Abstract
The nuclear regulator LaeA has been shown to govern production of multiple secondary metabolites in A. nidulans and A. fumigatus. Herein we examine the role of this protein in Aspergillus flavus. Similarly as in other Aspergilli, LaeA had a major effect on A. flavus secondary metabolism where ΔlaeA and over-expression laeA (OE::laeA) strains yielded opposite phenotypes resulting in decreased (increased) secondary metabolite production. The two mutant strains also exhibited striking morphological phenotypes in the loss (increase) of sclerotial production in comparison to wildtype. Growth on seed was marked by decreased (increased) conidial and aflatoxin production of the respective mutants; this was accompanied by decreased lipase activity in ΔlaeA, an enzymatic process correlated with seed maceration. Transcriptional examination of the mutants showed LaeA negatively regulates expression of its recently identified nuclear partner VeA, another global regulator of A. flavus secondary metabolites and sclerotia.
Keywords: Aflatoxin, LaeA, AflR, VeA, sclerotia, mycotoxin, secondary metabolism
Introduction
The genus Aspergillus represents a large, worldwide family of fungi with estimates nearing 200 species (Samson, 1992). Whereas most of the members of this genus are saprobes capable of thriving on plant, animal and manmade wastes, a few are potent pathogens of plants and animals. The two most frequently isolated pathogens from these diverse hosts are A. flavus and A. fumigatus, respectively. A. flavus is notorious for the production of the carcinogenic and mutagenic secondary metabolites commonly known as aflatoxins during growth on seed (Payne and Brown, 1998). In the US alone, agricultural economic losses due to aflatoxin contamination of food and feed are estimated to be $270 million annually (Richard and Payne, 2003). However, the effects are more keenly felt in developing countries where, in addition to contributing to enhanced cancer rates (Bressac et al., 1991), high aflatoxin contamination of human food also contributes to fatal toxicoses (Probst et al., 2007). This species is also associated with invasive aspergillosis (IA) in immunocompromised individuals (Hedayati et al., 2007). By far the majority of IA is attributed to A. fumigatus infection with instances of 50 to 90% mortality (Latge, 1999).
In common with all Aspergilli sequenced to date, both opportunistic pathogens have been found to contain a plethora of clustered genes devoted to the production of secondary metabolites or natural products. Many of these clusters were found to be regulated by a single nuclear protein called LaeA in A. fumigatus and the genetic model A. nidulans (Bok and Keller, 2004; Perrin et al., 2007). Deletion of A. nidulans laeA resulted in the reduction of secondary metabolite gene expression concomitant with loss/decrease in the production of the encoded metabolites, whereas overexpression of laeA yielded opposite results (Bok and Keller, 2004; Bok et al., 2006b). Deletion of the gene in two A. fumigatus strains resulted in decreased virulence in the mouse pulmonary model, associated with reduced killing of neutrophil cells (Bok et al., 2005; Sugui et al., 2007a) thus strongly supporting a role for LaeA mediated toxin production in IA development by A. fumigatus.
Here we examined the role of LaeA in the seed colonizing fungus A. flavus. Predictably, the ΔlaeA mutant was crippled in secondary metabolite production whereas the overexpression strain overproduced several metabolites including the carcinogen aflatoxin. Furthermore, ΔlaeA showed decreased lipase activity and was less able to colonize peanut and maize seed. Unexpectedly, however, ΔlaeA was unable to produce sclerotia, overwintering structures important in fungal survival. Sclerotial and secondary metabolism loss have also been reported in ΔveA mutants of A. flavus (Duran, R. M., et al., 2007). VeA and LaeA have recently been identified as members of a nuclear complex in A. nidulans (Bayram et al., 2008) and regulation of veA expression by LaeA as shown here may represent an internal feedback mechanism to maintain a nuclear complex linking morphology and secondary metabolism in the aspergilli.
Results
Identification and disruption of A. flavus laeA
The predicted sequence for the A. flavus laeA ortholog was obtained from GenBank (AY883016) and by conducting a BLAST search (http://www.ncbi.nlm.nih.gov/blast/) with the A. nidulans sequence against the A. flavus genome scaffolds (http://www.aspergillusflavus.org/genomics/) and then selecting the matching genomic region as well as 4 kb of up and downstream of the predicted laeA ORF using the gene prediction program FGENESH (http://www.bio.net/bionet/mm/bio-www/1999-January/000775.html). The resulting two exon gene was similar in structure to A. nidulans and A. fumigatus laeA. The A. flavus laeA gene is 100% identical to a putative A. oryzae laeA and its 5' exon aligns well with the sequences from A. nidulans and A. fumigatus with the most variation in the 5’ region (data not shown). A. flavus LaeA protein shows 75.6% identity to A. nidulans LaeA and contains a conserved s-adenosyl methionine binding (SAM) motif found to be critical for LaeA function (Bok et al., 2006c). Inactivation of the laeA locus was obtained by replacement of laeA by pyrG (Supplemental Fig.1A). Four out of twenty transformants contained the 4 and 1 kb fragments of a SpeI digest expected for a laeA disruption event and were called TJW71.1, TJW71.3, TJW71.4 and TJW71.7, respectively (Supplemental Fig. 1B). Physiological and pathogenicity experiments as below revealed all ΔlaeA strains shared the same phenotype and TJW71.1 was selected for the majority of experimentation.
A niaD- auxotroph of TJW71.1 was created to generate strain MLRM8.1 which was then complemented to laeA and niaD prototrophy with pLRM11. Four transformants were confirmed to contain wild type laeA (Supplemental Fig. 1C and data not shown). Examination of a representative transformant strain, TJW79.13 containing at least two copies of laeA (Supplemental Fig. 1C), was selected for further experiments to examine effects of over expression laeA. This strain is referred to as OE::laeA in this study.
LaeA is required for aflatoxin production, aflR expression and normal veA expression
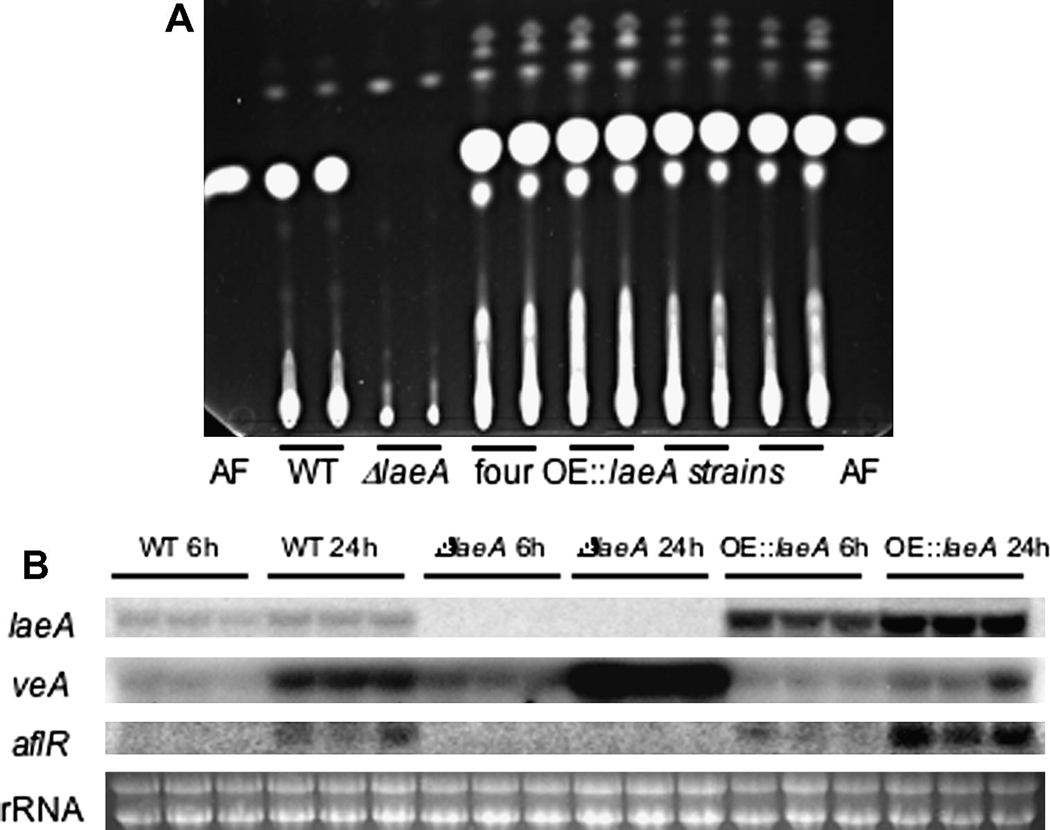
Drawing from observations of A. fumigatus and A. nidulans ΔlaeA strains, we predicted a loss of aflatoxin production mediated by loss of expression of the aflatoxin specific transcription factor aflR in the A. flavus ΔlaeA strain. As shown in Figure 1A and Supplementary Figure 2, while the ΔlaeA mutants did not show any detectable aflatoxin production, aflatoxin production was restored and in fact increased in TJW79.13 (and other complemented strains) as compared to wild type. Examination of laeA and aflR expression in wild type, TJW71.1 and TJW79.13 showed no detectable expression of either aflR and laeA in the ΔlaeA mutant but increased expression of both genes in TJW79.13 (Fig. 1B), thus correlating with TLC results.
Fig. 1.
Aflatoxin production and gene expression in ΔlaeA and OE::laeA mutants. (A) Extracts of the wild type A. flavus NRRL 3357, the ΔlaeA mutant (TJW71.1) and four OE::laeA strains were grown on YES media for 5 days at 29°C and extracts were separated on a thin layer chromatography plate. Each strain was extracted twice. Aflatoxin was visualized using long-wave (366 nm) UV light. Aflatoxin B1 standard is spotted on each side of the plate. (B) mRNA expression of laeA, veA and aflR in A. flavus NRRL 3357, ΔlaeA (TJW71.1) and OE::laeA (TJW79.13). All strains were cultured in liquid YEP medium and incubated at 250 rpm at 29°C. After 24 hours, mycelium was transferred to aflatoxin inducing medium, YES, for 6 and 24 hr. Each strain was grown three times.
In addition to aflR expression, we also examined expression of veA. VeA is a light regulated gene with similar global effects on secondary metabolite production in all the examined aspergilli (Stinnett et al., 2007). VeA is necessary for aflatoxin production and aflR expression in A. parasiticus (Calvo et al., 2004) and another isolate of A. flavus (Duran et al., 2007). Most recently VeA and LaeA, along with a third protein VelB, have been shown to function as a nuclear complex in A. nidulans (Bayram et al., 2008). As shown in Figure 1B, in comparison to wild type, the laeA deletion mutant TJW71.1 showed an increase and TJW79.13 a decrease in veA expression. These results suggest that veA is negatively regulated by LaeA.
Decreased conidial production and loss of sclerotia in ΔlaeA
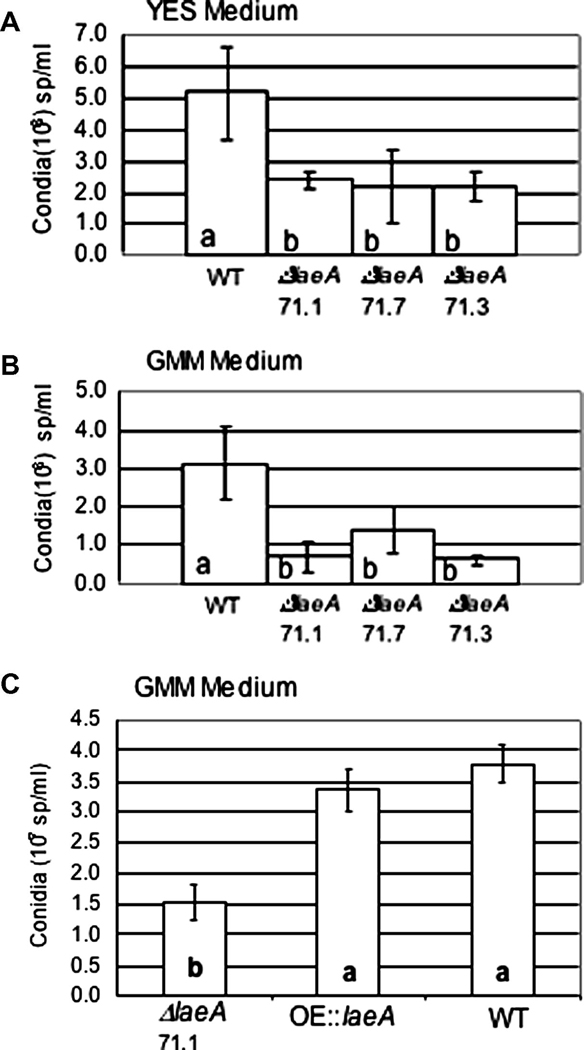
Similarly to A. fumigatus and A. nidulans ΔlaeA strains, the A. flavus ΔlaeA mutants presented a loss of pigmentation on the backside of GMM growth plates, although no difference in radial growth (data not shown). Quantitative analysis of conidial production showed an approximate two-fold decrease in spore production in the ΔlaeA mutant with no observable phenotype in the OE::laeA strain on GMM medium (Fig. 2A). This decrease was also observed in YES medium (Fig. 2B).
Fig. 2.
LaeA deletion results in reduced sporulation on growth medium. (A) The wild type A. flavus NRRL 3357 and three ΔlaeA mutants (TJW71.1, TJW71.7 and TJW71.3) were grown in YES medium for 5 days at 29°C. Columns with the same letter are not significantly different at a significance level of 0.05. (B) The wild type A. flavus NRRL 3357 and three ΔlaeA mutants (TJW71.1, TJW71.7 and TJW71.3) were grown on GMM medium for 5 days. Columns with the same letter are not significantly different at a significance level of 0.01. (C) The wild type NRRL 3357, ΔlaeA (TJW71.1) and OE::laeA (TJW79.13) where grown on GMM medium at 29°C for 5 days. Columns with the same letter are not significantly different at a significance level of 0.05. For all tests, conidia were counted using a hemocytometer. Values are a mean of 4 replicates.
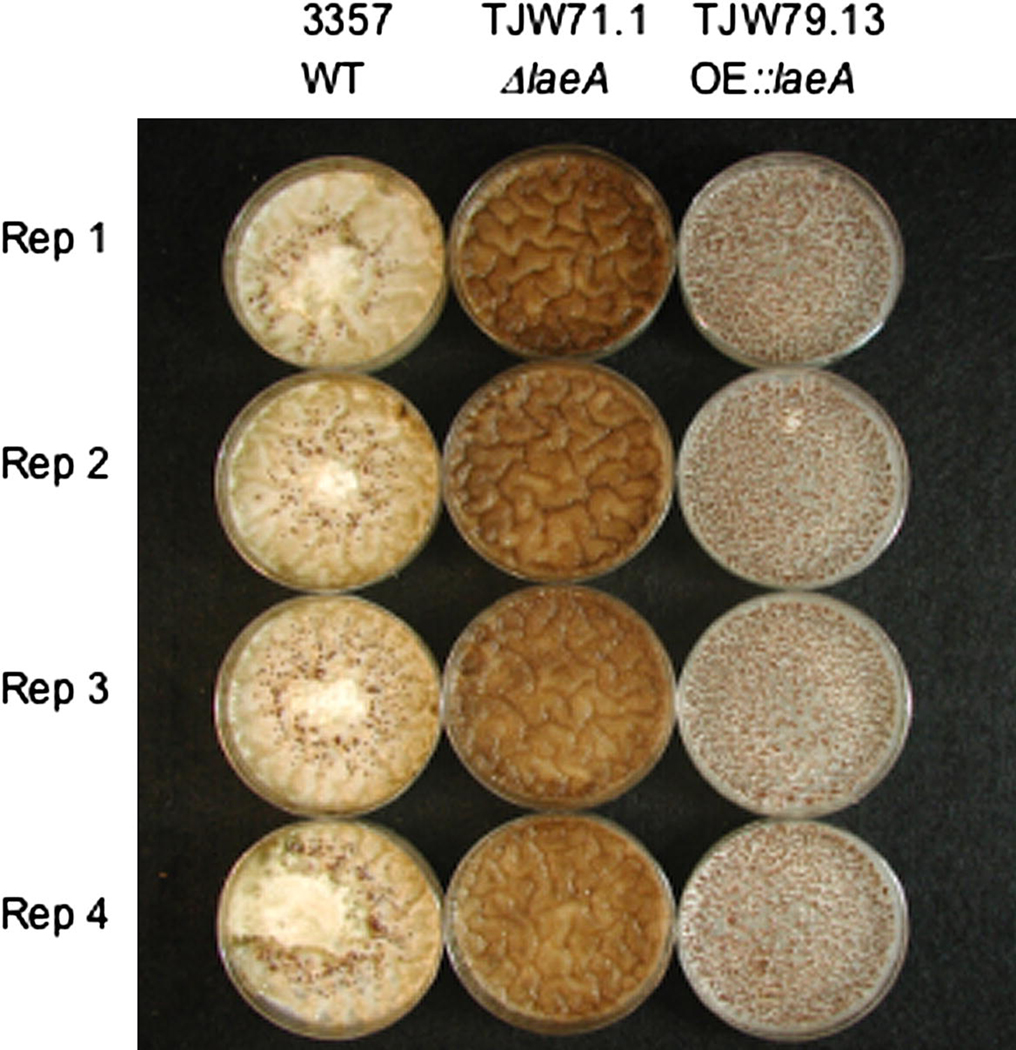
Sclerotial production has been linked to aflatoxin synthesis in several studies including the loss of both processes in ΔveA (Calvo et al., 2004; Duran et al., 2007). To determine if loss or overexpression of laeA resulted in aberrations in sclerotial production, the strains were grown on sclerotial inducing medium (GMM medium supplemented with 2% sorbitol). Striking phenotypes were observed for both mutants where the ΔlaeA strain did not produce any sclerotia and the OE::laeA strain produced statistically increased sclerotial numbers compared to wild type (Fig. 3).
Fig. 3.
Sclerotial production in ΔlaeA and OE::laeA mutants. The wild type NRRL 3357, ΔlaeA (TJW71.1) and OE::laeA (TJW79.13) where grown on GMM plus 2% sorbitol medium at 29° for 5 days in the dark. Plates were sprayed with ethanol to wash off conidia to visualize sclerotia. Sclerotia were absent in the ΔlaeA strain and overproduced in the OE::laeA strain as compared to wildtype. The average number of sclerotia produced per petri plate by the OE::laeA strain (1540 ± 90) was statistically greater (P ≤ 0.01) than the average number produced by wild type (217 ± 70).
laeA mutants are altered in host colonization
Although aflatoxin is not considered a virulence factor and alterations in its production would not a priori be considered important in seed colonization, we considered it possible that other changes wrought by deletion or overexpression of laeA could affect colonization of host seed. To examine this possibility, peanut seed were inoculated with all three strains of A. flavus.
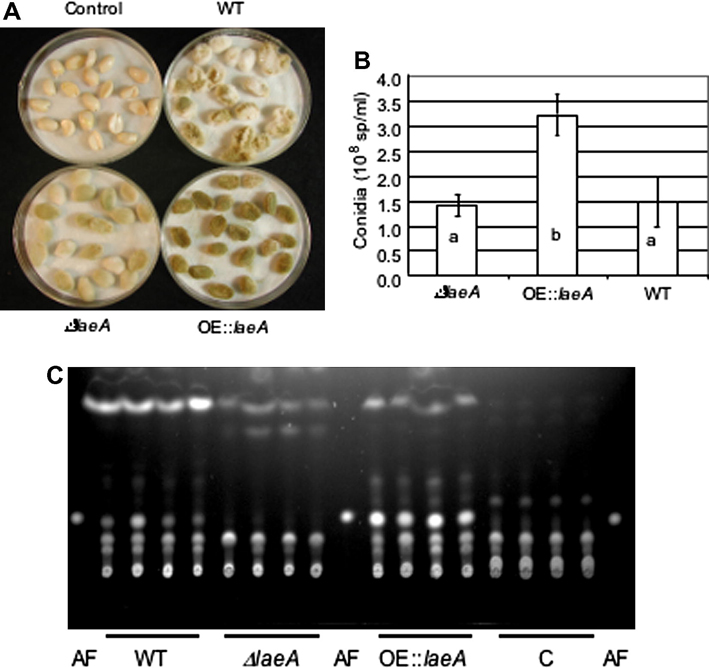
Macroscopically, the ΔlaeA strain appeared to grow less vigorously than wild type on peanut, whereas the OE::laeA strain showed an early enhanced ability to colonize peanut seeds (Fig. 4A), however, by day three any macroscopic difference was largely obscured (data not shown). Visual observations were correlated with conidial production by the OE::laeA strain on seed. On both day 2 and 3, the OE::laeA strain produced statistically more conidia than both wild type and ΔlaeA (Fig. 4B and data not shown). Although there was no decrease in conidial production by ΔlaeA on peanut seed as assessed by these assays, there was on maize seed (Supplemental Fig. 3).
Fig. 4.
Growth and aflatoxin production on peanut seed. The wild type NRRL 3357 ΔlaeA (TJW71.1) and OE::laeA (TJW79.13) were grown on peanut seed for 2 days at 29°C. (A) Representative picture of inoculated seed. (B) Conidia were counted from 4 replicates of seed as pictured in A. Columns with the same letter are not significantly different at a significance level of 0.05. (C) Aflatoxin production on peanut seed as examined by thin layer chromatography. C is control (seed mock inoculated with water). AF is aflatoxin B1 standard.
Figure 5C shows that aflatoxin production on the peanut seed reflected that of production on medium (Fig. 1A), namely that ΔlaeA did not produce aflatoxin on seed whereas the OE::laeA produced more than wild type. A similar aflatoxin profile was seen on maize seed (where only ΔlaeA strains were examined, Supplemental Fig. 3).
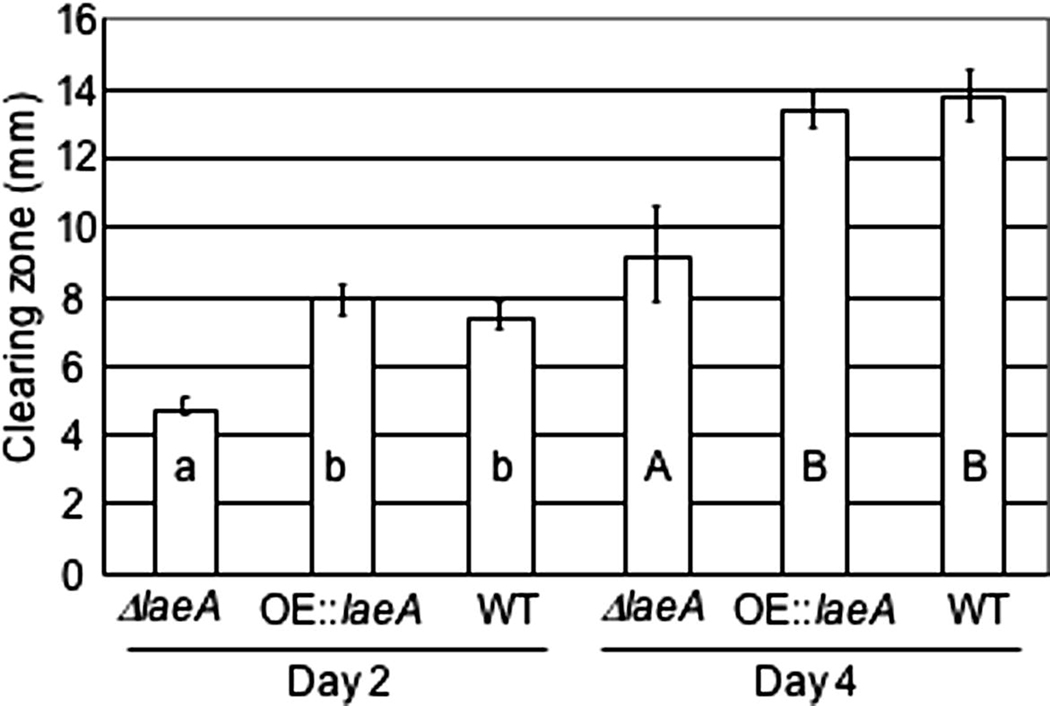
Fig. 5.
Lipase activity in ΔlaeA and OE::laeA mutants. The wild type NRRL 3357, ΔlaeA (TJW71.1) and OE::laeA (TJW79.13) where grown on lipase assay medium where increased clearing zone indicates increased lipase-like activity. Values are a mean of 4 replicates. Columns with the same letter are not significantly different at a significance level of 0.05. Day 2 and day 4 were analyzed separately.
Several enzyme activities have been associated with Aspergillus pathogenesis of seed, including lipase activity (Tsitsigiannis and Keller, 2006). We assessed potential lipase activity for all three strains. As shown in Figure 6, ΔlaeA showed lower lipase activity than either wild type or the OE::laeA strain at both day 2 and 4.
Global regulation of secondary metabolites in LaeA mutants
LaeA regulates production of several secondary metabolites in A. fumigatus and A. nidulans (Bok and Keller, 2004; Perrin et al., 2007). Here we examined the production of known compounds produced by wild type A. flavus NRRL 3357 (Frisvad et al., 1987) in comparison to both laeA mutants. The profile of these secondary metabolites depended on the growth substrate (Table 1). Two striking observations where made. First, the ΔlaeA mutant did not produce any secondary metabolites, with the exception of kojic acid on DG18 agar and aspergillic acid on TGY agar. Second, the OE::laeA strain produced additional metabolites not observed in wild type. These were all known sclerotial metabolites including paspaline/paspalicine, aflatrem and aflavinines and their production was attributed to the high production of sclerotia in this strain. The relative amounts of aflatoxins were slightly higher in the OE::laeA strain than in the wild type on CYA and DG18 agar, while the opposite was the case for YES agar. The amount of kojic acid was 3–10 times higher for the wild type than the OE::laeA strain on YES agar, but the opposite was the case for DG18 agar.
Table 1.
Production of secondary metabolites by wild type (NRRL 3357), ΔlaeA TJW71.1) and OE::laeA (TJW79.13) as measured by HPLC-DAD (diode array detection). The log to the area of the peaks is given as mAU (milli absorption units) as measured at 210 nm (ND means non detectable at 210 nm).
|
A. flavus strains |
Aflatoxin B1 RI 835 |
Aflatoxin B2 RI 810 |
Cyclopiazonic Acid RI 1094 |
Kojic Acid RI 570 |
Aspergillic Acid RI 1976 |
Paspaline / paspalinine RI 1261 / 1479 |
Aflatrem RI 1522 |
Aflavinines 1 |
Oryzachlorin RI 850 |
|---|---|---|---|---|---|---|---|---|---|
| 3357 CYA |
02.5 34 LU2 |
ND 32 LU |
2.8 | 0 | 0 | 0 | 0 | 0 | 0 |
| ΔlaeA CYA |
0 | ND | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
OE::laeA CYA |
2.8 43 LU |
ND 41 LU |
2.8 | 0 | 0 | 1.8 / 2.0 | 1.9 | 2.3 / 0 / 1.6 / 2.0 / 2.0 |
0 |
| 3357 YES |
3.1 84 LU |
ND 102 LU |
2.5 | 4.0 | 0 | 0 | 0 | 0 | 0 |
| ΔlaeA YES |
0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
OE::laeA YES |
2.9 58 LU |
ND 90 LU |
2.4 | 3.0 | 0 | 0 | 0 | 0 | 0 |
| 3357 WATM |
2.0 26 LU |
ND 27 LU |
2.7 | 0 | 0 | 0 | 0 | 0 | 0 |
| ΔlaeA WATM |
0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
OE::laeA WATM |
2.5 26 LU |
ND 28 LU |
2.5 | 0 | 0 | 0 / 1.8 | 1.6 | 0 / 1.6 / 2.0 / 1.9 / 1.9 |
0 |
| 3357 TGY |
0 | 0 | 0 | 0 | 2.1 | 0 | 0 | 0 | 0 |
| ΔlaeA TGY |
0 | 0 | 0 | 0 | 2.0 | 0 | 0 | 0 | 0 |
|
OE::laeA TGY |
0 | 0 | 0 | 0 | 1.9 | 0 | 0 | 0 | 0 |
| 3357 DG18 |
1.6 LU 2 |
ND LU 0 |
0 | 4.0 | 0 | 0 | 0 | 0 | 1.9 |
| ΔlaeA DG18 |
0 | 0 | 0 | 3.3 | 0 | 0 | 0 | 0 | 0 |
|
OE::laeA DG18 |
2.6 LU 29 |
ND LU 37 |
1.9 | 3.5 | 0 | 0 | 0 | 0 | 0 |
The aflavinines had retention indices (RI) 967, 1146, 1185, 1610, 1650
Aflatoxin B1 and B2 as measured by a fluorescence detector (LU units, excitation 230 nm, emission 450 nm)
Discussion
A critical attribute in LaeA function is its role in transcriptional regulation of Aspergillus secondary metabolite gene clusters. Genes required for expression of these biologically active small molecules are typically arranged in a contiguous manner and inherited as a single genetic locus (Hoffmeister and Keller, 2007). LaeA activity allows for directed expression of many of these loci with loss of laeA resulting in gene silencing (Bok et al., 2006b; Perrin et al., 2007). Although LaeA cellular mechanism is yet unknown, accruing data suggests LaeA may play a role in epigenetic regulation of such clusters (Bok et al., 2006c; Shwab et al., 2007), possibly as a member of a nuclear complex with VeA (Bayram et al., 2008). LaeA governs expression of multiple gene clusters in the soil saprophyte A. nidulans (Bok et al., 2006b). Feeding choice preferences of a fungivore demonstrated that laeA provides both a protective shield for the fungus as well as negatively impacts egg laying in the consuming springtails (Rohlfs et al., 2007). Similarly, LaeA function is critical for production of several toxins in A. fumigatus, including gliotoxin, a molecule injurious to mammalian cells and possibly important in the severity of IA as determined by the animal models and cellular response (Bok et al., 2006a; Spikes, S., et al., 2008; Sugui et al., 2007b). These studies have strongly implicated loss of metabolite production as a key factor contributing to the reduced virulence of ΔlaeA strains (Bok et al., 2005; Sugui et al., 2007a).
To address an impact on LaeA in A. flavus physiology and pathogenicity, we examined both ΔlaeA and OE::laeA strains. Our analysis showed reduced (no) expression of cyclopiazonic acid, kojic acid, oryzaechlorin and asperfuran in addition to aflatoxin in the ΔlaeA mutant, whereas several metabolites were up regulated in the OE::laeA strain, particularly those associated with sclerotial production, e.g. paspaline, aflatrem and aflavinines (Table 1). Although not produced by ΔlaeA in most media, kojic acid was produced when the strain was grown on DG18 medium. The production of aspergillic acid was approximately the same in all three isolates and appears not to be controlled by laeA. Both aspergillic acid and kojic acid are putative chelation agents and, as such, possibly play an essential role in general fitness of the fungus, which may impact the degree of regulation by LaeA of these metabolites. Interestingly, one metabolite, oryzachlorin, a epidithiodiketopiperazine similar in structure to gliotoxin, was down regulated in both mutants.
Loss of metabolite production has been correlated with morphological differentiation in fungi (Calvo et al., 2002), including reduced conidiospore formation (Wilkinson et al., 2004) and sclerotial loss (Calvo et al., 2004) in various Aspergilli. Although aflatoxin is not correlated with disease severity, sclerotial production has been associated with pathogenicity in some Aspergilli including A. flavus (Chang et al., 2001). The loss and increased sclerotial production of the two laeA mutants, and associated metabolites, was striking (Fig. 3 and Table 1) and reminiscent of the ΔveA phenotype. Deletion of veA in A. parasiticus (Calvo et al., 2004) and another strain of A. flavus (Duran et al., 2007) also show loss of aflR expression (and consequently aflatoxin production) coupled with loss of sclerotial formation. Examination of veA expression in ΔlaeA and OE::laeA strains supports a LaeA mediated negative regulatory mechanism governing veA expression. A coupling of all studies would suggest that both veA and laeA are required for aflatoxin and sclerotial formation, perhaps through formation of their joint regulatory complex in the nucleus (Bayram et al., 2008). Possibly regulation of veA by LaeA represents an internal mechanism to balance stoichiometry of this complex.
We also noted, based on visual observations and softness of infected seed, that ΔlaeA appeared less aggressive in tissue maceration compared to wild type or the OE::laeA strain (Fig. 4A). As one indicator of maceration potential, we assessed overall lipase/esterase activity of these mutants, a technique successfully employed in other studies to assess fungal degradative potential (Tsitsigiannis and Keller, 2006). Degradative enzymes are well known to play a significant role in fungal infections of plants, with lipases particularly important in pathogenesis of several hosts including seed in the A. flavus pathosystem (Berto et al., 1999; Commenil et al., 1995). The decreased lipase activity of the ΔlaeA mutant may indicate lipase activity contributes to pathogenesis through tissue degradation in this strain (Fig. 5). The differences in conidial production of this mutant on peanut and maize seed may reflect host differences or could be an attribute of timing of assessment and/or method of inoculation.
In summary, results from this study demonstrate the conserved function of laeA as a global regulator of secondary metabolism in the Aspergilli with varying impact on Aspergillus morphology. We speculate the development of chemical regulation by LaeA may have evolved as a protective device (Rohlfs et al., 2007) and, by chance, also provides considerable aggressive characteristics in disease development by opportunistic fungi. Regardless of the role of LaeA in fungal biology, our findings represent an advance in identification of a shared mechanism in Aspergillus resulting in damage of both plant and animal hosts.
Materials and methods
Strains and growth conditions
All strains were maintained as glycerol stocks and grown on potato dextrose agar (PDA) or glucose minimal medium (GMM, Shimizu and Keller, 2001) for spore production at 29°C. Media were supplemented with 10 mM uridine and 10 mM uracil as needed. Strains used in this study included Aspergillus flavus NRRL 3357 (prototroph), NRRL 3357.5 (pyrG auxotroph) and the transformants described below.
Vector construction
The laeA replacement vector was constructed as follows. A 1.4 kb fragment upstream of the laeA coding region was amplified with forward primer 5’ TGTGTCGACACTGCCCAGACATCTATA adding a SalI site as indicated by underline and reverse primer 5’ GTAGTACGAGTCGTGTGGTGGTGCGGCCGCCGC adding a NotI site as indicated. This fragment was digested with NsiI (which was internal to the SalI site) and NotI and ligated into the NsiI and NotI sites of pLMH26 (Maggio-Hall and Keller, unpublished data) upstream of the A. fumigatus pyrG. Next, a 1.5 kb fragment downstream of the laeA coding region was amplified with forward primer 5’ CAGCCGCGGACGATGCACTGAGCTGCCT adding a SacII site and reverse primer 5’ CCTCGCCAGCAACGGCCGAGACC approximately 20 bp downstream of a SacII site. This fragment was digested with SacII and ligated into pLMH26 linearized with SacII to yield pLRM5. The correct orientation of the downstream fragment was confirmed by PstI digestion and the correct candidates were then confirmed by digests with NotI and NsiI. The schematic of this replacement vector is depicted in Figure 1A. The laeA complementation vector was constructed by amplifying a 4.3 kb fragment containing the 2 kb laeA coding region and 2.3 kb upstream with forward primer 5’ GGGATCCTCCACAAAGCCTTTCGTAAAA and reverse primer 5’ TATCTAGAAGCACAGGCATGCGGCCGCA. This fragment was ligated into pCR-Blunt-II Topo (Invitrogen) to create intermediate vector pLRM9. A 3 kb HindIII fragment containing the niaD gene was cut from pGAPN-2 (Liang et al., 1997) and ligated into the HindIII site of pLRM9 to create the laeA complementation vector pLRM11.
Fungal transformation
Aspergillus protoplasts were produced and transformed using the modified polyethylene glycol method (Bok and Keller, 2004). To generate laeA::pyrG (i.e. ΔlaeA) strains, A. flavus NRRL 3357.5 protoplasts were transformed with 2 µg of a 5 kb pLRM5 PCR product generated using the forward primer 5’ CCTTGTATGATGTATGTATGATGAGC and the reverse primer 5’ TCTTGGGTCATTGGGTGGGCGG. Transformants were screened for uridine and uracil prototrophy followed by Southern analysis. The resulting laeA deletion strains used for further experiments were named as TJW71.1, TJW71.3, TJW71.4 and TJW71.7 respectively.
To generate nitrate auxotrophs of the laeA deletion strain, 106 conidia from TJW71.1 were spread onto GMM containing 750 mM chlorate and 1.6 mM ammonium chloride. Colonies that arose were transferred to fresh plates and single spores were purified. The resulting colonies were screened for mutation of niaD as described in Cove, where growth on different nitrogen sources allowed for classification of mutations in niaD, niiA and other genes required for nitrate utilization (Cove, 1966). One niaD auxotroph strain, MLRM8.1, was transformed with 10 µg of pLRM11 to yield the complemented strains, TJW79 series.
Physiology experiments
Conidial production, sclerotial formation and relative colony diameter were recorded for wild type, ΔlaeA and complemented laeA strains. Diameter growth was measured from point inoculation of 5 µl of a 106 spores/ml suspension of A. flavus conidia on GMM media. For quantitative analysis of conidial production, YES medium (2% yeast extract, 6% sucrose, pH 5.8) and GMM were overlaid with 5 ml of a 106 spores/ml suspension of A. flavus conidia in molten agar. Cultures were grown for 5 days at 29°C in light. Three 1.5-cm diameter cores were harvested from the center of each plate and homogenized in 3 ml of distilled water. 2 µl were removed, diluted 1:500, and conidia were counted using a hemocytometer. Sclerotial production was observed on the sclerotial inducing medium GMM +2% sorbitol. GMM + 2% sorbitol was overlaid with 3 ml of a 106 spores/ml suspension of A. flavus conidia in molten agar. Cultures were grown for 5 days at 29°C in dark conditions. Plates were then sprayed with 70% ethanol to kill and wash away conidia to aid in enumeration of sclerotia.
Seed infections
Peanut (Arachis hypogaea)
Mature peanut seeds were prepared by removing the brown exterior peanut layer (testa) using the fingers. The two cotyledons of each seed were separated and the embryo carefully removed without damaging any of the cotyledon tissue. Then, cotyledons were surface sterilized by placing them in a tea ball infuser and dipping them in a beaker containing 0.05% sodium hypochlorite in sterile water for 3 minutes. The tea ball was transferred to a new beaker containing sterile distilled water for 30 seconds (wash step), followed by a 5-second wash with 70% ethanol in a new beaker (additional sterilization step) and one more 30-second wash with sterile distilled water while shaking the tea ball. The cotyledons were drained completely and placed in a petri dish until the time of infection. All the steps were aseptically performed in a biosafety hood.
Peanut cotyledons were inoculated with a 105 spores/ml. Cotyledon treatments included water control (mock inoculation) and infection with fungal strains. For all treatments, 20 peanut cotyledons were immersed in 20 ml of sterile distilled water (control) or sterile distilled water with fungal conidia in 50 ml centrifuge tubes while shaking for 30 minutes in a rotary shaker at 50 rpm. Cotyledons were placed in petri dishes lined with 3 pieces of moist filter paper (to create a humidity chamber) and a water reservoir (lid of a 50 ml centrifuge tube containing 2ml of sterile water) to maintain high humidity. Cotyledons were incubated for 3 days at 29°C in dark conditions. The filter paper was moistened daily.
Maize (Zea mays)
Untreated maize seeds (X516WX raw) obtained from Kaltenberg Seed Farms, Inc. (Waunakee, WI) were inoculated similarly to peanut seed with the following differences. Seeds were wounded by creating a small hole near the embryo where they were inoculated with 5 µl of a 106 spores/ml suspension of A. flavus conidia. Ten seeds per replicate were transferred to the humidity chamber and incubated in the dark at 29°C for 3 days. The filter paper was moistened daily and any germinating seeds were removed and discarded. Analysis was performed on the same number of seed for each treatment and replication (e.g. if 3 seed germinated from one humidity chamber, then only 7 seed were examined for each treatment and replication).
All seed experiments were repeated three to four times.
Aflatoxin analysis from seed
Three days after infection, peanut cotyledons were collected in 50 ml centrifuge tubes with the addition of 3 ml of 0.01% Tween 80 (v/v in water) and vortexed vigorously for 1 minute. 1 ml was removed from each sample for conidia counting prior to aflatoxin extraction. For extraction, 5 ml of acetone was added to the samples followed by shaking for 10 minutes in a rotary shaker at 150 rpm. Samples were allowed to stand for 5 minutes at room temperature and then 5 ml of chloroform was added to each sample followed by shaking for 10 minutes at 150 rpm. Samples were allowed to stand for an additional 10 minutes at room temperature, vortexed briefly and centrifuged for 15 min at 2000 rpm to collect the organic lower phase. Samples were then dried out completely. The presence of abundant seed lipids in the samples hampered the clear observation of aflatoxin on TLC plates and a second extraction-purification was carried out as follows. Samples were re-suspended in 5 ml of 0.1M NaCl methanol:water (55:45) and 2.5 ml of hexane and vortexed vigorously at high speed for 1 minute. Samples were centrifuged at 2000 rpm for 5 minutes. The hexane layer was collected and the fatty acid inter-phase layer was discarded. The remaining aqueous phase was washed an additional time with 2.5 ml hexane as described above. The hexane extracts were combined, allowed to dry and then re-suspended in 500 µl of chloroform before 10 µl of each extract was separated on a silica gel TLC plate using a chloroform:acetone (95:5 [vol/vol]) solvent system.
Maize seed were extracted similarly but without the needed extra hexane extraction for peanut seed.
Lipase assays
To test for lipase activity, GMM medium was overlaid with 10 ml of a 106 spores/ml suspension of A. flavus conidia in molten agar. Cultures were grown for 4 days at 29°C in light conditions. Then, one 0.7-cm diameter agar plug was harvested from the center of each plate and added to a sterile test tube containing lipase medium (0.5% mycological peptone, 0.3% yeast extract in 1% agar containing 0.1% glyceryl tributyrate, Paterson et al., 1994). Tubes were incubated at 29°C in light conditions. Measurements of the clearing zone, indicative of lipase activity, were taken at days 2 and 4.
Aflatoxin production in media
YES and GMM media were overlaid with 5 ml of a 106 spores/ml suspension of A. flavus conidia in molten agar. Cultures were grown for 5 days at 29°C. Three 1.5-cm diameter cores were harvested from the center of each plate and homogenized in 3 ml of distilled water. 3 ml each of acetone and chloroform were added and the mixture was vortexed for 1 minute. The mixture was incubated at room temperature for 1 hour, was vortexed again and centrifuged for 10 minutes. The lower organic layer was removed and evaporated. Residue was resuspended in 100 µl of chloroform and 10 µl of the suspension were spotted onto a TLC plate (Whatman Ltd., Kent, England). This experiment was done in triplicate. Aflatoxin was resolved in a chloroform and acetone (95:5) solvent system and standards were purchased from Sigma-Aldrich (St. Louis, MO). Aflatoxin was visualized using long-wave (366 nm) UV light and digital photographs were taken.
Comprehensive secondary metabolite profiles
Three isolates A. flavus NRRL 3357, A. flavus TJW71.1 (Δ laeA) and A. flavus TJW79.13 (OE::laeA) were inoculated on the media CYA, YES (Frisvad and Samson, 2004), DG18 (Dichloran 18% glycerol agar, http://www.cbs.knaw.nl/cbs_home/cbs_home.html?http://www.cbs.knaw.nl/food/media.htm~main), WATM (Wickerhams Antibiotic test medium, (Raper and Thom, 1949)), and TGY (tryptone glucose yeast extract agar, http://www.cbs.knaw.nl/cbs_home/cbs_home.html?http://www.cbs.knaw.nl/food/media.htm~main) (in three points). The media were incubated for 7 days in the dark and 5 agar plugs were taken for HPLC analysis. The agar plugs were extracted with ethyl acetate / dichloromethane / methanol (3:2:1) with 1% formic acid added to this mixture and the extract ultrasonicated for 50 minutes. The extract was transferred to a new vial, and the organic solvents evaporated. The dry extract was re-dissolved in 0.5 ml methanol and filtered through a 0.45 µm filter before analysis. 3 µl of extract were injected into the chromatograph. The method used was based on Smedsgaard (Smedsgaard, 1997), but a Luna C18 (II) (Phenomenox, USA) column was used and the running time was 25 minutes. Retention indices were calculated for each compound and the retention indices and UV spectra were compared with authentic standards (Frisvad and Thrane, 1987).
Northern blot analysis of laeA, aflR and veA
Fifty ml liquid YEP medium (6% peptone, 2% yeast extract) was inoculated with 106 conidia/ml of A. flavus NRRL 3357, TJW71.1, or TJW79.13 in 50 ml flasks, incubated with shaking at 250 rpm at 29°C. After 24 hours, the mycelium was collected and incubated in the aflatoxin-stimulating YES medium for 6 and 24 hr (220 rpm, 29°C). Mycelia were harvested and total RNA was isolated using the Trizol (source: Invitrogen) method. The Northern blot was hybridized with a 670 bp HindIII/BamHI aflR fragment from pTMH52.1 (McDonald et al., 2005), a 4.3 kb laeA fragment from pLRM11 and a 1.7 kb veA fragment generated by PCR using forward primer 5’ CTAGCTGGTCATTATTTGATCTCG and reverse primer 5’ GTTGTAGAGTGGACGATCATCATG from genomic DNA.
Statistical analysis
Statistical differences in spore numbers were calculated by ANOVA tests using Minitab software (Penn State University).
Supplementary Material
Disruption and overexpression of the laeA gene in A. flavus. (A) Schematic diagram of the laeA open reading frame and how it was replaced with the pyrG selectable marker to generate the ΔlaeA mutant, TJW71. (B) Southern analysis of wild type (WT) strain and the laeA deletion strains. Genomic DNA was digested by SpeI. A 4 kb PCR product containing laeA was obtained with the forward primer 5’CCTTGTATGATGTATGTATGATGAGC and the reverse primer 5’ TCTTGGGTCATTGGGTGGGCGG and was used as a laeA probe (indicated by bar in panel A). Expected hybridization band patterns: wild type strain, 2.5 kb and 1.7 kb bands; ΔlaeA strains, 4.0 and 1 kb bands. The arrowheads at the top of the figure indicate the correct knock-out mutants, (C) Strain TJW1.1 was transformed with a full length laeA gene. Strain TJW79.13, an overexpression (OE) strain, containing two copies of laeA, was chosen for further studies. Genomic DNA was digested by HindIII. Expected hybridization band patterns: wild type strain, 5.4 and 5.6 kb bands (overlapping two bands); ΔlaeA, 4.5, 3.2 and 1.7 kb bands and OE::laeA contained the ΔlaeApattern plus additional copies of laeA.
Loss of aflatoxin production in ΔlaeA mutants. Extracts of the wild type A. flavus NRRL 33357 and three ΔlaeA mutants (TJW71.1, TJW71.7 and TJW71.3) grown on YES media for 5 days at 29°C were separated on a thin layer chromatography plate. Lanes 1–3 represent replicates for each strain. Aflatoxin was visualized using long-wave (366 nm) UV light. Aflatoxin B1 standard is spotted on each side of the plate.
LaeA deletion results in lower aflatoxin and spore production during maize infection. (a) Maize seed inoculated with A. flavus NRRL 3357 or three ΔlaeA mutants (TJW71.1, TJW71.7 and TJW71.3) were incubated in the dark at 29°C for 3 days and conidial production counted as described in text. Values represented are a mean of 4 replicates. Columns with the same letter are not significantly different at a significance level of 0.01. (b) Thin layer chromatography of extracts from the same maize seed show an absence of aflatoxin production in the seed infected with TJW71.1, TJW71.7 or TJW71.3. Aflatoxin was visualized using long-wave (366 nm) UV light. Lanes 1–4 represent replicates for each strain. The lane marked C represents the control seed mock inoculated with water.
Acknowledgements
Our thanks to Dr. Gary A. Payne for access to A. flavus genome data and for providing us with strains. This research was funded by NIH (MBRS) grant 5SO6GM08008 to S.P.K., and in part by NSF MCB-0236393, USAID Peanut Collaborative Research Support Program Grant LAG-G-00-96-90013-00 to N.P.K. and NIH R01 AI065728-01A1 to N.P.K. and J.C.F. JCF also thanks the Danish Research Agency for financial support (Center for Microbial Biotechnology).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bayram Ö, et al. VelB/VeA/LaeA coordinate light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Berto P, et al. Occurrence of a lipase in spores of Alternaria brassicicola with a crucial role in the infection of cauliflower leaves. FEMS Microbiol. Lett. 1999;180:183–189. doi: 10.1111/j.1574-6968.1999.tb08794.x. [DOI] [PubMed] [Google Scholar]
- Bok JW, et al. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell. 2005;4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, et al. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 2006a;74:6761–6768. doi: 10.1128/IAI.00780-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, et al. Genomic mining for Aspergillus natural products. Chem. Biol. 2006b;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, et al. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 2006c;61:1636–1645. doi: 10.1111/j.1365-2958.2006.05330.x. [DOI] [PubMed] [Google Scholar]
- Bressac B, et al. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–431. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- Calvo AM, et al. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, et al. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, et al. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia. 2001;153:41–48. doi: 10.1023/a:1015211915310. [DOI] [PubMed] [Google Scholar]
- Commenil P, et al. Purification and properties of an extracellular lipase from the fungus Botrytis cinerea. Lipids. 1995;30:351–356. doi: 10.1007/BF02536044. [DOI] [PubMed] [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta. 1966;113:51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- Duran RM, et al. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007;73:1158–1168. doi: 10.1007/s00253-006-0581-5. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA. Polyphasic taxonomy of Penicillium subgenus Penicillium - A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology. 2004:1–173. [Google Scholar]
- Frisvad JC, Thrane U. Standardized high-performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone retention indices and UV-VIS spectra (diode array detection) J Chromatogr. 1987;404:195–214. doi: 10.1016/s0021-9673(01)86850-3. [DOI] [PubMed] [Google Scholar]
- Hedayati MT, et al. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Latge JP. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SH, et al. Analysis of mechanisms regulating expression of the ver-1 gene, involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 1997;63:1058–1065. doi: 10.1128/aem.63.3.1058-1065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald T, et al. Inverted repeat transgenes silence mycotoxin production in Aspergillus and Fusarium species. Mol. Plant Microbe Interact. 2005;18:539–545. doi: 10.1094/MPMI-18-0539. [DOI] [PubMed] [Google Scholar]
- Paterson RRM, et al. Biochemical Techniques for Filamentous Fungi. Wallingford: CAB International; 1994. [Google Scholar]
- Payne GA, Brown MP. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- Perrin RM, et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007;3:e50. doi: 10.1371/journal.ppat.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst C, et al. Outbreak of an acute aflatoxicosis in Kenya in 2004: identification of the causal agent. Appl. Environ. Microbiol. 2007;73:2762–2764. doi: 10.1128/AEM.02370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper KB, Thom C. A Manual of the Penicillia. Baltimore: Williams & Wilkins Co.; 1949. [Google Scholar]
- Richard JL, Payne GA. Mycotoxins: Risks in plant, animal, and human systems. Ames, IA: Council for Agricultural Science and Technology; Task Force Report No. 139. 2003:xvi–199.
- Rohlfs M, et al. Secondary chemicals protect mould from fungivory. Biol. Lett. 2007;3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA. Current taxonomic schemes of the genus Aspergillus and its teleomorphs. In: Klich JWBMA, editor. Aspergillus: The biology and industrial applications. Butterworth Publishers; 1992. pp. 353–388. [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab E, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedsgaard J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J Chromatogr. A. 1997;760:264–270. doi: 10.1016/s0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- Spikes S, et al. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis. 2008;197:479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- Stinnett SM, et al. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 2007;63:242–255. doi: 10.1111/j.1365-2958.2006.05506.x. [DOI] [PubMed] [Google Scholar]
- Sugui JA, et al. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell. 2007a;6:1552–1561. doi: 10.1128/EC.00140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, et al. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell. 2007b;6:1562–1569. doi: 10.1128/EC.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis DI, Keller NP. Oxylipins act as determinants of natural product biosynthesis and seed colonization in Aspergillus nidulans. Mol Microbiol. 2006;59:882–892. doi: 10.1111/j.1365-2958.2005.05000.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson HH, et al. Increased conidiation associated with progression along the sterigmatocystin biosynthetic pathway. Mycologia. 2004;96:1190–1198. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disruption and overexpression of the laeA gene in A. flavus. (A) Schematic diagram of the laeA open reading frame and how it was replaced with the pyrG selectable marker to generate the ΔlaeA mutant, TJW71. (B) Southern analysis of wild type (WT) strain and the laeA deletion strains. Genomic DNA was digested by SpeI. A 4 kb PCR product containing laeA was obtained with the forward primer 5’CCTTGTATGATGTATGTATGATGAGC and the reverse primer 5’ TCTTGGGTCATTGGGTGGGCGG and was used as a laeA probe (indicated by bar in panel A). Expected hybridization band patterns: wild type strain, 2.5 kb and 1.7 kb bands; ΔlaeA strains, 4.0 and 1 kb bands. The arrowheads at the top of the figure indicate the correct knock-out mutants, (C) Strain TJW1.1 was transformed with a full length laeA gene. Strain TJW79.13, an overexpression (OE) strain, containing two copies of laeA, was chosen for further studies. Genomic DNA was digested by HindIII. Expected hybridization band patterns: wild type strain, 5.4 and 5.6 kb bands (overlapping two bands); ΔlaeA, 4.5, 3.2 and 1.7 kb bands and OE::laeA contained the ΔlaeApattern plus additional copies of laeA.
Loss of aflatoxin production in ΔlaeA mutants. Extracts of the wild type A. flavus NRRL 33357 and three ΔlaeA mutants (TJW71.1, TJW71.7 and TJW71.3) grown on YES media for 5 days at 29°C were separated on a thin layer chromatography plate. Lanes 1–3 represent replicates for each strain. Aflatoxin was visualized using long-wave (366 nm) UV light. Aflatoxin B1 standard is spotted on each side of the plate.
LaeA deletion results in lower aflatoxin and spore production during maize infection. (a) Maize seed inoculated with A. flavus NRRL 3357 or three ΔlaeA mutants (TJW71.1, TJW71.7 and TJW71.3) were incubated in the dark at 29°C for 3 days and conidial production counted as described in text. Values represented are a mean of 4 replicates. Columns with the same letter are not significantly different at a significance level of 0.01. (b) Thin layer chromatography of extracts from the same maize seed show an absence of aflatoxin production in the seed infected with TJW71.1, TJW71.7 or TJW71.3. Aflatoxin was visualized using long-wave (366 nm) UV light. Lanes 1–4 represent replicates for each strain. The lane marked C represents the control seed mock inoculated with water.