Abstract
DHR38 is the only Drosophila member of the NR4A subclass of vertebrate nuclear receptors, which have been implicated in multiple biological pathways, including neuronal function, apoptosis, and metabolism. Although an earlier study identified three point mutations in DHR38, none of these were shown to be a null allele for the locus, leaving it unclear whether a complete loss of DHR38 function might uncover novel roles for the receptor. Here we show that a specific DHR38 null allele, DHR38Y214, leads to fully penetrant pharate adult lethality, similar to the most severe phenotype associated with the EMS-induced mutations. DHR38Y214 mutants display minor effects on ecdysone-regulated transcription at the onset of metamorphosis. In contrast, cuticle gene expression is significantly reduced in DHR38Y214 mutant pupae. These studies define the essential functions of DHR38 and provide a genetic context for further characterization of its roles during development.
Keywords: nuclear receptor, transcription, cuticle, metamorphosis, ecdysone
Introduction
Nuclear receptors (NRs) comprise a family of ligand-regulated transcription factors that play central roles in growth, metabolism, and development (Chawla et al., 2001). They are defined by a highly conserved DNA-binding domain that consists of two zinc-fingers, and a less conserved ligand binding domain (LBD) that can regulate receptor activity, often in response to small lipophilic compounds. We use the fruit fly, Drosophila melanogaster, as a model system to characterize the regulation and function of NRs during development. Drosophila offers a number of unique advantages in these studies (King-Jones and Thummel, 2005). First, the Drosophila genome encodes only 18 canonical NRs in contrast to the 48 receptors found in humans and 284 receptors in C. elegans (Maglich et al., 2001; Escriva Garcia et al., 2003). In spite of their small number, the Drosophila NRs are distributed among all major subclasses of vertebrate NRs, often with a single fly receptor representing multiple vertebrate paralogs. Furthermore, in contrast to the complexity of vertebrate hormone signaling pathways, Drosophila has only one known physiologically active steroid hormone, 20-hydroxyecdysone (referred to here as ecdysone), which directs the major developmental transitions in the life cycle, including molting and metamorphosis. Ecdysone initiates transcriptional hierarchies through binding and activating an NR heterodimer comprised of the Ecdysone Receptor (EcR) and its RXR partner, Ultraspiracle (USP) (Riddiford et al., 2000). The late larval ecdysone pulse triggers puparium formation and the prepupal stage of development, terminating the juvenile growth phase and initiating adult maturation. A second ecdysone pulse approximately 10 hours after pupariation triggers adult head eversion and marks the prepupal-to-pupal transition. Detailed functional studies have shown that ecdysone exerts its effects through multiple primary-response transcription factors. These include the zinc finger proteins encoded by the Broad-Complex (BR-C), the E74 ETS-domain proteins, and the E75A nuclear receptor (Burtis et al., 1990; Segraves and Hogness, 1990; DiBello et al., 1991). These transcription factors, in turn, regulate batteries of downstream secondary-response effector genes that direct the appropriate stage- and tissue-specific biological responses to the hormone (Thummel, 2001; Henrich, 2005).
Here we characterize the Drosophila DHR38 NR, which has three vertebrate orthologs, NGFI-B, Nurr1, and NOR1, all members of the NR4A class of NRs. Studies in cultured cells and mouse models have shown that these receptors are widely expressed and function in multiple biological pathways (Maxwell and Muscat, 2006). These include critical roles in the central nervous system (Zetterstrom et al., 1997; Saucedo-Cardenas et al., 1998; Ponnio and Conneely, 2004), inflammatory responses (Pei et al., 2006a), T-cell apoptosis (Hsu et al., 2004), metabolism (Pei et al., 2006b), vascular disease (Bonta et al., 2007), and cancer (Mullican et al., 2007). Although X-ray crystallographic studies have shown that the Nurr1 and DHR38 LBDs lack a classic hydrophobic ligand-binding pocket, they remain subject to post-translational control by cofactor recruitment and/or post-translational modifications (Baker et al., 2003; Wang et al., 2003). DHR38, NGFI-B, and Nurr1 can heterodimerize with RXR (USP in Drosophila) to exert their regulatory functions (Sutherland et al., 1995; Maxwell and Muscat, 2006).
Our understanding of NR4A function has been complicated by redundancy between the three vertebrate family members (Maxwell and Muscat, 2006). This complexity, however, is overcome in C. elegans or Drosophila, where only a single NR4A receptor is present. In C. elegans, nhr-6 is expressed in chemosensory neurons and the developing somatic gonad, and plays a critical role in fertility through its effects on spermatheca and spermathecal valve development (Gissendanner et al., 2008). The molecular basis for these functions, however, remains unknown.
The DHR38 gene in Drosophila is ∼30 kb in length and consists of at least two mRNA isoforms, defined by the cTK11 and cTK61 cDNAs (Sutherland et al., 1995; Kozlova et al., 1998) (Fig. 1). DHR38 appears to be expressed at a relatively constant low level throughout development, requiring the use of riboprobes on northern blots or qRT-PCR for mRNA detection (Fisk and Thummel, 1995; Kozlova et al., 1998). Three EMS-induced point mutations have been used to define DHR38 functions during development (Kozlova et al., 1998). Homozygous mutants for each allele survive normally through early stages, but die late in development. DHR3843 and DHR3857 homozygotes and hemizygotes display rupturing of the cuticle at leg joints and consequent hemolymph leakage during the first 12 hours of adult life, followed by death a few hours later. A few homozygotes can survive as adults for several days, but are weak and cannot be maintained as a stock. A more severe phenotype is seen in DHR3856 hemizygotes, which develop normally until the end of metamorphosis. These mutants then display cuticle rupturing and hemolymph leakage inside the pupal case as the adult fly begins the movements that would normally lead to eclosion. Both DHR3843 and DHR3857 behave as hypomorphic alleles, with less severe phenotypes seen in homozygous mutants than when the mutation is combined with a deficiency for the DHR38 locus. In contrast, the nature of the DHR3856 allele cannot be determined because homozygotes die during larval stages (Kozlova et al., 1998). Although this early lethality might be due to a second-site mutation on the chromosome, it also raises the possibility that a defined DHR38 null mutation might uncover novel functions for this receptor prior to metamorphosis.
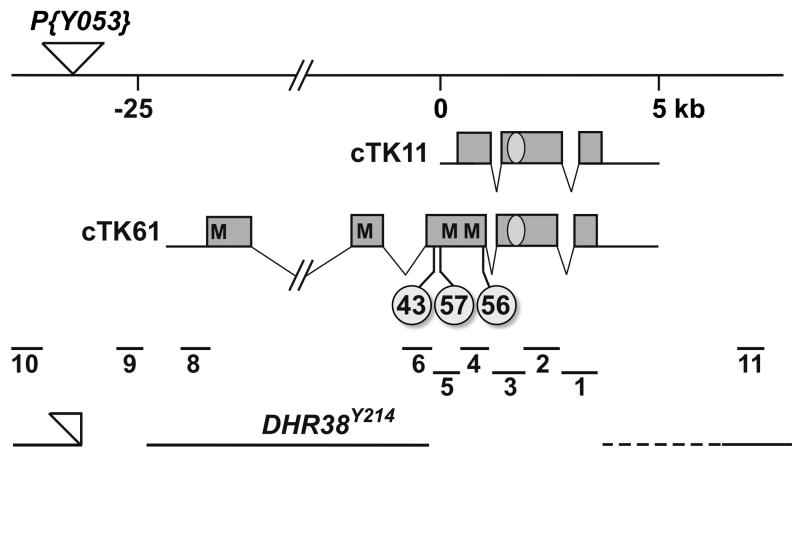
Fig. 1.
Genomic organization of the DHR38 locus and molecular nature of the mutations. The DHR38 genomic region is adapted from Kozlova et al. (1998). Distances are given in kilobases (kb) relative to the 5′ end of the cTK11 cDNA clone (Sutherland et al., 1995). The site of the P{Y053} viable P-element insertion is marked with an inverted triangle. The maps of cTK11 and cTK61 (Kozlova et al., 1998) are depicted below, with protein coding regions represented by boxes, untranslated regions by solid lines, and introns by angled thin lines. The DNA-binding domain is denoted by an oval, and the positions of the in-frame methionine codons in cTK61 are marked (M) as well as the locations of the three EMS-induced mutations, DHR3843, DHR3856, and DHR3857, labeled with circles. The locations of the ten PCR-amplified regions that were used to map the imprecise excision mutations are shown below the cDNA maps. At the bottom is depicted the molecular structure of DHR38Y214, with the genomic sequences present in the mutant represented by solid lines, deletions by an absence of the line, and uncertainty of the mapping by a dotted line. Part of the original P{Y053} insertion, bearing the w+ marker, is still present in this mutant (open triangle).
Here we describe the isolation of a specific null mutation in DHR38 and demonstrate that it has a lethal phenotype identical to that of DHR3856 hemizygotes. Consistent with this, we map the EMS-induced lesions in each DHR38 mutant allele and show that DHR3856, but not DHR3843 or DHR3857, disrupts sequences common to both DHR38 mRNA isoforms, accounting for the relative strength of its phenotype. The DHR38 null mutation has only a minor effect on the expression of ecdysone-regulated genes at the onset of metamorphosis. In contrast, a number of cuticle gene transcripts are significantly reduced in DHR38 mutant pupae, consistent with the lethal phenotype of these animals. These studies define an essential role for DHR38 in cuticle gene expression during pupal stages and adult cuticular integrity.
Results and Discussion
Molecular Mapping of EMS-Induced Mutations in DHR38
Each of the three EMS-induced point mutations in DHR38 was molecularly mapped in an effort to define their effect on gene function. PCR was used to amplify the DHR38 protein coding regions in genomic DNA isolated from homozygous or hemizygous mutants, and each fragment was subjected to DNA sequencing. These studies revealed premature stop codons upstream from sequences encoding the DNA-binding domain in all three alleles (Fig. 1). In DHR3843, the codon encoding glutamine 309 of the cTK61 isoform (Kozlova et al., 1998) is converted into a stop codon (CAG>TAG). In DHR3857, the codon encoding glutamine 318 of cTK61 is converted into a stop codon (CAA>TAA). Finally, in DHR3856, the codon encoding tyrosine 607 in cTK61 is converted into a stop codon (TAT>TAA). It is interesting to note that although all three mutations map to the same general location in DHR38, only the DHR3856 allele disrupts coding sequences common to both DHR38 transcripts (Fig. 1). The other two alleles affect only the longer DHR38 mRNA isoform. In addition, it is possible that DHR38 translation can re-initiate from either of two methionine codons that are located downstream from the DHR3843 and DHR3857 nonsense mutations (Fig. 1). These observations are consistent with the report that DHR3856 mutants display a more severe phenotype than either DHR3843 or DHR3857 mutants (Kozlova et al., 1998).
Generation and Characterization of a DHR38 Null Mutation
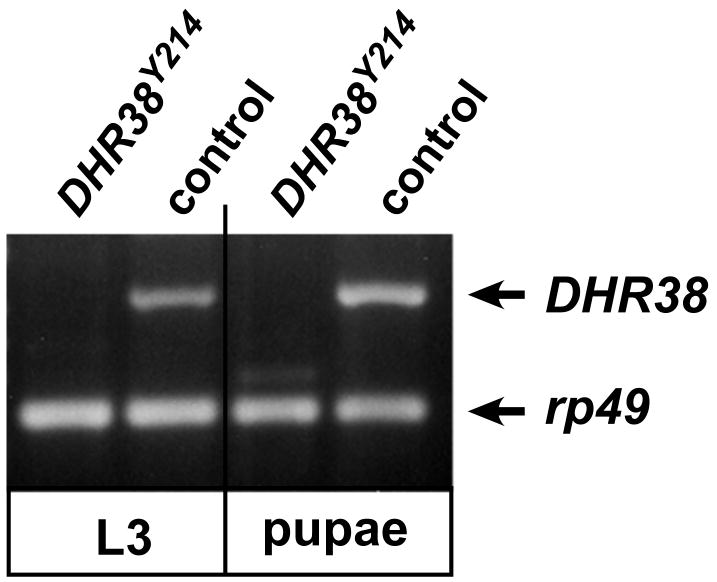
Although the DHR3856 mutant has a more severe phenotype than the other two EMS-induced mutations, it remains unclear whether it represents a true null allele (Kozlova et al., 1998). To address this issue, we used P-element imprecise excision to generate deletion mutations in DHR38. A homozygous viable P-element insertion, P{Y053}, was selected for this purpose, which maps about 2 kb upstream from the 5′ end of the cTK61 cDNA (Fig. 1). This P-element was mobilized and potential deletion mutants were recovered by scoring for lethality in combination with Df(2)KetelRX32, a large deletion that removes the 38D1-38E6 interval encompassing the DHR38 locus. Several resulting lethal mutations were characterized in more detail by PCR and DNA sequencing, using genomic DNA isolated from homozygous or hemizygous mutants. One mutation was identified, DHR38Y214, that lacks all protein coding sequences contained within cTK11, including sequences encoding the entire DNA-binding and ligand binding domains of the receptor, while sequences 3′ of DHR38 appear to be unaffected (Fig.1). In addition, DHR38Y214 carries a small deletion that removes part of the P{Y053} P-element and adjacent 3′ sequences that lie immediately upstream from the 5′ end of cTK61 (Fig.1). Part of the original P-element, however, remains in place, including the w+ marker gene along with genomic sequences that lie upstream from DHR38. This mutation thus appears to specifically affect the DHR38 locus and is a good candidate for a complete loss of function allele. Consistent with this, the DHR38Y214 allele fully complements lethal mutations that map on either side of DHR38, l(2)38Ea, l(2)38Eb and Ketel (Kozlova et al., 1998). In addition, whereas the coding sequence for the C-terminal 113 amino acids of the DHR38 protein could be detected by RT-PCR using RNA isolated from control third instar larvae or early pupae, no RNA was detectable in DHR38Y214 mutants (Fig. 2). This region is shared by all known DHR38 mRNA isoforms (Fisk and Thummel, 1995; Sutherland et al., 1995; Kozlova et al., 1998), supporting the conclusion that DHR38Y214 represents a null allele.
Fig. 2.
No DHR38 expression is detected in DHR38Y214 mutants. RT-PCR was used to detect DHR38 transcripts in homozygous DHR38Y214 mutants or y, w control animals, as wandering third instar larvae (L3) or 1-2 day old pupae (pupae). RT-PCR to detect the rp49 ribosomal protein transcript was used as a control for amplification and loading.
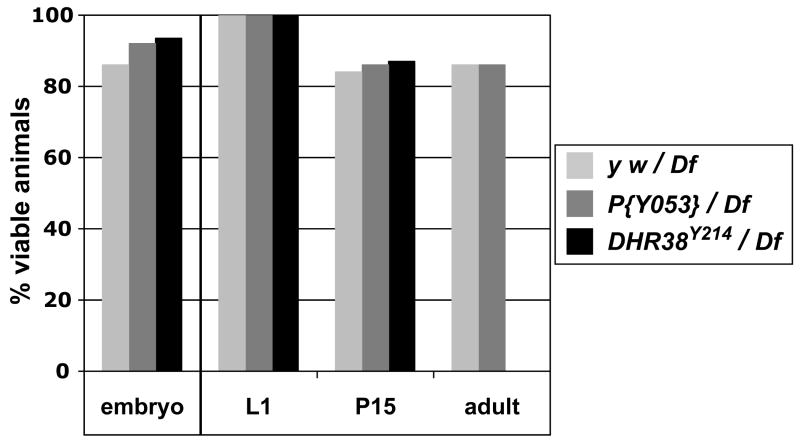
We next determined the lethal phenotype of the DHR38Y214 mutation in combination with the Df(2)KetelRX32 deficiency (Fig. 3). Approximately the same number of DHR38Y214/Df(2)KetelRX32 mutant embryos hatched into first instar larvae as either y w/Df(2)KetelRX32 or P{Y053}/Df(2)KetelRX32 controls, indicating that there is no embryonic lethality associated with this mutation (Fig. 3). Similarly, equivalent numbers of control or DHR38Y214/Df(2)KetelRX32 mutants collected as first instar larvae developed normally through the end of pupal development (P15). All mutants, however, died as pharate adults, within the pupal case prior to eclosion. DHR38Y214/Df(2)KetelRX32 mutant pupae have differentiated adult structures and appear normal except for a cuticle melanization phenotype and hemolymph leakage, indistinguishable from that seen in DHR3856 mutants (Kozlova et al., 1998). Penetrant pharate adult lethality was also observed for DHR38Y214 homozygotes and for DHR3856/DHR38Y214 mutants (data not shown). These observations indicate that DHR38Y214 represents a specific null allele for the locus.
Fig. 3.
DHR38Y214 mutants die as pharate adults. Males of three genotypes, y w, y w; P{Y053}/CyO, or y w; DHR38Y214/CyO were crossed with females carrying a deletion for the 38E interval, y w; Df(2)KetelRX32/CyO (Df), and viability was scored for each genotype at the indicated developmental stages. Embryonic viability was assayed by using a GFP-marked CyO balancer chromosome and determining the percentage of GFP− embryos that hatched into first instar larvae. Later viability was scored by using a CyO y+ balancer chromosome in a y w genetic background and scoring for the percent of y− first instar larvae (L1) that survived to either a late pupal stage (P15) or adulthood (adult).
Analysis of Ecdysone-Regulated Transcription in DHR38 Mutants
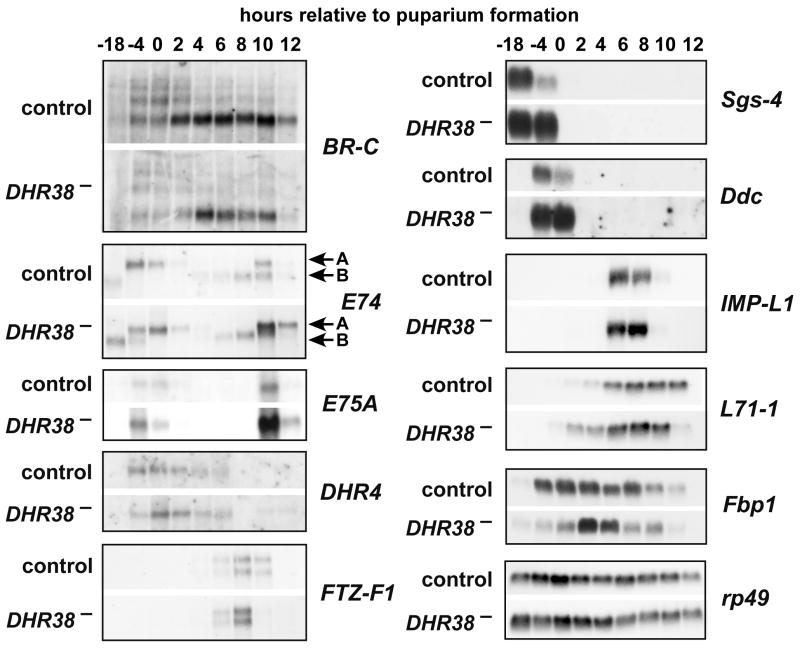
To date, only one gene has been shown to be dependent on DHR38 for its proper expression, Acp65A (Kozlova et al., 1998; Bruey-Sedano et al., 2005; Davis et al., 2007). This is, most likely, because hypomorphic alleles were used for those studies. In an effort to identify more DHR38 target genes, we examined the transcriptional cascade triggered by the steroid hormone ecdysone at the onset of metamorphosis. DHR38 has been proposed to impact ecdysone signaling through its interaction with USP (Sutherland et al., 1995). In addition, some mutations that result in pharate adult lethality are known to have a role in the ecdysone transcriptional cascade at puparium formation (e.g. E74A, Fletcher and Thummel, 1995). We thus decided to examine the effect of the DHR38 null allele on genes regulated by the late larval and prepupal pulses of ecdysone. RNA was isolated from both control and DHR38Y214/Df(2)KetelRX32 mutants staged as either mid-third instar larvae (-18 hrs), late third instar larvae (-4 hrs), newly formed prepupae (0 hr), or at two hour intervals thereafter until 12 hours after puparium formation. These RNA samples were analyzed by northern blot hybridization to detect the expression of genes that are induced directly by the hormone/receptor complex, BR-C, E74, E75A, DHR4, FTZ-F1, and Fbp1, as well as ecdysone-inducible secondary-response genes Sgs-4, Ddc, IMP-L1, L71-1 (Fig. 4). No significant effect was seen on E74, E75A, DHR4, or FTZ-F1 transcription. The apparent increase in E74A and E75A mRNA in 10 hour mutant prepupae is likely due to a sampling artifact caused by a few animals that had experienced the brief high titer prepupal ecdysone pulse (Richards, 1981). Similarly, there is little or no effect on Sgs-4, IMP-L1, L71-1, and Fbp1 expression in the DHR38 mutant. The increased level of Sgs-4 mRNA in -4 hr mutant larvae is likely due to the inaccuracy of staging third instar larvae by gut clearance (Andres and Thummel, 1994). L71-1 mRNA displays a 2-4 hour temporal shift to earlier times in the DHR38 mutant, while Fbp1 displays a more narrowed peak of expression in mid-prepupae. These results are consistent with our observation that developmental events associated with puparium formation and the prepupal-to-pupal transition, such as the destruction of the larval midgut and salivary glands, occur normally in DHR38 mutants (data not shown).
Fig. 4.
The DHR38 mutation has minor effects on ecdysone-regulated gene expression at the onset of metamorphosis. RNA was isolated from DHR38Y214/CyO and Df(2)KetelRX32/CyO heterozygotes (control) or DHR38Y214/Df(2)KetelRX32 mutants (DHR38−) and analyzed by northern blot hybridization. RNA was extracted from mid-third instar or late third instar larvae (-18 or -4 hrs relative to puparium formation), newly-formed prepupae (0 hrs), or at two hour intervals after puparium formation until 12 hrs. Northern blots were probed to detect BR-C, E74A, E74B, E75A, DHR4, FTZ-F1, Sgs-4, Ddc, IMP-L1, L71-1, and Fbp1. Hybridization to detect rp49 mRNA was performed as a control for loading and transfer.
In contrast, the BR-C and Ddc appear to display significant changes in expression level in the DHR38 mutant. All BR-C mRNA isoforms are reduced in the mutant (Fig. 4). This is interesting because ectopic expression of BR-C can lead to misregulation of pupal cuticle genes, although no effect of BR-C has been reported on cuticle function in adult flies (Zhou and Riddiford, 2002). In addition, levels of Ddc mRNA appear to be elevated in DHR38 mutants. Whereas the -4 hr time point is difficult to interpret, as mentioned above, puparium formation (0 hrs) provides an accurate time to synchronize the developmental stage of both control and mutant animals. This apparent effect on Ddc expression is of interest because this gene encodes dopa decarboxylase, and mammalian NR4A family members play a critical role in dopaminergic neuronal development (Zetterstrom et al., 1997; Saucedo-Cardenas et al., 1998). In addition, the increased expression of Ddc mRNA in DHR38 mutants is consistent with a recent paper by Davis et al. (2007), which showed that ectopic DHR38 expression can down-regulate Ddc promoter activity. This may not, however, be a specific effect because we have found that ectopic DHR38 expression at puparium formation can efficiently repress a number of ecdysone-inducible genes, including E74A, E75A, and DHR4 (data not shown). Down-regulation of these genes might be an indirect effect caused by ectopic DHR38 protein removing USP from functional EcR/USP receptor complexes (Sutherland et al., 1995). Further work is required to determine the significance of these effects of the DHR38 mutation on BR-C and Ddc expression.
Reduced Levels of Cuticle Gene Expression in DHR38 Mutant Pupae
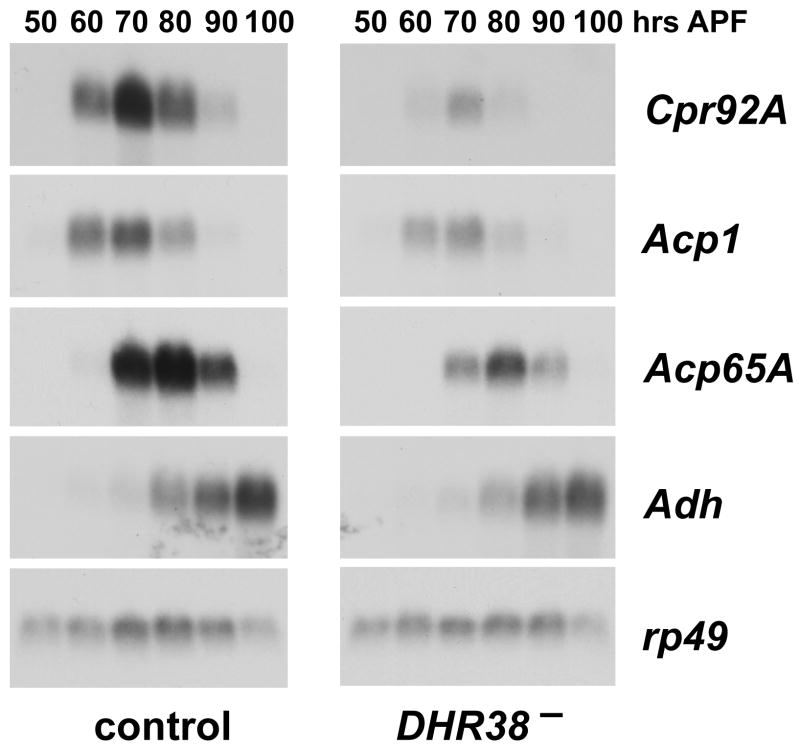
As part of a microarray study that will be reported elsewhere, three cuticle genes were identified among the most significantly down-regulated genes in DHR38 mutant pupae: Cpr92A (CG6240), Cpr49Ae (CG8505), and Acp65A (CG10297) (A.-F. Ruaud, T.K. and G.L., unpublished data). Northern blot analysis was used to validate these results. RNA was isolated from control and DHR38Y214/Df(2)KetelRX32 mutant pupae staged at 10 hour intervals during the final half of metamorphosis, and equal amounts of RNA were analyzed by northern blot hybridization to detect Cpr92A and Acp65A transcription (Fig. 5). We also examined the expression of Acp1 (CG7216), which was reduced in some, but not all of the DHR38 mutant datasets. This study revealed that these three cuticle genes are transiently expressed in late stage control pupae, preceding the deposition of adult cuticle. The expression of all three genes is also significantly reduced in the DHR38 mutant (Fig. 5). The earliest effect is seen at 60 hours after puparium formation, more than a day before the mutants die as pharate adults with ruptured cuticle and hemolymph leakage. In contrast, another gene expressed in late pupae, the adult-specific isoform of Adh, is only slightly affected in the mutant.
Fig. 5.
Cuticle gene expression is reduced in DHR38 mutant pupae. RNA was isolated from DHR38Y214/CyO and Df(2)KetelRX32/CyO heterozygotes (control) or DHR38Y214/Df(2)KetelRX32 mutants (DHR38−) staged at either 50, 60, 70, 80, 90, or 100 hrs after puparium formation (APF). Northern blot hybridization was used to detect transcripts from the Cpr92A, Acp1, and Acp65A cuticle genes, as well as the adult-specific Adh mRNA. Hybridization to detect rp49 mRNA was performed as a control for loading and transfer.
These results are consistent with earlier work by Bruey-Sedano et al. (2005), who showed that Acp65A expression is reduced in the hypomorphic DHR3802306 mutant (Kozlova et al., 1998). Acp65A is expressed in a specific pattern in the adult epidermis, underlying regions of flexible cuticle at the joints, suggesting that it is a critical target for DHR38 function (Bruey-Sedano et al., 2005). Like Acp65A, Cpr49Ae also encodes a protein with an RR-1 motif, associated with flexible cuticle (Karouzou et al., 2007). In contrast, Cpr92A encodes a protein with the RR-2 motif, associated with more rigid cuticle (Karouzou et al., 2007), and Acp1 appears to encode rigid cuticle that provides strength to the exoskeleton of the animal (Qiu and Hardin, 1995). DHR38 thus appears to regulate genes that direct the deposition of both flexible and rigid components of the exoskeleton. An attempt to rescue the lethality of DHR38Y214 mutants by epidermal expression of wild type DHR38 using the GAL4/UAS system was unsuccessful, due to the lethality associated with DHR38 overexpression (Kozlova et al., 1998) (A.-F. Ruaud, unpublished results). Future studies that attempt to control DHR38 expression more precisely may allow us to test the hypothesis that the essential functions for this gene reside in its expression in the epidermis of late pupae.
In this paper we demonstrate that a complete loss of DHR38 function results in pharate adult lethality, apparently as a result of reduced cuticle gene expression in late pupae and consequent loss of cuticular integrity, with death by exsanguination. This observation eliminates the possibility that a DHR38 null mutation might uncover an earlier essential function for the receptor. In addition, it provides an ideal genetic context for further characterization of DHR38 activity. Given that this receptor is the only member of the NR4A subclass in Drosophila, further studies of DHR38 mutants should provide insights into the ancestral roles of these receptors during development, with implications for better understanding the regulatory functions of NR4A family members in all higher organisms.
Experimental Procedures
Generation and Characterization of DHR38 Mutants
A homozygous viable P-element insertion was molecularly mapped ∼2 kb upstream from the 5′ end of the cTK61 cDNA clone (Genbank AJ002073) (Fig. 1). This P-element was mobilized using standard genetic protocols and imprecise excision mutations were recovered by scoring for lethality in combination with the Df(2)KetelRX32 deletion. Lethal excisions were molecularly mapped by extracting genomic DNA from 10-20 homozygous or hemizygous mutant third instar larvae (in combination with Df(2)KetelRX32) and scoring for the presence or absence of eleven short regions spanning the DHR38 locus by PCR and DNA sequencing (Fig. 1).
DHR38 transcripts were detected by RT-PCR analysis as described (Kozlova et al., 1998) using primers that are specific to a 407 bp region present in all known DHR38 mRNA isoforms (5′-ACATCATGGAGTTCAGCCGCA-3′ and 5′-GGAGGCATAACTTAGGGGCTA-3′). PCR was performed in 30 seconds steps at 94°, 61° and 72°C for 10 cycles without rp49 primers, and 22 cycles after rp49 primers were added.
Embryonic lethality was assayed by crossing stocks that carried the mutations over a CyO {actin-GFP} balancer chromosome and selecting for embryos that did not express GFP. A CyO, y+ balancer chromosome in a y w genetic background was used to assay for later lethal phases. Approximately 200–400 eggs were collected on grape juice-agar plates and allowed to develop for 24 hr at 25°C. First instar y− larvae were selected, transferred to vials with yeast paste, and allowed to develop. The sample size for each genotype was at least 100 animals.
Northern Blot Analysis
Total RNA was isolated from third instar larvae grown on food containing 0.05% bromophenol blue, selecting for either blue gut (-18 hrs relative to puparium formation) or clear gut (-4 hrs) animals (Andres and Thummel, 1994). Prepupae were staged by collecting newly-formed white prepupae (0 hrs) and harvesting animals at two hour intervals. Late pupae were staged by collecting animals that pupariated within a 1 hour interval and allowed to develop to the desired stage at 25°C. RNA was extracted, fractionated by formaldehyde gel electrophoresis, transferred to nylon membranes, and hybridized with radioactively labeled probes, as described (Andres et al., 1993).
Acknowledgments
We are grateful to K. Bates for help with sequencing the DHR38 mutant alleles, A. Beaton for providing the P{Y053} fly stock, J.-P. Charles for communicating results prior to publication, J. Willis for helpful discussions on cuticle genes, and A.-F. Ruaud for critical comments on the manuscript. This work was supported by the NIH (R01 DK075607) and the Howard Hughes Medical Institute.
References
- Andres AJ, Fletcher JC, Karim FD, Thummel CS. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev Biol. 1993;160:388–404. doi: 10.1006/dbio.1993.1315. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Baker KD, Shewchuk LM, Kozlova T, Makishima M, Hassell A, Wisely B, Caravella JA, Lambert MH, Reinking JL, Krause H, Thummel CS, Willson TM, Mangelsdorf DJ. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/s0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- Bonta PI, Pols TW, de Vries CJ. NR4A nuclear receptors in atherosclerosis and vein-graft disease. Trends Cardiovasc Med. 2007;17:105–111. doi: 10.1016/j.tcm.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Bruey-Sedano N, Alabouvette J, Lestradet M, Hong L, Girard A, Gervasio E, Quennedey B, Charles JP. The Drosophila ACP65A cuticle gene: deletion scanning analysis of cis-regulatory sequences and regulation by DHR38. Genesis. 2005;43:17–27. doi: 10.1002/gene.20150. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Davis MM, Yang P, Chen L, O'Keefe SL, Hodgetts RB. The orphan nuclear receptor DHR38 influences transcription of the DOPA decarboxylase gene in epidermal and neural tissues of Drosophila melanogaster. Genome. 2007;50:1049–1060. doi: 10.1139/g07-084. [DOI] [PubMed] [Google Scholar]
- DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriva Garcia H, Laudet V, Robinson-Rechavi M. Nuclear receptors are markers of animal genome evolution. J Struct Funct Genomics. 2003;3:177–184. [PubMed] [Google Scholar]
- Fisk GJ, Thummel CS. Isolation, regulation, and DNA-binding properties of three Drosophila nuclear hormone receptor superfamily members. Proc Natl Acad Sci U S A. 1995;92:10604–10608. doi: 10.1073/pnas.92.23.10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC, Thummel CS. The Drosophila E74 gene is required for the proper stage- and tissue-specific transcription of ecdysone-regulated genes at the onset of metamorphosis. Development. 1995;121:1411–1421. doi: 10.1242/dev.121.5.1411. [DOI] [PubMed] [Google Scholar]
- Gissendanner CR, Kelley K, Nguyen TQ, Hoener MC, Sluder AE, Maina CV. The Caenorhabditis elegans NR4A nuclear receptor is required for spermatheca morphogenesis. Dev Biol. 2008;313:767–786. doi: 10.1016/j.ydbio.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich VC. The ecdysteroid receptor. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science. Oxford: Elsevier; 2005. pp. 243–286. [Google Scholar]
- Hsu HC, Zhou T, Mountz JD. Nur77 family of nuclear hormone receptors. Curr Drug Targets Inflamm Allergy. 2004;3:413–423. doi: 10.2174/1568010042634523. [DOI] [PubMed] [Google Scholar]
- Karouzou MV, Spyropoulos Y, Iconomidou VA, Cornman RS, Hamodrakas SJ, Willis JH. Drosophila cuticular proteins with the R&R Consensus: annotation and classification with a new tool for discriminating RR-1 and RR-2 sequences. Insect Biochem Mol Biol. 2007;37:754–760. doi: 10.1016/j.ibmb.2007.03.007. [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS. Nuclear receptors--a perspective from Drosophila. Nat Rev Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Kozlova T, Pokholkova GV, Tzertzinis G, Sutherland JD, Zhimulev IF, Kafatos FC. Drosophila hormone receptor 38 functions in metamorphosis: a role in adult cuticle formation. Genetics. 1998;149:1465–1475. doi: 10.1093/genetics/149.3.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich JM, Sluder A, Guan X, Shi Y, McKee DD, Carrick K, Kamdar K, Willson TM, Moore JT. Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol. 2001;2:RESEARCH0029. doi: 10.1186/gb-2001-2-8-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13:730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol. 2006a;20:786–794. doi: 10.1210/me.2005-0331. [DOI] [PubMed] [Google Scholar]
- Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006b;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- Ponnio T, Conneely OM. nor-1 regulates hippocampal axon guidance, pyramidal cell survival, and seizure susceptibility. Mol Cell Biol. 2004;24:9070–9078. doi: 10.1128/MCB.24.20.9070-9078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Hardin PE. Temporal and spatial expression of an adult cuticle protein gene from Drosophila suggests that its protein product may impart some specialized cuticle function. Dev Biol. 1995;167:416–425. doi: 10.1006/dbio.1995.1038. [DOI] [PubMed] [Google Scholar]
- Richards G. The radioimmune assay of ecdysteroid titres in Drosophila melanogaster. Mol Cell Endocrinol. 1981;21:181–197. doi: 10.1016/0303-7207(81)90013-7. [DOI] [PubMed] [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci U S A. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Sutherland JD, Kozlova T, Tzertzinis G, Kafatos FC. Drosophila hormone receptor 38: a second partner for Drosophila USP suggests an unexpected role for nuclear receptors of the nerve growth factor-induced protein B type. Proc Natl Acad Sci U S A. 1995;92:7966–7970. doi: 10.1073/pnas.92.17.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NP, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- Zhou X, Riddiford LM. Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development. 2002;129:2259–2269. doi: 10.1242/dev.129.9.2259. [DOI] [PubMed] [Google Scholar]