Abstract

RNA oxidation is important in the etiology of disease and as a tool for studying the structure and folding kinetics of this biopolymer. Nucleobase radicals are the major family of reactive intermediates produced in RNA exposed to diffusible species such as hydroxyl radical. The nucleobase radicals are believed to produce direct strand breaks by abstracting hydrogen atoms from their own and neighboring ribose rings. By independently generating the formal C5-hydrogen atom addition product of uridine in RNA, we provide the first chemical characterization of the pathway for direct strand scission from a RNA nucleobase radical. The process is more efficient under anaerobic conditions. The preference for strand scission in double-stranded RNA over single-stranded RNA suggests that this chemistry may be useful for analyzing the secondary structure of RNA in hydroxyl radical cleavage experiments if they are carried out under anaerobic conditions.
Historically, homopolymers of RNA were used as model substrates in studies on nucleic acid damage by γ-radiolysis.1 More recently, RNA oxidation has become important in its own right due to its association with disease and its use as a tool for probing RNA folding and structure.2-6 Hydroxyl radical (HO•) is a primary damaging agent and direct strand scission is proposed to result from as many as 40% of its reactions with RNA. A variety of experiments established that π-bond addition is the major reaction pathway for HO• (and H•) with RNA and the resulting nucleobase radicals account for as much as 90% of the reactions.1,7 Consideration of strand break efficiency and hydroxyl radical reactivity suggest that nucleobase radicals and/or their respective peroxyl radicals abstract hydrogen atoms from ribose rings because direct strand scission requires formation of sugar radicals.1,8 Characterization of these pathways using HO• is complicated by the unselective nature by which diffusible reactive species react with nucleic acids. Our understanding of DNA oxidation has been greatly improved by independently generating reactive intermediates within synthetic oligonucleotides.9,10 We now report the reactivity of the first independently synthesized nucleobase RNA radical and its role in direct strand scission.
Hydroxyl radical and hydrogen atom preferentially add to the C5-position of pyrimidines (Scheme 1). We independently generated the monomeric form of the C5-hydrogen atom adduct of uridine, 5,6-dihydrouridin-6-yl radical (1) via Norrish Type I photocleavage of the nucleoside 2.11 Oligonucleotides containing 2 were prepared via automated solid phase oligonucleotide synthesis.12 Direct strand break formation following photolysis (350 nm, 4 h) of 2 was examined in single and double stranded RNA under aerobic and anaerobic conditions. Direct strand scission under aerobic conditions was less than 1/7th the amount when 1 was produced in a duplex (4) in the absence of O2. The photocleavage of monomeric 2 is independent of O2, suggesting that the observed trend is indicative of the effect of O2 on one or more steps in the mechanism for strand scission from 1.
Scheme 1.

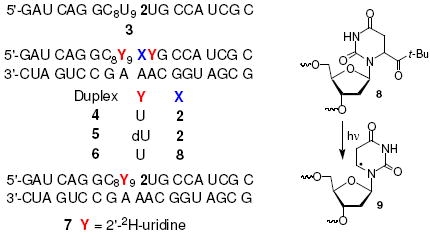
Under anaerobic conditions direct strand scission products corresponding to oxidation at the nucleotide at which 1 (or 9) was generated (“Intra”) and at the 5’-adjacent uridine (“Inter”) in 5’-32P-3 and 5’-32P-4 were observed, although more efficiently overall in the duplex (4, Table 1, Figure 1). The ratio of strand scission at the sites depended upon whether 1 was produced in single- or double-stranded RNA, although it was independent of photolysis time for a particular substrate. Strand scission occurred at the 5’-adjacent uridine >85% of the time when 5,6-dihydrouridin-6-yl radical (1) was produced in double-stranded RNA (4) but strand scission at the site where 1 is generated accounted for >80% of strand breaks in single-stranded substrate (3). Products containing 3’- or 5’-phosphate termini were identified via enzymatic dephosphorylation and comparison with independently synthesized markers. The 5’-termini consisted solely of phosphate groups. However, 3’-termini contained nucleotide fragments in addition to phosphate end groups. The structure of these other 3’-end groups have not yet been determined.
Table 1.
Direct strand scission under anaerobic conditions.a
| Substrate | Absolute Yield (%) |
% Interb | ||
|---|---|---|---|---|
| Inter | Intra | Total | ||
| 3 | 3.3 ± 0.7 | 17.3 ± 7.2 | 20.7 ± 7.2 | 17.9 ± 8.4 |
| 4 | 38.9 ± 3.0 | 6.0 ± 2.2 | 45.0 ± 4.8 | 86.8 ± 3.7 |
| 5 | 3.6 ± 1.6 | 13.2 ± 3.6 | 16.8 ± 5.2 | 20.7 ± 4.2 |
| 6 | 25.6 ± 3.5 | 4.0 ± 2.0 | 29.6 ± 1.5 | 86.3 ± 7.1 |
| 7 | 4.4 ± 0.5 | 10.9 ± 1.6 | 15.3 ± 1.6 | 29.0 ± 4.0 |
5’-32P-substrates photolyzed for 4 h.
% of total direct strand scission produced at “Inter” position.
Note: Each value is the average of at least 3 experiments. Each experiment consists of 3 replicates.
Figure 1.

Denaturing PAGE analysis of direct strand scission upon photolysis of (A) 5’-32P-3, (B) 5’-32P-4, (C) 5’-32P-7 under anaerobic conditions.
Qualitatively, the level of direct strand scission from 1 is significantly higher than from the analogous radical, 5,6-dihydro-2’-deoxyuridin-6-yl (9) in oligodeoxynucleotides.13,14 We speculated that the presence of the 2’-hydroxyl group, which results in a significant reduction in the C2’-carbon-hydrogen bond dissociation energy played a vital role in facilitating transfer of the radical from the nucleobase to the sugar.15 The importance of the 2’-hydroxyl in the ribose ring on direct strand scission was verified by replacing the 5’- and 3’-adjacent uridine nucleotides with 2’-deoxyuridine. Photolysis of 5’-32P-5 resulted in a more than 10-fold reduction in strand scission at the 5’-adjacent nucleotide and a 2-fold increase at the position where 1 was generated. In contrast, the amount of direct strand scission at the 5’-adjacent nucleotide when 5,6-dihydro-2’-deoxyuridin-6-yl (9) was flanked by uridine (5’-32P-6) was restored to a level comparable to that observed in 4.
These observations and examination of molecular models (Spartan ’02) led us to propose that the major pathway for direct strand scission from 5,6-dihydrouridin-6-yl radical (1) in duplex RNA under anaerobic conditions involves C2’-hydrogen atom abstraction from the 5’-adjacent uridine (Scheme 2). This hypothesis was supported by photolysis of 5’-32P-7 in which the 5’-adjacent uridine was deuterated at C2’. Formation of 1 in 5’-32P-7 resulted in more than an 8-fold reduction in the absolute amount of direct strand scission at the 5’-adjacent C2’-deuterated uridine compared to what is produced in 5’-32P-4. Although the level of strand scission at the nucleotide where 1 is generated increased in the deuterated substrate, the increase did not fully compensate for the diminution in strand scission at the 5’-adjacent nucleotide. Hence, we cannot rule out an increased contribution to the radical’s overall reactivity at a position within 1 or an adjacent nucleotide that does not result in direct strand scission (e.g. C1’) when the C2’-hydrogen of the 5’-adjacent uridine is deuterated.
Scheme 2.
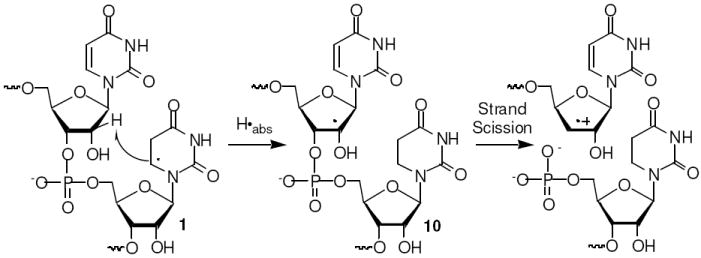
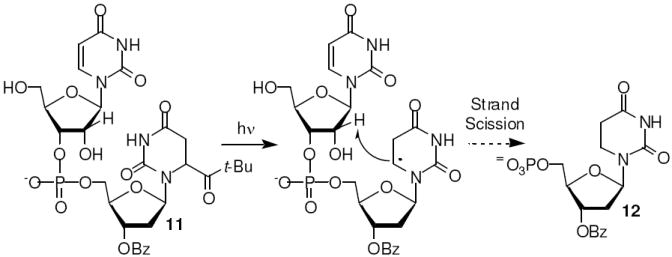
Formation of the C2’-radical (10) was expected to give rise to direct strand scission via elimination of the β-phosphate analogous to what was first proposed by von Sonntag for DNA cleavage from the respective C4’-radical.16,17 HPLC analysis of dinucleotide 11 (Scheme 3) that was photolyzed under anaerobic conditions was consistent with this mechanism. The dinucleotide produced the independently synthesized, anticipated elimination product, 12, in 14% yield. How the respective cation radical in RNA (Scheme 2) is ultimately converted into the fragment containing a 3’-phosphate terminus requires further investigation.
Scheme 3.
The kinetics of the strand scission process were probed using β-mercaptoethanol (BME) as a competitor. Direct strand scission in 5’-32P-4 was quenched by micromolar concentrations of the thiol. The thiol can prevent strand cleavage by trapping 1 and/or the subsequently formed ribose radical(s) (e.g. 10). The amount of RNA radical trapped by the thiol was defined as the difference in the amount of strand scission with and without BME. Plotting the ratio of thiol trapped radical versus strand scission as a function of BME concentration yielded a straight line whose slope represents the ratio of rate constants for thiol trapping and the rate determining step in strand scission.12 Depending upon whether we use the estimated rate constant for BME trapping of monomeric 1 (2.6 × 106 M-1s-1)11 or a generic rate constant for thiol reaction with an alkyl radical (8 × 106 M-1s-1)18, these data indicate that the rate limiting step in strand scission is between 29 and 90 s-1. Further investigation is required in order to determine whether hydrogen atom abstraction by 1 or phosphate elimination is the rate determining step in strand scission.
In summary, we have characterized the mechanism by which a RNA nucleobase radical is transformed into a direct strand break under anaerobic conditions. The major pathway in duplex RNA involves internucleotidyl hydrogen atom abstraction from the C2’-position of the 5’-adjacent nucleotide. The thiol competition experiment indicates that direct strand scission from 1 is too slow to compete with the millimolar concentration of thiol present in cells. However, the more efficient direct strand break formation in duplex RNA compared to single stranded substrate under anaerobic conditions may prove useful for extracting additional structural information from hydroxyl radical cleavage experiments.
Supplementary Material
Acknowledgments
We are grateful for generous support from the National Institute of General Medical Sciences (GM-054996). M.J.E.R. thanks the NIGMS for a Research Supplement to Promote Diversity in Health-Related Research.
Footnotes
Supporting Information. Procedures for the synthesis of all compounds and other experiments. ESI-MS of modified oligonucleotides. Sample phosphorimages of enzymatic end group analysis, deuterium effect on strand scission, and thiol trapping. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; London: 1987. [Google Scholar]
- 2.Moreira PI, Nunomura A, Nakamura M, Takeda A, Shenk JC, Aliev G, Smith MA, Perry G. Free Radical Biol Med. 2008;44:1493–1505. doi: 10.1016/j.freeradbiomed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Martinet W, DeMeyer GRY, Herman AG, Kockz MM. European Journal of Clinical Investigation. 2004;34:323–327. doi: 10.1111/j.1365-2362.2004.01343.x. [DOI] [PubMed] [Google Scholar]
- 4.Adilakshmi T, Bellur DL, Woodson SA. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB. Nature Protocols. 2008;3:1395–1401. doi: 10.1038/nprot.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tullius TD, Greenbaum JA. Curr Opin Chem Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Deeble DJ, Schulz D, Von Sonntag C. Int J Radiat Biol. 1986;49:915–926. doi: 10.1080/09553008514553151. [DOI] [PubMed] [Google Scholar]
- 8.Hildenbrand K, Schulte-Frohlinde D. Int J Radiat Biol. 1989;55:725–738. doi: 10.1080/09553008914550781. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg MM. Org Biomol Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 10.Gates KS. Chem Res Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman CA, Resendiz MJE, Sczepanski JT, Greenberg MM. J Org Chem. 2009;74:7007–7012. doi: 10.1021/jo9012805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.See Supporting Information.
- 13.Hong IS, Carter KN, Sato K, Greenberg MM. J Am Chem Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]
- 14.Carter KN, Greenberg MM. J Am Chem Soc. 2003;125:13376–13378. doi: 10.1021/ja036629u. [DOI] [PubMed] [Google Scholar]
- 15.Li M-J, Liu L, Wei K, Fu Y, Guo Q-X. J Phys Chem B. 2006;110:13582–13589. doi: 10.1021/jp060331j. [DOI] [PubMed] [Google Scholar]
- 16.Dizdaroglu M, Von Sonntag C, Schulte-Frohlinde D. J Am Chem Soc. 1975;97:2277–2278. doi: 10.1021/ja00841a051. [DOI] [PubMed] [Google Scholar]
- 17.Giese B, Dussy A, Meggers E, Petretta M, Schwitter U. J Am Chem Soc. 1997;119:11130–11131. [Google Scholar]
- 18.Newcomb M. Tetrahedron. 1993;49:1151–1176. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


