60.8 |
| PAGO4:AGO4 IP versus PAGO4:AGO9 IP | 22.1 | 40.9 | 63.0 |
| PAGO4:AGO6 IP versus PAGO4:AGO9 IP | 18.0 | 41.2 | 59.2 |
| Comparison of total and mutant data sets | |||
| Floral versus ago4-3 | 30.5 | 40.4 | 71.0 |
| Floral versus nrpe1/drd3-1 | 44.4 | 26.9 | 71.4 |
| ago4-3 versus nrpe1/drd3-1 | 17.1 | 49.3 | 66.4 |
| ago4-3 versus ago4-3 PAGO4:AGO4D660A | 18.7 | 47.8 | 66.5 |
Biological replicate data sets were taken into account whenever possible.
Note that segments with a significant level of sRNAs are considered loci (see Supplemental Methods online). For example, of all the segments defined within the Arabidopsis genome, 67.6% of the segments have sRNA levels sufficient to be considered loci in the floral replicates. Of this 67.6%, 4.7% of these loci are differentially represented among the floral replicates.
This estimate is a comparison of the three 454 FLAG AGO4 IPs (GSM415800-GSM415802) versus the Illumina FLAG AGO4 IP (GSM415893).
These comparisons were done using the Illumina FLAG AGO4 IP (GSM415893).
This AGO4IP comparison was done using the AGO4IP from aerial tissue (GSM415787).
The distinctness of the AGO9-associated sRNAs was also apparent from a display of the sRNAs that exactly match the Arabidopsis genome. As observed in previous studies, when using all sequence reads (unique and nonunique), there is a concentration of sRNAs in the centromeric regions in all data sets (see Supplemental Figure 3 online). However, when displaying unique reads only, certain sRNA loci are more highly represented in the AGO9 IP data than in the AGO4 and AGO6 IP data sets (see Supplemental Figure 4 online).
The locus representation among the sRNAs bound to AGO4, AGO6, and AGO9 could be influenced by biochemical properties of these proteins. Alternatively, it could be influenced by differential expression of the AGO4 group genes and the sRNA loci: an sRNA locus would be represented to an extent depending on the coincident expression of the genes for the sRNAs and the AGO4 group proteins.
To test these possibilities, we assembled promoter exchange constructs in which FLAG-tagged AGO6 and AGO9 are expressed from the AGO4 promoter (PAGO4). The constructs were transformed into wild-type Arabidopsis plants, the AGO4 group proteins were immunopurified from mixed inflorescence tissue using FLAG antibody (see Supplemental Figure 5A online), and the sRNA locus representation was assessed as described above. The biochemical property hypothesis would be favored if the sRNA locus representation is the same for AGO6 or AGO9 irrespective of whether these proteins were expressed from their native or from the AGO4 promoter. Alternatively, the coincident expression hypothesis would be favored if use of the AGO4 promoter caused the AGO6- and AGO9-sRNA loci to resemble those in the AGO4 IP sample.
The sRNAs purified from PAGO4:AGO6 and PAGO4:AGO9 were predominantly 24 nucleotides in length and had an extremely high proportion of 5′ nucleotide being A (>95%) (see Supplemental Figure 5B online). The corresponding sRNA loci were more similar to the AGO4-sRNA loci than the native promoter AGO6- and AGO9-sRNA loci (Table 1). Therefore, the differences between sRNA populations bound to the AGO4 group proteins are due to a large extent on their different expression patterns, and the data are consistent with the coincident expression hypothesis. However, there must be other factors involved because the sRNA locus representation when AGO6 and AGO9 were expressed under the AGO4 promoter was not identical to the representation in the AGO4 IP data set (Table 1). The biochemical properties of the AGO proteins must also have an effect.
Tissue-Specific Expression of AGO Proteins
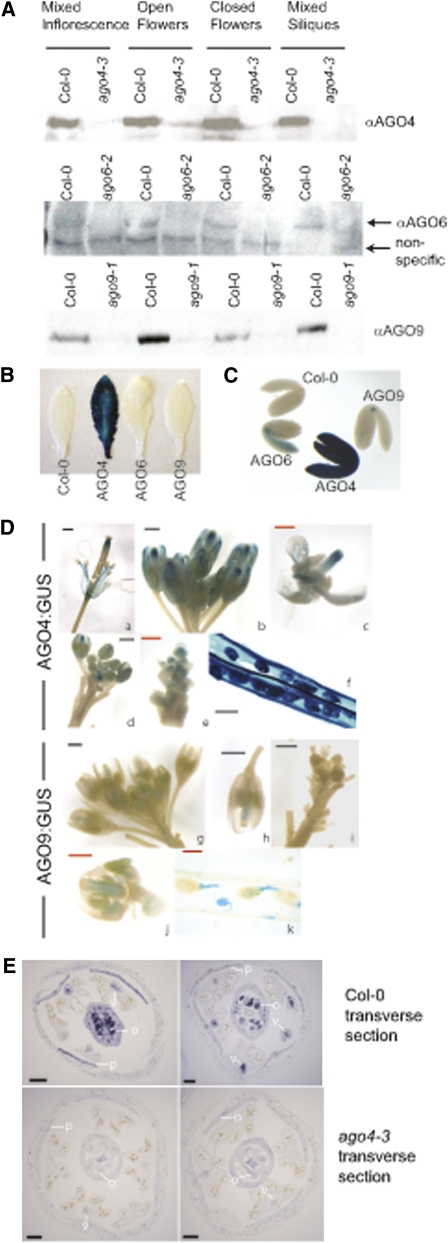
The coincident expression hypothesis requires that the AGO4 group proteins are differentially expressed. Immunoblotting with specific AGO antipeptide antibodies detected all three proteins in flower and silique samples, but the resolution of this method was not sufficient to detect differential expression (Figure 3A). We therefore generated AGO promoter(P): β-glucuronidase (GUS) Columbia-0 (Col-0) lines and used histochemistry to detect the extent of expression in a tissue-specific manner. This approach revealed that all three AGOs are expressed in embryos and mature plants (Figures 3B to 3D) but in different cell types. PAGO4:GUS showed widespread expression in embryos; leaves of mature plants (Figure 3B); all stages of flower development, including the vascular tissue of the sepals and the stamens and in the stigma and at the tip of the style; and in the siliques (Figures 3D and 3E). PAGO6:GUS expression was restricted to the shoot and root growing points and the vascular tissue connecting these and PAGO9:GUS was expressed in the embryonic shoot apex region (Figure 3C) and developing ovules from stage 8 flowers (Figure 3D).
Figure 3.
The AGO4 Group Proteins Accumulate in a Tissue-Specific Fashion.
(A) Immunoblots indicate that the AGO4 group proteins accumulate in both floral and silique tissue.
(B) Only PAGO4:GUS was detectable in leaf tissue; Col-0 lacks the GUS transgene.
(C) PAGO4:GUS, PAGO6:GUS, and PAGO9:GUS show differential expression patterns within embryos.
(D) PAGO4:GUS and PAGO9:GUS show differential expression patterns within mixed-stage floral tissue and siliques. (a) to (e) and (g) to (j) are floral tissue; (f) and (k) are siliques. Black bars = 500 μm; red bars = 250 μm.
(E) Immunohistochemistry on transverse sections of mixed-stage floral tissue shows specific AGO4 protein expression in developing ovules and vascular tissue. Only background staining was observed with the AGO4 antipeptide antibody on ago4-3 floral tissue. v, vascular tissue; o, ovules; p, petals. Bar = 100 μm.
The PAGO9:GUS expression patterns are consistent with in situ localizations using AGO9(pup1) probes (Scutt et al., 2003), and for AGO4, they correspond to immunolocalization with AGO4 antipeptide antibody on cross sections of mixed staged inflorescence tissue (Figure 3E). We detected AGO4 protein localization by this method in the developing ovules as well as the vascular tissue (Figure 3E). The low-level expression and restricted expression of PAGO6:GUS constructs is in line with microarray expression profiling (see Supplemental Figure 6 online), indicating that the corresponding RNA only accumulates at low levels. Combined, these results confirm that there is differential expression of the AGO4 group proteins as required by the coincident expression hypothesis.
We then asked whether expression patterns alone were enough to account for the functional differences of the AGO4 group proteins affecting sRNA accumulation and epigenetic modification. To test this possibility, we transformed the PAGO4:AGO6 and PAGO4:AGO9 constructs into ago4-3 (see Supplemental Figure 5A online) and assayed for complementation of sRNA and DNA methylation phenotypes at AtSN1, SIMPLEHAT2, and AtREP2. We reasoned that, if expression was sufficient to account for the functional differences of the AGO4 group genes, the chimeric constructs would complement ago4-3. However, if the encoded proteins are functionally distinct, then complementation would not occur or be incomplete.
All three AGO proteins were expressed at the same levels in the ago4-3 mutant background (Figure 4A) from the chimeric AGO4 promoter constructs. As expected, the sRNA and DNA methylation phenotypes at all loci tested were complemented by the AGO4 coding sequence construct, as monitored by RNA gel blot analyses and bisulfite sequencing, respectively (Figures 4B and 4C). At SIMPLEHAT2 and AtSN1, PAGO4:AGO9 only conferred partial complementation of sRNA production, whereas PAGO4:AGO6 was unable to complement sRNA accumulation of these loci (Figures 4B and 4C). By contrast, at AtREP2, both PAGO4:AGO9 and PAGO4:AGO6 complement sRNA accumulation (Figure 4B). However, only PAGO4:AGO9 could fully complement the loss of asymmetric DNA methylation. PAGO4:AGO6 partially complemented this phenotype (Figure 4C). Both PAGO4:FLAG AGO6 and PAGO4:FLAG AGO9 can bind sRNAs from these loci when the promoter exchange constructs were expressed in wild-type plants (see Supplemental Figure 7 online).
These data with promoter fusion constructs therefore reinforce the importance of coincident expression of AGO proteins and sRNA loci. However, they also confirm, as with the genome-wide analysis of sRNA loci (Table 1), that other factors related to the biochemistry of the AGO proteins are also required for the functional differences in these proteins.
Biochemical Properties of AGO4, AGO6, and AGO9
Biochemical differences affecting function of AGO4 group proteins are unlikely be related to the ability to bind sRNAs of different length and 5′ nucleotide because these proteins all bind 24-nucleotide sRNAs with predominantly 5′ A (see Supplemental Figure 5B online) even when they are expressed from the AGO4 promoter. It seemed likely therefore that any biochemical differences could involve interactions of these AGO proteins with other proteins or factors associated with chromatin loci.
As AGO4 and the POLV CTD have been shown to associate in vivo and in vitro (Li et al., 2006; El-Shami et al., 2007), we tested the possibility that variation in this POLV interaction with AGO6 or AGO9 might explain why these proteins expressed from the AGO4 promoter could not fully complement sRNA accumulation and methylation at AtSN1 and SIMPLEHAT2. The assay for the interaction of these proteins was based on immunoprecipitation of the AGO proteins using the FLAG epitope followed by immunoblotting with a POLV antibody. The results showed that all three AGO proteins bound to POLV with AGO6 binding more strongly than the other two AGO4 group proteins (Figure 5).
Figure 5.
The AGO4 Group Proteins Are Capable of Interacting with the Largest Subunit of POLV.
Each AGO was immunopurified using the FLAG epitope, and equal amounts of AGO protein were present on the agarose beads (αFLAG beads panel). The αPOLV input panel shows an equal input of the POLV largest subunit, except in the nrpe1/drd3-1 mutant. The αPOLV co-IP panel contains the FLAG-AGO recombinant protein and any interacting proteins. This was subjected to immunoblotting using the POLV antibody. PAGO4:FLAG AGO6 repeatedly showed the greatest degree of co-IP interaction.
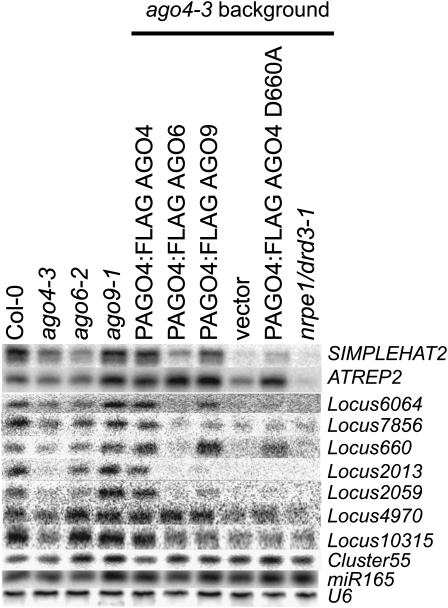
We also explored the possibility that the slicing activity of AGO4 group proteins could explain their functional differences. The slicing ability of AGO4 is required for sRNA production on a locus-specific basis (Qi et al., 2006), so we tested whether it correlated with the complementation ability of the AGO4 group proteins. First, we recreated the AGO4 slicer mutant, PAGO4:FLAG AGO4D660A, with a mutation in a conserved residue that is required for slicer function (Qi et al., 2006). We then used RNA gel blot analyses to assess this mutant's ability to complement sRNA phenotypes of ago4-3. If slicing ability is the factor that determines whether AGO6 or AGO9 could complement AGO4-dependent loci, we predicted that the phenotype of PAGO4:AGO6 or PAGO4:AGO9 would resemble that of AGO4D660A.
The mutant PAGO4:FLAG AGO4D660A had the same phenotype reported previously in that it complemented sRNA production at AtREP2 but not SIMPLEHAT2 (Qi et al., 2006) (Figure 6). In this respect, AGO4D660A was similar to AGO6 and AGO9. However, this similarity broke down when additional AGO4-dependent sRNA loci were tested. These additional loci resembled SIMPLEHAT2, AtREP2, and AtSN1 in that their sRNA levels were lower than the wild type in ago4-3, but there was otherwise no consistent pattern when ago4-3 was complemented by AGO4D660A, PAGO4:FLAG AGO6, or PAGO4:FLAG AGO9. The levels of sRNA from Locus 660 were complemented by AGO4D660A and PAGO4:FLAG AGO9 but not PAGO4:FLAG AGO6; Locus 2059 was not complemented by any of these constructs; Locus 4970 was complemented by PAGO4:FLAG AGO6 and PAGO4:FLAG AGO9 but not AGO4D660A.
Figure 6.
Locus-Specific sRNA Accumulation by AGO6/AGO9 Expression Does Not Correlate with the AGO4D660A Mutant.
AGO4-dependent loci were tested by RNA gel blot analyses and are denoted by locus numbers. All loci required NRPE1/DRD3 for sRNA accumulation. Most require an AGO4 catalytic PIWI domain. Genomic locations for each locus are listed in Supplemental Table 2 online.
A locus-specific effect of slicer activity is also reinforced by our analysis of sRNA content of ago4-3, the POLV mutant, nrpe1/drd3-1, and an ago4-3 PAGO4:FLAG AGO4D660A transgenic line using high-throughput sequencing. The results, summarized in Table 1, were consistent with the RNA gel blot analyses presented in Figure 6 and showed that the nrpe1/drd3-1 mutant affects sRNA accumulation at 44% of the predicted sRNA loci (Table 1), which is in agreement with previous studies (Mosher et al., 2008). The ago4-3 mutation had a weaker effect involving only 30% of the loci (Table 1, Figure 6). However, a strong correlation exists between loci affected by the nrpe1/drd3-1 and the ago4-3 mutation since only 17% of the loci were differentially represented in samples from these two mutants (Table 1). Similarly, only 18% of the loci were estimated to be differentially represented between ago4-3 and ago4-3 PAGO4:FLAG AGO4D660A. This suggests that sRNA accumulation at most AGO4-dependent loci is not complemented by the catalytic mutation; therefore, sRNA accumulation most often requires the enzymatic cleavage of a target RNA (Table 1).
These analyses of POLV interactions and slicer mutation confirm that there are indeed biochemical differences between AGO4 group proteins that could contribute to the functional differences of these proteins. However, there is not a simple relationship between the various phenotypes tested here and these biochemical properties, and we conclude that there may be other locus-specific factors involved that may act in combination with slicer activity or POLV interaction to influence the functional diversity of the AGO4 group proteins.
DISCUSSION
Coincident Expression of AGO4 Group Proteins and Their Bound RNA
In this article, we have investigated functional diversification of the AGO4 group proteins in Arabidopsis. Our results rule out that these three AGO proteins have different functions because they bind different types of sRNA: they all bind 24-nucleotide sRNAs with a 5′ A (Figure 1). Instead, we show that at least two other factors must be involved of which the first is associated with the expression pattern of the different AGO4 group genes. The evidence for this first factor is from our findings that their genes are differentially expressed (Figure 3) and that the genomic sRNA loci are not equally represented among the sRNAs bound to the AGO4 group proteins (Table 1). However, when the AGO4 group proteins were each expressed under the AGO4 promoter, they bound to populations of sRNAs that are more similar both quantitatively and qualitatively to those bound by AGO4 (Table 1).
Other eukaryotic AGOs and PIWI proteins accumulate in a tissue-specific fashion (Vagin et al., 2006; Farazi et al., 2008; Hock and Meister, 2008; Malone et al., 2009), suggesting that differential expression is a general strategy for their functional diversification. One of the clearest examples is with the PIWI proteins that are expressed in or the near the germline of animal cells where they associate with piRNAs, which are thought to silence transposable elements (Vagin et al., 2006; Malone et al., 2009). However, in all of these examples, the differentially expressed AGO proteins would not bind to different populations of sRNAs if the corresponding sRNA loci are constitutively expressed: there must also be variation in the expression of these loci between cell types. According to this idea, the population of sRNAs bound to any AGO protein would be determined by the coincidence of the AGO protein and the expression of an sRNA locus in any particular cell type.
The piRNAs also conform to this coincidence mechanism because, like their partner PIWI proteins, they are expressed specifically in germ line cells. In plants, which do not sequester their germline in the same manner as animals, some sRNA accumulation has been shown to be tissue specific as is required by the coincidence model. For example, some transposable element–derived sRNAs accumulate in the vegetative nucleus of the pollen (Slotkin et al., 2009). Correspondingly, many of these same types of sRNAs are uniparentally expressed in the endosperm and their accumulation peaks 5 d after fertilization (Mosher et al., 2009).
The AGO4 group proteins, including AGO4 and AGO6, have been associated with epigenetic modifications and, in particular, with genome defense through the silencing of transposable elements. If genome defense is the main role of these proteins, their expression should be restricted to gametes or cells that generate gametes. However, from our promoter fusion analysis (Figure 3), the expression of the AGO4 group genes cannot be explained simply in terms of genome defense because they are expressed in somatic cell types. Therefore, in addition to a role in genome defense, it seems likely that these proteins may have various roles associated with genetic and epigenetic control in leaves and other organs. As most of the sRNAs bound to the AGO4 group proteins are associated with repeats or transposable elements, this possibility has a further implication for the biological effects of transposable elements. It suggests that they may influence genetic and epigenetic control in plants via sRNAs and AGO4 group proteins, perhaps acting redundantly with other epigenetic control mechanisms. This idea is an extension of the pioneering controlling element concept of McClintock because it implies that transposons may have a role in genetic control.
Additional Factors Affecting Functional Diversification of AGO4 Group Proteins
However, coincidence of AGO protein and sRNA locus expression is not the only mechanism of functional divergence among the AGO4 group: there must be additional factors. Evidence for these factors is from our observation that AGO6 and AGO9 are incapable of fully complementing sRNA accumulation and asymmetric DNA methylation to the same extent as AGO4 even when they are expressed from the AGO4 promoter (Figure 6, Table 1). From these data it seems that there are locus-specific factors that determine whether sRNA accumulation and its effects are dependent on AGO4, AGO6, or AGO9.
Locus-specific effects could be explained if the AGO proteins are present at the corresponding target loci. This hypothesis is consistent with experiments in S. pombe demonstrating that tethering of RITS, which includes S. pombe AGO1, is sufficient to induce heterochromatin formation (Buhler et al., 2006). The recent findings that AGO4 in Arabidopsis is present at target loci are also consistent with this hypothesis (Daxinger et al., 2008; Wierzbicki et al., 2008, 2009).
In this study, we tested two possible explanations of the locus-specific factor. In one assay, we asked whether there was a correlation between the ability of AGO to slice target RNAs and the locus specificity. This possibility was ruled out because a slicer defective AGO4D660A exhibited locus specificity in its ability to complement ago4-3, but this specificity did not correlate with the complementation activity of either AGO6 or AGO9 (Figure 6). Thus, we ruled out that slicer activity is a prime determinant of locus specificity.
We also assayed the ability of the AGO4 group proteins to interact with POLV (Figure 5). The strength of this interaction could affect the targeting mechanism if it varies between the different AGO4 group proteins. Our results revealed that, of the three AGO4 group proteins, AGO6 interacted most strongly with POLV (Figure 5). However, the AGO6-dependent sRNA loci are not necessarily those that also had a strong POLV dependency, as indicated by the phenotype of drd3-1. Therefore, it is unlikely that the POLV interaction alone can explain the locus specificity of the AGO4 group proteins.
Other proteins that may influence the locus specificity of AGO4 group protein action include a, SNF2 helicase homolog DRD1 and a hinge domain protein DMS3 that influences structural maintenance of chromosomes (Daxinger et al., 2008; Wierzbicki et al., 2008, 2009). These proteins both influence the role of POLV in the silencing pathway involving AGO4 and, presumably, AGO6 and AGO9. A recently identified protein, KTF1/SPT5-like/RDM3, that interacts with AGO4 via its WG/GW motifs and that is also present in the POLV complex (Bies-Etheve et al., 2009; He et al., 2009; Huang et al., 2009) could also be involved in the locus specificity of AGO4 group proteins.
The likelihood that KTF1 is present in separate AGO4 and POLV complexes raises the possibility of competitive interaction: KTF1 is homologous to a transcriptional elongation factor (El-Shami et al., 2007), so when bound to POLV, it might enhance the transcription of chromatin-associated RNAs that are targets of RNA silencing. The interaction with AGO proteins might compete with this interaction by sequestration of the transcription cofactor. The level of silencing at a locus would then be determined by the amount of POLV at the locus with additional factors being the levels of KTF1 and AGO. The relative strength of the interaction between KTF1 POLV and the different AGO proteins would also be a factor.
Thus, we propose that AGO4, AGO6, and AGO9 participate in the RdDM pathway but functionally diverge in terms of their ability to promote sRNA accumulation and DNA methylation. Their effector contributions can be differential, but the bound sRNAs are not necessarily different. These effector differences result from a combination of factors, which include the cells in which the AGOs and the sRNAs are expressed and also most likely from the proteins present or interacting with the AGO effector complex at the locus itself. Other factors may also be important, such as the subcellular compartment within which the AGO resides or when in the cell cycle it is expressed. Further experiments will reveal how epigenetic modifications are created by these AGO4 group proteins in a tissue-specific and locus-specific manner.
METHODS
Plant Materials
Mutants with the following alleles were used in this study: ago4-1 and gl1-1 (CS6364; Zilberman et al., 2003), ago4-3 (WiscDSLox338A06), ago6-2 (SALK_031553), ago9-1 (SALK_127358), nrpd1a-4 (SALK_083051), rdr2-2 (SALK_059661), dcl3-1 (SALK_005512), and nrpe1/drd3-1 (Kanno et al., 2005). All plants were grown under controlled conditions in 16 h light at 22°C. Mixed-stage floral and aerial tissue of ∼7 weeks maturity was collected.
Transgenic Constructs
All constructs are based on pGREEN vector 0229 (Basta resistant) except for vectors containing AGO4 promoter/terminator sequences, which reside in pGREEN 0179 (hygromycin resistant) (www.pgreen.ac.uk). The specific cloning steps used to create the constructs are described in Supplemental Methods online.
RNA Gel Blot and RT-PCR Analyses
For RNA gel blot and RT-PCR analyses, total RNA was extracted from mixed-stage inflorescence tissue using Trizol (Invitrogen). Approximately 25 μg total RNA was subjected to electrophoresis in 8 M urea 15% polyacrylamide gels and transferred to nylon membrane using capillary blotting. Oligos used for sRNA detection are listed in Supplemental Table 2 online.
RT-PCR was completed on mixed-stage inflorescence RNA with subsequent DNase treatment (TurboDNase; Ambion) following the manufacturer's instructions. First-strand cDNA was generated using oligo(dT). PCR was performed using these primers: ACTIN2 (ActinF2/ActinR), AGO1 (AGO1FW/AGO1RV), AGO4 (AGO4FW/AGO4RV), AGO6 (AGO6FW/AGO6RV), AGO9 (AGO9FW/AGO9RV), and ROS1 (DBO206/DBO207) (see Supplemental Table 2 online). If quantitative RT-PCR was completed, SYBR Green (Sigma-Aldrich) was used to measure amplification on a Chromo4 (Bio-Rad). Triplicate readings were averaged and standard errors were calculated after normalization to ACTIN2. Error bars represent a propagation of standard errors.
Methylation Analysis
DNA was isolated from mixed-stage floral tissue using Puregene Core Kit A (Qiagen). For McrBC analysis, 500 ng of DNA was mock or McrBC digested (NEB) for 12 h. Amplification of AtSN1 (ATS15/At SN1 F2) or ACTIN2 (ActinF2/ActinR) followed. Results were measured using quantitative PCR as above. For bisulfite analysis, 500 ng of DNA was converted using the EZ DNA Methylation-Gold Kit (Zymo Research). Subsequent PCR was completed for AtSN1 (JP1821/JP1822) (Henderson et al., 2006), SIMPLEHAT2 (DBO413/DBO414), and AtREP2 (DBO411/DBO412). The converted DNA was amplified using standard parameters, but with an extension temperature of 62°C. At least 16 clones per replicate were sequenced.
Peptide Antibodies and Protein Extraction
Antipeptides for the specific antibodies were as follows: AGO1 (VRKRRTDAPSEGGEGC-CONH2) (Qi et al., 2005; Baumberger et al., 2007), AGO2 (CGRKPQVPSDSASPSTST-CONH2), AGO4 (CRELKKRNP-NENGEFE-CONH2), AGO6 (H2N-IEPEQPSKRDYDITTC-CONH2), and AGO9 (DSDEPNGSGLPPPC-CONH2). The POLV CTD antipeptide antibody was described previously (Pontier et al., 2005; Huang et al., 2009). Detailed protocols of the protein extraction, immunoprecipitations, coimmunoprecipitation with POLV, and immunohistochemistry can be found in the Supplemental Methods online.
GUS Staining
GUS staining was performed as described previously (Galli et al., 2003). Siliques were dissected open and stained according to the above method followed by clearing in Hoyer's solution for 7 d (Liu and Meinke, 1998).
sRNA Cloning and Bioinformatic Analyses
For total sRNA samples, RNA was extracted using Trizol (Invitrogen) and isolated from the total RNA fraction using mirVana (Ambion). For AGO-associated sRNAs, after washing the IP beads, the AGO agarose bead complex was incubated in ∼500 μL Trizol and RNAs were extracted once with 0.2 (v/v) chloroform. sRNAs were precipitated in an equal volume of isopropanol. sRNAs were cloned as described previously (Chappell et al., 2005) with the minor modifications that (1) 15 cycles of PCR were used to amplify the library, and (2) adapter sequences were used that were appropriate for 454 or Illumina sequencing. The details of the computational analyses are in the Supplemental Methods online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AGO4 (At2g27040), AGO6 (At2g32940), and AGO9 (At5g21150). Analysis concerning the repetitive elements AtSN1, SIMPLEHAT2, and AtREP2 were based on the elements in the following locations: AtSN1 (Chrom3: 15794511..15794910), SIMPLEHAT2 (Chrom 5: 24819856..24820440), and At REP2 (Chrom 1: 540434..540986). The sRNA sequencing data sets have been deposited in the Gene Expression Omnibus, and accession numbers are as follows: Col-0 floral (GSM415783, GSM415784, and GSM415785), Col-0 aerial (GSM415786), 454 FLAG AGO4 IP (GSM415800, GSM415801, and GSM415802), Illumina FLAG AGO4 IP arial (GSM415787), Illumina FLAG AGO4 IP floral (GSM415788), FLAG AGO6 IP (GSM415789 and GSM415790), AGO9 IP (GSM415791 and GSM415792), PAGO4:FLAG AGO4 IP (GSM415793), PAGO4:FLAG AGO6 IP (GSM415794), PAGO4:FLAG AGO9 IP (GSM415795), nrpe1/drd3-1 (GSM415796), ago4-3 PAGO4:FLAG AGO4 D660A (GSM415797), Col-2 floral (GSM415798), and ago4-3 (GSM415799). A complete description of these data sets can be found in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization and Complementation of ago4-3.
Supplemental Figure 2. Complementation of ago6-2 by PAGO6:FLAG AGO6.
Supplemental Figure 3. Graphical Representation of Unique and Nonunique sRNA Frequency.
Supplemental Figure 4. Graphical Representation of Uniquely Matching sRNA Frequency.
Supplemental Figure 5. The Profile of AGO4-, AGO6-, and AGO9-Associated sRNAs When Expressed under the AGO4 Promoter.
Supplemental Figure 6. Organ-Specific Tissue Expression of AGO4, AGO6, and AGO9 as Measured by Publically Available Microarray Studies.
Supplemental Figure 7. Small RNAs Bound to Promoter Exchange Constructs.
Supplemental Table 1. High-Throughput Sequencing Data Sets.
Supplemental Table 2. Oligo Sequences Used in This Article.
Supplemental Methods. Transgenic Construct Construction, Protein Extraction, Immunoprecipitation, and Immunohistochemistry Protocols, and Detailed sRNA and Statistical Analysis Methods.
Acknowledgments
We thank Jodie Pike, Eric Kemen, and Michael Burrell at The Sainsbury Laboratory, James Hadfield, Nik Matthews, James Hadfield, Rory Stark, and Kevin Howe at the Cambridge Research Institute, and Kim Rutherford at the University of Cambridge for their help with sequencing and data processing the sRNA libraries. E.R.H. was supported by an EMBO long-term fellowship (ALTF 252-2005), Biotechnology and Biological Science Research Council Grant BB/C006739/2, and European Commission FP6 Integrated Project SIROCCO Contract LSHG-CT-2006-037900.
References
- Baumberger N., Baulcombe D.C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits micro RNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.-H., Lie M., Havecker E., Ziegler-Graff V., Baulcombe D.C. (2007). The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N., Pontier D., Lahmy S., Picart C., Vega D., Cooke R., Lagrange T. (2009). RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 10: 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Voinnet O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Buhler M., Verdel A., Moazed D. (2006). Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Chappell L., Baulcombe D., Molnar A. (2005). Isolation and cloning of small RNAs from virus-infected plants. Current Protocols in Microbiology, Coico D.R., Kowalik T., Quarles J.M., Stevenson B., Taylor R.K., Simon A.E., Downey T., (Hoboken, NJ: John Wiley & Sons; ), pp. 16H.2.1–16.H.2.17 [DOI] [PubMed] [Google Scholar]
- Daxinger L., Kanno T., Matzke M. (2008). Pol V transcribes to silence. Cell 135: 592–594 [DOI] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. (2007). Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21: 2539–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Farazi T.A., Juranek S.A., Tuschl T. (2008). The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135: 1201–1214 [DOI] [PubMed] [Google Scholar]
- Forstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D. (2007). Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M., Theriault A., Liu D., Crawford N.M. (2003). Expression of the Arabidopsis transposable element Tag1 is targeted to developing gametophytes. Genetics 165: 2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Zhu S.H., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.L., Wang C.S., Jin H.L., Zhu J.K. (2009). An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobson S.E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38: 721–725 [DOI] [PubMed] [Google Scholar]
- Hock J., Meister G. (2008). The Argonaute protein family. Genome Biol. 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.F., Jones A.M.E., Searle I., Patel K., Vogler H., Hubner N.C., Baulcombe D.C. (2009). An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 16: 91–93 [DOI] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willman M.R., Wu G., Yoshikawa M., Gutierrez-Nava M., Poethig R.S. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. (2005). Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Kim V.N. (2008). Sorting out small RNAs. Cell 133: 25–26 [DOI] [PubMed] [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W.L., Lagrange T., Pikaard C., Jacobsen S.E. (2006). An ARGONAUTE4-containing nuclear processing center colocalized with cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Liu C.M., Meinke D.W. (1998). The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J. 16: 21–31 [DOI] [PubMed] [Google Scholar]
- Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J. (2009). Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S.J., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5 ' terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Morel J.B., Godon C., Mourrain P., Beclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. (2008). PolIVb influences RNA-directed DNA-methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 3145–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C. (2004). Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18: 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. (2004). Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572 [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. (2005). Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A.M., Hannon G.J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qi Y., He X.H., Wang X., Kohany O., Jurka J., Hannon G.J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C. (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutt C.P., Vinauger-Douard M., Fourquin C., Ailhas J., Kuno N., Uchida K., Gaude T., Furuya M., Dumas C. (2003). The identification of candidate genes for a reverse genetic analysis of development and function in the Arabidopsis gynoecium. Plant Physiol. 132: 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzic M., Becker J.D., Feijo J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2006). Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 20: 759–771 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zhu J., Kapoor A., Zhu J.-K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]