Abstract
Uncontrolled TNF-α synthesis is known to play an important role in numerous inflammatory disorders, and multiple transcriptional and post-transcriptional regulatory mechanisms have therefore evolved to dampen the production of this important pro-inflammatory cytokine. By examining the anti-inflammatory properties of the vitamin B3 constituent nicotinamide, we have uncovered a novel regulatory pathway controlling TNF-α production. Exogenous nicotinamide inhibits TNF-α secretion through modulation of mRNA translation efficiency. Moreover, the capacity to produce TNF-α appears to be directly correlated with intracellular NAD levels, suggesting that a NAD-dependent biological event that can be inhibited by nicotinamide controls TNF-α synthesis in cells of the immune system. Sirtuins represent NAD-dependent deacetylases involved in regulation of gene expression in both mammals and yeasts, and are known to be inhibited by nicotinamide. We demonstrate herein that similarly to nicotinamide, structurally unrelated sirtuin inhibitors downregulate TNF-α secretion with minimal effect on TNF-α gene transcription. By over-expressing individual sirtuin members in cell lines transiently expressing TNF-α, we have identified SIRT6 as a sirtuin member able to upregulate TNF-α synthesis in vitro. In agreement with this finding, bone-marrow derived dendritic cells from SIRT6 KO mice display reduced TNF-α synthesis in response to CpG. Collectively, these data delineate a novel relationship between metabolism and the inflammatory response, by uncovering the role of SIRT6 in the control of TNF-α secretion.
INTRODUCTION
Nicotinamide adenine dinucleotide (NAD) is an essential coenzyme regulating numerous cellular metabolic pathways. In the last few decades, a growing number of studies have uncovered an additional, nonredox function of NAD in cellular physiology. In particular, NAD has been recognized as a substrate for a growing group of “NAD–dependent” enzymes with multiple roles in cellular regulation 1. NAD-dependent protein modifications are catalyzed by enzymes broadly defined as ADP-ribosyl transferases. Two enzyme families, known respectively as mono-ADP-ribosyltransferases (mARTs) and poly(ADP-ribose) polymerases (PARPs) have in particular been recently identified in mammals. mARTs represent a family of ectoenzymes including GPI-anchored membrane proteins and secreted forms, while PARPs (comprising 18 members to date 2) are intracellular proteins usually located within the nuclear compartment. NAD also represents a substrate for members of a third evolutionarily conserved family of proteins collectively known as “sirtuins” 34. These proteins, (for which seven members, SIRT1 to SIRT7 have been described) regulate a wide range of biological processes including gene silencing 5, aging 67, cell survival in response to stress 8,9, cell differentiation 10 and metabolism 11,12. The founding member of this family, Sir2 in yeasts and its mammalian homologue SIRT1, catalyzes a unique reaction in which the cleavage of NAD and the deacetylation of substrate are coupled to the formation of O-acetyl-ADP-ribose 13
A common feature of all these “NAD-dependent” enzymes is the cleavage of the ADP-ribose moiety of NAD with the concomitant release of nicotinamide (NAm), which therefore serves as an endogenous end product inhibitor 14. NAm, in conjunction with nicotinic acid, also represents an essential vitamin (Vitamin B3 or niacin) required as an exogenous precursor of NAD in most living organisms 15. Collectively, these observations have shifted interest in NAm from a simple nutrient (NAD precursor) to a pharmacological agent able to interfere with NAD-consuming enzymes. It is therefore presently believed that intracellular NAD and NAm levels may regulate numerous cellular reactions by modulating the activity of a large number of NAD-dependent enzymes 16. Recently, experiments performed in yeasts support this contention, and suggest an important role for the NAm/NAD ratio in regulating several biological processes 17.
Our own interest in the potential link between NAD biosynthesis and cell regulation derives from studies performed in immune cells that led to the identification of the gene encoding for the murine nicotinamide phosphoribosyltransferase (NAmPRT, EC 2.4.2.12) 18. This enzyme catalyzes the first step of the salvage pathway allowing NAD biosynthesis from NAm. NAmPRT levels were found upregulated upon stimulation of immune cells both in vivo and in vitro 19,20. Although elevated NAmPRT expression may simply represent a physiological response to meet the demand for increased metabolic resources in activated/proliferating cells, it may also represent a regulatory mechanism providing adequate intracellular NAD levels required for the effector function of one or several NAD-consuming enzymes important for immunoregulation.
Several lines of experimental evidence support a functional link between NAD metabolism and inflammation. NAm has been shown to inhibit the production of key inflammatory mediators such as NO 21 and cytokines 22,23. These observations have been generally interpreted in light of the recently uncovered role of PARP-1, in the regulation of an inflammatory response 24. Indeed (i) PARP-1 has been shown to act as a transcriptional activator of NF-kB 25 (ii) NAm and other structurally unrelated pharmacological PARP-1 inhibitors are of remarkable therapeutic efficacy in experimental models of inflammatory-related diseases 24 and finally (iii) PARP-1 −/− mice are protected from endotoxic shock 26,27. These and other observations engendered the view that PARP-1 may represent the molecular link between NAm, NAD biosynthesis and secretion of pro-inflammatory cytokines through regulation of NF-kB transcriptional activity.
The purpose of the present study was to evaluate the functional link between NAD metabolism and inflammation. The observations described in this study indicate a contrasting role for NAm and NAD in regulating the secretion of TNF-α. While adequate intracellular NAD levels are required for optimal TNF-α production, exogenous NAm inhibits TNF-α suggesting an important role for a NAD-dependent, NAm-inhibitable enzymatic activity in regulating the production of this potent pro-inflammatory mediator. However, and in contrast to a prevailing view, NAm appears to exert its anti-inflammatory properties in a PARP-1-independent fashion. Using a set of structurally unrelated pharmacological inhibitors and a genetic approach, we identify herein an important role for SIRT6, a member of the sirtuin family, in the regulation of TNF-α production during an inflammatory response.
RESULTS
NAm protects mice against an endotoxic shock
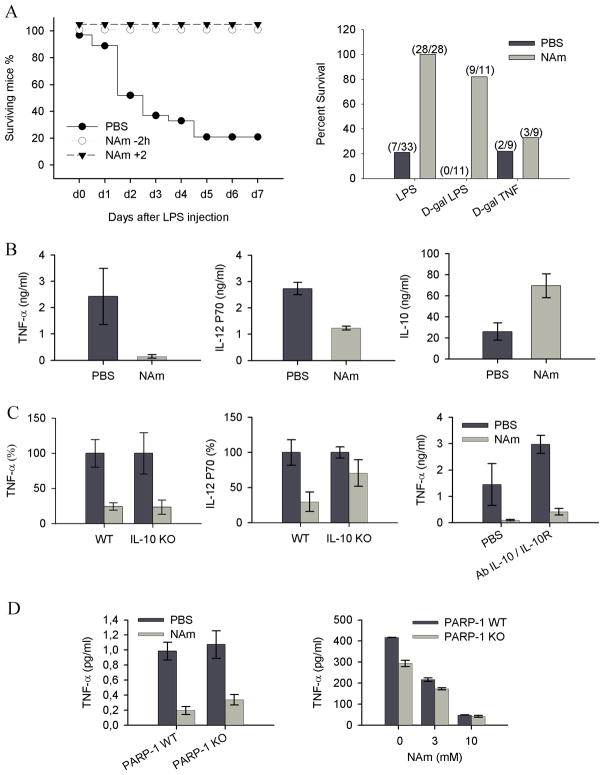
A well defined model of acute septic shock was used to evaluate the anti-inflammatory properties of NAm in vivo. Groups of mice were injected with a lethal dose of LPS (20 mg/kg) in combination with either saline or NAm (500 mg/kg) at various times before or after endotoxin administration. As shown in Figure 1A, NAm was found to protect mice against a lethal high dose of LPS even when injected 2 hours following endotoxin challenge.
Figure 1. In vivo anti-inflammatory properties of NAm.
(A) Naïve C57BL/6 mice were injected with LPS (20 mg/kg) or with a combination of a low dose of LPS (5 μg/kg) and D-galactosamine (0.75 g/kg) or with a combination of TNF (25 μg/kg) and D-galactosamine (0.75 g/kg). Survival rates were monitored daily as shown in the left panel, or determined 7 days post-injection (right panel). (B) Cytokine serum levels were determined by ELISA 90 min (TNF-α and IL-10) or 120 min (IL-12) following LPS (2 mg/kg) and PBS or NAm (500mg/kg) injection as indicated. (C) C57BL/6 mice or IL-10 KO syngenic mice were injected as in B. When indicated, mice were treated with a combination of antibodies to IL-10 (clone JES5-2A5) and to IL-10R (clone 1B1.2). (D) Wt and PARP-1 KO mice were treated as indicated in A. Serum TNF-α levels were determined 90 min after LPS injection and shown in the left panel. Purified splenic dendritic cells from wt and PARP-1 KO mice were stimulated in vitro with CpG (100 ng/ml) and TNF-α levels (right panel) determined in the supernatant of 16h cultures.
D-galactosamine (D-Gal) sensitizes animals to LPS by causing severe liver damage secondary to TNF-induced hepatocyte apoptosis. As shown in Figure 1A, NAm protected D-gal sensitized mice against an LPS, but not against a TNF-α challenge, indicating that this vitamin did not affect the late stages of the biological response to endotoxemia, but rather interfered with an early event of the inflammatory response, as confirmed by the analysis of the peak response of several cytokines released in the serum of treated animals. NAm administration led to a notable shift in the cytokine response, with marked reduced levels of TNF-α and IL-12, and increased levels of the anti-inflammatory cytokine IL-10 (Figure 1B). Since endogenous IL-10 is known to modulate the secretion of pro-inflammatory mediators during LPS challenge in vivo 28, we repeated these experiments in mice genetically deficient for IL-10. As shown in Figure 1C, NAm failed to inhibit IL-12 secretion in the absence of endogenous IL-10, while it strongly reduced LPS-induced, serum TNF-α levels in both mouse strains. To further confirm that NAm inhibited TNF-α production in an IL-10-independent fashion, wt animals were injected with a combination of blocking antibodies to IL-10 and IL-10R before LPS challenge. As expected, in vivo blocking of IL-10 signalling led to enhanced TNF-α secretion that however retained sensitivity to the inhibitory effects of NAm (Figure 1C). Finally, based on the ability of NAm to inhibit PARP-1 enzymatic activity (with an IC50 in the 30 μM range 29), we wished to examine the potential role of PARP-1 in mediating the anti-inflammatory properties of this vitamin. PARP-1 KO mice responded to LPS challenge by secreting control levels of serum TNF-α and were equally sensitive to the anti-inflammatory properties of NAm both in vivo and in vitro (Figure 1D). Collectively, these observations are best explained by assuming that the anti-inflammatory properties of NAm are due to the ability of this vitamin to block TNF-α secretion by activated cells in a PARP-1 independent fashion.
NAm inhibits TNF-α synthesis at the post-transcriptional level
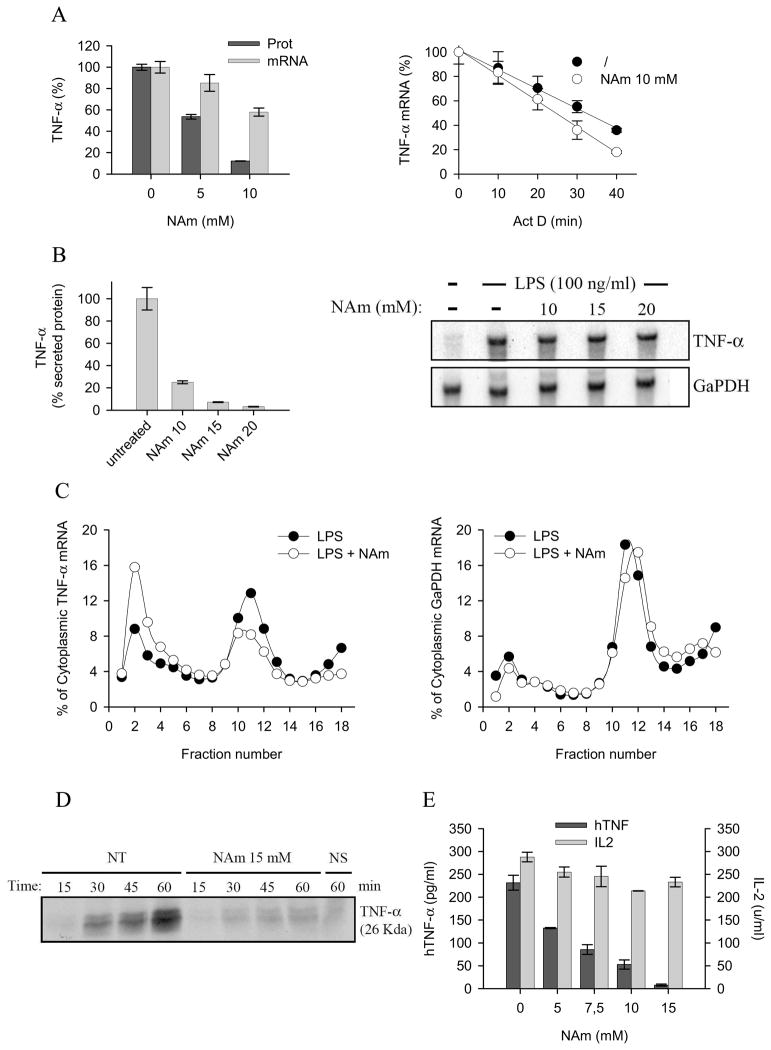
To better understand the mechanisms by which NAm affects TNF-α production, the well described mouse RAW264.7 macrophage cell line was used as responder cell. TNF-α secretion by LPS-stimulated RAW 246.7 was inhibited by exogenous NAm with minimal effect on mRNA accumulation (Figure 2A). mRNA stability was not or only marginally inhibited by NAm, suggesting that this vitamin inhibited TNF-α protein production by interfering with a post-transcriptional step (Figure 2A). To further support this contention, RAW 246.7 cells were stimulated with LPS for 2 hours, washed, and subsequently incubated in the presence of media or graded doses of NAm for an additional 30 minutes. Again, NAm strongly inhibited TNF-α protein secretion, with minimal effect on TNF-α transcripts as judged by ELISA and northern blot analysis, respectively (Figure 2B). Cytoplasmic extracts from LPS or LPS/NAm treated cells were than fractionated over sucrose gradients and TNF-α mRNA quantified by northern blot. LPS treatment led to the typical accumulation of TNF-α mRNA in the polysome containing, heavy fraction of the gradient (Figure 2C, fractions 10 to 13). NAm decreased the proportion of TNF-α transcripts that associate with polysomes, concomitant with a shift towards fractions containing smaller polysomes and monosomes (Figure 2C, fractions 1 to 4). Polysome containing GaPDH mRNA display the same profile in both control, and NAm-treated cells (Figure 2C, right panel). Finally, RAW 264.7 cells were incubated in medium containing radioactive amino acids and the synthesis of TNF-α in response to LPS was followed by immunoprecipitation and polyacrylamide gel electrophoresis. As shown in Figure 4C, NAm significantly reduced the accumulation of TNF-α protein in cell extracts, in keeping with the conclusion that it affected the translation efficiency of TNF-α mRNA. (Figure 2D). To eliminate any possible interference of NAm with the TLR signalling cascade regulating gene transcription and mRNA splicing, the human 293T cell line was transiently transfected with constructs expressing the complete human TNF-α cDNA or the murine IL-2 ORF under the control of a viral (CMV) promoter. As shown in Figure 2E, NAm selectively inhibited TNF-α secretion in this experimental setting with minimal effect on IL-2 production, strongly suggesting that it did not interfere with the general transcriptional or translational machinery. Finally, an ELISA assay detecting both soluble and cell-associated forms of TNF-α was used to exclude any interference of this vitamin with the intracellular traffic or maturation of the high molecular weight, transmembrane form of the TNF-α precursor protein. NAm inhibited TNF-α protein accumulation in this assay, confirming the inhibitory effect of NAm on TNF-α protein synthesis (Supplementary Fig. 1). Collectively, these observations suggest a role for NAm modulating the translational efficiency of TNF-α mRNA
Figure 2. NAm inhibits TNF-α protein synthesis.
(A) RAW 264.7 macrophages were treated or not with NAm and stimulated with 100 ng/ml of LPS. Culture media and cells were collected 2h after stimulation. TNF-α protein concentration in culture supernatant was measured by ELISA while TNF mRNA expression (normalized to GaPDH mRNA) was evaluated in cell extracts by quantitative RT-PCR (right panel). RAW264.7 cells were stimulated with 100 ng/ml LPS for 2 h. Transcription was then stopped by addition of 5 μg/ml actinomycin D (Act D) in the presence of NAm or left untreated. Cells were harvested at the time intervals shown, and TNF mRNA levels were quantified by quantitative RT-PCR. TNF/GaPDH RNA ratios were plotted as percentages of values at the time of Act D addition (left panel). (B) RAW 264.7 cells were stimulated for 2h with 100 ng/ml LPS, washed and treated for 30 min with graded doses of NAm. Relative expression of TNF-α protein levels (left panel) and mRNA (right panel) were determined respectively by ELISA and northern blot analysis. GaPDH mRNA levels were determined as internal control. (C) Polysome profile analysis of RAW 264.7 cells treated with 15 mM NAm. Densitrometric quantification of the relative RNA northern blot signal intensities across the gradient for TNF-α (left panel) and GaPDH (right panel) transcripts. Data are from one of three representative experiments. (D) RAW 264.7 cells were stimulated with LPS (100 ng/ml) for 1h, washed and incubated in cysteine- and methionine-free DMEM. 30 min later the cells were incubated in DMEM supplemented with [35S]-labeled cysteine and methionine, brefeldin A and with 15 mM NAm for the indicated time periods. Immunoprecipitated TNF-α was separated by SDS-PAGE and the gels dried and analyzed by autoradiography (NS: not stimulated by LPS). (E) 293T cells were co-transfected with plasmid encoding human TNF-α and with a plasmid encoding murine IL-2. Secreted cytokines levels were determined in the supernatant of transfected cells by ELISA. NAm was found to selectively inhibit the production of hTNF-α.
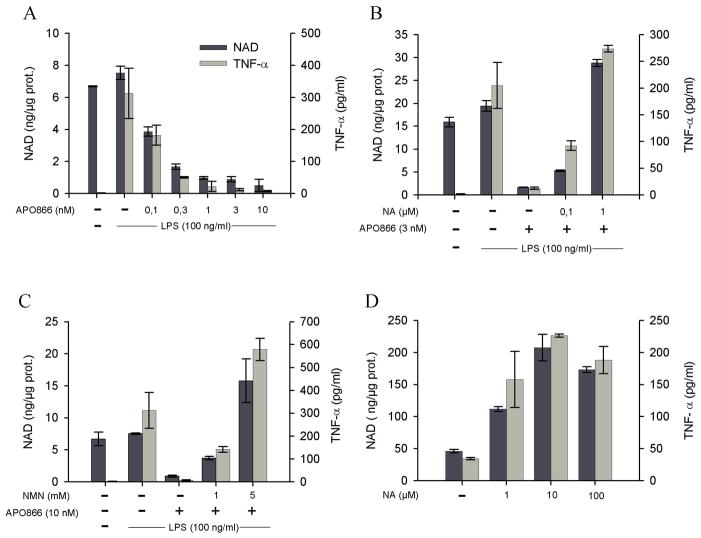
Figure 4. Intracellular NAD levels determine TNF-α production capacities.
(A) The human THP-1 cells line was incubated overnight in the presence of graded doses of the NAmPRT inhibitor APO866. Cells were than stimulated with LPS and levels of secreted TNF-α and intracellular NAD content determined 2 hours later, as indicated in the method section. (B), (C) The experiment was conducted as in (A), except that responder cells were incubated in media supplemented with the NAD precursors NA (B) or NMN (C). (D) THP-1 cells were incubated overnight in media supplemented with NA at the indicated doses and levels of secreted TNF-α and intracellular NAD content evaluated as in (A).
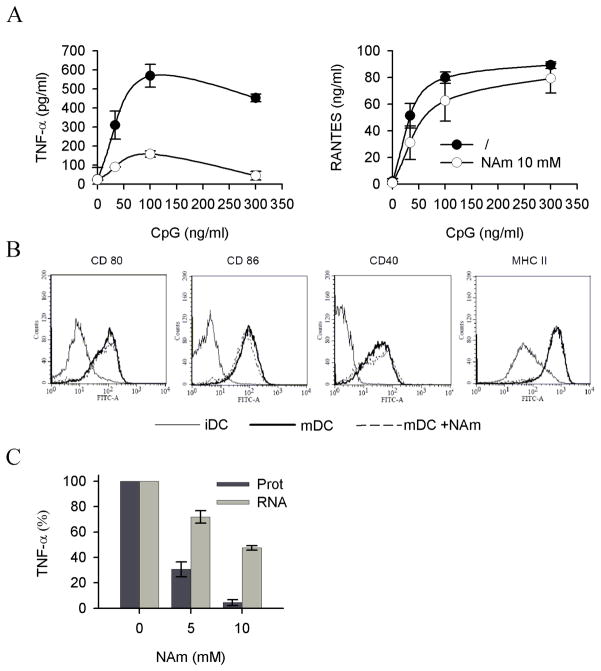
To evaluate the effect of exogenous NAm on TNF-α protein synthesis in a more physiological setting, freshly isolated, splenic murine dendritic cells were used in vitro as responder cells. These cells respond to microbial stimulation by undergoing a complex biological response (referred to as maturation) characterized by secretion of pro-inflammatory soluble mediators (cytokines and chemokines) and upregulation of cell surface markers including the co-stimulatory ligands/receptors CD40, CD80, CD86 and MHC class II. In keeping with previous observations, exogenous NAm inhibited TNF-α synthesis in response to CpG without affecting the capacity of these cells to secrete the chemokine RANTES (Figure 3A) or to upregulate CD40, CD80, CD86 and MHC class II molecules expression (Figure 3B). Noteworthy, and in keeping with previous studies, NAm preferentially affected protein synthesis over mRNA accumulation, as shown in Figure 3C.
Figure 3. Immunomodulatory properties of NAm on splenic dendritic cells.
(A) Purified splenic dendritic cells were stimulated with graded doses of CpG in the presence or absence of NAm (10mM) and TNF-α and RANTES levels determined in the supernatant of overnight cultures. (B) Purified dendritic cells were stained with the indicated antibodies at day 0 (immature DC, iDC) or after maturation in vitro in the absence (mDC) or in the presence of 10 mM NAm (mDC + NAm). (C) Splenic dendritic cells were stimulated with 100 ng/ml CpG and TNF-α protein concentration in culture supernatants was measured by ELISA while TNF mRNA expression (normalized to RPL-32 mRNA) was determined in cell extracts by quantitative RT-PCR.
Endogenous NAD levels determine TNF-α protein synthesis efficacy
Since NAm represents an endogenous feedback inhibitor of all NAD-dependent enzymes described to date, the previous observations suggest an important role for a NAD-dependent enzyme in regulating TNF-α protein synthesis. To directly evaluate this possibility, we designed an experiment aimed at manipulating the intracellular NAD levels in a TNF-α producing cell line. Eukaryotic cells can synthesise NAD from four distinct precursors including the amino acid tryptophan, the niacin (vitamin B3) components nicotinic acid (NA) and nicotinamide (NAm) and finally the recently identified intermediate nicotinamide riboside (NR 30, see Supplementary Fig. 2). It is noteworthy that only hepatocytes express high levels of tryptophan 2,3 dioxygenase 15 (TDO, EC 1.13.11.11), the first enzyme of the enzymatic pathway leading to NAD biosynthesis from tryptophan, indicating thus that most cells in the organism are dependent upon exogenous niacin (NAm or NA) for NAD biosynthesis. The human THP-1 macrophage cell line has been shown to express both nicotinamide phosphoribosyltranferase (NAmPRT, EC 2.4.2.12) and nicotinic acid phosorphorybosyl transferase (NAPRT, EC 2.4.2.1) rendering this cell line able to use both niacin components as NAD precursors31. Since conventional media formulations do not contain nicotinic acid nor nicotinamide riboside, NAm represents the only NAD precursor available under standard culture conditions (e.g. 8μM in RPMI and 33μM in DMEM). Moreover, NAD biosynthesis from NAm can be specifically inhibited by APO866, a recently described and potent low molecular weight inhibitor of NAmPRT (with no effect on NAPRT 32) with an IC50 in the nM range 31. THP-1 cells were incubated in control media (containing NAm as sole NAD precursor), or in media supplemented with graded doses of APO866. As expected, APO866 was able to strongly inhibit intracellular NAD concentrations upon overnight incubation with an approximate IC50 of 0.1 nM (Figure 4A). This decrease in intracellular NAD levels was mirrored by a reduced capacity to produce TNF-α in response to LPS (Figure 4A). Exogenous nicotinic acid (NA) restored intracellular NAD levels in APO866-treated cells, and concomitantly allowed control-level TNF-α secretion in response to LPS despite continuous exposure to the NAmPRT inhibitor (Figure 4B). Similarly, addition of nicotinamide mononucleotide (NMN), the product of the NAmPRT-catalyzed reaction (see Supplementary Fig. 2), restored both intracellular NAD levels and TNF-α production in THP-1 cells, confirming both the specificity of action of APO866 and the role of intracellular NAD in regulating TNF-α production (Figure 4C). Accordingly, culture of THP-1 cells in the presence of an exogenous source of NA led to a striking increase in intracellular NAD levels (400 % increase when compared to control cells) and supra-optimal TNF-α secretion capacity (Figure 4D). Note that neither APO866 nor NAD precursors affected cell viability in this experimental setting (not shown). Collectively, these observations establish a close relationship between intracellular NAD levels and control of TNF-α synthesis.
SIRT6 regulates TNF-α production
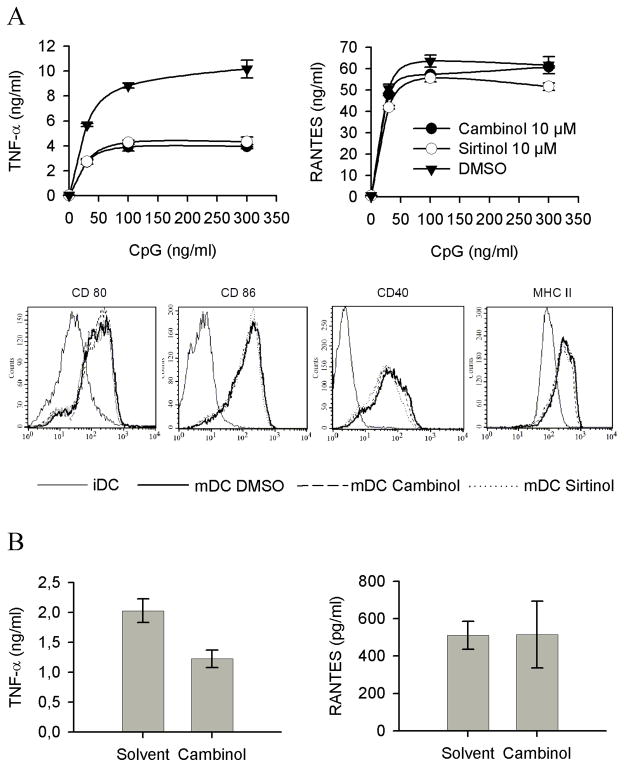
The previous observations suggested that TNF-α protein synthesis was under the control of a NAD-dependent, NAm-sensitive biological event. Recently, studies mostly performed in yeasts have identified sirtuins as important sensors of intracellular NAD and NAm levels, providing a molecular link between energy metabolism and signalling mediated by this family of NAD-dependent enzymes. Since TNF-α protein synthesis appeared to be under the positive and negative control of respectively NAD and NAm, we hypothesized that one or multiple members of the sirtuin family could exert a positive control on TNF-α production. To evaluate this hypothesis, murine dendritic cells and cell lines were incubated in the presence of a panel of structurally unrelated sirtuin inhibitors 33,34 and stimulated by CpG. All sirtuin inhibitors led to a marked reduction in TNF-α protein secretion, with minimal effect on RANTES synthesis and on expression of cell surface associated markers (Figure 5A). Similar observations have been performed using other responder cells lines and an additional sirtuin inhibitor (dyihydrocoumarin 35, data not shown). These observations suggest a role for a sirtuin member in regulating the efficacy of TNF-α synthesis in response to microbial stimulation, and led us to evaluate the possible anti-inflammatory properties of cambinol, known to display reduced toxicity in vivo 34, in a mouse model of acute septic shock In keeping with the in vitro data, cambinol displayed anti-inflammatory properties in vivo, as shown by its ability to significantly inhibit TNF-α secretion in response to acute LPS injection (Figure 5B). Note that RANTES synthesis in vivo was used as a control for possible non specific effect of the drug on the innate immune response. Finally, and in keeping with previous observations related to NAm, reduction of intracellular NAD levels and incubation in the presence of sirtuin inhibitors appeared to inhibit TNF-α protein synthesis with minimal effect on mRNA accumulation (see Supplementary Fig. 3).
Figure 5. Immunomodulatory properties of sirtuin inhibitors.
(A) Purified splenic dendritic cells were pretreated with 10 μM of the indicated sirtuin inhibitors and stimulated with graded doses of CpG in the continous presence of inhibitors. TNF-α and RANTES levels determined in the supernatant of overnight cultures. Purified dendritic cells were stained with the indicated antibodies at day 0 (immature DC, iDC) or after maturation in vitro in the presence of solvent (DMSO mDC) or 10 μM of indicated inhibitors (mDC Cambinol or mDC Sirtinol). (B) C57BL/6 mice were injected i.p. with cambinol (100 mg/kg) or its solvent (10%/10% ethanol/Cremophore solution) 1 hours before LPS treatment. The figure represents the TNF-α and RANTES serum levels of 4 individual mice in each group and is representative of three independent experiments.
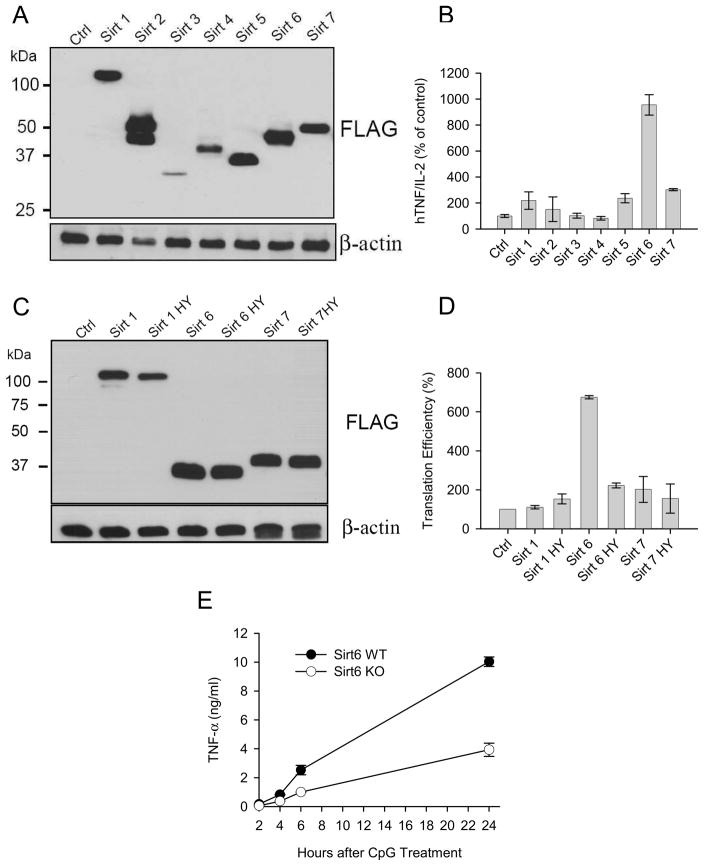
To positively identify the sirtuin member regulating TNF-α translation efficiency, we constructed plasmids encoding all murine sirtuins and transiently co-transfected them with a hTNF-α expression vector and a murine IL-2 reporter vector (included to normalize for extract concentrations, transfection efficiency and overall translation status) in the 293T human cell line. Despite some variations, all transfected cells lines expressed the corresponding sirtuin member as shown in Figure 6A. Analysis of the relative expression of TNF-α vs IL-2 proteins in the supernatants of transfected cells lines revealed a positive influence of SIRT6 on TNF-α protein synthesis in this experimental setting (Figure 6B). Since none of the mitochondrial (SIRT3, SIRT4 and SIRT5) or predominantly cytoplasmic (SIRT2) sirtuins (Figure 6B) affected TNF-α secretion in this assay, they were excluded from further analysis. The effect of nuclear sirtuins on TNF-α expression was analyzed in more details, as shown in Figures 6C and 6D. In this experiment, cells were transiently transfected with plasmid encoding wild type or catalytically inactive mutant versions of SIRT1, SIRT6 and SIRT7, and translational efficiency evaluated following a previously described approach 36 by comparing TNF-α and IL-2 mRNA levels to their protein counterpart. Overexpression of SIRT6 consistently resulted in increased TNF-α protein secretion relative to its mRNA levels (Figure 6D). In keeping with the pharmacological studies, the ability of SIRT6 to augment TNF-α translation efficiency required its enzymatic activity, since a mutant SIRT6 construct lacking enzymatic activity (data not shown) was unable to affect TNF-α production (Figure 6D). Finally, to confirm the role of SIRT6 in regulating TNF-α synthesis, bone marrow-derived dendritic cells from wt and SIRT6 KO mice 37 were stimulated in vitro with CpG and supernatants tested for TNF-α content. Lack of SIRT6 expression decreased the capacity of BMDC to produce TNF-α, confirming the role of this sirtuin member in regulating the production of this important pro-inflammatory cytokine (Figure 6E).
Figure 6. SIRT6 regulates TNF-α protein synthesis.
(A) The human cell line 293T was transiently transfected with plasmids encoding the indicated tagged forms of murine sirtuin members. Protein expression was determined by western blot using an anti-FLAG mAb. (B) Pools of 293T cells were transiently transfected with three constructs encoding respectively for human TNF-α, mouse IL-2 and a selected murine sirtuin as indicated. The results represent the relative content of human TNF-α compared to murine IL-2 released in the supernatant during 6 hours of culture. (C) The human cell line 293T was transiently transfected with plasmids encoding the indicated tagged forms of wt and mutant forms of selected murine sirtuin members. Protein expression was determined as in (A). (D) TNF-α translation efficiency of triple transfectants, as described in (B), was determined as detailed in the text (mean of two independent experiments). (D) Bone marrow derived dendritic cells from wt and SIRT6 KO mice were stimulated with 1 μg/ml CpG and TNF-α protein levels determined in the supernatant collected at the indicated timing.
DISCUSSION
Inflammation represents an essential host response to microbial challenge, allowing the recruitment and activation of leukocytes at the site of infection 38. Infection-mediated autoagression may however subsides even as the offending agents have been eliminated, stressing the importance of fine tuning the inflammatory response to achieve the appropriate level and duration of the response 39. Inadequate production of TNF-α is involved in numerous pathological situations 40 such as diabetes, Crohn’s disease and rheumatoid arthritis. Accordingly, numerous studies have revealed that expression of TNF-α is tightly controlled, both transcriptionally and post-transcriptionally by modulation of mRNA stability and translational efficiency 41.
The present study uncovers a close relationship between NAD metabolism and production of TNF-α, identifying a novel regulatory pathway controlling TNF-α synthesis by cells of the immune system. We demonstrate in this report that the TNF-α translation efficiency requires adequate intracellular NAD levels and is inhibited by exogenous NAm, revealing a potentially important link between cellular metabolites and inflammation.
The anti-inflammatory properties of NAm have been known for decades 22. However, despite the longstanding interest in the pharmacological properties of this vitamin, the mechanisms by which it affects an inflammatory response have not been clearly established. One the major difficulties in interpreting the biological properties of NAm relates to its wide range of putative targets 42. In general, the anti-inflammatory properties of NAm have been attributed to its ability to inhibit PARP-1, but experiments described herein (see Figure 1) and performed by others 23,27 fail to support this hypothesis. Studies performed mostly in yeasts have led to consider members of the sirtuin family as important sensors of the intracellular NAm/NAD status 43,44,45. The present work concurs with this hypothesis and suggest a link between NAD metabolism, sirtuin activity and regulation of TNF-α secretion. Experiments performed using APO866 (a potent inhibitor of the salvage pathway of NAD biosynthesis from NAm 31) and exogenous NAD precursors demonstrate a clear link between intracellular NAD levels and TNF-α production. All sirtuin inhibitors tested in this study (and encompassing structurally distinct molecules such as NAm, sirtinol and cambinol) displayed similar pharmacological properties in vitro, inhibiting TNF-α synthesis at a post-transcriptional step (see Supplementary Fig. 3) with minimal effect on RANTES secretion and expression of cell surface markers induced upon stimulation (Figures 3 and 5). To further strengthen the central role of sirtuins in sensing intracellular NAD levels in this model, we have been able to demonstrate the ability of APO866 to both reduce intracellular NAD levels (see Figure 4) and to modulate the activity of SIRT1 in a cellular model, as described in Supplementary Figure 5. Briefly, APO866 treatment led to the accumulation of the acetylated form of p53, a well known target for SIRT1 46. The first strategy that was used to identify the sirtuin member involved in TNF-α synthesis was based on a lentiviral-mediated short hairpin RNA system to individually knockdown all seven members of the sirtuin family. Although expression of each target gene was moderately suppressed using this approach, it did not lead to a reproducible and significant reduction in TNF-α synthesis (not shown). We therefore developed an in vitro assay allowing us to evaluate the effect of each sirtuin member on TNF-α synthesis. SIRT6 was the only sirtuin member found to positively regulate TNF-α secretion when co-transfected with a TNF-α encoding plasmid. In keeping with our pharmacological observations, a mutant form of SIRT6 lacking enzymatic activity 47,37 was found ineffective in this assay (see Figure 6). The role of SIRT6 in regulating TNF-α production was further confirmed using inflammatory cells derived from SIRT6 KO mice (Figure 6D). Note however that an in depth analysis of the role of SIRT6 in vivo awaits the development of a mouse strain expressing a conditional allele of SIRT6, since SIRT6 KO mice display multiple phenotypic abnormalities including uncontrolled cell death and an altered immune system that may non-specifically affect an inflammatory response in vivo 37.
Similarly to other sirtuin members, SIRT6 expresses both an ADP-ribosyltransferase and protein deacetylase activity, although the physiological substrates for these enzymatic reactions remain to be determined 37. In addition, SIRT6 is predominantly nuclear and further experiments are therefore warranted using cells of the immune system to evaluate a possible cytoplasmic location of this protein compatible with a role in translational regulation. Based on the observations reported herein, we would suggest a role for SIRT6 in a NAD-dependent, post-translational modification of a cellular substrate involved in regulation of the translational efficiency of selected mRNAs.
The present studies may also contribute to a better understanding of the in vivo pharmacological properties of NAm. NAm represents a potentially valuable pharmacological agent due to its low cost, non-proprietary status and well established safety profile in humans. The present study raises however some concern about its efficacy in vivo as an anti-inflammatory compound. Indeed, NAm represents both a feedback inhibitor of NAD-consuming reactions and an endogenous NAD precursor. As such, NAm can both inhibit and promote the enzymatic activity of NAD-requiring enzymes depending on dosage and timing of administration. Thus, while high doses of NAm in vivo inhibit TNF-α secretion when injected 1 hour before a microbial stimulus, they could in fact increase an inflammatory response at a later stage, by replenishing the intracellular NAD pool on immune cells. Collectively, these studies highlight the possible “double-edged sword” type of response induced by NAm in vivo, with both beneficial and detrimental effects on the inflammatory response depending on dose and treatment duration.
In marked contrast, the possibility to selectively inhibit sirtuin activity in vivo without altering the intracellular NAD status may represent a novel and promising strategy for treating inflammatory disorders. APO866 has been shown to represent a potent NAmPRT inhibitor with limited toxicity in vivo 31. Acute administration of this compound in vivo significantly inhibits TNF-α secretion in response to LPS (see Supplementary Fig. 5 and Busso et al, submitted for publication). Similarly, chronic treatment with this compound has been recently shown to protect mice against a collagen-induced arthritis, confirming the possibility to inhibit an inflammatory response in vivo by modulating NAD metabolism (Busso et al, submitted for publication). Based on the evidence provided herein, sirtuin inhibitors may represent novel valuable drug candidates. Most inhibitors available to date appear to affect the enzymatic activity of multiple sirtuin members and further studies are therefore warranted to identify a specific SIRT6 inhibitor able to modulate an in vivo inflammatory response.
In conclusion, the present study provides a novel example of “chemomodulation” in a complex organism, a mechanism whereby small-molecule metabolites involved in “energy metabolism” act as molecular regulators of gene expression, and paves the way to novel anti-inflammatory strategies of potential clinical value.
METHODS
Mice and in vivo experiments
C57BL/6 mice, were purchased form Harlan (Nederland). All mice were bred in our pathogen-free facility and used at 6–9 weeks of age. All experiments were performed in compliance with the relevant laws and institutional guidelines and have been approved by the local committee from the Institut de Biologie et Médecine Moléculaires from the Université Libre de Bruxelles (Gosselies, Belgium). IL-10 knockout mice were purchased from Jackson Laboratories (Bar Harbor, Maine). PARP-1 knockout mice were kindly provided by Dr. G. de Murcia (Ecole Supérieure de Biotechnologie de Strasbourg, Illkirch, France). Mice were injected intravenously (i.v.) into the lateral tail vein or intraperitoneally (i.p.) with indicated doses of LPS from E. coli (serotype 0111:B4), NAm (both from Sigma, St Louis, MO), APO866 (previously identified as FK866 31 was synthesized and provided by Astellas Pharma GmbH, Munich, Germany) or Cambinol. Control animals were injected with the same volume of PBS or solvent. NAm, LPS, D-galactosamine (D-gal, Sigma), TNF-α (eBioscience, San Diego, CA) and the IL-10 antibodies cocktail (anti-IL-10R mAb 1B1.2, anti-IL-10 mAb JES5-2A5, produced in our own laboratory) were diluted in pyrogen-free saline and injected as described.
Purification of dendritic cells
Splenic dendritic cells were purified as follows. Mice were injected s.c. with 106 B16 melanoma cells engineered to stably produce murine Fms-like tyrosine kinase 3 48 and spleens were harvested 9 days later. Spleen cells were digested with collagenase type 3 (Worthington Biochemicals, New Jersey, USA), further dissociated in Ca2+-free medium, and separated into low and high density fraction on a Nycodenz gradient (Nycomed, Oslo, Norway). The low density fraction was separated according to CD11c expression by incubation with anti-CD11c–coupled microbeads followed by one passage over a MACS column (Miltenyi Biotec, Bergisch-Gladbach, Germany). The CD11c-positive cells were cultured in RPMI 1640 containing 2% Ultroser HY (Life Technologies, Paisley, Scotland), and were induced to mature during overnight culture in the presence of the indicated dose of CpG-ODN 1826 (Eurogentec, Belgium). Bone marrow-derived dendritic cells (BMDC) were generated as follows. Frozen bone marrow cells were plated at 2×105 cells/ml in medium supplemented with 10 ng/ml GM-CSF in 6-well plates in a volume of 6 ml. At days 3 and 6, 4 ml of supernatant was replaced by 4 ml of fresh medium containing GM-CSF. At day 9, cells were collected, centrifuged and resuspended in fresh medium in the presence of the indicated dose of CpG-ODN.
Antibodies and Cytofluorometric analysis
Purified DCs were analyzed by flow cytometry with a FACSort™ (Becton Dickinson). The cells were labeled with PE-coupled N418 (anti-CD11c), 3/23 (anti-CD40), 16–10A1 (anti-CD80), GL-1 (anti-CD86), or 14.4.4 (anti-MHC II (I-Ed), all purchased from BD Biosciences (Mountain View, CA). The cells were gated based on characteristic forward and light scatter to eliminate dead cells and debris from analysis.
Cells lines, plasmids
293T cells (ATCC) and B16 melanoma were maintained in Dulbecco’s Modified Eagle Medium containing 10% heat-inactivated FBS, 1 mM sodium pyruvate and 0,05 Mm 2-ME. THP-1 and BMDC cells were cultured in RPMI 1640 medium containing 10% FBS, 1 mM sodium pyruvate, L-glutamine 2 mM, essential amino acids and 0.05 mM 2-ME. RAW264.7 cells originally obtained from American Type Culture Collection (Rockville, MD) were maintained in DMEM medium with 5% FBS and all additives.
All constructs were generated using a standard PCR-based cloning strategy, and the integrity of individual clones was verified through DNA sequencing. To construct C-terminal FLAG-tagged SIRTs expression vectors, the coding regions were amplified from different DNA templates using primer sets containing Flag sequence and cloned into pCMV-SPORT6 (templates and primers sequences are available in the supplementary data/methods).
Point mutants of SIRT6-Flag vectors (G52A, H113Y and double mutants G52A/H113Y) and SIRT7-Flag vectors (H188Y) were generated by site-directed mutagenesis using a standard PCR-based protocol. DNA transfection to 293T cell lines was performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. Cells were harvested 48h after transfection and cells were plated on 96-well plates and cultured for an additional 6h in fresh medium. Culture supernatants were tested for TNF-α and IL-2 content by ELISA. Cells were lysed and FLAG-tagged SIRTs expression were analysed by Western blotting or total RNA was extracted, and TNF/IL2 mRNA levels were analyzed by quantitative RT-PCR.
Cells lines Culture Assays
RAW264.7 cells were stimulated with LPS 100 ng/ml and were treated with the indicated dose of NAm. The culture supernatants were tested for TNF-α content by ELISA. THP-1 cells were cultured overnight with increasing doses of APO866, and then stimulated with LPS 100 ng/ml for 2h. In the experiments aimed at manipulating the intracellular NAD levels, cells were co-incubated with the indicated dose of nicotinic acid (NA) and nicotinamide mononucleotide (NMN, both from Sigma). The culture supernatants were tested for TNF-α content by ELISA and intracellular NAD levels were measured by an enzymatic assay (see below).
NAD and cytokine determinations
Intracellular NAD concentrations were determined by an enzymatic cycling assay. Cells were lysed in NAD extraction buffer containing 100 mM Na2CO3 and 20 mM NaHCO3. Samples (20μl) were mixed with a cycling buffer (160μl) containing 125 mM Tris HCl pH 8.8, 1.25 mM phenazine ethosulfate, 0.625 mM thiazolyl blue tetrazolium bromide (MTT), 0.25 mg/ml alcohol dehydrogenase and 1.25 % BSA (all from Sigma). The cycling reaction was initiated by adding 20 μl of ethanol 6 M. The samples were incubated at 37°C and the optical density at 570 nm was measured after 5, 10, 15 and 20 minutes using an ELISA plate reader. Serial dilutions of NAD were used as a standard. NAD concentrations were normalized to protein concentrations, determined for each sample using the micro-BCA kit (Pierce). Cytokines levels in culture media and mouse serum were determined using ELISA kits and/or antibody pairs from eBioscience (TNF-α); R&D Systems (Minneapolis, MN) (RANTES) and BD Bioscience (IL-2, IL-10, and IL-12).
RNA extraction, Quantitative RT-PCR and Northern blotting
Total cellular RNA was isolated from cells with TRIZOL reagent (Invitrogen), according to the manufacturer’s instructions. RNA was reverse-transcribed by M-MLV reverse transcriptase, using oligo(dT) primers. Specific mRNAs were quantitated by SYBR Green (Eurogentec, Belgium) based RT-PCR using ABI Prism 5700 sequence detection system (Applied Biosystems). Primers used for quantitative RT-PCR are listed in the supplementary data. The results for TNF quantitative RT-PCR assays were normalized to those obtained for the corresponding GaPDH or RPL-32 mRNA, providing a relative quantification value. For Northern blot, the quality of the RNA samples was verified by agarose gel electrophoresis. A measure of 5 μg of total RNA was loaded on a 1.5% agarose gel and Northern blot was performed as described 49. Blots were hybridized with antisense [α-32P]UTP labelled riboprobes.
TNF mRNA stability
The effect of NAm on the stability of TNF mRNAs was evaluated using the transcriptional inhibitor, actinomycin D (5 μg/ml, Calbiochem). Actinomycin D was added 2 h after LPS stimulation with or without NAm. Cells were harvested at various time-points following actinomycin D treatment, and RNA was isolated. Quantitative RT-PCR was performed to determine the levels of TNF mRNAs. The house-keeping gene GAPDH was used as internal control.
Polysome analysis
RAW cells stimulated for 2h with 100 ng/ml LPS were washed and treated for 30 min with 15 mM NAm. Cells were washed with ice-cold PBS supplemented with cycloheximide (150 μg/ml) and resuspended in polysome extraction buffer (10 mM Tris pH 7.2, 140 mM KCl, 10 mM MgCl2, 100 U/ml RNasin, Heparin 0.5 μg/ml, 10 mM dithiothreitol, cycloheximide 150 μg/ml, 0.5% NP-40). Nuclei and the majority of mitochondria were sedimented by centrifugation for 10 min at 10,000g at 4°C. Extracts were separated over linear sucrose gradients (15%–50% w/w in 25 mM Tris-HCl (pH 7.5), 25 mM NaCl, 10 mM MgCl2, Heparin 0.5 μg/ml, cycloheximide 150 μg/ml, 10 mM dithiothreitol) by ultracentrifugation. (39,000 rpm for 2 hr at 4°C in a SW40 swing out rotor). Fractions of 0.5 ml were collected with continuous monitoring of absorbance at 254 nm. For RNA isolation the fractions were mixed in Trizol reagent (Life Technologies Ltd, Paisley, UK), and RNA were extracted according to the manufacturer’s protocol. The RNA from each fraction was electrophoresed through 1.5 % agarose gels and subjected to Northern blot analysis as described above.
Metabolic labeling
Cells were stimulated with LPS (100 ng/ml) for 1h, washed and incubated in cysteine- and methionine-free DMEM. 30 min later, the cells were incubated in DMEM supplemented with [35S]-labeled cysteine and methionine (Amersham Bioscience), brefeldin A (eBioscience) and 15 mM NAm for the indicated time periods, followed by immunoprecipitation using polyclonal rabbit anti mouse TNF-α (ImmunoSource, Zoersel, Belgium). Immunoprecipitated samples were separated by SDS-PAGE and the gels were dried and analyzed by autoradiography after 24h at −80°C
Western blotting
Cells were lysed in radioimmunoprecipitation lysis buffer (RIPA) containing protease inhibitors (Roche EDTA-free cocktail), centrifuged for 10 min at 10,000g and the insoluble debris were discarded. Samples (2 μg protein) were separated on 10% polyacrylamide gels and transferred to a 0.45 μm PVDF membrane (Millipore, Bedford MA). The membrane was blocked with TBS containing 5% BSA, 0.1% Tween-20. FLAG-tagged SIRTs were detected using anti-FLAG M2 mAb, while actin was detected using rabbit polyclonal antiserum to actin. Bound antibodies were revealed with horseradish peroxidase-conjugated anti-mouse or protein A by ECL reagent (Amersham Bioscience, Amersham, UK). All antibodies and secondary reagents were from Sigma.
Supplementary Material
Acknowledgments
The authors wish to thank Dr. G de Murcia for providing the PARP-1 KO mouse strain and Dr. V. Sartorelli for providing the pHan-SIRT1 vector. This work was supported by The Belgian Program in Interuniversity Poles of Attraction Initiated by the Belgian State, Prime Minister’s office, Science Policy Programming and by a Research Concerted Action of the Communauté française de Belgique and by APOXIS SA. F. V. G. and M.G. have been supported by Research Grants from the FNRS, Belgium. The scientific responsibility is assumed by the authors.
Footnotes
Competing financial interests: T. De Smedt is an Apoxis employee
References
- 1.Ziegler M. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem. 2000;267:1550–64. doi: 10.1046/j.1432-1327.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 2.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 3.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–74. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 4.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–21. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 7.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell Postprint. 2007 doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–62. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 10.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 11.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 12.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders BD, Zhao K, Slama JT, Marmorstein R. Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes. Mol Cell. 2007;25:463–72. doi: 10.1016/j.molcel.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rongvaux A, Andris F, Van Gool F, Leo O. Reconstructing eukaryotic NAD metabolism. Bioessays. 2003;25:683–90. doi: 10.1002/bies.10297. [DOI] [PubMed] [Google Scholar]
- 16.Yang T, Sauve AA. NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. Aaps J. 2006;8:E632–43. doi: 10.1208/aapsj080472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauve AA, Moir RD, Schramm VL, Willis IM. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol Cell. 2005;17:595–601. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 18.Rongvaux A, et al. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–34. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 19.Jia SH, et al. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–27. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye SQ, et al. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med. 2005;171:361–70. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- 21.Chen CF, et al. The protective effect of niacinamide on ischemia-reperfusion-induced liver injury. J Biomed Sci. 2001;8:446–52. doi: 10.1007/BF02256606. [DOI] [PubMed] [Google Scholar]
- 22.Fukuzawa M, et al. Inhibitory effect of nicotinamide on in vitro and in vivo production of tumor necrosis factor-alpha. Immunol Lett. 1997;59:7–11. doi: 10.1016/s0165-2478(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 23.Ungerstedt JS, Blomback M, Soderstrom T. Nicotinamide is a potent inhibitor of proinflammatory cytokines. Clin Exp Immunol. 2003;131:48–52. doi: 10.1046/j.1365-2249.2003.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuzzocrea S. Shock, inflammation and PARP. Pharmacol Res. 2005;52:72–82. doi: 10.1016/j.phrs.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–53. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver FJ, et al. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. Embo J. 1999;18:4446–54. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhnle S, Nicotera P, Wendel A, Leist M. Prevention of endotoxin-induced lethality, but not of liver apoptosis in poly(ADP-ribose) polymerase-deficient mice. Biochem Biophys Res Commun. 1999;263:433–8. doi: 10.1006/bbrc.1999.1393. [DOI] [PubMed] [Google Scholar]
- 28.Gerard C, et al. Interleukin 10 reduces the release of tumor necrosis factor and prevents lethality in experimental endotoxemia. J Exp Med. 1993;177:547–50. doi: 10.1084/jem.177.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rankin PW, Jacobson EL, Benjamin RC, Moss J, Jacobson MK. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. J Biol Chem. 1989;264:4312–7. [PubMed] [Google Scholar]
- 30.Bieganowski P, Brenner C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell. 2004;117:495–502. doi: 10.1016/s0092-8674(04)00416-7. [DOI] [PubMed] [Google Scholar]
- 31.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–42. [PubMed] [Google Scholar]
- 32.Khan JA, Tao X, Tong L. Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat Struct Mol Biol. 2006;13:582–8. doi: 10.1038/nsmb1105. [DOI] [PubMed] [Google Scholar]
- 33.Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–43. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- 34.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–77. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 35.Olaharski AJ, et al. The flavoring agent dihydrocoumarin reverses epigenetic silencing and inhibits sirtuin deacetylases. PLoS Genet. 2005;1:e77. doi: 10.1371/journal.pgen.0010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–18. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 38.Nathan C. Points of control in inflammation. Nature. 2002;420:846–52. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 39.Henson PM. Dampening inflammation. Nat Immunol. 2005;6:1179–81. doi: 10.1038/ni1205-1179. [DOI] [PubMed] [Google Scholar]
- 40.Feldmann M, Maini RN. Lasker Clinical Medical Research Award. TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat Med. 2003;9:1245–50. doi: 10.1038/nm939. [DOI] [PubMed] [Google Scholar]
- 41.Stoecklin G, Anderson P. Posttranscriptional mechanisms regulating the inflammatory response. Adv Immunol. 2006;89:1–37. doi: 10.1016/S0065-2776(05)89001-7. [DOI] [PubMed] [Google Scholar]
- 42.Szabo C. Nicotinamide: a jack of all trades (but master of none?) Intensive Care Med. 2003;29:863–6. doi: 10.1007/s00134-003-1737-8. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J. Are poly(ADP-ribosyl)ation by PARP-1 and deacetylation by Sir2 linked? Bioessays. 2003;25:808–14. doi: 10.1002/bies.10317. [DOI] [PubMed] [Google Scholar]
- 44.Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–12. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 46.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 47.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–20. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 48.Mach N, et al. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000;60:3239–46. [PubMed] [Google Scholar]
- 49.Kruys V, Thompson P, Beutler B. Extinction of the tumor necrosis factor locus, and of genes encoding the lipopolysaccharide signaling pathway. J Exp Med. 1993;177:1383–90. doi: 10.1084/jem.177.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.