Abstract
The condensation level of chromatin is controlled by epigenetic modifications and associated regulatory factors and changes throughout differentiation and cell cycle progression. To test whether changes of chromatin condensation levels per se affect access and binding of proteins, we used a hypertonic cell treatment. This shift to hyperosmolar medium increased nuclear calcium concentrations and induced a reversible chromatin condensation comparable to the levels in mitosis. However, this condensation was independent of mitotic histone H3 serine 10 phosphorylation. Photobleaching and photoactivation experiments with chromatin proteins—histone H2B-GFP and GFP-HP1α—before and after induced chromatin condensation demonstrated that hypercondensation reduced their dissociation rate and stabilized their chromatin binding. Finally, measuring the distribution of nucleoplasmic proteins in the size range from 30 to 230 kDa, we found that even relatively small proteins like GFP were excluded from highly condensed chromatin in living cells. These results suggest that structural changes in condensed chromatin by themselves affect chromatin access and binding of chromatin proteins independent of regulatory histone modifications.—Martin, R. M., Cardoso, M. C. Chromatin condensation modulates access and binding of nuclear proteins.
Keywords: fluorescence microscopy, GFP, mitosis, photoactivation, photobleaching
The cell nucleus is a highly organized organelle for storing and translating genetic information. Although there are no substructures separated by membranes, the nucleus is compartmentalized for different functions in nucleic acid metabolism (1). The nuclear DNA is organized together with structural proteins into dynamic higher-order chromatin structures, which reflect and control gene expression during the cell cycle and cellular differentiation (2, 3). As a consequence of a complex and not yet understood interplay between chromatin condensation state, transcriptional activity, and modifications of chromatin-organizing proteins, chromatin subsets are termed euchromatin and heterochromatin (4, 5). Euchromatin in interphase cells is actively transcribed and less condensed, while heterochromatin is transcriptionally inactive, with a condensation level similar to mitotic chromosomes (6). During mitotic chromatin condensation, most processes associated with DNA (e.g., transcription and replication) stop and only resume after chromatin decondensation in early G1 phase. However, association of RNA polymerase I components to rDNA loci has been shown to occur already during mitosis (7). The condensation of chromatin is characterized by a reduction of volume due to a spatial organization into densely packed higher-order structures (8). Specific histone modifications, e.g., histone H1 and H3 phosphorylation, occur at mitosis and contribute to the individualization and condensation of chromosomes. The consequences of this compaction into condensed chromosomes at mitosis are reduced free volumes and less exposed surface of the chromatin substructures on which molecules could interact (9, 10). However, it is unclear whether and how changes in the chromatin condensation state and/or histone modifications affect the distribution and access of nucleoplasmic proteins and the mobility of chromatin organizing proteins.
Interphase chromatin has been shown to be accessible to macromolecules (11, 12), chromatin proteins (13), and neutral inert proteins (14), even at the single-molecule level (15). Furthermore, certain transcription and chromatin factors have been shown to still have access to chromosomes in mitosis when the DNA metabolism stops (7). Therefore, the relationship between nuclear protein distribution, chromatin accessibility, and how changes in chromatin condensation over the cell cycle affect DNA metabolism remains unclear. In this study, we have manipulated chromatin condensation in interphase cells to test whether the level of chromatin condensation independently of mitotic histone modifications can affect mobility and accessibility of nuclear proteins.
MATERIALS AND METHODS
Plasmids
The histone H2B-mRFP plasmid was constructed by cutting out CFP from pH2B-CFP (16) with BamHI + MfeI and ligating in the mRFP cDNA cut with EcoRI + BamHI from pRSET-mRFP1 (17). The photoactivatable HP1α construct was created by inserting the human HP1α cDNA derived from pBCHGN-HP1α (18) cut with HindIII and BamH I into the HindIII and BglII of pPAGFP-C1 (19). The plasmid constructs EGFP (Clontech, Heidelberg, Germany), GFP-PCNAL2 (20), GFP-DNA Ligase I (21), and GFP-Dnmt1 (22) were described before.
Cell culture, transfections, and chromatin condensation
Human HeLa and stable HeLa H2B-GFP cells (23) were grown in DMEM with 10% FCS, 5 mM l-glutamine, and 5 g/ml gentamicin. For live cell microscopy, the cells were plated in 8-well Ibidi chambers (Ibidi, Munich, Germany). Single or double transfection of cells was carried out by the CaPO4 precipitation method (21) or using Transfectin reagent (Bio-Rad, Munich, Germany). Hyperosmolar chromatin condensation was induced by supplying the cells with a dilution of 10× PBS in growth medium to yield 4× PBS, equivalent to a 500 mM saline solution. The Fluo 3 (Invitrogen, Paisley, UK) labeling and measurements were performed as described before (24).
Immunofluorescence
For immunofluorescence staining, cells were washed in PBS, and in case of hyperosmolar treatment, they were incubated for 15 min in 4× PBS, fixed in 3.7% formaldehyde diluted in PBS or 4× PBS, respectively, and permeabilized with 0.25% Triton X-100 in PBS. The primary anti-phospho H3 rabbit polyclonal antibody (catalog no. 06-570; Millipore, Schwalbach, Germany) was diluted in PBS containing 0.2% fish-skin gelatin together with 2 U DNase I (Sigma, St. Louis, MO, USA) in DNase buffer containing 60 mM Tris HCl, 0.66 mM MgCl2, and 1 mM mercaptoethanol to increase chromatin penetration by the antibody and detected using goat anti-rabbit IgG (Jackson, Newmarket, UK). Immunofluorescence staining for proliferating cell nuclear antigen (PCNA) was performed as described earlier (25).
Fluorescence microscopy
Live-cell microscopy was performed with a Zeiss LSM510Meta confocal setup on a Zeiss Axiovert 200 M inverted microscope with an ×63 phase-contrast plan-apochromat oil objective, NA 1.4 (Carl Zeiss, Jena, Germany). The microscope was housed in a chamber heated to 37°C (Okolab, Ottaviano, Italy). For all acquisition settings, the main beam splitter was HFT UV/488/543/633; GFP was excited by 488-nm Ar ion laser and detected with a 500- to 530-nm bandpass filter. RFP was excited by a 543-nm HeNe laser and was detected using a 585- to 615-nm bandpass filter. Photobleaching and photoactivation were performed with a short intense pulse of a 488-nm Ar ion laser beam in selected areas of interest, as shown in Fig. 3. Acquisition settings were identical for individual cells in repeated imaging experiments.
Figure 1.
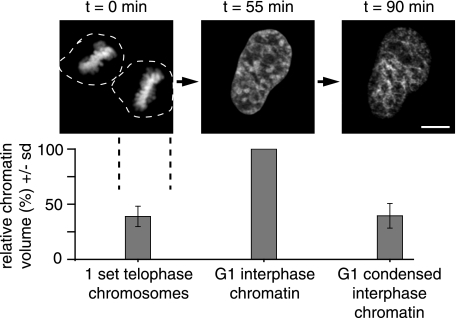
Manipulation and quantification of chromatin condensation states. Individual HeLa cells expressing H2B-GFP as a marker for chromatin (23) were visualized starting in mitosis (left panel) into G1 (middle panel), and following incubation in hyperosmolar medium (right panel). At each time point, complete high-resolution confocal z stacks were acquired; projections are shown. Scale bar = 5 μm. Chromatin volume calculations were derived from complete z stacks. Bar diagram summarizes chromatin volume data as means ± sd (n=10 cells) relative to the value for G1 interphase chromatin in the same cell.
Figure 2.
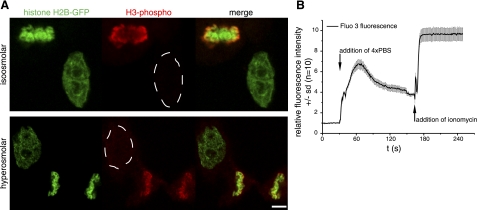
Different pathways of chromatin condensation in mitosis and hyperosmolar condensation. A) Histone H3 is modified by phosphorylation at serine 10 during mitosis and was detected in formaldehyde-fixed HeLa H2B-GFP cells using anti-phospho H3S10-specific antibodies. Top panel: phosphorylated histone H3 signal (red) colocalizes with histone H2B-GFP-labeled chromosomes in the mitotic cell, but it is not detected in the interphase cell. Bottom panel: chromatin was condensed by hyperosmolar treatment to induce an interphase chromatin volume comparable to mitotic chromosomes (see Fig. 1). As in untreated cells, mitosis-specific histone H3 phosphorylation is not detectable in the interphase nucleus, and only in the mitotic chromosomes, suggesting a different pathway of mitotic and hyperosmolar chromatin condensation. B) Intranuclear calcium levels in HeLa cells (n=10), measured by time-lapse microscopy using the fluorescent calcium sensor Fluo 3. For untreated cells, initial Fluo 3 fluorescence intensity was normalized to 1 and, following incubation in hyperosmolar 4× PBS solution, increased 7-fold. To test for saturation effects and dynamic range of the Fluo 3 measurement conditions, the calcium ionophore ionomycin was added subsequently to the cells to induce maximum calcium uptake. This displays the whole range of fluorescence intensity measurable by the calcium sensor under the same imaging conditions and further suggests that the change in chromatin condensation after 4× PBS incubation involves a significant change in calcium level. Scale bar = 5 μm.
Figure 3.
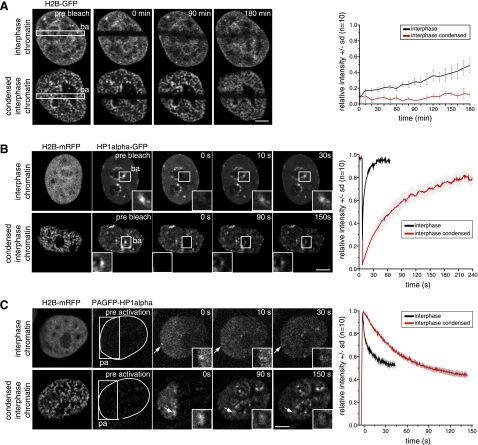
Distribution and mobility of chromatin proteins in different chromatin condensation states. HeLa cells stably expressing H2B-GFP, as well as HeLa cells, transiently transfected with plasmids coding for H2B-mRFP and either GFP-HP1α or PAGFP-HP1α were used in photobleaching or photoactivation experiments of the respective chromatin proteins, as indicated. For all experiments, areas photobleached (ba) and photoactivated (pa) are indicated by squares; times after photobleaching/activation are given in the images. A) Photobleaching experiment shows histone H2B-GFP dynamics in interphase HeLa cells. Top row: untreated interphase chromatin. Bottom row: hyperosmolar condensed chromatin. B) Photobleaching experiments with HP1α-GFP for cells in interphase (top panel) and with hyperosmolar condensed interphase chromatin (bottom panel). Corresponding fluorescence recovery curves are at right; means ± sd (n=10 cells). C) Photoactivation experiments with HP1α fused to PAGFP. Dotted lines in images before fluorescence activation encircle nucleus and chromosomes, visualized independently with H2B-mRFP fusion. Graphs at right show dissociation of HP1α from chromatin, measured by loss of photoactivated fusion protein from interphase chromatin; means ± sd (n=10 cells). Insets: magnified view of accumulations of HP1α at heterochromatin (arrows; B, C). Scale bars = 5 μm.
Image analysis
Image analysis was performed with the Zeiss LSM Image Examiner software for fluorescence intensity and colocalization measurements. The latter included the Pearson correlation coefficient (R), measured by selecting the nuclear areas, excluding the nucleoli or for mitotic cells an oval area, including the chromosomes and cytoplasm. Chromatin volume calculations and data for photobleaching and photoactivation experiments were processed in Image J (U.S. National Institutes of Health, Bethesda, MD, USA). Images were Gauss filtered and background subtracted. For the calculation of chromatin volumes, the image stacks were corrected for bleaching during image acquisition along the z axis. The stacks were first analyzed by line scans to set the boundaries of the chromatin signal intensity and accordingly thresholded to include all euchromatin and heterochromatin in G1 cells and the mitotic chromosomes. The voxel counter tool in Image J was used to calculate the respective volume of the thresholded chromatin. Photobleaching and photoactivation data were corrected for cell translation and rotation in the stack_reg plug-in. The acquired data sets were analyzed, evaluated, and displayed in Origin 7 (OriginLab, Northampton, MA, USA). The photobleaching and photoactivation redistribution data were double normalized for the loss or increase of signal by the bleach or activation laser pulse and photobleaching or photoactivation by image acquisition, respectively, and averaged according to Phair et al. (26). The fluorescence recovery after photobleaching (FRAP) data curves were fitted with a biexponential decay model in Origin 7, and the half equilibrium times t1/2 were derived from the fit curves. The mobile fraction of HP1α molecules was calculated with data from the FRAP fit curves according to the formula Mf = (F∞ − Fo)/ (Fi − Fo), where Fi = fluorescence before bleaching, Fo = fluorescence after bleaching, and F∞ = fluorescence after full recovery (27).
RESULTS
Manipulation and quantitative evaluation of chromatin condensation mechanisms
Maximal chromatin condensation takes place at every mitosis in cycling cells and results in a cessation of DNA metabolism. The latter could be the result of M-phase-specific modifications of chromatin proteins (e.g., histone modifications) directly preventing the access of factors to the genome and/or structural changes of chromatin hindering access by increased compaction. To distinguish between these possibilities, we made use of an approach to induce hypercondensation of chromatin during interphase (28).
We first compared the chromatin volumes of individual living HeLa cells labeled with histone H2B-GFP fusion proteins in different cell cycle stages and after chromatin-condensing treatment. The same cells were imaged beginning in mitosis and through reentry into G1, followed by induced chromatin condensation by incubation in hyperosmolar medium for 5 min. We then measured the chromatin volumes in the three conditions to test whether the treatment reduced interphase chromatin volumes to the level of mitotic chromosome volumes (Fig. 1). The chromatin volume measurements for mitosis and hyperosmolar condensation in G1 showed similar levels (mean±sd) of 39 ± 9 and 40 ± 11%, respectively, relative to G1 chromatin in the same cells before treatment (Fig. 1). This hypercondensation of chromatin halted DNA-associated processes, such as transcription and replication, and was fully reversible within 10 min of returning to normal osmolar medium (28). From the results of our volume measurements, we concluded that this approach generated condensation of chromatin in interphase nuclei to the same extent as mitotic chromatin.
Next, we assessed the occurrence of typical chromatin modifications in mitotic chromosomes and hypercondensed interphase chromatin. At mitosis, phosphorylated histone H3 at serine 10 is associated with the individualization and condensation of chromosomes. Although we found similar levels of chromatin condensation (Fig. 1), no mitosis-specific histone H3 phosphorylation was detected (Fig. 2A). Hence, the process of hyperosmolar chromatin condensation relies on a different pathway, which does not involve cell cycle-specific histone modifications but rather profound structural changes.
Because we had previously reported that cellular energy depletion using Na azide leads to interphase chromatin condensation with increased calcium level in the nucleus (24), we measured the levels of this cation using the fluorescent calcium sensor Fluo 3 before and after hypertonic treatment. Our data demonstrate that the condensation of chromatin by hyperosmolarity is accompanied by a 7-fold rise in the intranuclear calcium level (Fig. 2B). Subsequent addition of the calcium ionophore ionomycin to the cells led to an even higher increase establishing that our measurements were not performed under saturating conditions.
Taken together, these results indicate that reversible hypercondensation of interphase chromatin occurs in the absence of mitotic histone modifications, likely as a result of increased intranuclear calcium levels, and leads to a stop in DNA metabolism.
Accessibility of proteins to different chromatin condensation states
Next, we assayed the effect of chromatin condensation levels on the accessibility of nuclear proteins.
A large number of nuclear proteins bind to or are incorporated into chromatin structures throughout the cell cycle. We, therefore, tested whether chromatin condensation level affects their access to and their dynamics at chromatin. We selected a nucleosome core histone (H2B) and a heterochromatin binding protein (HP1) as representative chromatin proteins and analyzed their chromatin access and mobility by photobleaching and photoactivation experiments.
The nucleosome protein H2B-GFP was not redistributed on hyperosmotic chromatin hypercondensation (Fig. 3A). FRAP analysis showed, in accordance to previous reports (13), that the half-time of recovery t1/2 in interphase chromatin for this core histone was ∼180 min. On hyperosmolar treatment, only ∼5% fluorescence recovery could be measured within the same time, indicating that the access to and/or release of the core histone H2B from fully condensed interphase chromatin was affected. By comparison, the reported recovery of H2B-GFP already reached the same value (5%) 20 min after bleaching in mitotic prophase and metaphase cells and was slightly faster in anaphase and telophase (13–17%). This faster exchange of core histones in late mitosis was suggested to be linked to transcriptional reactivation and remodeling of chromatin (7).
Chromatin proteins that are not core components of the nucleosome often associate less tightly to chromatin and DNA than histones. The heterochromatin protein HP1α binds to methylated histone H3 and accumulates in heterochromatin. We measured the half-recovery time of GFP-HP1α in interphase heterochromatin of HeLa cells and compared it with hypercondensed chromatin. In interphase heterochromatin, t1/2 was 2.3 s, and full recovery was reached after 50–60 s (Fig. 3B), which fits well with previously reported half-recovery times of 1–10 s (18, 29). The mobile fraction of HP1α molecules on interphase heterochromatin within the time frame of the experiment was calculated to be 97%. The immobile or very slow exchanging fraction of 3% is in agreement with the data reported before of 2.5–7% (29, 30). However, in hypercondensed interphase chromatin, HP1α recovery in the FRAP analysis was much slower, with t1/2 increased to 44 s and the mobile fraction reduced to 82%. Full recovery of HP1α was reached after 240 s and is thus slower than reported for mitotic chromatin, which was ∼60 s (29). The 15% reduction in the HP1α mobile fraction is consistent with previous data showing a 14–15% increased fraction of very slow HP1α molecules in pericentromeric heterochromatin of prometaphase and metaphase cells (29). Even higher immobile fractions were found on transcriptional inhibition by actinomycin D (25–40%) or in resting primary T cells (30%) (29, 31).
To further clarify whether HP1α was transiently trapped in the hypercondensed interphase chromatin, we performed photoactivation experiments and measured the dissociation of fluorescent PAGFP-HP1α molecules from the heterochromatin spots in the photoactivated area. Analysis of the fluorescence decay curves (Fig. 3C) revealed that HP1α t1/2 of dissociation from interphase heterochromatin was 8 s, compared to a 6.5-fold slower t1/2 of 52 s in hypercondensed interphase chromatin. These results indicate a reduced mobility and a transient trapping of HP1α.
Biochemical fractionation of chromatin proteins complexes by, e.g., gel filtration without the use of DNase treatment or other DNA disrupting agents to better approximate the whole cell situation typically resulted on detection of chromatin protein containing fractions throughout a broad range of high molecular masses. Thus, such an analysis would not realistically reflect the in vivo scenario.
Taken together, we could demonstrate that the hypercondensed interphase chromatin was less accessible and reduced the exchange of chromatin proteins like core histones and heterochromatin-binding proteins.
We then measured the access of nonchromatin proteins to the differently condensed chromatin. We chose GFP as a neutral probe protein and imaged the same cells before and after hyperosmolar treatment to directly compare the distribution of proteins relative to chromatin within the same cell. Furthermore, we compared the results to mitotic chromatin.
Figure 4 shows confocal optical sections of living cells displaying the distribution of the inert tracer protein GFP relative to H2B-mRFP-labeled chromatin. The relative distribution was then analyzed quantitatively first by plotting the GFP signal intensity vs. the histone-labeled chromatin along a line across the nucleus (line-scan analysis, Fig. 4B) and second by a correlation analysis (Fig. 4C). The latter included both the display of all red and green pixels within the nucleus, excluding the nucleoli, in intensity scatterplots, and the calculation of the Pearson’s coefficient R as a measure for correlation (+1), anticorrelation (−1), or no correlation (0) of two color channels in images.
Figure 4.
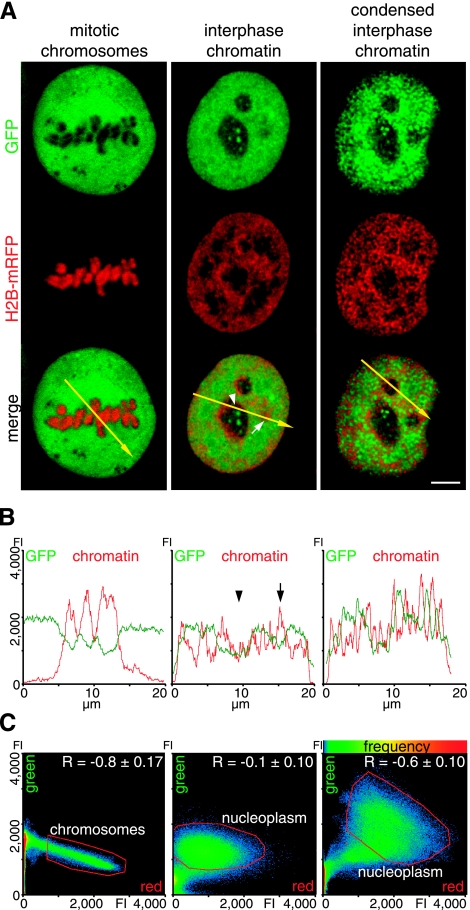
Effect of chromatin condensation state on protein accessibility. A) Distribution of the inert tracer protein GFP, from the plasmid construct EGFP (Clontech), and histone H2B-mRFP-labeled chromatin in representative optical sections of cells in mitosis (left panel), in interphase (middle panel), and in the same interphase cells again after induced chromatin condensation (right panel). B) Line-scan analysis displays pixel intensity distribution of GFP (green) and chromatin (red), along the yellow arrow in the respective merged image in A. Regions with reduced protein concentration correspond to heterochromatin (arrow) and nucleoli (arrowhead). C) Scatterplots display pixel of merged image, according to intensity values of red and green channels; frequency of red-green pixel combination is coded by color (hot colors represent higher frequencies). Red line demarcates pixels corresponding to mitotic chromosomes, as well as to interphase nucleus, excluding nucleoli (referred to as nucleoplasm). Pearson correlation coefficient R is mean ± sd of 10 midsections in individual cells. Scale bar = 5 μm.
In mitosis, the GFP was mostly excluded from chromatin, visible as dark areas where the chromosomes are located (Fig. 4A, left panel). The exclusion is displayed in the line scan by a drop of the GFP intensity and inverse progression of the chromatin intensity. The scatterplot showed a distribution in a sharp cone of moderate negative slope, with the lowest GFP intensities in the region occupied by the chromosomes. Finally, the Pearson’s correlation coefficient of r = −0.8 ± 0.2 clearly demonstrated an anticorrelation of GFP and mitotic chromosomes. In the interphase cell (Fig. 4A, middle panel), GFP showed an overall much more homogeneous distribution throughout the nucleus, with the exception of the nucleoli, corroborated further by the line-scan analysis depicting a reduction of GFP in the nucleoli (Fig. 4B, arrowhead). In some larger heterochromatin spots, we could observe some reduction of the GFP concentration (Fig. 4B, arrow). The scatterplot displayed the GFP and chromatin pixel intensities as a cloud in the center of the plot, and the R value of −0.1 ± 0.1, close to 0, indicated no correlation (Fig. 4C). After induced condensation of interphase chromatin to mitotic chromosome volume, the GFP redistributed mostly to the regions not occupied by chromatin, which corresponded to an enlarged interchromatin space (Fig. 4A, right panel). The GFP exclusion from condensed interphase chromatin was clearly shown by the inverse correlation of the line-scan data (Fig. 4B), and the shift toward a negative slope on the scatterplot and the negative R value of −0.6 ± 0.1 (Fig. 4C).
To test whether this exclusion of proteins from the compacted chromatin was a general phenomenon for nonchromatin-bound nuclear proteins, we extended this analysis to several nucleoplasmic proteins of increasing size up to 230 kDa (Supplemental Fig. 1). As for GFP, all proteins tested, independent of their size, showed a similar level negative correlation with mitotic as well as interphase hypercondensed chromatin.
DISCUSSION
In summary, making use of a hypertonic treatment, we could achieve in interphase cells a chromatin condensation level similar to mitotic chromosomes in the absence of the typical mitotic histone modifications and likely due to increased intranuclear calcium level. Increasing the chromatin condensation led to a slowed exchange of chromatin proteins. Furthermore, although, in general, interphase chromatin was accessible to nonchromatin proteins within a size range of 30–230 kDa, on hypercondensation, they redistributed away and exhibited an anticorrelated distribution to the same level than to mitotic chromatin. This reduction in concentration of nonchromatin proteins could, therefore, concomitantly with other chromatin modifications, be involved in the shutdown of DNA metabolism in mitosis. These data are also consistent with the results from our recent analysis of the mobility and access of single streptavidin proteins to heterochromatin, where these interphase chromatin domains exhibited quite some permeability to this neutral average-sized protein (15).
It has been debated over the past years whether and how chromatin can exclude nuclear factors and the contribution of this effect to DNA-associated processes and heterochromatin function. Different studies indicated that macromolecules have access to interphase chromatin (11, 14, 32, 33). However, these studies have used mainly carbohydrates like dextrans, which may behave differently from nuclear proteins (32, 33). In addition, access to chromatin was not analyzed at different condensation levels. Photodynamic studies showed that chromatin in interphase and mitosis is accessible to chromatin factors like histones and heterochromatin proteins (7, 29, 30). Our data also showed that condensed chromatin was accessible to nucleosomal proteins and other chromatin-associated factors that are involved in heterochromatin maintenance. On the other hand, our results indicate that the concentration of nucleoplasmic proteins that are not associated with nucleosomes is low in chromatin with a higher condensation level. We suggest that the partial exclusion of nucleoplasmic proteins is due to increased structural restrictions and dense environments occupied by condensed chromatin and concentrated chromatin-binding factors. In the condensed chromatin structures, the competition for available volume favors high-affinity chromatin binding factors and decreases the number of nucleoplasmic proteins with less or no affinity to those structures. In mitosis, the reduced access of proteins could, in addition to specific chromatin modifications (e.g., histone phosphorylation), reduce genome-wide accessibility and, thereby, contribute to the mitotic shutdown of transcription, replication, and other DNA-dependent processes. The modulation of protein access and consequently local protein concentration could constitute a general mechanism for the regulation of binding dynamics, enzymatic activities, and DNA metabolism.
Supplementary Material
Acknowledgments
The authors thank Sabine M. Görisch for many discussions, Jeffrey H. Stear for comments on the manuscript and Ulrike Ziebold [Max Delbrück Center for Molecular Medicine (MDC), Berlin, Germany] for the kind gift of anti-phospho H3 antibody. The authors are indebted to Heinrich Leonhardt for numerous discussions and helpful suggestions throughout the course of this project. This work was funded by grants of the German Research Council (DFG) to M.C.C.
References
- Cremer T, Kreth G, Koester H, Fink R H, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;10:179–212. [PubMed] [Google Scholar]
- Belmont A S, Dietzel S, Nye A C, Strukov Y G, Tumbar T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kupper K, Dietzel S, Fakan S. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol Cell. 2004;96:555–567. doi: 10.1016/j.biolcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Richards E J, Elgin S C. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Francastel C, Schubeler D, Martin D I, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Bermudez F, Ellenberg J. Measuring structural dynamics of chromosomes in living cells by fluorescence microscopy. Methods. 2007;41:158–167. doi: 10.1016/j.ymeth.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Belmont A S. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Daban J R. High concentration of DNA in condensed chromatin. Biochem Cell Biol. 2003;81:91–99. doi: 10.1139/o03-037. [DOI] [PubMed] [Google Scholar]
- Gorisch S M, Wachsmuth M, Toth K F, Lichter P, Rippe K. Histone acetylation increases chromatin accessibility. J Cell Sci. 2005;118:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- Verschure P J, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter A E, Belmont A S, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552–4564. doi: 10.1128/MCB.25.11.4552-4564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Cook P R. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack C, Saito K, Tamura M, Kinjo M. Microenvironment and effect of energy depletion in the nucleus analyzed by mobility of multiple oligomeric EGFPs. Biophys J. 2006;91:3921–3936. doi: 10.1529/biophysj.105.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Martin R M, Buschmann V, Bazett-Jones D, Leonhardt H, Kubitscheck U, Cardoso M. Probing intranuclear environments at the single molecule level. Biophys J. 2008;94:2847–2858. doi: 10.1529/biophysj.107.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Lippincott-Schwartz J, Presley J F. Dual-colour imaging with GFP variants. Trends Cell Biol. 1999;9:52–56. doi: 10.1016/s0962-8924(98)01420-2. [DOI] [PubMed] [Google Scholar]
- Campbell R E, Tour O, Palmer A E, Steinbach P A, Baird G S, Zacharias D A, Tsien R Y. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, McNairn A J, Jenuwein T, Gilbert D M, Singh P B, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Patterson G H, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Rahn H P, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso M C. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso M C, Joseph C, Rahn H P, Reusch R, Nadal-Ginard B, Leonhardt H. Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J Cell Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran H P, Schermelleh L, Leonhardt H, Cardoso M C. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1181–1186. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Sullivan K F, Wahl G M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- Martin R M, Gorisch S M, Leonhardt H, Cardoso M C. An unexpected link between energy metabolism, calcium, chromatin condensation and cell cycle. Cell Cycle. 2007;6:2422–2424. doi: 10.4161/cc.6.19.4738. [DOI] [PubMed] [Google Scholar]
- Easwaran H P, Leonhardt H, Cardoso M C. Distribution of DNA replication proteins in Drosophila cells. BMC Cell Biol. 2007;8:42. doi: 10.1186/1471-2121-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R D, Gorski S A, Misteli T. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 2004;375:393–414. doi: 10.1016/s0076-6879(03)75025-3. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, Yang S, Rohr K, Leonhardt H, Solovei I, Cremer C, Fakan S, Cremer T. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G K, Terjung S, Brown J P, Aucott R L, Baron-Luhr B, Singh P B, Georgatos S D. Plasticity of HP1 proteins in mammalian cells. J Cell Sci. 2007;120:3415–3424. doi: 10.1242/jcs.012914. [DOI] [PubMed] [Google Scholar]
- Festenstein R, Pagakis S N, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- Gorisch S M, Richter K, Scheuermann M O, Herrmann H, Lichter P. Diffusion-limited compartmentalization of mammalian cell nuclei assessed by microinjected macromolecules. Exp Cell Res. 2003;289:282–294. doi: 10.1016/s0014-4827(03)00265-9. [DOI] [PubMed] [Google Scholar]
- Verschure P J, Van Der Kraan I, Manders E M, Hoogstraten D, Houtsmuller A B, Van Driel R. Condensed chromatin domains in the mammalian nucleus are accessible to large macromolecules. EMBO Rep. 2003;4:861–866. doi: 10.1038/sj.embor.embor922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.