Abstract
Integrity of animal biomembranes is critical to preserve normal cellular functions and viability. Phosphatidylcholine, an indispensible membrane component, requires the enzyme CCTα for its biosynthesis. Nuclear expression of CCTα is needed for expansion of the nuclear membrane network, but mechanisms for CCTα nuclear import are unknown. Herein, we show that in epithelia, extracellular Ca2+ triggers CCTα cytoplasmic-nuclear translocation. CCTα nuclear import was associated with binding to 14-3-3ζ, a key regulator of protein trafficking. 14-3-3ζ was both sufficient and required for CCTα nuclear import. Helix G within the 14-3-3ζ binding groove interacts with a putative molecular signature within the CCTα carboxyl-terminal phosphoserine motif (residues 328–343). 14-3-3ζ was critically involved in preserving phosphatidylcholine synthesis and cell viability in a model of Pseudomonas aeruginosa infection where Ca2+ concentrations increase within epithelia. Thus, 14-3-3ζ controls CCTα nuclear import in response to calcium signals, thereby regulating mammalian phospholipid synthesis. Agassandian, M., Chen, B. B., Schuster, C. C., Houtman, J. C. D., Mallampalli, R. K. 14-3-3ζ escorts CCTα for calcium-activated nuclear import in lung epithelia.
Keywords: membrane, homeostasis, lipid
Animal membranes are enriched with phosphatidylcholine (PtdCho), a zwitterionic phospholipid that serves as a major component of various secretory products, including bile, high-density lipoproteins, and pulmonary surfactant. PtdCho synthesis requires three enzymes: choline kinase (EC 2.7.1.32), which catalyzes the first committed step; CTP:phosphocholine cytidylyltransferase (CCT) (EC 2.7.7.15), which is rate-limiting and rate-regulatory; and choline phosphotransferase (EC 2.7.8.2), which catalyzes the final reaction within the CDP-choline pathway (1). CCTα, the predominant species in lung epithelia, comprises 367 aa with 4 functional domains: a basic residue NH2-terminal nuclear localization signal (NLS), a catalytic core (C), a membrane-binding domain (M), and a carboxyl-terminal phosphorylation domain (P) (1). CCTα is also an amphitrophic enzyme; thus, it can switch between an inactive soluble or cytoplasmic form to an active, membrane-bound species. Notably, the ability of CCTα to reversibly translocate to nuclear or endoplasmic reticulum membranes after stimulation by lipid activators is a predominant, well-recognized regulatory mechanism for enzyme activation (1, 2). Accordingly, in many cell types, CCTα is localized to the nucleus in association with the nuclear envelope (3,4,5); in lung epithelia, the enzyme is distributed within the cytoplasm and nucleus (6, 7). Not surprisingly, the canonical NLS within the CCTα NH2 terminus is required for nuclear import (3). However, the elucidation of the repertoire of adaptor proteins that direct nuclear import of CCTα has not been investigated and yet might improve our understanding by which eukaryotic cells maintain membrane phospholipid homeostasis.
In mammals, the 14-3-3 protein family has emerged as an important class of ubiquitously expressed biomolecules that regulate protein trafficking and function (8). 14-3-3 includes 7 evolutionarily conserved members that function as adaptors or scaffolding proteins interacting with a multitude of cellular partners to regulate diverse processes, such as metabolism, signal transduction, apoptosis, and malignant transformation (8). Although the brain contains very high levels of 14-3-3, lung epithelial cells, both normal and malignant, are enriched with 14-3-3ζ and 14-3-3ε isoforms (9). Monomeric 14-3-3 is functional, containing 9 α-helices; more often, 14-3-3 functions as dimers, requiring NH2-terminal α-helices for dimer formation (10). Crystal structures reveal that helices αC, αE, αG, and αI form a large ligand-binding groove for interaction with target proteins (10). 14-3-3 associates with most targets in a phospho-specific manner, recognizing RSXpSXP and RXXXpSXP motifs; however, studies also demonstrate that 14-3-3 members interact with binding partners in a phosphorylation-independent manner (10,11,12).
The interplay between 14-3-3, calcium (Ca2+) signals, Ca2+-binding ligands, and nuclear import proteins highlights the complexity by which cells regulate nuclear trafficking. For example, 14-3-3 promotes nuclear import of the cytoskeletal protein myopodin by modulating interaction of myopodin’s NLS with importin-α (13). 14-3-3 promotes nuclear import of its cargos, in part, by direct binding to the ubiquitous second messenger, Ca2+ (14). Ca2+ regulates nuclear entry of nuclear factor of activated T cells (15), myopodin (13), simian virus 40 (SV40) (16), and the NF-κB essential modulator (17). Many targets of Ca2+-signaling pathways are controlled by binding to calmodulin (CaM) the intracellular Ca2+ receptor that coordinates responses to extracellular stimuli. 14-3-3ε binds CaM, and CaM and 14-3-3 binding regulate nuclear retention of Ras-related small G proteins (18, 19).
We hypothesized that 14-3-3 is a molecular chaperone that escorts CCTα to the nucleus in a Ca2+-dependent manner. Here, we demonstrate that extracellular Ca2+ induces CCTα cytoplasmic-nuclear translocation, a process mediated by specific molecular sequence residues that direct association between 14-3-3ζ and CCTα. CCTα interacts with helix G of dimeric 14-3-3ζ, and binding to 14-3-3ζ requires a molecular recognition site within the enzyme’s M-binding domain and a portion of its carboxyl-terminal P domain. 14-3-3ζ was both required and sufficient to drive nuclear CCTα import. 14-3-3ζ was observed to maintain phosphatidylcholine synthesis and reduce apoptosis in a model of Pseudomonas aeruginosa infection where Ca2+ signals increase within epithelia. The results provide new insight into the molecular trafficking of a key regulatory enzyme and molecular mechanisms that preserve membrane phospholipid homeostasis.
MATERIALS AND METHODS
Materials
The murine lung epithelial (MLE) cell line was obtained from American Type Culture Collection (Manassas, VA, USA). 14-3-3ζ plasmids were a kind gift from Dr. Xiaoping Du (Department of Pharmacology, University of Illinois, Urbana, IL, USA; ref. 20). Rabbit CCTα and lysophosphatidylcholine acyltransferase (LPCAT) polyclonal antiserum raised against synthetic peptides were generated by Covance Research Products. (Richmond, CA, USA). The 14-3-3ζ antibody was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). PARP antibodies were from Cell Signaling (Danvers, MA, USA), and the Caspase-Glo and Cell Titer-Glo kits were from Promega (Madison, WI, USA). Recombinant 14-3-3ζ (YWHAZ) was obtained from GenWay Biotech (San Diego, CA, USA). Rabbit IgG TrueBlot kits were from eBioscience (San Diego, CA, USA). The ECL plus Western blotting detection system was purchased from Amersham Biosciences (Piscataway, NJ, USA). A23187 was from Calbiochem (La Jolla, CA, USA). The TnT reticulocyte assay system was from Promega. The talon metal affinity resin was from Clontech Laboratories, Inc. (Valencia, CA, USA). Amylose resin was purchased from New England BioLabs (Ipswich, MA, USA). The QuikChange site-directed mutagenesis kit was from Stratagene (La Jolla, CA, USA). The Geneclean2 Kit was obtained from Bio101 (Carlsbad, CA, USA). Synthetic peptides used for binding were made by CHI Scientific (Maynard, MA, USA). The pCR4-TOPO plasmids and Escherichia coli TOP10 competent cells were obtained from Invitrogen (Carlsbad, CA, USA). The Fugene 6 transfection reagent was purchased from Roche Diagnostics (Indianapolis, IN, USA). Nuclear extraction kits were from Chemicon International (Temecula, CA, USA). All DNA sequencing was performed by the University of Iowa DNA Core Facility.
Cell culture
MLE cells were maintained in Hite’s medium with 2% FBS at 37°C in 5% CO2. After reaching 80% confluence, the cells were harvested using 0.25% trypsin and 0.1% EDTA and plated onto appropriate culture dishes. After incubation overnight, the medium was replaced in serum-free medium alone (control) or with added Ca2+ (0.4–2.4 mM) for various times. In some experiments, cells were exposed to A23187 (100 nM) for 10 min prior to addition of Ca2+. P. aeruginosa was used as a probe to increase cell Ca2+ and was grown in tryptic soy broth and added to cells as described previously (21). Determination of cell viability and caspase activity was performed using the CellTiter Glo Luminescent Cell Viability and Caspase-GloAssay kits following the manufacturer instructions.
Isolation of cellular fractions
Isolation was performed according to the manufacturers’ instructions provided in the nuclear extraction kit. Cells were resuspended in 5-cell pellet volumes of ice-cold cytoplasmic lysis buffer, incubated on ice for 15 min, and centrifuged at 250 g for 5 min at 4°C. The cell pellet was resuspended in cytoplasmic lysis buffer and subjected to centrifugation, and the remaining pellet was resuspended in the nuclear extraction buffer, followed by rotation for 30–60 min at 4°C. The nuclear extract was contained in the supernatant after centrifugation of the mixture at 160,000 g for 5 min. The residual pellet after the spin represented the membrane fraction.
Immunofluorescence microscopy
MLE cells were cultured on 35-mm glass-bottom dishes to 50% confluence. Culture medium was removed and replaced with serum-free medium alone (control), or in combination with Ca2+ (1.0 mM) or A23187 (10 nM), as described above, and incubated for 10 to 60 min. Cells were then fixed in 4% paraformaldehyde and 0.5% glutaraldehyde, and permeabilized with 0.25% Triton X-100 for 15 min at room temperature. Cells were treated with 10% BSA in PBS for 1 h before probing with CCTα, LPCAT, and 14-3-3ζ primary antibodies (1 h), followed by Alexa 488/568 fluorescent probes (1 h). Cells were washed 3× with PBS, and viewed using a combination laser-scanning microscope system (LSM510/ConfoCor2; Zeiss, Jena, Germany). For CFP-CCTα, cells were cotransfected with CFP constructs with Fugene 6 transfection reagent for 24 h. After washing 3 times, cells were incubated in serum-free medium with 1 mM CaCl2 for 1 h. CCTα localization was detected at the single-cell level using the laser-scanning microscope. For quantitation of fluorescence, 10 individual cells for each condition were analyzed. After subtraction of background, regions of interest were selected in the nucleus and in the cytosol of each cell, and the average intensity was measured and calculated by using ImageJ analysis software (U.S. National Institutes of Health, Bethesda, MD, USA).
Enzyme activity
CCT activity assays were performed with or without lipid activator in reactions, as described previously (22).
PtdCho synthesis
Cells were pulsed with 5 μCi of [methyl-3H] choline in the presence or absence of Ca2+ for 10–60 min. Cellular lipids were extracted, lipids were analyzed by thin-layer chromatography, and PtdCho was quantitated as described previously (22).
Fluorescence resonance energy transfer (FRET) analysis
A 1050-bp CCTα fragment was cloned and ligated into pAmCyan-Cl (Clontech) to generate CFP-CCTα, as described previously (7). To construct YFP-14-3-3, total cellular RNA was isolated from mouse type II cells and used as an RT template. Murine primary type II cells were isolated as described (23). Resulting cDNAs were then amplified using the primers 5′-ACTAGATCTATGGGGGATCGAGAGCAG-3′ (forward primer) and 5′-ACTGTCGACTCAGTTGCCTTCTCCTGCTT-3′ (reverse primer) to produce a 741-bp fragment encoding 14-3-3ζ. The forward and reverse primers contain sites for BglII plus a 3-bp, or a SalI site plus a 3-bp overhang, respectively. The PCR product was purified using Gene Clean, digested with SalI and BglII, and ligated into the YFP vector.
To analyze the interaction between CCTα and 14-3-3ζ by FRET, cells were cultured on a collagen-coated 2-chamber coverglass system as described above. Cells were cotransfected with YFP-14-3-3ζ and CFP-CCTα with Fugene 6 transfection reagent for 24 h. After washing 3 times, cells were incubated in serum-free medium with 1 mM CaCl2 for 1 h. The interaction between CCTα and 14-3-3ζ was detected at the single-cell level using a combination laser-scanning microscope system (LSM510/ConfoCor2) as described previously (24).
Immunoblot analysis
Immunoblotting was performed as described previously (7, 22). Immunoreactive proteins were probed with an ECL Plus Western blotting detection system. The dilution factor for the CCTα was 1:1000, for 14-3-3ζ and 1 μg/ml for V5 antibody.
Immunoprecipitation of CCTα and 14-3-3ζ
Immunoprecipitation was performed with the rabbit IgG TrueBlot system per the manufacturer’s instructions. Cell lysates were precleared with anti-rabbit IgG beads (1 h) and incubated with primary antibody to CCTα, 14-3-3ζ, normal rabbit IgG, or preimmune serum for 1.5 h. Lysates were incubated overnight with anti-rabbit IgG beads at 4°C, and washed with TrueBlot lysis buffer (3×); beads were boiled for 5 min in Laemmli buffer, and proteins were finally separated by SDS-PAGE prior to CCTα and 14-3-3ζ immunoblotting.
Construction of CCTα carboxyl-terminal mutants and CFP chimeras
Full-length CCTα, an internal deletion mutant lacking residues 231–251 (CCTd21), an NH2-terminal truncated CCTα variant (CCTN40), and CCTα carboxyl-terminal deletion mutants CCT260, CCT280, and CCT300 were constructed previously (7, 22). Additional CCTα mutants that varied in carboxyl-terminal extent were generated by site-directed mutagenesis using similar PCR-based strategies with appropriate forward and reverse primers using pCR4-CCT as a template (7, 22). These mutants include CCT285, CCT287, and CCT288. The plasmids were transformed into E. coli TOP10 competent cells for large-scale plasmid preparation.
A full-length CCTα variant (CCTS288A) harboring a point mutation where S288 was substituted with Ala was generated using the QuickChange site-directed mutagenesis kit. Several additional constructs were similarly generated that all contain a S288 point mutation but vary in carboxyl-terminal extent and/or the presence of additional mutations at Ser phosphoacceptor sites. For example, CCT327 and CCT340 each harbor a S288A point mutation but only contain the first 327 or 340 residues of CCTα, respectively. CCTS288A was used as a template in PCR for generation of CCT327 and CCT340 and also variants with multiple mutations at carboxyl-terminal Ser residues: Ser-329, Ser-331 and Ser-339. Thus, CCT339, CCT329/339, CCT331/339, and CCT329/331/339 all contain 367 residues with Ser to Ala substitutions at these sites in addition to Ser-288. The CCTα internal deletion mutant, termed CCTΔ327/336, which lacks residues 328–335 (RSPSPSFR), was generated by site-directed mutagenesis, also using CCTS288A as a template. The CFP-CCTα was generated as described previously (7). The CCT329/339 construct was digested with BgIII and SalI, purified, and then ligated into pECFP-C1 (previously digested with the same restriction enzymes) using T4 ligase.
Construction of R18 plasmid
V5-tagged R18 was constructed as follows, using 5′-CACCCCTCATTGTGTCCCTAGAGATCTTTCTTGGCTTGATCTTGAAGCTAATATGTGTTTACCT-3′ (forward DNA fragment) and 5′-AGGTAAACACATATTAGCTTCAAGATCAAGCCAAGAAAGATCTCTAGGGACACAATGAGG-3′ (reverse complementary DNA fragment that encodes the R18 peptide). The forward DNA fragment contains a CACC overhang. The two DNA fragments were combined, heated at 95°C for 2 min, and cooled, followed by directional cloning into a pcDNA3.1D/V5-his expression vector.
siRNA and transfectional analysis
For expression of desired plasmids or siRNA, MLE cells were transfected with test plasmids (2–4 μg/100-mm dish) using Fugene 6 transfection reagent. Twenty-four hours after transfection, cells were incubated in serum-free medium for 12–15 h, and then Ca2+ (0.4–2.4 mM) was added for various times prior to cell isolation. For siRNA, MLE cells were plated on 35-mm glass-bottomed tissue culture dishes at 105 cells/dish, transfected with 14-3-3 siRNA (100 nM), or scrambled RNA (100 nM) using Fugene 6. Twenty-four hours after transfection, cells were treated with or without Ca2+ (1.0 mM) in serum-free DMEM/F12 at 37°C for 1 h. Cells were fixed and processed for CCTα immunostaining and counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Thermo Scientific, Waltham, MA, USA) to visualize the nucleus. In rescue studies, cells were transfected with 14-3-3ζ wild type or a 14-3-3ζ mutant for an additional 24 h prior to exposure to Ca2+ (1.0 mM) in serum-free DMEM/F12 at 37°C for 1 h.
In vitro transcription and translation (TnT) and 14-3-3ζ pulldown assays
CCTα cDNAs cloned into pCR4-TOPO4 were added (2 μg/reaction) directly to the rabbit reticulocyte lysate (TnT-coupled reticulocyte lysate system) and incubated with T7 RNA polymerase in a 50-μl reaction containing [35S]-methionine (20 μCi/reaction) for 90 min at 30°C per the manufacturer’s instructions. Ten microliters of reaction products was boiled for 5 min in Laemmli buffer and stored at −80°C. The remaining 40 μl of reaction products was rotated with 14-3-3ζ resin at 4°C overnight. After incubation, resin was washed 3 times with ice-cold PBS, and proteins were eluted by boiling for 5 min in Laemmli buffer. Proteins eluted from the resin, and products of in vitro translation (10 μl) were resolved by SDS-PAGE, gels were dried, and autoradiography was performed.
Purification of 14-3-3ζ mutants
Wild type and mutants of 14-3-3ζ in a pmalC2 vector were expressed in BL21Star E. coli (20). Overnight cultures of BL21Star E. coli expressing 14-3-3ζ mutants were used to inoculate large-scale cultures. Once cultures reached an absorbance of 1 at 600 nm, IPTG was added to a final concentration of 0.1 mM, and cultures were allowed to grow to stationary phase. Cultures were harvested by centrifugation, and cell pellets were stored at −80°C. Thawed cells were then suspended in Buffer A (7), sonicated, and centrifuged; the resultant supernatant was used for protein purification. Recombinant fusion proteins with an NH2-terminal maltose-binding protein (MBP) tag were purified by affinity chromatography using amylose resin high flow. Fusion proteins were then eluted with 10 mM maltose and used for binding to CCTα resins.
Preparation of protein-talon resins
14-3-3ζ or CCTα resin was prepared using recombinant NH2 terminally His-tagged 14-3-3ζ or affinity-purified His-tagged CCTα and talon metal affinity resin. 14-3-3ζ or CCTα (0.05 mg) was rotated in binding buffer (150 mM NaCl, 20 mM Tris-HCl and 10 mM imidazole, pH 7.9) with talon metal affinity resin for 1.5 h, at 4°C.
14-3-3ζ or CCTα resin binding assays
Fifteen microliters of 14-3-3ζ or CCTα resin was incubated with either cell lysates or cellular fractions that were isolated as described previously (22). Resins were also incubated with purified CCTα (1–2 μg), purified 14-3-3ζ fusion proteins, or products generated from in vitro translation for 1–2 h or overnight at 4°C. After incubation, resins were washed 3 times with buffer containing 300 mM NaCl and 40 mM imidazole. Proteins were released from resin by boiling for 5 min in Laemmli buffer and separated using SDS-PAGE prior to immunoblotting with appropriate detection reagents or autoradiography to visualize the products.
Isothermal titration calorimetry
The affinities of 14-3-3ζ for specific CCTα-binding sites was determined by isothermal titration calorimetry. This method determines the affinity and thermodynamic parameters of the interaction of two molecules by measuring the heat generated on the mixing of two compounds. For these experiments, 20 injections of commercially prepared peptides harboring phosphorylated residues (S288, S329, and S339) were injected in the 14-3-3ζ using an iTC200 (MicroCal, Piscataway, NJ, USA). Experiments were conducted at 25°C in phosphate buffer (pH 7.4). The concentration of 14-3-3 ζ in the cell was 10 μM, while the concentration of peptides in the syringe was 100 μM. The titration curve was fit to determine the binding stoichiometry (n), binding constant (Kd), and thermodynamic parameters (enthalpy and entropy changes) using the Origin software provided by the manufacturer.
Statistical analysis
Statistical analysis was performed by 1-way ANOVA or Student’s t test. Data are presented as means ± se.
RESULTS
Ca2+ induces nuclear import of CCTα and 14-3-3ζ
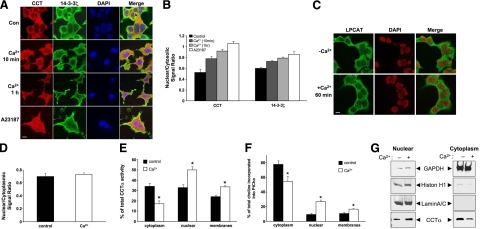
CCTα exhibits cytoplasmic-nuclear shuttling, and 14-3-3 proteins regulate cellular trafficking. The 14-3-3ζ isoform is detected in high levels in lung epithelia (9). Thus, we first tested whether 14-3-3ζ might colocalize with CCTα in MLE cells in response to extracellular Ca2+ using fluorescent microscopy. Under control conditions, CCTα and 14-3-3ζ were both detected within the cytoplasm (Fig. 1A, B). Although the kinetics for nuclear entry differed somewhat between CCTα and 14-3-3ζ, incubation with extracellular Ca2+ led to nuclear translocation of both proteins in a time-dependent manner (Fig. 1A, B). A merged view of two fluorescent images at 10 and 60 min reveals colocalization of CCTα with 14-3-3ζ in the cytoplasm and nucleus. The Ca2+ ionophore, A23187, also increased nuclear expression of the proteins in Fig. 1B, where we used the nuclear marker, DAPI (Fig. 1A). As a specificity control, Ca2+ exposure did not trigger nuclear translocation of LPCAT, a related protein involved in phospholipid metabolism (Fig. 1C, D). Thus, the data support physical transport of CCTα to the nuclear compartment after Ca2+ stimulation.
Figure 1.
Ca2+ stimulates nuclear translocation of CCTα and 14-3-3ζ in lung epithelia. A–D) MLE cells were incubated in serum-free medium with (+Ca2+) or without (−Ca2+) calcium chloride (1.0 mM) for various times. Cells were also exposed to A23187 (10 nM) for 10 min. Cells were then fixed and processed for immunofluorescent staining using either 14-3-3ζ or CCTα primary antibodies followed by incubation with Alexa 488/568 secondary antibodies (Invitrogen) and DAPI (Thermo Scientific) (A, B), or LPCAT primary antibodies followed by incubation with FITC (Sigma, St. Louis, MO, USA) and DAPI antibodies (C, D). Subcellular localization of proteins was monitored using a confocal microscope. Bars represent quantitative measurements of relative nuclear/cytosolic intensities (B, D). E, F) Cells stimulated with (θ) or without (ν) calcium chloride (0.4 mM) were harvested after 10 min, and cytosolic, nuclear, and membrane fractions were isolated and processed for CCT activity (E) and PtdCho synthesis (F). Data represent values expressed as percentage of total enzyme activity (D) or [3H]-choline incorporation into cells (E). G) Subcellular fractions were confirmed using nuclear and cytosolic markers and correlated with CCTα translocation. Data in each panel represent n = 3 separate experiments. *P < 0.05 vs. control. Scale bars = 10 μm.
To investigate whether the cytoplasmic-nuclear shift of CCTα by Ca2+ was physiologically relevant, we assayed CCTα activity and PtdCho synthesis. Ca2+ did not alter total cellular CCTα activity (data not shown). There was a significant reduction in cytosolic CCTα activity coordinate with increases in nuclear and membrane-associated activity by 10 min in cells exposed to Ca2+ vs. control (Fig. 1E). PtdCho synthesis in total cell lysates was also not altered by Ca2+ (data not shown). However, PtdCho synthesis within cytoplasm was reduced at 10 min after Ca2+ exposure, concomitant with increased PtdCho synthesis within membrane and nuclear fractions (Fig. 1F). Immunoreactive CCTα also shifted in response to Ca2+ within subcellular fractions that were analyzed using appropriate markers (Fig. 1G). Overall, these data suggest that CCTα and 14-3-3ζ both relocate to the nucleus after a Ca2+ stress coordinate with a shift in enzyme function, perhaps as a means to preserve the enzyme’s critical role in maintaining phospholipid synthesis within the nuclear compartment (2).
Ca2+-induced interaction of CCTα and 14-3-3ζ
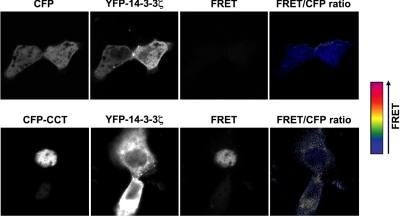
We used FRET analysis to more directly evaluate CCTα and 14-3-3ζ interaction (Fig. 2). In FRET, the interaction between two distinct molecules can be determined by labeling each with different fluorophores (the donor and the acceptor); interaction between these fluorophores results in energy transfer from the excited donor to the acceptor. This method is sensitive for determination of protein-protein interaction within the nanometer range (25). Cells were cotransfected with YFP-14-3-3 with either CFP (negative control) or CFP-CCTα. The panels show CFP, YFP, FRET, and the FRET:CFP ratio (pseudocolor); the latter represents intensity of the FRET signal. In cells coexpressing CFP and YFP-14-3-3, no FRET signal was detected (Fig. 2, top panel; FRET). In cells coexpressing CFP-CCTα and YFP-14-3-3, an intense FRET signal was detected, indicating protein interaction between CCTα and 14-3-3 (Fig. 2, bottom panel).
Figure 2.
Ca2+ stimulates association of CCTα and 14-3-3ζ in lung epithelia. FRET was used to assess protein interaction in cells cotransfected with YFP-1433 with either CFP (negative control) or CFP-CCT. Panels show CFP, YFP, FRET, and the FRET:CFP ratio (pseudocolor). Pseudocolor scale indicates the FRET:CFP ratio, which represents intensity of the FRET signal. In cells coexpressing CFP and YFP-14-3-3, no FRET signal was detected (top panel). In cells coexpressing CFP-CCT and YFP-14-3-3, an intense FRET signal was detected (bottom panel). Data represent n ≥ 3 separate experiments.
14-3-3ζ binds CCTα in a Ca2+-dependent manner
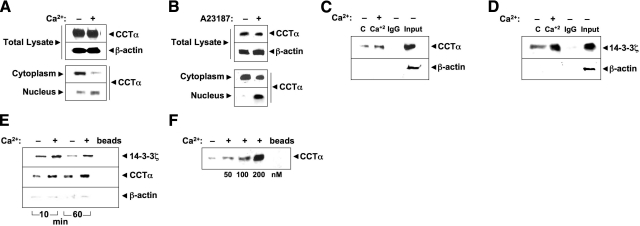
We next examined biochemical interaction of CCTα and 14-3-3ζ. First, cellular fractions isolated from cells treated with or without Ca2+ (Fig. 3A) or Ca2+ ionophore (Fig. 3B) and total lysates were probed with CCTα antibody. After Ca2+ or Ca2+ ionophore exposure, CCTα decreased in cytosol and increased in the nuclear isolates (Fig. 3A, B). Next, 14-3-3ζ was immunoprecipitated from lung cells, followed by CCTα immunoblotting (Fig. 3C); the antibodies were also switched in separate experiments (Fig. 3D). In either approach, CCTα and 14-3-3ζ were detected in association, an interaction increased by Ca2+; limited if any signals were detected using the negative control, normal rabbit IgG. Thus, endogenous CCTα binds 14-3-3ζ in lung epithelia, an interaction stimulated by Ca2+.
Figure 3.
Ca2+ stimulates CCTα binding to 14-3-3ζ. A, B) MLE cells were incubated in serum-free medium with (+) or without (−) calcium chloride (0.4 mM) for 1 h (A) or A23187 (100 nM) for 10 min (B). Cell lysates were harvested, subcellular fractions were isolated, and fractions were processed for CCTα immunoblotting. C, D) Coimmunoprecipitation. Cell lysates were harvested and incubated with primary antibody to CCTα or 14-3-3ζ for 1.5 h. Normal rabbit IgG was used as a negative control. Cell lysates were rotated overnight with anti-rabbit IgG beads at 4°C. The next day, immunoprecipitates were released from beads by 5 min of boiling in Laemmli buffer. Proteins were resolved by SDS-PAGE, followed by immunoblot analysis with CCTα (C) or 14-3-3ζ (D) and β-actin antibodies. E) His pulldown analysis. Cells were transfected with His-CCTα; after 24 h, cells were incubated in serum-free medium with (+) or without (−) calcium chloride (0.4 mM) for 1 h prior to harvest. Cellular lysates were purified on cobalt, and eluants processed for immunoblot analysis with 14-3-3ζ (above) or CCTα (below) antibodies, or β-actin antibodies to demonstrate the specificity of 14-3-3ζ-CCTα interaction. F) Purified CCTα was incubated with (+) or without (−) Ca2+ (0–200 nM) for 1 h, followed by 14-3-3ζ pulldown assays prior to CCTα immunoblotting. Data represent n = 3 separate experiments.
As a second approach to assess protein-protein interaction, we used pulldown assays using His-tagged CCTα. Cells were transfected with his-CCTα, lysates were purified by talon beads, and eluants were processed for 14-3-3ζ immunoblotting. As shown in Fig. 3E, Ca2+ increased association of 14-3-3ζ with overexpressed CCTα at 10 and 60 min, consistent with coimmunoprecipitation results. CCTα binding to 14-3-3ζ was specific, as other lipogenic proteins were not detected in this complex (Supplemental Fig. 1). As CCTα binding to 14-3-3ζ in lung epithelia may be indirect, we performed pulldown assays using recombinant 14-3-3ζ conjugated to affinity talon resin incubated with purified CCTα (Fig. 3F). The CCTα preparation effectively bound 14-3-3ζ beads in a Ca2+-dependent manner at low nM concentrations in vitro, indicating a direct molecular association between the two proteins. Thus, endogenous and overexpressed CCTα bind 14-3-3ζ in lung epithelia, a process that appears to be a direct molecular association regulated by Ca2+ availability.
Mapping of a 14-3-3ζ-binding site within CCTα
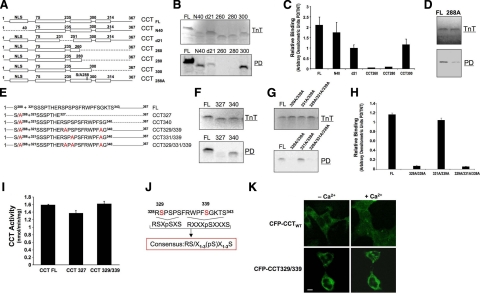
CCTα mutants were synthesized using in vitro translation followed by 14-3-3 pulldown assays and autoradiography (Fig. 4A–H). We used equal amounts of synthesized mutants and 14-3-3ζ resin to evaluate enzyme binding affinity to 14-3-3. The NLS may be indispensable for 14-3-3 binding to its targets, and 14-3-3 interacts with calmodulin (CaM)/CaM kinase (13, 26). Deletion of the NH2 terminus containing the NLS (CCTα N40) was not sufficient to abrogate enzyme binding to 14-3-3ζ (Fig. 4B, C). A catalytic-membrane hinge region (CCTα d21) internal deletion mutant containing a CCTα CaM/CaM kinase dock site still bound 14-3-3ζ, albeit with lower levels of binding. Moreover, CCTα carboxyl-terminal truncation mutants CCT260 and CCT280 did not bind 14-3-3ζ, whereas a distal carboxyl truncation lacking the CCTα phosphorylation domain (CCT300) still interacted with 14-3-3ζ (Fig. 4B, C). Thus, a putative 14-3-3ζ-binding site was identified within a span of 20 residues (280–300) within the CCTα membrane-binding domain. Further, a CCTα mutant containing only the first 285 or 287 residues lacked the ability to interact with 14-3-3ζ entirely, whereas a 288-truncated construct exhibited low-level 14-3-3ζ binding (Supplemental Fig. 2). Thus, Ser-288 appeared required but not sufficient for optimal binding of CCTα to 14-3-3ζ. Last, a full-length CCTα construct containing a point mutation at S288 has exhibited reduced binding to 14-3-3ζ (Fig. 4D). The data as a whole suggest existence of two docking regions within CCTα. The first motif spans 20 residues (280–300) with S288 as a potentially important site. A second region was also hypothesized to exist more downstream in the carboxyl-terminal region because of weaker binding of some constructs (CCT288 vs. full-length CCTα) and because 14-3-3 tends to bind Ser-enriched phosphorylation domains.
Figure 4.
Mapping of a 14-3-3ζ-binding site within CCTα. A) Map illustrates sequences of wild-type or individual CCTα mutants using deletional mutagenesis. Dashed lines represent deleted residues. Constructs tested include wild-type CCT (CCTFL), an NH2-terminal deletion mutant (CCTN40) devoid of the nuclear localization signal (NLS), and an internal deletion mutant lacking 21 residues within the catalytic-membrane binding hinge region (CCTd21). Other constructs include a series of deletion mutants progressively truncated within the CCTα carboxyl terminus (CCT260, CCT280, and CCT300) or point mutations (S288). B) TnT: individual mutants were synthesized in vitro using rabbit reticulocyte lysate in a reaction containing [35S]-methionine (20 μCi/reaction). Pulldown (PD): newly translated reaction products were incubated with 14-3-3ζ beads and sedimented, and pellets were processed for SDS-PAGE and autoradiography. C) Relative binding of constructs as shown by densitometry and expressed as ratios of synthesized/bound protein levels. D) Full-length CCTα construct harboring a point mutation at S288 was tested for 14-3-3ζ binding. E) Map illustrates sequences of wild-type CCTα (CCTFL), or 5 individual CCTα mutants all containing a Ser-288A point mutation with additional deletions or multiple Ser to Ala substitutions within the phosphorylation domain. Dashed lines represent deleted residues. CCT327 and CCT340 are carboxyl-terminal truncated variants (containing 327 and 340 residues, respectively). Remaining CCTα mutants are full-length constructs all harboring multiple-point mutations as candidate phosphoserine sites. F) Individual CCT327 and CCT340 mutants were synthesized in vitro using TnT as above; products were incubated with 14-3-3ζ beads and processed for SDS-PAGE and autoradiography as in B. G, H) Binding of phosphomimetic mutation constructs as shown by autoradiography (G) and densitometry (H). I) CCT activities of TnT-synthesized mutants, CCT327 and CCT329/339, that lacked ability to bind 14-3-3, were determined to assess folding of mutants. J) Sequence alignment. CCTα residues 328–343 are at top, the known consensus 14-3-3 binding motifs are at center (underscored), and a proposed modified motif for 14-3-3ζ binding as identified within CCTα by mapping is at bottom. K) MLE cells were transfected with CFP-CCTα or CFP-CCT329/339 plasmids in the presence or absence of Ca2+ as in Fig. 1A. Cells were then visualized by confocal microscopy for subcellular localization. Compared to nuclear and cytosolic CFP-CCTα, CFP-CCT329/339 was retained within the cytoplasm despite an exogenous Ca2+ stimulus. Scale bar = 10 μm. Data in immunoblot panels represent n = 3 separate experiments (except CCTN40 construct; n=2). Densitometric data represent means ± sd.
To optimally unveil a second binding motif within the CCTα carboxyl terminus, we tested constructs all containing a Ser-288A point mutation to minimize binding from the first site (Fig. 4E–H). The relative levels of binding between mutant CCT proteins and 14-3-3ζ was quantitated (Fig. 4H). Two truncated mutants within the phosphorylation domain, CCT327 and CCT340 (each containing the Ser-288A point substitution) exhibited differential levels for 14-3-3ζ binding, suggesting that a second binding domain resides between CCTα residues 327 and 340 of CCTα (Fig. 4F). Within this stretch of residues, we identified two sequences, 328RSPSPS 333 and 335RWPFSGK 341, similar to canonical high-affinity phosphorylation-dependent 14-3-3 -binding sites RSXpSXP (mode 1) and RXXXpSXP (mode 2), respectively. Of the Ser point-mutation constructs, only those CCTα variants that contained a minimum of triple amino acid substitutions at Ser-288, Ser-329, and Ser-339 displayed markedly reduced interaction with 14-3-3ζ (Fig. 4G, H). Enzymatic activities of these synthesized mutants (CCT327 and CCT329/339) that failed to bind 14-3-3 were comparable with wild-type CCTα, suggesting that these proteins may retain similar conformations to native enzyme essential for catalysis (Fig. 4I). Thus, these observations underscore the requirement of specific carboxyl-terminal CCTα Ser residues for 14-3-3ζ binding and the identification of a binding motif [(RSX1–3(pS)X1–3S (Fig. 4J)] within the enzyme’s P domain.
To estimate the relative contribution of the binding sites within CCTα, we used commercially prepared peptides corresponding to molecular signatures for 14-3-3ζ binding using ITC. All three specific 14-3-3ζ-binding peptides contained relevant sites (Ser-288, Ser-329, and Ser-339) that were phosphorylated at these residues. The titration curves of the solution containing 14-3-3ζ with Ser-288, Ser-329, and Ser-339 peptides, binding stoichiometry (n), dissociation constant (Kd), and thermodynamic parameters are represented in Supplemental Fig. 3. The data reveal that 14-3-3ζ binds to all three peptides with approximately equal affinity.
To assess the functionality of phosphomimetic mutations within the putative 14-3-3 recognition site, cells were transfected with CFP-CCTα and CFP-CCT329/339. CFP-CCTα is normally in the cytosol when transiently transfecting the plasmid on plated cells (Fig. 4K). Further, the fusion protein translocated to the nucleus, similar to endogenous CCTα, in response to Ca2+ (Figs. 1A and 4K). In contrast, the CFP-CCT329/339 fusion protein was detected in the cytoplasm, despite exogenous Ca2+ (Fig. 4K). Of note, while the CFP-CCTα chimera displays cytosolic localization using these transfection methods, nuclear-associated CFP-CCTα is often observed using high-efficiency transfection protocols in suspended cells or nucleofection (7).
Mapping of a CCTα-binding site within 14-3-3ζ
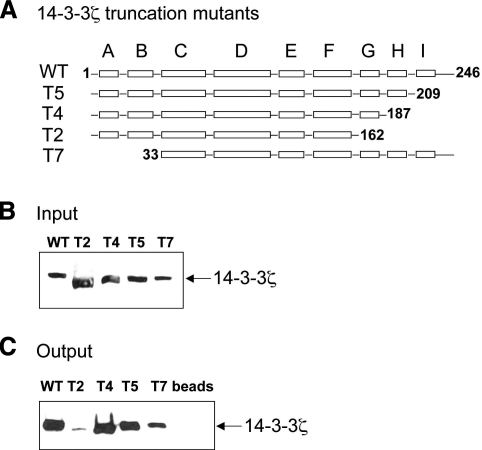
To identify a CCTα-binding site within 14-3-3ζ, we examined the binding of purified 14-3-3ζ MBP-truncation fusion protein mutants (T2, T4, T5, and T7) to CCTα-conjugated talon resin (20). The T2, T4, and T5 plasmids are carboxyl-terminal 14-3-3ζ truncation mutants devoid of helixes G, H, and I (T2), helixes H and I (T4), helix I (T5), and a 14-3-3ζ NH2-terminal truncation (T7) (Fig. 5A). Wild-type 14-3-3ζ MBP-fusion protein was used as a positive control and CCTα-conjugated talon resin was used as a negative control (Fig. 5B, C). The wild-type and 14-3-3ζ mutants expressed in E. coli were chromatographically purified prior to CCTα-conjugated resin pulldown assays and CCTα immunoblotting (Fig. 5B, C). The results show that removal of helix I (T5) or two helixes, I and H (T4) from the 14-3-3ζ carboxyl terminus does not alter binding to CCTα. Further truncation, including helixes I, H, and G dramatically reduced CCTα binding vs. full-length 14-3-3ζ (Fig. 5C). 14-3-3ζ was not detected in CCTα-conjugated resin that was used as a negative control. Hence, helix G (residues 162–187) of 14-3-3ζ is required for CCTα binding. As a whole, these results suggest that helix G of 14-3-3ζ binds to CCTα, possibly via distinct phosphoserines within the enzyme’s phosphorylation domain.
Figure 5.
14-3-3ζ helix G is required for interaction with CCTα. A) Map illustrates wild type (Wt) or individual 14-3-3ζ mutants lacking various α-helical domains (T2–T5). 14-3-3ζ mutant T7 lacks the NH2-terminal domains necessary for dimerization. Wild-type and 14-3-3ζ truncation mutants were expressed in BL21Star E. coli. After transformation into BL21 competent cells, bacteria were inoculated into large-scale cultures, harvested, and processed for protein purification by affinity chromatography. B) Eluants were processed for 14-3-3ζ immunoblotting (input) prior to application to CCTα resin for binding assays. C) Purified 14-3-3 wild-type and individual mutants were incubated with CCTα-beads for 2 h. Protein complexes from beads were eluted and processed for SDS-PAGE and 14-3-3ζ immunoblotting. Data represent n = 3 separate experiments.
14-3-3ζ is both required and sufficient for CCTα nuclear import
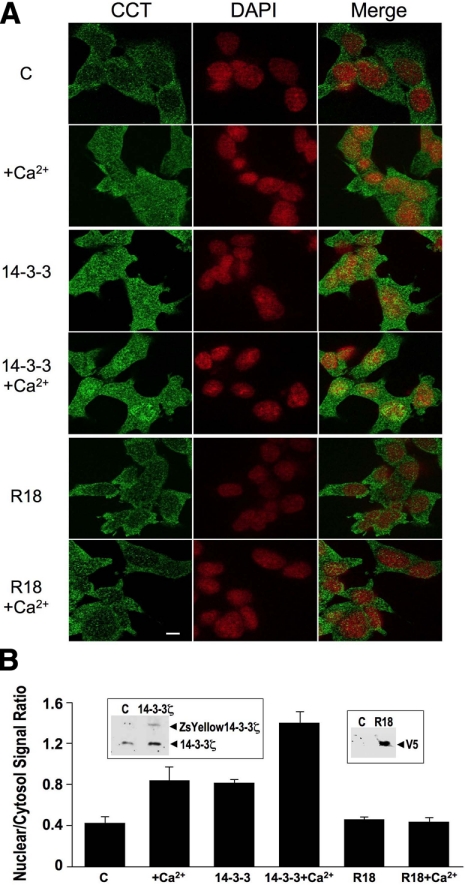
We overexpressed both wild-type 14-3-3ζ protein and a plasmid encoding a broad spectrum inhibitor, R18, which binds to the binding groove of 14-3-3 members, and processed cells for CCTα immunofluorescence (Fig. 6A) (27). Compared to control, 14-3-3ζ was sufficient to increase nuclear CCTα expression comparable to effects of Ca2+ alone; however, maximal nuclear levels of CCTα were achieved with stimulation of cells with Ca2+ after overexpression of 14-3-3ζ (Fig. 6B). In contrast, similar high-level nuclear expression of CCTα was not observed in cells that were transfected with the R18 construct, even in the presence of Ca2+ (Fig. 6).
Figure 6.
14-3-3 is sufficient and required for CCTα nuclear import. A) MLE cells were plated on 35-mm glass-bottom tissue culture dishes at 100,000 cells/dish, transfected with YFP-14-3-3 or V5-tagged R18 (2 μg/dish) using Fugene 6. At 24 h after transfection, cells were treated with or without calcium chloride (1.0 mM) in serum-free DMEM/F12 at 37°C for 1 h. Cells were fixed and immunostained with an antibody recognizing CCTα, followed by Alexa 488 fluorescent probes; cells were also counterstained with DAPI (to visualize the nucleus). Scale bar = 10 μm. B) Fluorescent signals within the cytosol and the nucleus were calculated and graphed and expressed as ratios of nuclear/cytoplasmic intensity. Insets: immunoblotting was performed to confirm overexpressed 14-3-3 (left) and R18 levels (right) vs. untransfected cells [control (C)], the latter detected using V5 antibodies.
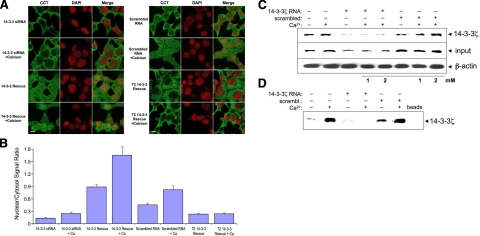
In other loss-of-function studies, we transfected cells with 14-3-3ζ siRNA to evaluate its role on CCTα nuclear translocation using immunocytochemical (Fig. 7A, B) and biochemical (Fig. 7C, D) approaches. Figure 7C demonstrates that 14-3-3ζ expression was effectively reduced with 14-3-3 siRNA (input). Moreover, exogenous Ca2+ failed to translocate CCTα to the nucleus in cells transfected with 14-3-3 siRNA compared to cells exposed to scrambled RNA (Fig. 7A, B, D). 14-3-3ζ overexpression in cells with or without exogenous Ca2+ restored CCTα nuclear import (Fig. 7A, B). Exogenous Ca2+ increased intracellular Ca2+ despite using an L-type channel inhibitor, suggesting that the signal originates from intracellular stores (Supplemental Fig. 4). Overexpression of a 14-3-3ζ mutant (T2) lacking critical helices for CCTα binding was unsuccessful in rescue assays, even in presence of Ca2+ (Fig. 7A, B). Thus, 14-3-3ζ is necessary and yet sufficient to transport CCTα to the nucleus of lung epithelia.
Figure 7.
Loss of function and rescue with 14-3-3ζ regulate CCTα nuclear translocation. A) MLE cells were plated and transfected with 14-3-3 siRNA (100 nM) or scrambled RNA (100 nM). At 24 h after transfection, cells were treated with or without calcium chloride (1.0 mM) in serum-free medium for 1 h. One set of these cells was then fixed and immunostained with an antibody recognizing CCTα, followed by Alexa 488 fluorescent probes; cells were also counterstained with DAPI (to visualize the nucleus). Other sets were transfected with 14-3-3ζ WT and a 14-3-3ζ mutant (T2) for an additional 24 h prior to calcium chloride (1.0 mM) exposure for 1 h. Cells were then fixed and immunostained as above. Scale bars = 10 μm. B) Fluorescent signals within the cytosol and the nucleus were calculated and graphed and expressed as ratios of nuclear/cytoplasmic intensities. C) Cells were transfected with 14-3-3 siRNA (100 nM) and scrambled RNA (100 nM) and treated with or without calcium chloride as in A. Total cell lysates were processed for immunoblotting with 14-3-3ζ (input; middle) or β-actin antibody (bottom) as a loading control. The other portion of total cell lysates was processed for pulldown assays using CCTα beads followed by 14-3-3ζ immunoblotting (top) to assess protein interaction. D) Nuclear fraction isolated from total cell lysates was processed for pulldown assays with 14-3-3ζ beads followed by CCTα immunoblotting to analyze enzyme translocation to the nucleus. Data represent n = 3 separate experiments.
14-3-3ζ regulates phospholipid homeostasis in a bacterial sepsis model
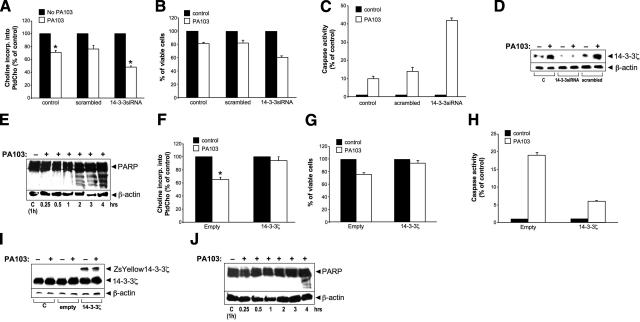
P. aeruginosa inhibits PtdCho synthesis by inhibiting CCTα activity; loss of CCTα function and impaired PtdCho synthesis are linked to the apoptotic program (28, 29). P. aeruginosa also increases lung epithelial cell Ca2+ (ref. 30 and unpublished results) and triggers nuclear translocation of CCTα (Supplemental Fig. 5). Thus, we assessed the biological relevance of 14-3-3ζ for lipid synthesis within the context of an inflammatory cell injury model. The combination of P. aeruginosa infection and treatment of lung epithelia with 14-3-3ζ siRNA reduced PtdCho production (Fig. 8A) and increased cell death (Fig. 8B), caspase activity (Fig. 8C), and PARP cleavage (a marker of apoptosis, Fig. 8D, E) to a greater extent than bacterial infection of cells alone. Conversely, overexpression of 14-3-3ζ lessened or blocked adverse effects of P. aeruginosa infection on PtdCho production and cell viability (Fig. 8F–G) and lessened apoptosis (Fig. 8H–J). Hence, 14-3-3ζ plays a key role in lung epithelial homeostasis during P. aeruginosa infection.
Figure 8.
Loss and gain of function of 14-3-3ζ regulate phospholipid synthesis and apoptosis in lung epithelia after P. aeruginosa (PA103) infection. MLE cells were transfected with 14-3-3 siRNA, control (scrambled) RNA (A–E), or ZsYellow14-3-3 ζ plasmid (F–J) for 24 h prior to infection with P. aeruginosa (MOI=5) for 2–3 h (A–D, F–I). Cells were harvested for assays of PtdCho synthesis (A, F), cell viability (B, G) and caspase activity (C, H). Levels of 14-3-3ζ and β-actin were assayed by immunoblot analysis (D, I). PARP cleavage as a marker of apoptosis was detected using immunoblotting (E, J).
DISCUSSION
The molecular factors that regulate nuclear-cytoplasmic shuttling of CCTα remain elusive. The new observation in this study is that calcium stimulates CCTα nuclear import via binding to 14-3-3ζ. We show that specific molecular determinants within CCTα and 14-3-3ζ mediate interaction between these two proteins. Notably, we uncovered a conserved 14-3-3 binding motif (328RSPSPS 333 and 335RWPFSGKTS 343) within the CCTα carboxyl-terminal phosphoserine domain that was required to drive nuclear translocation of the enzyme. Our data suggest that additional CCTα-14-3-3ζ elements further upstream (residues 280–300) also partake in 14-3-3 binding. Our data suggest that CCTα trafficking in response to inflammatory signals involving a Ca2+ → 14-3-3ζ → CCTα pathway might be one component of a more elaborate homeostatic control mechanism to recruit CCTα molecules to the nucleus. Because bacterial pathogens such as P. aeruginosa degrade CCTα by actions of cytosolic proteinases, we speculate that during inflammatory stress, nuclear shift of a subpopulation of CCTα molecules by this pathway represents a dynamic mechanism whereby cells can preserve the enzyme. Consistent with this, 14-3-3ζ knockdown led to impaired phospholipid synthesis and accelerated the apoptotic program underscoring the physiological relevance of 14-3-3ζ in mediating CCTα transport (Fig. 8).
Our data are consistent with those of others where modest increases in intracellular Ca2+ concurrent with activation of cell survival pathways, sequestration of 14-3-3 with proapoptotic effectors, appears to be a protective mechanism in cells (31). In these studies, related Ca2+ signaling molecules, including CaM and CaMK kinase, also play important transport roles. As stated above, nuclear retention of CCTα might also utilize similar elements as a survival mechanism by sequestering the enzyme from sites of active proteolysis. For example, long-term (18 h) exposure to Ca2+ ionophore stimulates CCTα degradation and reduces PtdCho synthesis by calpains (32). Thus, recruitment of nuclear-bound CCTα molecules could be an early-phase stress response to evade calpains activated by P. aeruginosa and augment nuclear membrane expansion (28); nuclear CCTα could also be held in reserve as a sequestered, inactive form.
We used extracellular Ca2+ as a stimulus because it triggers intracellular elevations of Ca2+ that are observed during alveolar epithelial cell stress (33). Of note, the extracellular concentrations of Ca2+ used here are well below the physiological levels detected in lung alveolar lining fluid (34, 35). Our results are consistent with studies by DeLong et al. (4), where CCTα was exclusively nuclear after cells were exposed to high concentrations (250 mM) of Ca2+ during calcium phosphate transfection. On the basis of imaging data, perhaps as little as 10 min of cellular exposure is sufficient to incite 14-3-3ζ binding to CCTα; this interaction triggers translocation of this complex to the nucleus leading to nuclear enzyme activation and increased nuclear membrane phospholipid synthesis (Fig. 1). The results suggest that 14-3-3ζ together with Ca2+-regulated effectors act as a rapid mechanism for nuclear import when other modes of regulation, such as increased CCTα gene transcription or stabilization of enzyme turnover may be more delayed.
Our data as a whole suggest the existence of at least two docking regions within CCTα for 14-3-3. The first motif spans 20 residues (280–300) with S288 as a potentially important site, because unlike full-length CCTα, two carboxyl-terminal truncation mutants, CCT260 and CCT280, did not bind 14-3-3, and point mutagenesis of S288 reduced 14-3-3 binding (Fig. 4B, D). Thus, a putative 14-3-3-binding region of ∼20 residues was identified within the CCTα M domain. Further, one distal carboxyl truncation lacking the CCTα phosphorylation domain (CCT300) still bound 14-3-3 at lower levels compared to full-length CCTα; a CCTα mutant containing only the first 285 or 287 residues lacked the ability to interact with 14-3-3 entirely. Thus, Ser-288 appears to be required but not sufficient for optimal binding of CCTα to 14-3-3.
The general mechanism of action of 14-3-3 proteins appears to involve the formation of multiple protein complexes, with its targets often depending on the phosphorylation state of the 14-3-3 binding partner. A second motif for CCTα interaction with 14-3-3ζ was identified from 328RSPSPSFRWPFSGKTS 343 residing in the P domain (Fig. 4). The relative binding affinities of the motifs (280–300 and 328–343) were comparable by ITC analysis (Supplemental Fig. 3). Other residues might interact with 14-3-3, perhaps including sites within the M domain. In this regard, an internal deletion mutant (d21) lacking a CaM-binding site (7) also exhibited significantly reduced 14-3-3ζ binding (Fig. 4B). This suggests that CaM-CaM kinase docking within this region could facilitate CCTα phosphorylation to recruit 14-3-3ζ within the complex. This requires further investigation. The 328RSPSPSFRWPFSGKTS 343 sequence has similarities to two canonical high-affinity 14-3-3-phosphorylation-dependent binding motifs, RSXpSXP (mode 1) and RXXXpSXP (mode 2), where pS represents a phosphoserine (8). Recently, a novel 14-3-3ζ-binding site, RX1–2(pS)X2–3S, was identified within the cytoplasmic tail of platelet glycoprotein Iba (36). The stretch of residues 328–343 within the CCTα phosphorylation domain contains Ser at positions found in mode 1 and mode 2 binding motifs and the glycoprotein Iba-binding site (Fig. 4). Hence, the CCTα binding motif (RS/X1–3(pS)X1–3S) represents a slight modification of known 14-3-3 consensus binding motifs.
CCTα phosphorylation might serve as a mechanism to regulate its enzymatic localization. Conserved Ser residues within 14-3-3-binding motifs recognized by kinases are often critical determinants of protein localization (37). All 16 Ser within the CCTα P domain are candidates for kinase phosphorylation; subsequent docking by 14-3-3 at a recognition signature within this domain was supported by using a phosphoSer 14-3-3-binding motif antibody (data not shown). Thus, it is likely that Ser in the 14-3-3-binding motif within CCTα are phosphorylated, and these residues may be phosphorylated in vivo (38). Ser-315 is phosphorylated by Ca2+-activated extracellular signal regulating kinases (ERK1/2) (22). However, Ca2+ did not activate ERK1/2 nor did cellular expression of MEK1, an upstream kinase that elicits ERK1/2 phosphorylation and activation, induce 14-3-3ζ binding to CCTα (data not shown). Alternatively, the presence of Arg residues at −3 and −4 positions within the mode 1 and mode 2 signatures of CCTα appear to serve as attractive targets for CaMK (8). Preliminary data suggest that CaMKI effectively phosphorylates CCTα in vitro and its inhibition attenuates CCTα nuclear entry (data not shown). CaM was also detected in association with the 14-3-3ζ-CaMKI-CCTα enzyme complex but binds more proximally at Glu 243 to stabilize the enzyme (7). Additional studies would be needed to determine whether CaMKI phosphorylates CCTα in vivo and whether CaM is required to direct 14-3-3ζ to its docking site within CCTα and direct enzyme trafficking.
The 14-3-3 NH2 terminus harbors domains critical for its dimerization, whereas regions involved in ligand binding are present in both the carboxyl- and NH2-terminal regions and within the amphipathic α-helical groove (10). Although 14-3-3 monomers can bind a phosphopeptide independent of dimerization, our data indicate that helix G of 14-3-3ζ (residues 162–187) and 14-3-3ζ dimerization are needed for optimal interaction with CCTα (39). Whereas monomeric 14-3-3 can bind proteins containing high-affinity sites, dimeric 14-3-3 can interact with partners containing two or more low-affinity 14-3-3-binding sites (40, 41). Indeed, simultaneous binding of dimeric 14-3-3 to two distinct sites may be necessary to achieve stable association with target proteins (40). Compared to a single binding site, two tandem binding sites on one phosphopeptide increases 14-3-3 binding affinity for its ligand 30-fold (42). Because 14-3-3 mutants lacking the dimerization domain (NH2-terminal helices A and B) bind CCTα at lower levels (Fig. 5B, C), perhaps both high- and low-affinity motifs within the enzyme participate in interaction with 14-3-3ζ dimers. In this respect, 14-3-3 can bind to dephosphorylated proteins. Further, phosphorylation-dependent binding to 14-3-3 facilitates secondary phosphorylation-independent interactions (43). This pattern of ligand recognition would fit 14-3-3ζ interaction with CCTα, where one binding pocket of dimeric 14-3-3 could engage the phosphoserine-containing motif, and the second half of the dimer interacts with dephosphorylated epitopes or an adaptor molecule bound to the membrane-binding target surface. This dimeric association together with binding to divalent cations could stabilize the complex facilitating translocation of CCTα to the nucleus. Further structural studies will be needed to corroborate interactions between the amphipathic groove of 14-3-3ζ, Ca2+, and the α-helix of CCTα, including Ser-288 and the phosphoserine-binding motif. Our data do not exclude the possibility that binding of these elements alters the enzyme’s conformation, resulting in unmasking of its NLS to promote nuclear import; conversely, molecular interference of nuclear signals by 14-3-3 has been described elsewhere (44). The data as a whole implicate a potentially important role for 14-3-3ζ binding within distinct molecular sites of CCTα that controls Ca2+-regulated nuclear trafficking under calcium stress.
Supplementary Material
Acknowledgments
This work was supported by a Merit Review Award from the U.S. Department of Veteran’s Affairs, and U.S National Institutes of Health R01 grants HL097376, HL081784, and HL068135 (to R.K.M).
References
- Jackowski S, Fagone P. CTP:phosphocholine cytidylyltransferase: paving the way from gene to membrane. J Biol Chem. 2005;280:853–856. doi: 10.1074/jbc.R400031200. [DOI] [PubMed] [Google Scholar]
- Gehrig K, Cornell R B, Ridgway N D. Expansion of the nucleoplasmic reticulum requires the coordinated activity of lamins and CTP:phosphocholine cytidylyltransferase α. Mol Biol Cell. 2008;19:237–247. doi: 10.1091/mbc.E07-02-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, MacDonald J I, Kent C. Identification of the nuclear localization signal of rat liver CTP:phosphocholine cytidylyltransferase. J Biol Chem. 1995;270:354–360. doi: 10.1074/jbc.270.1.354. [DOI] [PubMed] [Google Scholar]
- DeLong C J, Qin L, Cui Z. Nuclear localization of enzymatically active green fluorescent protein-CTP:phosphocholine cytidylyltransferase alpha fusion protein is independent of cell cycle conditions and cell types. J Biol Chem. 2000;275:32325–32330. doi: 10.1074/jbc.M004644200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sweitzer T D, Weinhold P A, Kent C. Nuclear localization of soluble CTP:phosphocholine cytidylyltransferase. J Biol Chem. 1993;268:5899–5904. [PubMed] [Google Scholar]
- Ridsdale R, Tseu I, Wang J, Post M. CTP:phosphocholine cytidylyltransferase alpha is a cytosolic protein in pulmonary epithelial cells and tissues. J Biol Chem. 2001;276:49148–49155. doi: 10.1074/jbc.M103566200. [DOI] [PubMed] [Google Scholar]
- Chen B B, Mallampalli R K. Calmodulin binds and stabilizes the regulatory enzyme, CTP:phosphocholine cytidylyltransferase. J Biol Chem. 2007;282:33494–33506. doi: 10.1074/jbc.M706472200. [DOI] [PubMed] [Google Scholar]
- Dougherty M K, Morrison D K. Unlocking the code of 14-3-3. J Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Qi W, Liu X, Qiao D, Martinez J D. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer. 2005;113:359–363. doi: 10.1002/ijc.20492. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee W H, Sobott F, Papagrigoriou E, Robinson C V, Grossmann J G, Sundstrom M, Doyle D A, Elkins J M. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci U S A. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Weller J L, Ho A, Chemineau P, Malpaux B, Klein D C. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci U S A. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson M L, Francis M S, Peden A, Aili M, Stefansson K, Palmer R, Aitken A, Hallberg B. A nonphosphorylated 14-3-3 binding motif on exoenzyme S that is functional in vivo. Eur J Biochem. 2002;269:4921–4929. doi: 10.1046/j.1432-1033.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- Faul C, Huttelmaier S, Oh J, Hachet V, Singer R H, Mundel P. Promotion of importin alpha-mediated nuclear import by the phosphorylation-dependent binding of cargo protein to 14-3-3. J Cell Biol. 2005;169:415–424. doi: 10.1083/jcb.200411169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal G S, Huber J L, Huber S C. Biological significance of divalent metal ion binding to 14-3-3 proteins in relationship to nitrate reductase inactivation. Plant Cell Physiol. 1998;39:1065–1072. doi: 10.1093/oxfordjournals.pcp.a029303. [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Frauwirth K A. Reciprocal NFAT1 and NFAT2 nuclear localization in CD8+ anergic T cells is regulated by suboptimal calcium signaling. J Immunol. 2007;179:3734–3741. doi: 10.4049/jimmunol.179.6.3734. [DOI] [PubMed] [Google Scholar]
- Li P P, Nguyen A P, Qu Q, Jafri Q H, Aungsumart S, Cheng R H, Kasamatsu H. Importance of calcium-binding site 2 in simian virus 40 infection. J Virol. 2007;81:6099–6105. doi: 10.1128/JVI.02195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold C M, Wu Z H, Huang T T, Miyamoto S. Calcium-dependent regulation of NEMO nuclear export in response to genotoxic stimuli. Mol Cell Biol. 2007;27:497–509. doi: 10.1128/MCB.01772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H C, Wu W L, So S C, Chung Y W, Tsang L L, Wang X F, Yan Y C, Luk S C, Siu S S, Tsui S K, Fung K P, Lee C Y, Waye M M. Modulation of the Ca2+-activated Cl(−) channel by 14-3-3epsilon. Biochem Biophys Res Commun. 2000;270:581–587. doi: 10.1006/bbrc.2000.2454. [DOI] [PubMed] [Google Scholar]
- Beguin P, Mahalakshmi R N, Nagashima K, Cher D H, Ikeda H, Yamada Y, Seino Y, Hunziker W. Nuclear sequestration of beta-subunits by Rad and Rem is controlled by 14-3-3 and calmodulin and reveals a novel mechanism for Ca2+ channel regulation. J Mol Biol. 2006;355:34–46. doi: 10.1016/j.jmb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Gu M, Du X. A novel ligand-binding site in the zeta-form 14-3-3 protein recognizing the platelet glycoprotein Ibα and distinct from the c-Raf-binding site. J Biol Chem. 1998;273:33465–33471. doi: 10.1074/jbc.273.50.33465. [DOI] [PubMed] [Google Scholar]
- Agassandian M, Miakotina O L, Andrews M, Mathur S N, Mallampalli R K. Pseudomonas aeruginosa and sPLA2 IB stimulate ABCA1-mediated phospholipid efflux via ERK-activation of PPARα-RXR. Biochem J. 2007;403:409–420. doi: 10.1042/BJ20061364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agassandian M, Zhou J, Tephly L A, Ryan A J, Carter A B, Mallampalli R K. Oxysterols inhibit phosphatidylcholine synthesis via ERK docking and phosphorylation of CTP:phosphocholine cytidylyltransferase. J Biol Chem. 2005;280:21577–21587. doi: 10.1074/jbc.M412409200. [DOI] [PubMed] [Google Scholar]
- McCoy D M, Fisher K, Ryan A J, Mallampalli R K. Transcriptional regulation of lung cytidylyltransferase in developing transgenic mice. Am J Respir Cell Mol Biol. 2006;35:394–402. doi: 10.1165/rcmb.2005-0401OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B B, Mallampalli R K. Masking of a nuclear signal motif by monoubiquitination leads to mislocalization and degradation of the regulatory enzyme, CCTα. Mol Cell Biol. 2009;29:3062–3075. doi: 10.1128/MCB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuweiler H, Sauer M. Using photoinduced charge transfer reactions to study conformational dynamics of biopolymers at the single-molecule level. Curr Pharm Biotechnol. 2004;5:285–298. doi: 10.2174/1389201043376896. [DOI] [PubMed] [Google Scholar]
- Obsilova V, Vecer J, Herman P, Pabianova A, Sulc M, Teisinger J, Boura E, Obsil T. 14-3-3 Protein interacts with nuclear localization sequence of forkhead transcription factor FoxO4. Biochemistry (Mosc) 2005;44:11608–11617. doi: 10.1021/bi050618r. [DOI] [PubMed] [Google Scholar]
- Wang B, Yang H, Liu Y C, Jelinek T, Zhang L, Ruoslahti E, Fu H. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry (Mosc) 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- Zhou J, Wu Y, Henderson F, McCoy D M, Salome R G, McGowan S E, Mallampalli R K. Adenoviral gene transfer of a mutant surfactant enzyme ameliorates pseudomonas-induced lung injury. Gene Ther. 2006;13:974–985. doi: 10.1038/sj.gt.3302746. [DOI] [PubMed] [Google Scholar]
- Cui Z, Houweling M, Chen M H, Record M, Chap H, Vance D E, Terce F. A genetic defect in phosphatidylcholine biosynthesis triggers apoptosis in Chinese hamster ovary cells. J Biol Chem. 1996;271:14668–14671. doi: 10.1074/jbc.271.25.14668. [DOI] [PubMed] [Google Scholar]
- Jacob T, Lee R J, Engel J N, Machen T E. Modulation of cytosolic Ca2+ concentration in airway epithelial cells by Pseudomonas aeruginosa. Infect Immun. 2002;70:6399–6408. doi: 10.1128/IAI.70.11.6399-6408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S, Tokumitsu H, Soderling T R. Calcium promotes cell survival through CaM-K kinase activation of the protein-kinase-B pathway. Nature. 1998;396:584–587. doi: 10.1038/25147. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ryan A J, Medh J, Mallampalli R K. Oxidized lipoproteins inhibit surfactant phosphatidylcholine synthesis via calpain-mediated cleavage of CTP:phosphocholine cytidylyltransferase. J Biol Chem. 2003;278:37032–37040. doi: 10.1074/jbc.M304316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M, Bertocchi C, Jennings P, Haller T, Mair N, Singer W, Pfaller W, Ritsch-Marte M, Dietl P. Ca2+ entry is essential for cell strain-induced lamellar body fusion in isolated rat type II pneumocytes. Am J Physiol Lung Cell Mol Physiol. 2004;286:L210–L220. doi: 10.1152/ajplung.00332.2003. [DOI] [PubMed] [Google Scholar]
- Cooley J, McDonald B, Accurso F J, Crouch E C, Remold-O'Donnell E. Patterns of neutrophil serine protease-dependent cleavage of surfactant protein D in inflammatory lung disease. J Leukoc Biol. 2008;83:946–955. doi: 10.1189/jlb.1007684. [DOI] [PubMed] [Google Scholar]
- Hirche T O, Crouch E C, Espinola M, Brokelman T J, Mecham R P, DeSilva N, Cooley J, Remold-O'Donnell E, Belaaouaj A. Neutrophil serine proteinases inactivate surfactant protein D by cleaving within a conserved subregion of the carbohydrate recognition domain. J Biol Chem. 2004;279:27688–27698. doi: 10.1074/jbc.M402936200. [DOI] [PubMed] [Google Scholar]
- Mangin P, David T, Lavaud V, Cranmer S L, Pikovski I, Jackson S P, Berndt M C, Cazenave J P, Gachet C, Lanza F. Identification of a novel 14-3-3zeta binding site within the cytoplasmic tail of platelet glycoprotein Ibα. Blood. 2004;104:420–427. doi: 10.1182/blood-2003-08-2881. [DOI] [PubMed] [Google Scholar]
- Paroni G, Cernotta N, Dello R C, Gallinari P, Pallaoro M, Foti C, Talamo F, Orsatti L, Steinkuhler C, Brancolini C. PP2A regulates HDAC4 nuclear import. Mol Biol Cell. 2008;19:655–667. doi: 10.1091/mbc.E07-06-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R B, Kalmar G B, Kay R J, Johnson M A, Sanghera J S, Pelech S L. Functions of the C-terminal domain of CTP:phosphocholine cytidylyltransferase. Effects of C-terminal deletions on enzyme activity, intracellular localization, and phosphorylation potential. Biochem J. 1995;310:699–708. doi: 10.1042/bj3100699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian R R, Masters S C. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Tzivion G, Shen Y H, Zhu J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene. 2001;20:6331–6338. doi: 10.1038/sj.onc.1204777. [DOI] [PubMed] [Google Scholar]
- Cahill C M, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. 2001;276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- Yaffe M B, Rittinger K, Volinia S, Caron P R, Aitken A, Leffers H, Gamblin S J, Smerdon S J, Cantley L C. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Shen Y H, Godlewski J, Bronisz A, Zhu J, Comb M J, Avruch J, Tzivion G. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol Biol Cell. 2003;14:4721–4733. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin A J, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.