Abstract
Immune responses to autoantigens are in part controlled by deletion of autoreactive cells through genetically regulated selection mechanisms. We have directly analyzed peripheral CD4+ proinsulin (PI) 76–90 (SLQPLALEGSLQKRG)-specific T cells using soluble fluorescent major histocompatibility complex class II tetramers. Subjects with type I diabetes and healthy controls with high levels of peripheral proinsulin-specific T cells were characterized by the presence of a disease-susceptible polymorphism in the insulin variable number of tandem repeats (INS-VNTR) gene. Conversely, subjects with a ‘protective' polymorphism in the INS-VNTR gene had nearly undetectable levels of proinsulin tetramer-positive T cells. These results strongly imply a direct relationship between genetic control of autoantigen expression and peripheral autoreactivity, in which proinsulin genotype restricts the quantity and quality of the potential T-cell response. Using a modified tetramer to isolate low-avidity proinsulin-specific T cells from subjects with the susceptible genotype, transcript arrays identified several induced pro-apoptotic genes in the control, but not diabetic subjects, likely representing a second peripheral mechanism for maintenance of tolerance to self antigens.
Keywords: autoimmune diabetes, insulin, proinsulin, INS-VNTR, genotype, tetramer
Introduction
Insulin is an important antigen in both murine1 and human2 autoimmune diabetes (type 1A diabetes (T1D)). T-cell reactivity to a major proinsulin epitope, PI(73–90), occurs as an early marker of disease progression in the pre-diabetic stage among genetically high-risk subjects.2, 3 Direct detection of these proinsulin-specific T cells was recently achieved using soluble, fluorescent human leukocyte antigen (HLA)-peptide tetramers containing the PI(73–90) epitope to bind to peripheral CD4 T cells proliferating in response to proinsulin peptide stimulation.4 In humans, expression levels of insulin are regulated in part by a direct transcriptional influence of a polymorphic insulin gene variable number of tandem repeats (INS-VNTR) genetic element associated with the proinsulin gene promoter region. Higher expression levels of proinsulin, due to the presence of the VNTR III haplotype in the thymus, are associated with a 3- to 4-fold relative protection from diabetes.5, 6, 7 Using proinsulin-specific HLA tetramers to detect CD4 T cells in subjects with either the VNTR III protective haplotype or the allelic VNTR I diabetes susceptibility haplotype, we found markedly lower frequencies of high-avidity T cells in VNTR III-positive subjects, consistent with a genetically controlled mechanism of immune tolerance based on polymorphic antigen expression levels involved in thymic selection of autoreactive lymphocytes.
In human autoimmune diseases many epitopes recognized by autoreactive T cells are of moderate or low avidity,8, 9, 10 which may allow these cells to escape deletional tolerance mechanisms during thymic development. Expression of proinsulin, similar to other tissue-specific protein antigens, occurs in thymic medullary epithelial cells influenced by the transcriptional regulator AIRE (autoimmune regulator),11 and NOD mice deficient in thymic insulin expression have accelerated development of T1D, consistent with escape of high-avidity autoreactive T cells and a prominent role for central tolerance in protection from insulin autoimmunity.12, 13 Transcriptional levels of proinsulin in the human thymus are under genetic control of the polymorphic INS promoter region, which carries a variable number of tandem repeats of a 14-bp consensus element, classified as VNTR I (short, 26–63 repeats), VNTR II (63–140 repeats) or VNTR III (long, 141–209 repeats). European ancestry populations express either VNTR I or III,14 and insulin transcription in thymic tissue is higher in VNTR I/III heterozygotes or VNTR III/III homozygotes when compared with VNTR I/I homozygotes.15, 16
Allelic variation at INS-VNTR confers differential susceptibility to T1D. Subjects with the VNTR III genotype have a lower frequency of anti-insulin autoantibodies,17 more preserved β-cell function18 and a lower concordance rate of diabetes among twins19 when compared with subjects having the VNTR I/I genotype. Overall, the INS-VNTR genotype is associated with an odds ratio for T1D susceptibility of 2.2,20 and the presence of the VNTR III genotype can moderate disease risk, even in populations that carry high-risk HLA genotypes.21
Results and discussion
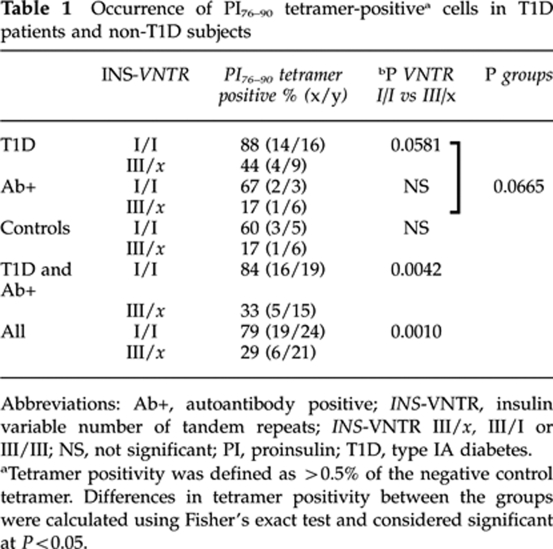
One postulated mechanism to account for the T1D protective effect of VNTR III is that higher thymic expression may lead to deletion of proinsulin-reactive T cells during development. To directly test this hypothesis, we analyzed peripheral blood lymphocytes for recognition of HLA class II-proinsulin complexes in subjects with VNTR I/I or VNTR III/x (where x=I or III) genotypes. CD4+CD25– peripheral T lymphocytes from 24 Caucasian VNTR I/I and 21 VNTR III/x subjects matched for HLA-DRB1*0401 were cultured with autologous antigen-presenting cells in the presence of PI(76–90),22 to allow for expansion of antigen-specific cells, and subsequently incubated with soluble DRB1*0401-PI(76–90) complexes conjugated to a phycoerythrin fluorochrome. Out of 45 HLA-DRB1*0401-positive subjects analyzed, 6 were homozygous for DRB1*0401, 12 had in addition DRB1*0301 and the remaining 27 had some other DRB1* allele. The PI 73–90 epitope is immunodominant in both humans and in humanized HLA-DR4/CD4 double-transgenic mice immunized with human proinsulin.23 Of VNTR I/I subjects, 79% (19/24) had detectable tetramer-positive T cells, compared with 29% (6/21) of the VNTR III/x subjects (P<0.0010; Table 1). As shown in Figure 1a, the difference in overall distribution of tetramer positivity was highly significant between groups (P<0.0004) and, with two exceptions, all the VNTR III/x subjects were at or near background levels of tetramer detection.
Table 1.
Occurrence of PI76–90 tetramer-positivea cells in T1D patients and non-T1D subjects
Figure 1.
Relationship between INS-VNTR genotype and detection of CD4 T cells, which bind PI76–90 tetramers. Subjects with VNTR class I alleles have high levels of peripheral blood T cells recognizing soluble DRB1*0401-PI76–90 tetramer compared with the subjects having the protective class III alleles (a). The same T-cell samples were analyzed using the PI76–90 9S modified tetramer; no difference between the two groups of subjects was detected, implying presence of low-avidity PI-reactive T cells in both (b). In addition, no difference in tetramer positivity between the two groups of subjects was found when tetramers specific for another T1D autoantigen, GAD555–567, or foreign antigen, HA306–318, were used, indicating proinsulin specificity of the VNTR association (c, d). ***P=0.0004, differences in the level of tetramer-reactive T cells were calculated using Mann–Whitney U test. Solid horizontal lines represent means. Dashed horizontal lines indicate 0.5% binding of negative control tetramer. $INS-VNTR III/x where x=I or III. Typing for HLA-DRB1*04 subtypes was performed using sequence-specific primers and probes in conjunction with real-time PCR.27 A total of 45 HLA-DRB1*0401-positive subjects were analyzed out of which 6 were homozygous for DRB1*0401, 12 had in addition DRB1*0301 and the remaining 27 had in addition some other DRB1* allele. T1D patients of INS-VNTR I/I as well as INS-VNTR I/III genotype were of comparable age (average 25.8±8.9 years vs 26.2±11.6 years, respectively) and had similar duration of the disease (3.4±1.9 years vs 5.5±1.5 years, respectively) as were autoantibody-positive subjects (36.3±10.4 vs 42±12.8) and healthy controls (40.4±14.5 vs 50.5±14.6). The VNTR genotype was assigned based on the genotype at the −23 A/T single-nucleotide polymorphism (SNP) at the INS promoter.14, 28 Peripheral blood mononuclear cells were isolated from heparinized blood and CD4+CD25− T cells enriched in two steps. In the first step CD4+ T cells were separated from all non-CD4 cells using CD4+ T cell isolation kit (Miltenyi Biotec, Auburn, CA, USA). In the second step CD25+ T cells were depleted using anti-CD25-coated micro beads. CD4+CD25− T cells were stimulated in vitro for 12–14 days in 48-well tissue culture plates with autologous adherent non-CD4+ cells prepulsed with either PI peptides, hemagglutinin (HA) peptide (10 μg ml−1) or without peptides. At days 6, 8 and 10, human recombinant IL-2 (10 IU ml−1, Chiron Corporation, Emeryville, CA, USA) was added to the cultures.
To improve the detection of low-avidity CD4+ self-reactive T cells in T1D, HLA class II tetramer reagents have been developed with agonist peptides containing amino acid substitutions, which improve binding of the tetramer to the antigen-specific regulatory T cells. Yang et al.4 recently described substitution of lysine to serine at peptide position p9 within the PI(76–90) peptide to stabilize binding to DRB1*0401 in soluble tetramers, resulting in enhanced detection of a proinsulin-positive CD4 T-cell population. When the same T-cell samples were analyzed using this ‘9S' tetramer, higher levels of tetramer-positive cells were detected, in both VNTR I/I and VNTR III/x subjects (Figure 1b), and there was no difference between groups. Thus, low-avidity T cells are present in both populations, independent of genotype, whereas the presence of CD4 T cells with higher avidity, capable of recognizing the native proinsulin peptide, is under INS genetic control.
Parallel tetramer assays were performed using either another T1D autoantigen, GAD65,24 or an antigen from the hemagglutinin of influenza virus.25 As shown in Figures 1c and d, no differences in tetramer positivity for these antigens were found among subjects classified by INS-VNTR genotype, indicating specificity for the proinsulin immune response.
The lymphocyte samples for these assays come from a mixture of T1D subjects, autoantibody-positive, nondiabetic ‘at-risk' subjects and non-autoimmune controls. INS-VNTR genetic control of T-cell detection using proinsulin tetramers was similar in all three groups, as summarized in Table 1. In other words, the correlation between INS-VNTR and the presence of peripheral proinsulin tetramer-binding T cells was found in all groups based on genotype alone, irrespective of disease status. Overall, combining VNTR I/I and VNTR III/x subjects, there was also a trend for higher tetramer positivity in the T1D group when compared with the non-diabetic groups, consistent with the presence of pre-existing in vivo expansion of autoreactive cells in the diabetic population (Supplementary Figure S1). Alternatively, the in vitro culture conditions of the assay may allow for preferential expansion of these autoreactive populations.
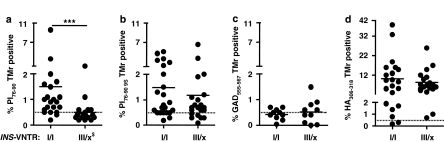
As tetramer-binding studies with the modified ‘9S' peptide allowed us to detect low-avidity PI(76–90)-binding T cells, we used HLA-DRB1*0401–9S tetramers for sorting of the proinsulin-specific CD4 T cells by flow cytometry from INS-VNTR I/I subjects. The recovery of cells was limited because of the low frequency of antigen-specific T cells, but in four cases (two controls and two T1D) we purified sufficient tetramer-positive cells for transcript array analysis. Figure 2 shows the flow cytometry profiles used for cell sorting: parallel tetramer sorting of the influenza hemagglutinin-positive T cells and parallel transcript arrays from the same samples used as controls.
Figure 2.
Flow cytometry profiles of CD4 T cells of T1D and control subjects used for RNA isolation and subsequent transcript arrays, showing tetramer binding with the PI 76–90 9S modified tetramer. Parallel HA306–319 tetramer-positive binding (foreign control antigen) and an irrelevant control tetramer, outer surface protein A (OspA) 163–175, are also shown. The numbers in the upper right quadrant represent percentages of the tetramer-positive cells in the CD4+CD25− T-cell subset after 14 days of culture. Major histocompatibility complex (MHC) class II tetramers were produced by constructing the expression vectors and generating soluble biotinylated HLA-DRB1*0401 molecules, as described elsewhere.25 HLA molecules were loaded with peptides by detergent-facilitated exchange and dialyzed to remove the unbound fractions. Tetramers were obtained by incubation with phycoerythrin-labeled streptavidin. PI76–90 immunodominant epitope (SLQPLALEGSLQKRG),22 a PI76–90 9S substitute peptide (SLQPLALEGSLQSRG), in which substitution of K88 to S in peptide position 9 (9P) enhanced its binding affinity and agonistic activity,4, 22 and two irrelevant control peptides, an influenza A hemagglutinin, HA306–318 (KYVKQNTLKLA)25 as a positive control and Borrelia burgdorferi OspA163–175 (KSYVLEGTLTAEK) as a negative control were each loaded into HLA-DR4 molecules assembled into tetramers. After 12–14 days of culture, T cells were stained with tetramers for flow cytometry analysis as described.29
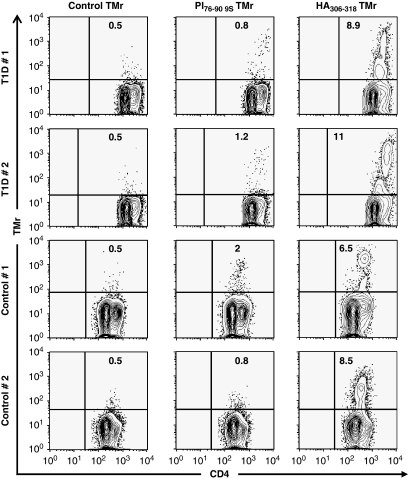
Of the 84 transcripts selected for cytokines and candidate immunologic markers, 6 clearly distinguished between the proinsulin tetramer-positive cells of the normal control subjects and T1D subjects (Figure 3). Four of these markers are closely related, representing pro-apoptotic transcripts associated with peripheral tolerance mediated by activation-induced cell death: FASLG (Fas ligand (TNF superfamily, member 6)), interleukin-2 (IL-2), tumor necrosis factor and TNFSF11 (TNF (ligand) superfamily, member 11). These markers were observed in both normal and T1D subjects responding to a foreign antigen, but only in the normal subjects responding to proinsulin. The failure to upregulate these transcripts in the T1D T cells activated by proinsulin may indicate a functional resistance to this pathway of peripheral tolerance.
Figure 3.
Partial expression profile of PI-specific, tetramer-sorted cells. Upregulated transcripts for FASLG, tumor necrosis factor (TNF), TNFSF11 and IL-2, which are commonly associated with peripheral tolerance, distinguish T1D subjects (a) from control subjects (b). Similar associations were not observed for control HA306–318-specific cells (c, d). Relative transcript expression was calculated as fold up- or downregulation in peptide-stimulated cultures relative to autologous non-stimulated cultures. Gene expression profiling was performed on tetramer-positive cells isolated by flow cytometry using a BD FACSVantage cell sorter (BD Biosciences, San Jose, CA, USA). From the sorted tetramer-positive cells, total RNA was extracted using a MagMAX Total RNA Isolation Kit (Applied Biosystems, Inc., Foster City, CA, USA). First-strand complementary DNA was synthesized with the RT2 first-strand kit (SABiosciences, Corp., Frederick, MD, USA) and samples plated on RT2 profiler PCR arrays for common human cytokines (SABiosciences), followed by the amplification by real-time PCR on a StepOne Plus or an ABI7000 sequence detection system (Applied Biosystems, Inc.). Amplification was carried out in a total volume of 25 μl for 40 cycles of 15 s at 95 °C and 1 min at 60 °C each. Expression of each gene was determined by normalizing the expression to five housekeeping genes. Relative gene expression for each subject was calculated by comparing peptide-stimulated samples to an unstimulated autologous sample to calculate a fold-change value.
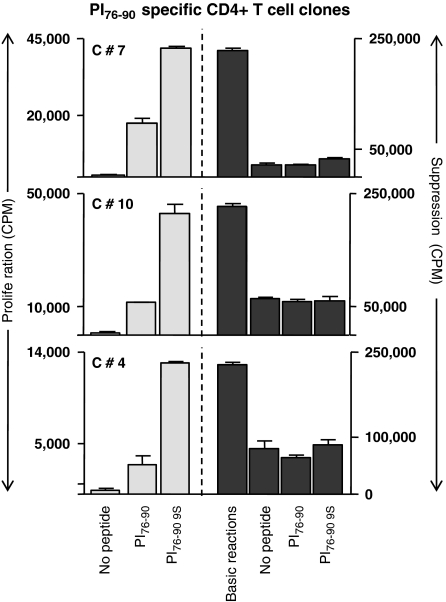
In addition, there were two marker transcripts, IL-10 and IL-7, that were upregulated in the T1D subjects but not in the normal controls responding to proinsulin. The presence of IL-10 transcripts in the T cells from T1D subjects responding to proinsulin is of interest because this is reminiscent of previous studies in which peripheral blood stimulation with proinsulin peptides was predominantly associated with IL-10 release.22, 26 Figure 4 illustrates mixed T-cell proliferation assays, in which three T-cell clones purified from the sorted ‘9S' proinsulin tetramer-positive population were added to autologous responder peripheral blood mononuclear cells. In each case, suppression of the bystander T cells, irrespective of additional antigen, was observed. It is possible that IL-10 and IL-7 are associated with cell survival and anergy in this low-avidity autoreactive population, contributing to their relative resistance to apoptotic tolerance mechanisms.
Figure 4.
PI-specific T cells exert bystander suppression. Shown are three representative clones that proliferated specifically to native and 9S PI peptides (10 μg ml−1). Addition to responder T cells suppressed proliferation, irrespective of antigen exposure. The s.d. bars represent average of three replicate wells. PI76–90-specific T-cell clones were isolated from CD4+ T cells of a diabetic subject, as described previously.22 They were further characterized in a 3H thymidine incorporation assay to confirm their PI specificity using both wild-type PI76–90 peptide and PI76–90 S9 substitute peptide.2, 26 For suppression assays, 20 000 clone T cells per well were activated with irradiated DRB1*0401 PBMCs (60 000 per well) as antigen-presenting cells (APCs) pulsed with either of the PI peptides or alone. Activated cells were then added to a responder cell culture consisting of CD25+-depleted CD4+ T cells of a DRB1*0401 donor activated by 5 μg ml−1 soluble anti-CD3 (UCHT1; BD Biosciences Pharmingen, San Diego, CA, USA), 5 μg ml−1 soluble anti-CD28 (CD28.2; Pharmingen) and irradiated autologous APCs (non-CD4 cells). The ability of each T-cell clone to suppress proliferation of the responder cells was determined by 3H thymidine incorporation during the final 16 h of the 5-day assay.
These studies illustrate simultaneous and dual mechanisms of antigen-specific tolerance to diabetes-associated autoantigens. The first, most fundamental level of control is a genetically determined threshold for central tolerance through deletion of high-avidity autoreactive T cells. The relationship between specific INS-VNTR alleles and levels of proinsulin tetramer-binding CD4 T cells, corresponding directly with the previously observed levels of thymic antigen expression, clearly identifies this developmental checkpoint as a fundamental determinant of T1D susceptibility. This is the first direct demonstration in human disease of this important central control of antigen-specific autoimmunity, but it is functionally incomplete. Using the modified ‘9S' agonist tetramer, we also identified low-avidity autoreactive T cells in the peripheral circulation, which were survivors that had escaped central thymic deletion. In our control, non-diabetic subjects, transcript profiling of these cells suggested a second peripheral tolerance mechanism characterized by upregulation of pro-apoptotic genes. Thus, both central and peripheral deletional checkpoints are invoked to control T-cell autoreactivity to proinsulin, manifest through genetic control of antigen expression and the avidity threshold for antigen-specific response.
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by grants from the Juvenile Diabetes Research Foundation and the Washington State Life Sciences Discovery Fund. We thank Drs William Kwok and Eddie James for production of tetramers and Drs Junbao Yang, John Gebe and Carla Greenbaum for helpful discussions.
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
References
- Nakayama M, Abiru N, Moriyama H, Babaya N, Liu E, Miao D, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinovic-Bello I, Boehm BO, Ziegler AG. Predominantly recognized proinsulin T helper cell epitopes in individuals with and without islet cell autoimmunity. J Autoimmun. 2002;18:55–66. doi: 10.1006/jaut.2001.0566. [DOI] [PubMed] [Google Scholar]
- Durinovic-Bello I, Hummel M, Ziegler AG. Cellular immune response to diverse islet cell antigens in IDDM. Diabetes. 1996;45:795–800. doi: 10.2337/diab.45.6.795. [DOI] [PubMed] [Google Scholar]
- Yang J, Danke N, Roti M, Huston L, Greenbaum C, Pihoker C, et al. CD4+ T cells from type 1 diabetic and healthy subjects exhibit different thresholds of activation to a naturally processed proinsulin epitope. J Autoimmun. 2008;31:30–41. doi: 10.1016/j.jaut.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- Sabater L, Ferrer-Francesch X, Sospedra M, Caro P, Juan M, Pujol-Borrell R. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J Autoimmun. 2005;25:312–318. doi: 10.1016/j.jaut.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Undlien DE, Bennett ST, Todd JA, Akselsen HE, Ikaheimo I, Reijonen H, et al. Insulin gene region-encoded susceptibility to IDDM maps upstream of the insulin gene. Diabetes. 1995;44:620–625. doi: 10.2337/diab.44.6.620. [DOI] [PubMed] [Google Scholar]
- Deng L, Mariuzza RA. Recognition of self-peptide-MHC complexes by autoimmune T-cell receptors. Trends Biochem Sci. 2007;32:500–508. doi: 10.1016/j.tibs.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijonen H, Elliott JF, van Endert P, Nepom G. Differential presentation of glutamic acid decarboxylase 65 (GAD65) T cell epitopes among HLA-DRB1*0401-positive individuals. J Immunol. 1999;163:1674–1681. [PubMed] [Google Scholar]
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Brimnes MK, Jensen T, Jorgensen TN, Michelsen BK, Troelsen J, Werdelin O. Low expression of insulin in the thymus of non-obese diabetic mice. J Autoimmun. 2002;19:203–213. doi: 10.1006/jaut.2002.0616. [DOI] [PubMed] [Google Scholar]
- Thebault-Baumont K, Dubois-Laforgue D, Krief P, Briand JP, Halbout P, Vallon-Geoffroy K, et al. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J Clin Invest. 2003;111:851–857. doi: 10.1172/JCI16584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt BJ, Payne F, Lowe CE, Hermann R, Healy B, Harold D, et al. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes. 2004;53:1884–1889. doi: 10.2337/diabetes.53.7.1884. [DOI] [PubMed] [Google Scholar]
- Pugliese A, Zeller M, Fernandez A, Jr, Zalcberg LJ, Bartlett RJ, Ricordi C, et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- Walter M, Albert E, Conrad M, Keller E, Hummel M, Ferber K, et al. IDDM2/insulin VNTR modifies risk conferred by IDDM1/HLA for development of type 1 diabetes and associated autoimmunity. Diabetologia. 2003;46:712–720. doi: 10.1007/s00125-003-1082-z. [DOI] [PubMed] [Google Scholar]
- Nielsen LB, Mortensen HB, Chiarelli F, Holl R, Swift P, de Beaufort C, et al. Impact of IDDM2 on disease pathogenesis and progression in children with newly diagnosed type 1 diabetes: reduced insulin antibody titres and preserved beta cell function. Diabetologia. 2006;49:71–74. doi: 10.1007/s00125-005-0042-1. [DOI] [PubMed] [Google Scholar]
- Hermann R, Laine AP, Veijola R, Vahlberg T, Simell S, Lahde J, et al. The effect of HLA class II, insulin and CTLA4 gene regions on the development of humoral beta cell autoimmunity. Diabetologia. 2005;48:1766–1775. doi: 10.1007/s00125-005-1844-x. [DOI] [PubMed] [Google Scholar]
- Concannon P, Chen WM, Julier C, Morahan G, Akolkar B, Erlich HA, et al. Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the type 1 Diabetes Genetics Consortium. Diabetes. 2009;58:1018–1022. doi: 10.2337/db08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzo C, Contu D, Cordell HJ, Lampis R, Congia M, Marrosu MG, et al. Heterogeneity in the magnitude of the insulin gene effect on HLA risk in type 1 diabetes. Diabetes. 2004;53:3286–3291. doi: 10.2337/diabetes.53.12.3286. [DOI] [PubMed] [Google Scholar]
- Durinovic-Bello I, Rosinger S, Olson JA, Congia M, Ahmad RC, Rickert M, et al. DRB1*0401-restricted human T cell clone specific for the major proinsulin73–90 epitope expresses a down-regulatory T helper 2 phenotype. Proc Natl Acad Sci USA. 2006;103:11683–11688. doi: 10.1073/pnas.0603682103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congia M, Patel S, Cope AP, De Virgiliis S, Sonderstrup G. T cell epitopes of insulin defined in HLA-DR4 transgenic mice are derived from preproinsulin and proinsulin. Proc Natl Acad Sci USA. 1998;95:3833–3838. doi: 10.1073/pnas.95.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijonen H, Novak EJ, Kochik S, Heninger A, Liu AW, Kwok WW, et al. Detection of GAD65-specific T-cells by major histocompatibility complex class II tetramers in type 1 diabetic patients and at-risk subjects. Diabetes. 2002;51:1375–1382. doi: 10.2337/diabetes.51.5.1375. [DOI] [PubMed] [Google Scholar]
- Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104:R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinovic-Bello I, Schlosser M, Riedl M, Maisel N, Rosinger S, Kalbacher H, et al. Pro- and anti-inflammatory cytokine production by autoimmune T cells against preproinsulin in HLA-DRB1*04, DQ8 type 1 diabetes. Diabetologia. 2004;47:439–450. doi: 10.1007/s00125-003-1315-1. [DOI] [PubMed] [Google Scholar]
- Gersuk VH, Nepom GT. A real-time PCR approach for rapid high resolution subtyping of HLA-DRB1*04. J Immunol Methods. 2006;317:64–70. doi: 10.1016/j.jim.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinovic-Bello I, Jelinek E, Schlosser M, Eiermann T, Boehm BO, Karges W, et al. Class III alleles at the insulin VNTR polymorphism are associated with regulatory T-cell responses to proinsulin epitopes in HLA-DR4, DQ8 individuals. Diabetes. 2005;54 Suppl 2:S18–S24. doi: 10.2337/diabetes.54.suppl_2.s18. [DOI] [PubMed] [Google Scholar]
- James EA, LaFond R, Durinovic-Bello I, Kwok W. Visualizing antigen specific CD4+ T cells using MHC class II tetramers. J Vis Exp. 2009. p. 25. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.