Abstract
Recent genetic studies have led to identification of numerous loci that are associated with susceptibility to autoimmune diseases. The strategy of using information from these studies has facilitated the identification of novel juvenile idiopathic arthritis (JIA) susceptibility loci, specifically, PTPN22 and IL2RA. Several novel autoimmune susceptibility loci have recently been identified, and we hypothesise that single-nucleotide polymorphisms (SNPs) within these genes may also be JIA susceptibility loci. Five SNPs within the genes AFF3, IL2/IL21, IL7R, CTLA4 and CD226, previously associated with multiple autoimmune diseases were genotyped, in a large data set of Caucasian JIA patients and controls, and tested for association with JIA. We identified two susceptibility loci for JIA, AFF3 and the IL2/IL21 region and additional weak evidence supporting an association with the CTLA4 and IL7R genes, which warrant further investigation. All results require validation in independent JIA data sets. Further characterisation of the specific causal variants will be required before functional studies can be performed.
Keywords: autoimmune, juvenile idiopathic arthritis, CTLA4, AFF3, IL2
Introduction
Autoimmune diseases are caused by dysregulation of the immune system leading to an immune response to self-tissue. Autoimmune diseases are complex genetic diseases and in the last few years great progress has been made in the search for susceptibility loci.1, 2 As more confirmed autoimmune disease susceptibility loci are identified, an interesting story is emerging in that many of these loci predispose to more than one autoimmune disease. This confirms the hypothesis that shared alleles contribute to a spectrum of diseases and suggests that common immunological pathways are involved in susceptibility to these phenotypically distinct diseases.3
Juvenile idiopathic arthritis (JIA) is another complex genetic autoimmune disease characterised by chronic inflammatory disease in children. It is a group of heterogeneous disorders but encompasses all forms of arthritis of unknown aetiology that starts before the age of 16 and which persists for at least 6 weeks.4 The strategy of using information from autoimmune disease genome-wide association studies or candidate gene studies have facilitated the search for novel JIA susceptibility loci. Indeed two recently identified confirmed JIA susceptibility loci, PTPN225 and IL2RA6 are putative autoimmune susceptibility genes as they also show association with rheumatoid arthritis (RA),5, 7 type I diabetes (T1D)7, 8 and Graves' disease.9, 10 In addition, using a strategy of examining confirmed RA susceptibility loci in JIA, we have recently reported evidence for association of two further loci (TRAF1/C5 and STAT4) with JIA susceptibility.11
Several novel putative autoimmune susceptibility loci have recently been identified with association with multiple autoimmune diseases. These include the IL2/21 region on chromosome 4q2312, 13 and the genes encoding IL7R,14, 15, 16 CTLA4,17 AFF314 and CD226.18 We hypothesise that these genes may also confer susceptibility to JIA and, therefore, the aim of this study was to determine whether single-nucleotide polymorphisms (SNPs) within these genes are also associated with susceptibility to JIA.
Results and discussion
In this study, using a strategy of examining previously associated autoimmune loci in JIA, we have identified association of two loci with JIA susceptibility (Table 1). First, we show association of a SNP (rs1160542) in the 5′ region of the AFF3 gene (Table 1), a gene that is preferentially expressed in lymphoid cells and has a potential regulatory role in lymphoid development.19 This SNP has been associated with RA20 and a perfect proxy SNP (r2=1), rs9653442 has been associated with T1D14 with similar odds ratio and allele frequencies to that observed in this study of JIA.
Table 1.
Association analysis results for those SNPs associated with multiple autoimmune diseases in a cohort of patients with JIA
| Marker | Chr | Genea | HWE controls | Major allele/minor allele (1/2) | MAF cases | MAF controls | Genotype frequency cases (%)b | Genotype frequency controls (%)c | Trend test P-valued | Allelic OR (95% CI) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 12 | 11 | 22 | 12 | 11 | |||||||||
| rs1160542 | 2 | AFF3 | 0.32 | A/G | 0.5 | 0.45 | 217 (23.7) | 483 (52.8) | 215 (23.5) | 574 (19.3) | 1493 (50.3) | 900 (30.3) | 2.05 × 10−5 | 1.25 (1.13–1.39) |
| rs3087243 | 2 | CTLA4 | 0.74 | G/A | 0.43 | 0.46 | 180 (19.7) | 428 (46.8) | 306 (33.5) | 634 (20.8) | 1523 (50.0) | 892 (29.3) | 0.05 | 0.9 (0.81–1.0) |
| rs6822844 | 4 | IL2/IL21 | 0.19 | G/T | 0.15 | 0.18 | 22 (2.3) | 232 (24.8) | 683 (72.9) | 125 (3.6) | 1003 (29.0) | 2326 (67.3) | 0.0006 | 0.78 (0.67–0.9) |
| rs6897932 | 5 | IL7R | 0.06 | C/T | 0.27 | 0.29 | 62 (6.6) | 377 (40.0) | 504 (53.4) | 267 (7.6) | 1482 (42.3) | 1756 (50.1) | 0.06 | 0.9 (0.8–1.01) |
| rs763361 | 18 | CD226 | 0.84 | C/T | 0.48 | 0.46 | 222 (23.5) | 464 (49.2) | 257 (27.3) | 745 (21.2) | 1750 (49.9) | 1012 (28.9) | 0.13 | 1.08 (0.98–1.2) |
Abbreviations: Chr, chromosome; HWE, P-value statistic for Hardy–Weinberg equilibrium test; JIA, juvenile idiopathic arthritis; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
A Bonferroni correction of five was applied to correct for the number of loci studied, resulting in a P-value threshold of 0.01 for claims of significance.
Genotyping was performed using the Sequenom iPLEX platform. A 90% sample quality control rate and 90% SNP genotyping success rate was imposed on the analysis.
The gene name refers to the nearest gene in the region although SNPs are not necessarily intra-genic.
UK Caucasian JIA patients (n=1054) from three sources. The British Society for Paediatric and Adolescent Rheumatology (BSPAR) National Repository of JIA (n=654), a cohort of UK Caucasian patients with long-standing JIA (n=201), described previously29 and a third cohort collected as part of the Childhood Arthritis prospective Study (CAPS), a prospective inception cohort study of JIA cases from five centres across United Kingdom (n=199).30
Healthy Caucasian control DNA samples were available from five centres in the United Kingdom as described previously31: Manchester, 924 controls (including 228 in 1958 birth cohort controls); Sheffield, 995 controls; Leeds 532 controls; Aberdeen 862 controls; Oxford 536 controls, total control sample size=3531.
Genotype and allele frequencies were compared between cases with JIA and controls using the Cochrane–Armitage trend test implemented in PLINK32 and allelic odds ratios (ORs) and their 95% confidence intervals (CIs) calculated.
Second, we found strong association of a SNP (rs6822844) mapping within the IL2–IL21 locus on chromosome 4q27, which has previously been associated with RA, T1D and coeliac disease (Table 1).12, 13 The SNP lies approximately 24 kb 5′ of the IL21 gene. The SNP lies within a block of high linkage disequilibrium, which contains four genes, KIAA1109, TENR, IL2 and IL21. As for the other diseases, it is the common allele, which predisposes to JIA. This association confirms a recently published association of the IL2_IL2 region with JIA susceptibility.21 We performed a meta-analysis of the two studies, which yielded highly significant evidence for association (odds ratio 0.77 95% confidence interval 0.69–0.87, P=1 × 10−5) with no evidence for heterogeneity between the two cohorts (P=0.81). This finding is interesting in light of the previous confirmed association of the IL2RA gene with JIA6 and may suggest that the IL2 pathway is particularly important in JIA susceptibility.
We found a weak trend toward association of a SNP in the IL7R gene with JIA (Table 1), in line with the previous association of this SNP with RA, T1D14 and multiple sclerosis15, 16 the common allele of the SNP was increased in cases compared with controls, although this did not achieve statistical significance. However, this study was under-powered with only 18% power to detect an effect (Supplementary Table 2). Therefore, additional independent studies and meta-analyses of this SNP will be required to confirm it as associated with JIA susceptibility. The SNP is a non-synonymous SNP within exon 6 of IL7R and has a functional effect on gene expression, resulting in altered ratios of soluble and membrane-bound isoforms of the protein.15
SNPs within the CTLA4 gene, previously associated with T1D and autoimmune thyroid disease17 have previously been examined in JIA with conflicting results.22, 23 This may reflect true genetic heterogeneity at this locus or may be due to the modest sample sizes used in previous investigations. We found a weak association of the CTLA4 CT60 SNP (rs3087243) with UK JIA cases (Table 1), although this study only had 53% power to detect an effect (Supplementary Table 2). However, no evidence for association of this SNP with JIA was detected in a recent large study of US JIA families and controls.23 We used the Cochran–Mantel–Haenszel test to perform a meta-analysis combining data from this study and the Prahalad study; this yielded weak but statistically significant evidence for association (odds ratio 0.92 95% confidence interval 0.84–1.0, P=0.05) with no evidence for heterogeneity (by Breslow–Day test) between the two cohorts (P=0.44). Further analysis of this SNP in independent data sets followed by meta-analysis will be essential to robustly determine whether CTLA4 represents a JIA susceptibility locus. It is obviously a good candidate as an autoimmune susceptibility locus because of its role as a negative regulator of T-cell activation.17 Furthermore, the CT60 SNP is found within the 3′ untranslated region, in which the G allele is associated with susceptibility to several autoimmune diseases and also has a functional effect of lower mRNA levels of the soluble CTLA4 isoform.17
Finally, a non-synonymous SNP, rs763361, in exon 7 of the CD226 gene has recently been associated with multiple autoimmune diseases including T1D, multiple sclerosis and possibly autoimmune thyroid disease and RA.18 In our total JIA analysis, we found no significant association of the SNP with JIA (Table 1). However, we only had 24% power to detect an effect (Supplementary Table 2).
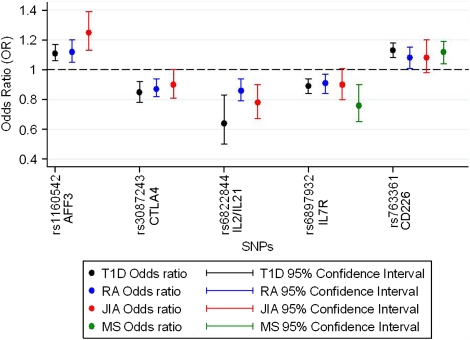
Figure 1 shows a comparison between the association analysis results in T1D, RA, multiple sclerosis and JIA. For all the SNPs tested, the same allele was associated with JIA as was associated with the other autoimmune diseases and effect sizes are similar. Hence, the failure to confirm association with CTLA4 and IL7R at the corrected threshold could be due to a lack of statistical power (53 and 18%, respectively) (Supplementary Table 2). It has not always been the case for the overlapping autoimmune disease susceptibility loci, that the same allele is associated. For example in PTPN22, the minor allele of the R620W SNP is associated with greater risk of developing RA, JIA, T1D and SLE but is protective for Crohn's disease.24, 25 There is also emerging data suggesting that one of the associated SNPs at the IL2RA locus confers differing risk and protective effects for T1D and multiple sclerosis.26, 27
Figure 1.
Plot of odds ratios for minor allele for SNPs previously associated with autoimmune disease, comparison with JIA. Plots of odds ratios and 95% confidence intervals for the association analysis of all SNPs, results in T1D (black dots and lines), in RA (blue dots and lines), in JIA (red dots and lines) and in MS (green dots and lines). References12, 14, 16, 17, 18 and Barton (2009) submitted.
JIA is a phenotypically heterogeneous disease and can be classified into more clinically homogeneous diseases using the ILAR classification criteria (Supplementary Table 1).28 However, comparing each of the ILAR subtypes separately against controls would result in a large number of hypothesis tests. Therefore, we first examined whether there was evidence of a difference in allele frequencies between the seven ILAR subtypes. Differences between subtypes were assessed using χ2 tests on the 7 × 2 tables. Only when a difference was found (P<0.05) were separate odds ratios and 95% confidence intervals calculated for the subgroups. In all cases, this was not significant (P>0.05) (data not shown). Therefore, further stratification by ILAR subtype was not performed. Larger sample sizes will be required to fully examine subgroup differences.
In conclusion, adopting the strategy of targeting loci with previous evidence for association in multiple autoimmune diseases has identified two novel JIA loci, AFF3 and the IL2/IL21 locus.
Conflict of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by the Arthritis Research Campaign: arc grant reference no: 17552. We thank David Strachan for facilitating access to the 1958 birth cohort. We acknowledge use of genotype data from the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02.
Appendix
Childhood Arthritis Prospective Study (CAPS): Arc Epidemiology Unit, University of Manchester—Kimme Hyrich, Mark Lay, Sham Lal, Paul Gilbert, Peter Ward; Alderhey—Eileen Baildam, Carol Lydon, Lynsey Brown; Glasgow—Joyce Davidson, Janet Gardner-Medwin, Vicki Price, Jane Sim, Maureen Todd; Great Ormond Street—Lucy Wedderburn, Alexandra Meijer, Julie Jones; Newcastle—Helen Foster, Mark Friswell, Michael Eltringham; Manchester—Alice Chieng, Joanne Buckley; Other—Tauny Southwood
UKRAG Consortium: University of Manchester1: Stephen Eyre, Anne Hinks, Laura J Gibbons, John Bowes, Edward Flynn, Paul Martin, Xiayi Ke, Rachelle Donn, Wendy Thomson, Anne Barton, Jane Worthington
University of Leeds2: YEAR Consortium†, Stephen Martin, James I Robinson, Ann W Morgan, Paul Emery
University of Sheffield3: Anthony G Wilson
University of London4: Sophia Steer
University of Aberdeen5: Lynne Hocking, David M Reid
University of Oxford6: Pille Harrison, Paul Wordsworth
1arc-Epidemiology Unit, Stopford Building, The University of Manchester, Manchester, UK
2Leeds Institute of Molecular Medicine, Section of Musculoskeletal Disease, University of Leeds, Leeds LS9 7TF, UK
3School of Medicine & Biomedical Sciences, The University of Sheffield, Sheffield S10 2JF, UK
4Clinical and Academic Rheumatology, Kings College Hospital NHS Foundation Trust, Denmark Hill, London SE5 9RS, UK
5Bone Research Group, Department of Medicine & Therapeutics, University of Aberdeen, Aberdeen AB25 2ZD, UK
6University of Oxford Institute of Musculoskeletal Sciences, Botnar Research Centre, Oxford OX3 7LD, UK
YEAR Consortium: Management Team—Professor Paul Emery1, Professor Philip Conaghan1, Dr Mark Quinn2, Dr Ann W Morgan1, Dr Anne-Maree Keenan1, Dr Elizabeth Hensor1, Julie Kitcheman1. Consultants—Dr Andrew Gough3, Dr Michael Green2,3, Dr Richard Reece4, Dr Lesley Hordon5, Dr Philip Helliwell16, Dr Richard Melsom6, Dr Sheelagh Doherty7, Dr Ade Adebajo8, Dr Andrew Harvey9, Dr Steve Jarrett9, Dr Gareth Huson1, Dr Amanda Isdale2, Dr Mike Martin1, Dr Zunaid Karim9, Prof Dennis McGonag1e10, Dr Colin Pease1, Dr Sally Cox1. SpRs—Dr Victoria Bejarano1, Dr Jackie Nam1. Nurses—Claire Brown1, Christine Thomas1, David Pickles1, Alison Hammond1, Beverley Neville3, Alan Fairclough4, Caroline Nunns4, Anne Gill2, Julie Green2, Belinda Rhys-Evans1, Barbara Padwell1, Julie Madden10, Lynda Taylor10, Sally Smith1, Heather King1, Jill Firth6, Jayne Heard7, Linda Sigsworth6. Lab Staff—Diane Corscadden1, Karen Henshaw1, Lubna-Haroon Rashid1, Stephen G Martin1, James I Robinson1
1Section of Musculoskeletal Disease, LIMM, Leeds, UK
2York District Hospital, York, UK
3Harrogate District Hospital, Harrogate, UK
4Huddersfield Royal Infirmary, Huddersfield, UK
5Dewsbury District and General Hospital, Dewsbury, UK
6St Luke's Hospital, Bradford, UK
7Hull Royal Infirmary, Hull, UK
8Barnsley District General Hospital, Barnsley, UK
9Pinderfields General Hospital, Wakefield, UK
10Calderdale Royal Hospital, Halifax, UK
British Society of Paediatric and Adolescent Rheumatology (BSPAR) study group: M Abinum, MD, M Becker, MD, A Bell, MD, A Craft, MD, E Crawley, MD, J David, MD, H Foster, MD, J Gardener-Medwin, MD, J Griffin, MD, A Hall, MD, M Hall, MD, A Herrick, MD, P Hollingworth, MD, L Holt, MD, S Jones, MD, G Pountain, MD, C Ryder, MD, T Southwood, MD, I Stewart, MD, H Venning, L Wedderburn, MD, P Woo, MD and S Wyatt, MD.
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
References
- Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17 R2:R116–R121. doi: 10.1093/hmg/ddn246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Todd JA. Genetic analysis of autoimmune disease. Cell. 1996;85:311–318. doi: 10.1016/s0092-8674(00)81110-1. [DOI] [PubMed] [Google Scholar]
- Ravelli A, Martini A. Juvenile idiopathic arthritis. Lancet. 2007;369:767–778. doi: 10.1016/S0140-6736(07)60363-8. [DOI] [PubMed] [Google Scholar]
- Hinks A, Worthington J, Thomson W. The association of PTPN22 with rheumatoid arthritis and juvenile idiopathic arthritis. Rheumatology (Oxford) 2006;45:365–368. doi: 10.1093/rheumatology/kel005. [DOI] [PubMed] [Google Scholar]
- Hinks A, Ke X, Barton A, Eyre S, Bowes J, Worthington J, et al. Association of the IL2RA/CD25 gene with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:251–257. doi: 10.1002/art.24187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CE, Cooper JD, Brusko T, Walker NM, Smyth DJ, Bailey R, et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- Brand OJ, Lowe CE, Heward JM, Franklyn JA, Cooper JD, Todd JA, et al. Association of the interleukin-2 receptor alpha (IL-2Ralpha)/CD25 gene region with Graves' disease using a multilocus test and tag SNPs. Clin Endocrinol (Oxf) 2007;66:508–512. doi: 10.1111/j.1365-2265.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, Howson JM, et al. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53:3020–3023. doi: 10.2337/diabetes.53.11.3020. [DOI] [PubMed] [Google Scholar]
- Hinks A, Eyre S, Ke X, Barton A, Martin P, Flynn E, et al. Overlap of disease susceptibility loci for rheumatoid arthritis (RA) and juvenile idiopathic arthritis (JIA) Ann Rheum Dis 2009. e-pub ahead of print 11 August 2009 [DOI] [PMC free article] [PubMed]
- van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallstrom E, Khademi M, et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 2008;10:5–10. doi: 10.1038/gene.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Staudt LM. LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood. 1996;87:734–745. [PubMed] [Google Scholar]
- Barton A, Eyre S, Ke X, Hinks A, Bowes J, Flynn E, et al. Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum Mol Genet. 2009;18:2518–2522. doi: 10.1093/hmg/ddp177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers HM, Kurreeman FA, Stoeken-Rijsbergen G, Brinkman DM, Kamphuis SS, Van Rossum MA, et al. Association of the autoimmunity locus 4q27 with juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:901–904. doi: 10.1002/art.24296. [DOI] [PubMed] [Google Scholar]
- Suppiah V, O'doherty C, Heggarty S, Patterson CC, Rooney M, Vandenbroeck K. The CTLA4+49A/G and CT60 polymorphisms and chronic inflammatory arthropathies in Northern Ireland. Exp Mol Pathol. 2006;80:141–146. doi: 10.1016/j.yexmp.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Prahalad S, Bohnsack JF, Whiting A, Clifford B, Jorde LB, Guthery SL, et al. Lack of association of functional CTLA4 polymorphisms with juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:2147–2152. doi: 10.1002/art.23602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier LM, Lowe CE, Cooper J, Downes K, Anderson DE, Severson C, et al. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 2009;5:e1000322. doi: 10.1371/journal.pgen.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcina A, Fedetz M, Ndagire D, Fernandez O, Leyva L, Guerrero M, et al. IL2RA/CD25 gene polymorphisms: uneven association with multiple sclerosis (MS) and type 1 diabetes (T1D) PLoS ONE. 2009;4:e4137. doi: 10.1371/journal.pone.0004137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International league of associations for rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–392. [PubMed] [Google Scholar]
- Packham JC, Hall MA. Long-term follow-up of 246 adults with juvenile idiopathic arthritis: functional outcome. Rheumatology (Oxford) 2002;41:1428–1435. doi: 10.1093/rheumatology/41.12.1428. [DOI] [PubMed] [Google Scholar]
- Adib N, Hyrich K, Thornton J, Lunt M, Davidson J, Gardner-Medwin J, et al. Association between duration of symptoms and severity of disease at first presentation to paediatric rheumatology: results from the childhood arthritis prospective study. Rheumatology (Oxford) 2008;47:991–995. doi: 10.1093/rheumatology/ken085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.