A role for B cell receptor (BCR) signaling in lymphomagenesis has been inferred by studying immunoglobulin genes in human lymphomas1,2 and by engineering mouse models3, but genetic and functional evidence for its oncogenic role in human lymphomas is needed. Here we describe a form of “chronic active” BCR signaling that is required for cell survival in the activated B cell-like (ABC) subtype of diffuse large B cell lymphoma (DLBCL). The signaling adapter CARD11 is required for constitutive NF-kB pathway activity and survival in ABC DLBCL4. Roughly 10% of ABC DLBCLs have mutant CARD11 isoforms that activate NF-kB5, but the mechanism that engages wild type CARD11 in other ABC DLBCLs has been elusive. An RNA interference genetic screen revealed that a BCR signaling component, the kinase BTK, is essential for survival of ABC DLBCLs with wild type CARD11. As well, knockdown of proximal BCR subunits (IgM, Igκ, CD79A, CD79B) killed ABC DLBCLs with wild type CARD11 but not other lymphomas. The BCRs in these ABC DLBCLs formed prominent clusters in the plasma membrane with low diffusion, similar to BCRs in antigen-stimulated normal B cells. Somatic mutations affecting the ITAM signaling modules6 of CD79B and CD79A were detected frequently in ABC DLBCL biopsy samples but rarely in other DLBCLs and never in Burkitt’s or MALT lymphomas. Remarkably, 18% of ABC DLBCLs mutated one functionally critical residue of CD79B, the first ITAM tyrosine. These mutations increased surface BCR expression and attenuated LYN kinase, a feedback inhibitor of BCR signaling. These findings establish chronic active BCR signaling as a new pathogenetic mechanism in ABC DLBCL, suggesting several therapeutic strategies.
DLBCL is a heterogeneous diagnostic category consisting of molecularly distinct subtypes that differ in gene expression, oncogenic aberrations and clinical outcome7,8. The ABC DLBCL subtype relies on constitutive NF-kB signaling to block apoptosis but the germinal center B cell-like (GCB) subtype does not9. Recurrent CARD11 mutations in ABC DLBCL provided genetic evidence that NF-kB signaling is central to its pathogenesis5. However, most ABC DLBCLs have wild type CARD11 yet nonetheless rely upon CARD11 to activate NF-kB signaling4,9.
In normal B cells, CARD11 is engaged upon antigenic stimulation of BCR signaling. Antigen specificity of the BCR is provided by surface immunoglobulin, but signaling is mediated by two associated proteins, CD79A (Ig-α) and CD79B (Ig-β)10. The CD79A/B heterodimer is a scaffold for the assembly and membrane expression of the BCR and also initiates downstream signaling to the NF-kB, PI3 kinase, ERK MAP kinase and NF-AT pathways. Engagement of the BCR by antigen induces SRC-family kinases to phosphorylate tyrosines in the ITAM motifs of CD79A and CD79B. The tyrosine kinase SYK is activated by binding to the phosphorylated ITAMs, triggering a signaling cascade that involves the tyrosine kinase BTK, phospholipase Cγ, and protein kinase C β (PKCβ). PKCβ phosphorylates CARD11, causing it to recruit BCL10 and MALT1 into a multiprotein “CBM” complex that activates IκB kinase (IKK), thereby initiating NF-kB signaling.
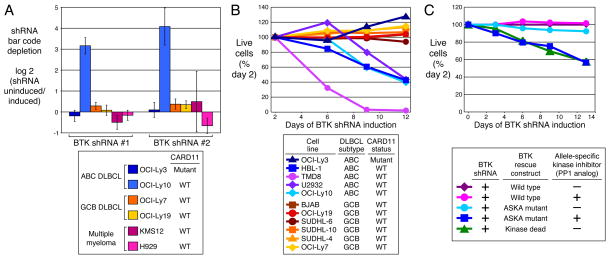
A potential role for BCR signaling in ABC DLBCLs with wild type CARD11 was revealed by an RNA interference screen. Two small hairpin RNAs (shRNAs) targeting the BCR pathway component BTK were highly toxic for an ABC DLBCL line with wild type CARD11 (OCI-Ly10) but not for one with mutant CARD11 (OCI-Ly3), nor for GCB DLBCL and multiple myeloma lines (Fig. 1A; Supplemental Fig. 1). In subsequent survival assays, a BTK shRNA was toxic for four ABC DLBCL lines with wild type CARD11 but not for OCI-Ly3 or six GCB DLBCL lines (Fig. 1B). BTK kinase activity was required to rescue ABC DLBCL lines from the toxicity of BTK knockdown (Fig. 1C).
Figure 1. BTK is a critical kinase for survival of ABC DLBCL cells.
A. RNA interference screen in lymphoma and multiple myeloma cell lines. An shRNA library targeting 442 kinases was screened in the indicated cell lines as described4. Shown is the selective toxicity of two BTK shRNAs after 3 weeks in culture. Bar values are mean +/− s.d. of four independent transductions. B. Selective toxicity of a BTK shRNA for ABC DLBCLs with wild type CARD11. DLBCL cell lines were infected with a retrovirus that expresses BTK shRNA #1 together with GFP. Shown is the fraction of GFP+ cells relative to the GFP+ fraction on day 2. C. BTK kinase activity is required for survival of ABC DLBCL cells. OCI-Ly10 cells were transduced with cDNAs encoding wild type or mutant BTK (kinase-dead allele or analog-sensitive kinase allele (ASKA)29). Wild type but not kinase-dead BTK rescued cells with endogenous BTK knockdown. The ASKA isoform-specific kinase inhibitor 1-NM-PP1 (2 mM) killed cells bearing the BTK ASKA allele.
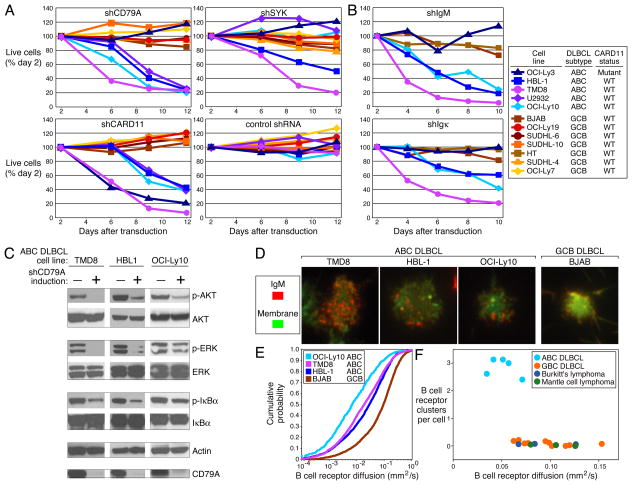
The role of BTK in BCR signaling prompted us to investigate the reliance of ABC DLBCLs on other BCR pathway components. A CD79A shRNA killed all four ABC DLBCL lines with wild type CARD11 but not the one with mutant CARD11 or the GCB DLBCL lines (Fig. 2A). In contrast, a CARD11 shRNA killed all ABC DLBCL lines and a control shRNA was non-toxic. In HBL-1, the knockdown of surface CD79A expression by different shRNAs caused a proportional decrease in surface IgM, implying that the toxicity of CD79A knockdown was due to loss of surface BCR (Supplemental Fig. 2A). CD79B shRNAs were also toxic to ABC DLBCLs, and the degree of CD79B knockdown was proportional to the decrease in surface BCR and to toxicity (Supplemental Fig. 2B, C). To investigate the role of the immunoglobulin receptor, we developed shRNAs targeting IgM and Igκ (Supplemental Fig. 3). These shRNAs were also selectively toxic to ABC DLBCLs with wild type CARD11, establishing a direct role for immunoglobulin in this signaling (Fig. 2B).
Figure 2. Chronic active BCR signaling in ABC DLBCL lines.
A. Survival of DLBCL cell lines following shRNA-mediated knockdown of BCR signaling components CD79A, SYK, and CARD11. B. Knockdown of immunoglobulin heavy or light chain is toxic for ABC DLBCLs with chronic active BCR signaling. C. Phosphoproteins in multiple signaling pathways depend upon chronic active BCR signaling. Indicated ABC DLBCL cell lines were transduced with an shRNA targeting CD79A and phosphorylated or total proteins were assessed by Western blotting before and after 48-hour shRNA induction. D. Clustering of IgM in the plasma membrane was observed only in ABC DLBCL lines with chronic active BCR signaling, using TIRF microscopy. Plasma membrane density was revealed by membrane dye R18. E. Reduced diffusion of surface IgM in ABC DLBCL lines with chronic active BCR signaling as compared to the GCB DLBCL line, as quantified by TIRF microscopy. F. Immobile BCR clusters are characteristic of lines representing ABC DLBCL but not other lymphoma types.
The NF-kB pathway is activated by BCR signaling in ABC DLBCLs since knockdown of BTK, CD79A, CD79B and CARD11 decreased expression of NF-kB target genes and inhibited IKK (Supplemental Fig. 4). BCR signaling also activates the PI3 kinase and ERK MAP kinase pathways in these cells since CD79A knockdown inhibited phosphorylation of AKT and ERK in addition to IκBα (Fig. 2C).
A subsequent focused shRNA screen suggested that other BCR signaling components contribute to chronic active BCR signaling, including SYK, BLNK, PLCG2, and PRKCB (Supplemental Fig. 5). A SYK shRNA killed two ABC DLBCL lines with wild type CARD11 (HBL-1, TMD8) but not two others (OCI-Ly10, U2932), and also had no effect on OCI-Ly3 or GCB DLBCL lines (Fig. 2A), despite comparable knockdown (Supplemental Fig. 6A). Not only was OCI-Ly10 insensitive to SYK knockdown but it also died upon ectopic expression of wild type but not kinase-dead SYK (Supplemental Fig. 6B). A previous study using a SYK inhibitor, R406, concluded that most DLBCLs rely upon tonic BCR signaling11. However, R406 killed SYK-independent GCB and ABC DLBCL lines (including OCI-Ly10), suggesting that its toxicity in these lines may be due to inhibition of other kinases and not BCR signaling (Supplemental Fig. 6C).
We next used total internal reflection fluorescence (TIRF) microscopy to visualize BCRs on the surface of lymphoma lines. In normal mouse B cells, TIRF microscopy revealed that antigen exposure causes BCRs to form clusters with reduced diffusion, leading to BCR signaling12. All 5 ABC DLBCL lines displayed prominent BCR clusters that were not present in 16 other lines derived from GCB DLBCL, Burkitt’s lymphoma or mantle cell lymphoma (Fig. 2D, F). BCR clusters were also observed in biopsies from 3 patients with ABC DLBCL (Supplemental Fig. 7A). Moreover, the BCRs in ABC DLBCLs diffused less rapidly than those in other lymphoma lines (Fig. 2E, F). We observed a correlation between BCR clusters and phosphotyrosine accumulation in ABC DLBCL lines suggesting that these structures may be actively signaling (Supplemental Fig. 7B).
Together, these findings establish an ongoing requirement for proximal BCR signaling in ABC DLBCLs with wild type CARD11. Since these lines also depend upon CARD11, like antigen-activated normal B cells, we refer to this phenomenon as “chronic active” BCR signaling. We wish to distinguish chronic active BCR signaling from “tonic” BCR signaling. Tonic BCR signaling promotes cell survival in all mature mouse B cells13,14, but mice deficient in CBM components have relatively normal numbers of mature follicular B cells15. It thus appears likely that CARD11 is not essential for tonic BCR signaling but is required for chronic active BCR signaling. Moreover, chronic active BCR signaling is characterized by BCR clustering, which is not observed in resting mouse B cells that depend on tonic BCR signaling12.
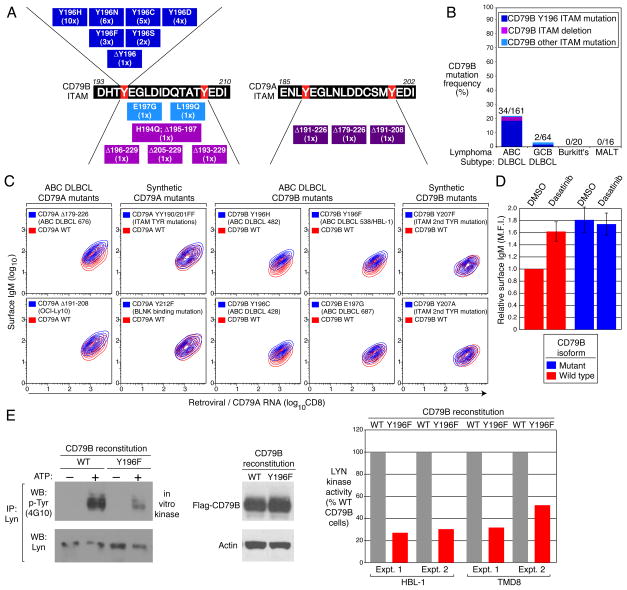
To provide genetic evidence of BCR signaling in the pathogenesis of ABC DLBCL, we resequenced genes in the BCR pathway in DLBCL cell lines and biopsies. We identified missense mutations affecting the first tyrosine of the CD79B ITAM motif in two cell lines, HBL-1 (Y196F) and TMD8 (Y196H) (Fig. 3A). Both lines were heterozygous for this mutation, but >90% of the CD79B mRNA in HBL-1 was derived from the mutant allele (data not shown). These mutations prompted us to resequence the CD79B ITAM region in 225 DLBCL biopsies. Remarkably, in 18% (29/161) of ABC DLBCLs the first ITAM tyrosine was replaced by a variety of amino acids due to point mutations and in one case, this residue was removed by a 3 base pair deletion (Fig. 3A, B). Less common were missense mutations in other ITAM residues and deletions that disrupted all or part of the motif. Of 64 GCB DLBCLs, only one had a mutation affecting the first ITAM tyrosine and one other had a different ITAM mutation (L199Q). Overall, the frequency of CD79B ITAM mutations was significantly higher in ABC DLBCL (21.1%) than in GCB DLBCL (3.1%) (p=8.9 × 10−4). CD79B ITAM mutations were not present in 20 Burkitt’s lymphoma and 16 gastric MALT lymphoma biopsies. For 6 ABC DLBCL cases, analysis of non-malignant tissue established that the CD79B mutations were somatically acquired by the malignant cells (Supplemental Fig. 8).
Figure 3. CD79A and CD79B ITAM mutations in ABC DLBCL.
A. CD79B and CD79A ITAM mutations in DLBCL biopsies and lines (case number in parenthesis). B. CD79B ITAM mutation frequencies in lymphoma biopsies. C. Mutant CD79A and CD79B isoforms increase surface IgM. The GCB DLBCL line BJAB was reconstituted with either wild type or mutant CD79A/B proteins. Surface IgM is depicted relative to CD79 RNA levels, estimated using bicistronic expression of CD8. “Synthetic” mutants were not observed in patient samples. D. CD79B mutations prevent down-modulation of surface BCR by BCR signaling. The ABC DLBCL line HBL-1 was reconstituted with wild type or Y196H mutant CD79B and treated for 24 hours with DMSO or dasatinib, a BCR signaling inhibitor. Surface IgM (mean fluorescence intensity; M.F.I.) is depicted relative to the levels in cells with wild type CD79B treated with DMSO. Error bars depict +/− s.e.m.; 2 experiments. E. CD79B mutations inhibit LYN kinase activity in ABC DLBCLs. The indicated ABC DLBCL lines were reconstituted with wild type or Y196F mutant CD79B. LYN kinase activity in immunoprecipitates was estimated by densitometric analysis of Western blots as phospho-LYN (using anti-phospho-tyrosine antibody 4G10) relative to total LYN.
The CD79A ITAM region of the ABC DLBCL line OCI-Ly10 has a splice donor site mutation16 causing an 18 amino acid deletion that removes most of the ITAM, including the second tyrosine. Though OCI-Ly10 was heterozygous for this mutation, >90% of the CD79A mRNA was mutated (data not shown). One ABC DLBCL biopsy had a similar splice site mutation and another had mutations that deleted the entire CD79A ITAM (Fig. 3A). CD79A mutations were rare among ABC DLBCLs, occurring in 2.9% (2/68) of biopsies.
In mouse B cells, mutations in the CD79A or CD79B ITAM tyrosines elevate surface BCR expression by inhibiting receptor internalization17. Indeed, GCB DLBCL cells reconstituted with CD79A or CD79B mutants derived from ABC DLBCLs had more surface IgM expression than cells with wild type isoforms, but this was not the case for CD79 ITAM mutations that were not observed in patient samples (Fig. 3C). Likewise, ABC DLBCL cells reconstituted with mutant CD79B had higher surface BCR expression than those reconstituted with wild type CD79B (Fig. 3D). Interruption of chronic active BCR signaling with the kinase inhibitor dasatinib (see below) elevated surface BCR expression in ABC DLBCL cells with wild type but not mutant CD79B (Fig. 3D). Hence, one function of the CD79 mutations is to maintain surface BCR expression in the face of chronic active BCR signaling.
We hypothesized that the CD79B mutations might be genetically selected in ABC DLBCLs for their ability to circumvent negative regulatory circuits that attenuate BCR signaling. Whereas several SRC-family tyrosine kinases can initiate BCR signaling, LYN is unique in mediating negative feedback on BCR signaling18. Indeed, LYN-deficient mice succumb to an autoimmune disease that has been traced to BCR hyperactivity19. Interestingly, LYN is required for BCR internalization20,21, suggesting that CD79 mutations might elevate surface BCR expression by inhibiting LYN. To test this, we knocked down endogenous CD79B expression in HBL-1 and TMD8 cells, both of which harbor a CD79B mutation, and complemented them with exogenous wild type or mutant CD79B cDNAs. Immunoprecipitation of LYN followed by an in vitro kinase assay demonstrated greater LYN kinase activity in cells reconstituted with wild type CD79B (Fig. 3E). These data suggest a model in which CD79B mutations are selected in ABC DLBCLs to attenuate negative autoregulation by LYN during chronic active BCR signaling.
The CD79 mutants are not loss-of-function mutants since they prevented death of ABC DLBCL cells caused by knockdown of endogenous CD79 isoforms (Supplemental Fig. 9). However, the CD79 mutants were not functionally superior to their wild type counterparts in this assay (Supplemental Fig. 9), and did not spontaneously activate NF-kB when introduced into GCB DLBCL cells, unlike CARD11 mutants5 (data not shown). We therefore propose that the CD79 ITAM mutations may be selected early in the genesis of the malignant clone, perhaps to allow it to respond abnormally well to a self or foreign antigen (Supplemental Fig. 10). Interestingly, mutations that impair CD79A or CD79B ITAM function in mouse B cells lead to exaggerated antigen responsess17,22,23. Future research should investigate the potential role of antigenic stimulation in chronic active BCR signaling and in the spontaneous BCR clustering that characterizes ABC DLBCLs. BCR clustering does not depend upon the CD79B mutations (Supplemental Fig. 11), suggesting that other mechanisms contribute to this aspect of chronic active BCR signaling.
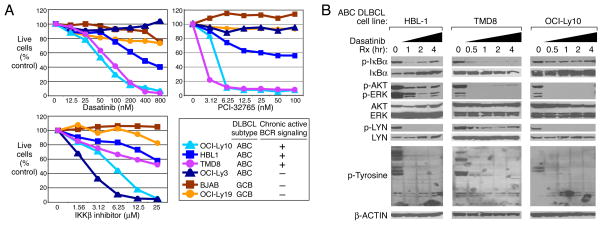
We considered therapeutic strategies to exploit chronic active BCR signaling in ABC DLBCL. Dasatinib, a BCR-ABL inhibitor approved for the treatment of chronic myelogenous leukemia, also inhibits other SRC-family kinases and BTK24. Dasatinib killed ABC DLBCL lines that rely upon chronic active BCR signaling but not the BCR-independent line OCI-Ly3 or GCB DLBCL lines (Fig. 4A). A selective BTK inhibitor, PCI-3276525, was also selectively toxic to cell lines with chronic active BCR signaling (Fig. 4A). By contrast, all ABC DLBCL lines were sensitive to an IKKβ inhibitor. In BCR-dependent lines, dasatinib reduced IκBα, AKT, ERK, and LYN phosphorylation, as well as total protein tyrosine phosphorylation and IKK activity (Fig. 4B; Supplemental Fig. 12). Dasatinib toxicity may thus be due to NF-κB inhibition, which causes apoptosis, and AKT/mTOR inhibition, which causes “metabolic catastrophe”26. Indeed, rapamycin, an mTOR inhibitor, synergized with an IKKβ inhibitor in killing ABC DLBCL lines with chronic active BCR signaling (Supplemental Fig. 13). Our studies suggest that the position of molecular lesions in the BCR and NF-kB signaling pathways could be used to guide therapy of ABC DLBCL. ABC DLBCLs with wild type CARD11 and chronic active BCR signaling might respond to a BTK inhibitor, such as PCI-32765, and possibly to inhibitors of SRC-family kinases, PKCβ, or SYK, in some cases. By contrast, CARD11-mutant tumors would need to be treated with agents that target downstream components of the NF-kB pathway such as IKK27. A precise delineation of which ABC DLBCL cases depend on chronic active BCR signaling awaits the development of predictive biomarkers and on the results of clinical trials involving BCR signaling inhibitors.
Figure 4. Therapeutic strategies to target chronic active BCR signaling.
A. Viability of DLBCL lines assessed by MTT assay after 4 days of treatment with varying doses of dasatinib, the BTK inhibitor PCI-32765 (compound 13 in ref.25), or an IKKβ inhibitor27. B. Effect of dasatinib on phospho-protein levels in ABC DLBCL cells. Three ABC DLBCL lines were treated with dasatinib (50 nM) for the indicated times and analyzed by Western blotting.
Methods Summary
Cell lines possessing the ecotropic retroviral receptor and the tetracycline repressor were generated and used in RNA interference library screening, shRNA toxicity assays and cDNA complementation studies as described4. DLBCL cell lines were assigned to the ABC or GCB subtypes by gene expression profiling (ref.4; Supplemental Fig. 14). shRNA screening results are given in Supplemental Tables 1 and 3, and shRNA sequences are listed in Supplemental Tables 2 and 3. Specific shRNA-mediated mRNA and protein knockdown was documented (Fig. 2C; Supplemental Fig. 6A, 15). IKK reporter lines were engineered to express an IκBα-Photinus luciferase fusion and Renilla luciferase27. TIRF imaging of the BCR was based on previously described techniques12.
Tumor biopsies were obtained prior to treatment from patients with de novo DLBCL28, gastric MALT lymphoma, and Burkitt’s lymphoma. All samples were studied according to a protocol approved by the National Cancer Institute Institutional Review Board.
Methods
Cell lines
All cell lines were maintained in a humidified 5% CO2 incubator at 37° C. Cell lines were grown in RPMI 1640 medium supplemented with glutamine, beta-mercaptoethanol, penicillin/streptomycin, and 10% fetal bovine serum, except for OCI-Ly3 and OCI-Ly10, which were maintained in Iscove’s modified Dulbecco’s medium supplemented with beta-mercaptoethanol, penicillin/streptomycin, and 20% heparinized human plasma. All cell lines had been previously modified to express an ecotropic retroviral receptor, as well as a fusion protein of the Tet repressor and the blasticidin resistance gene, as previously described4.
Retroviral transductions
Retroviral supernatants were prepared as previously described5. In brief, Lipofectamine 2000 (Invitrogen) was used to transfect 293T producer cells with a plasmid mixture for gag and pol, a mutant ecotropic env, and each particular retrovirus. After two days, supernatant was passed through a 0.45 μM filter, mixed with polybrene (8 μg/mL), and used for centrifugal transduction of target cells expressing the ecotropic retroviral receptor. In some instances a second infection was performed, using fresh supernatant collected 3 days after transfection of producer cells.
shRNA library screening
shRNA library screening was performed as described4. The shRNA library was constructed in a pMSCV-based retroviral vector (pRSMX_Puro) with two expression cassettes, one for constitutive expression of a selectable marker (conferring puromycin resistance) and the other for inducible expression of shRNA, after release of binding of the bacterial tetracycline repressor by doxycycline (20 ng/mL).
For the focused screen of BCR pathway genes (Supplemental Fig. 5), the same procedure was applied except that the abundance of each shRNA was enumerated using an Illumina GAII sequencer. Comparison was made between an shRNA-uninduced sample from day 0 and an shRNA-induced sample from day 24 of culture. The magnitude and standard error of the shRNA effect was calculated by using a logistic regression model to estimate the relative probability that an shRNA was found in the shRNA-induced vs. uninduced samples. This model included a normalization factor that was a constant for all genes from a given shRNA pool in a given experiment and including normal random effects representing gene by experiment interactions.
Other shRNA vectors and shRNA sequences
Modifications were made to the puromycin resistance gene cassette in the pMSCV-based retroviral shRNA vector, including fusion of the puromycin resistance gene to EGFP (pRSMX_PuroGFP) as previously described4 or replacement of the puromycin resistance gene with the E. coli IMPDH gene to allow selection in mycophenolic acid. shRNA sequences used in this study are presented in Supplemental Tables 2 and 3.
Expression vectors and cDNA mutagenesis or modification
Retroviral vectors for inducible cDNA expression were either pBMN-based (http://www.stanford.edu/group/nolan/plasmid_maps/pmaps.html), or pMSCV-based with the cDNA expressed from a doxycycline-inducible CMV promoter in which a binding site for the bacterial tetracycline repressor is inserted at the transcription start site (derived from pCDNA4/TO (Invitrogen)). All mutagenesis was performed using the QuikChange kit from Stratagene and verified by dye termination sequencing. Modification of the CD79A and CD79B cDNAs for the surface expression experiments shown in Fig. 4C, D, and E included insertion of a FLAG peptide coding sequence just downstream of the signal peptide cleavage site, and fusion to EGFP at the 3′ end, after removal of the stop codon and addition of a 6 amino acid spacer as previously described30. 3′ GFP fusions were also constructed for SYK (Supplemental Fig. 6B).
shRNA toxicity and complementation assays
The toxicity of individual shRNA sequences was chiefly tested with the PuroGFP vector as previously described4. In brief, following infection with an shRNA- and GFP-expressing retrovirus, FACS was used to determine the fraction of live cells that were GFP+ two days after transduction. Doxycycline was then added to induce shRNA expression and the fraction of GFP+ live cells was determined at various intervals during subsequent culture. Parallel cultures were prepared using a vector expressing a control shRNA. The GFP+ fraction from the test shRNA cultures were normalized both to the GFP+ fraction from the control shRNA culture on the same day and to the GFP+ fraction on Day 2. In the case of CD79B (Supplemental Fig. 2B, C), adequate knockdown by shRNA required selection of shRNA- and GFP-expressing single cell clones by limiting dilution. Toxicity assays of these clones were performed by mixing the clones with untransduced cells and determining an initial GFP+ fraction prior to the addition of doxycycline to induce shRNA expression. The fraction of GFP+ cells at various time points were normalized to this initial value.
For complementation studies with CD79A (Supplemental Fig. 9), OCI-Ly10 cells were infected with retroviruses expressing wild type or mutant CD79A coding regions along with Lyt2 (mouse CD8). Subsequently, cells were infected with a retrovirus expressing a CD79A shRNA (targeting the 3′UTR) and GFP and the fraction of Lyt2+/GFP+ cells was monitored over time as above.
For complementation studies with CD79B (Supplemental Fig. 9), HBL-1 cells were infected with a retrovirus expressing a CD79B shRNA (targeting the 3′UTR) and GFP, selected in puromycin, and single cell cloned. A clone was selected with the best doxycycline-inducible knockdown of CD79B as assessed by FACS. This clone was infected with a retrovirus expressing a wild type or mutant CD79B coding region along with Lyt2 (mouse CD8). These GFP+ cells were mixed with unmodified HBL-1 cells and the fraction of GFP+/Lyt2+ cells was monitored over time, as above.
To analyze the influence of the CD79B mutations on chronic active BCR signaling in ABC DLBCLs (Fig. 4D, E), HBL1 or TMD8 cells were infected with a retroviral vector with doxycycline-inducible expression of an shRNA directed at the CD79B 3′UTR-directed and an IMPDH drug selection marker. After selection in mycophenolic acid, single cell clones were assayed for doxycycline-inducible knockdown of CD79B by FACS. Clones were subsequently infected with a retrovirus expressing wild type of mutant (Y196F) CD79B coding region cDNAs from the MSCV LTR together with a hygromycin resistance gene. After selection in hygromycin, the CD79B shRNA was induced for 3–5 days prior to assay.
BTK ASKA assay
BTK was modified by a “gatekeeper” T474A mutation, as well as a second S538A mutation used previously to construct a BTK ASKA mutant31. Wild type, kinase-dead (K430R), and ASKA forms of BTK were introduced as 3′GFP fusions without a selectable marker, and then the mixed (GFP- and GFP+) population was infected with a retrovirus expressing an shRNA targeting the BTK 3′ UTR and selected with puromycin. The starting proportion of GFP+ cells was determined by FACS with bead quantitation, and then shRNA expression was induced with doxycycline in the presence or absence of the ASKA inhibitor 1NM-PP1 (2 μM; Sigma) for continued culture and periodic quantitation. The increase in GFP+ cells in the various cultures was normalized to the increase observed for WT BTK without 1NM-PP1.
FACS assays for protein level
For quantitation of most surface markers, FACS was performed by staining unfixed cells on ice. For quantitation of CD79A, cells were fixed in 1.6% paraformaldehyde, permeabilized in methanol, and stained with an antibody recognizing a peptide sequence in the intracellular domain (BD Biosciences). For analysis of total and surface-expressed levels of exogenous mutant CD79A or CD79B in the BJAB model system, as well as their effect on surface IgM (Fig. 4C), BJAB cells were first prepared to express an shRNA targeting either endogenous gene, using selected single clones in the case of CD79B, and then infected with bicistronic expression vectors containing 5′FLAG- and 3′GFP-tagged versions (described above) of the respective genes and an IRES-Lyt2 (murine CD8a) cassette. After induced knockdown of the endogenous proteins, 3-color FACS assays were performed in which GFP fluorescence was used as a measure of total exogenous protein, an Alexa-647 antibody against Lyt2 (BD Biosciences, excited with a He/Ne laser) was used as a marker of exogenous mRNA, and a PE-conjugated antibody was used to detect surface expression of IgM (direct) or FLAG (by secondary detection of unlabeled mouse anti-FLAG, M2 from Sigma).
Western blotting
Control or doxycycline-treated cells were lysed in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% NP-40, 2 mM EDTA) supplemented with a cocktail of protease inhibitors (Roche) and phosphatase inhibitors (Sigma) for 30 min. Lysates were cleared by centrifugation at 15,000 × g at 4° C for 10 min and protein concentrations were determined by BCA protein assay (Pierce). 80–100 ug of lysates were subjected to electrophoresis through a 4–12% Bis-Tris gel (Invitrogen) and immobilized on the nitrocellulose membranes. Proteins were detected by using the following antibodies: CD79A, SYK, IκBα (Santa Cruz Biotechnology), BTK (ref.32; kindly provided by Donn Stewart), p-tyrosine (4G10, Milipore), AKT, ERK, p-AKT (S473), p-ERK (p44-T202, p42-Y204), β-ACTIN, p-LYN (Y507), and p-Iκ B α (S32) (Cell Signaling Technology).
Lyn Immunoprecipitation and in Vitro Kinase Assay
Cells were suspended at 107 cells/ml in PBS and lysed by mixing 1:1 (v/v) with RIPA buffer (0.5% Triton X-100, 0.5% deoxycholate, and 0.05% SDS) in lysis buffer. Cells were lysed for 10 min on ice and then clarified by microcentrifugation (12,000 × g) for 20 min at 4 °C to yield a postnuclear supernatant (PNS). Lyn was immunoprecipitated from 0.5–1 ml of detergent extracts by incubation with 2 μg anti-Lyn mouse monoclonal antibody H6 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 1–2 h on ice and then rotated for 45 min with 35 μl of ImmunoPure Immobilized Protein A (Pierce) at 4 °C. Immunoprecipitates were washed twice with lysis buffer without detergent and then once with kinase assay buffer (20 mM Tris, pH 7.6, 10 mM MgCl2, and 1 mM Na3VO4). After washing, Lyn immunoprecipitates were subjected to in vitro kinase assays by adding 200 μl of kinase assay buffer containing either 1 mM ATP or no ATP. Samples were then incubated at 37 °C for samples for 15 min. Reactions were quenched by the addition of 50 μl of 5× non-reducing SDS sample buffer followed immediately by boiling.
MTT assays
Cells in 96-well plates were treated with dasatinib, rapamycinand/or IKKβ inhibitor for 4 or 8 days with replacement of fresh medium supplemented with 1× concentration of drugs every 2 days. At harvesting, cells were treated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) for 0.5–1 hour and the produced colored substrate was solubilized in isopropanol and 1% hydrochloric acid. The absorbance of the colored supernatant at 570-nm wavelength was measured by a spectrophotometer with subtraction of background absorbance at 630-nm wavelength.
TIRF microscopy
TIRF imaging of the BCR was based on previously described techniques12. Briefly, surface IgM was labeled by staining with anti-IgM-Cy5 Fab (Jackson Research) for 15 min on ice in Hank’s balanced salt solution containing 1% calf serum. Cells were then incubated with 1 mM R18 dye (Invitrogen) at room temperature for 1 min, washed and allowed to adhere to coverslips coated with planar lipid bilayers consisting of 1,2-dioleoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids). Cells were imaged live at 37°C, or alternatively, fixed with 2% paraformaldehyde, permeabilized with 0.1% Triton-X100 and stained with anti-phosphotyrosine antibodies (4G10-FITC, Upstate). Colocalization of the BCR and phosphotyrosine signal was quantified by Pearson correlation coefficient using ImageJ (National Institutes of Health). For measurements of single molecule diffusion of the BCR, surface IgM was labeled with 2 ng/ml anti-IgM-Cy3 Fab for 10 min at room temperature and tracks of single BCR molecules were recorded at 35 ms resolution. Images were bandpass-filtered and the position of the BCR in each point of the track was determined by two-dimensional Gaussian fitting using Matlab (Mathworks) scripts. Short-range diffusion coefficients from individual BCR tracks were calculated as described12.
IKK reporter assay
As previously described for OCI-Ly34, stable clones of TMD8 were constructed with separate vectors to express a fusion between IκBα and Photinus luciferase (from pGL3, Promega) as the reporter, and Renilla luciferase (from phRL-TK, Promega) for normalization. Clones responsive to an IKKβ small molecule inhibitor were identified and used with the OCI-Ly3 reporter clone for short-term (4 hr) incubation with dasatinib (Supplemental Fig. 12), or, after infection with pRSMX_Puro and selection, shRNA induction for 1–4 days (Supplemental Fig. 4C). After development with the Dual-Glo luciferase assay system (Promega), the ratio of IkBα-Photinus to Renilla luminescence was normalized to that in untreated or uninduced cultures.
Patient samples
Tumor biopsy specimens were obtained prior to treatment from 223 patients with de novo DLBCL that have previously been analyzed by gene expression profiling28, 16 patients with Mucosa-associated lymphoid tissue (MALT) lymphoma, and 20 patients with Burkitt’s lymphoma. All samples were studied according to a protocol approved by the National Cancer Institute Institutional Review Board.
PCR amplification and sequencing
Genomic DNA from cell lines and patient samples was extracted with the DNeasy Tissue kit (Qiagen) according to the manufacturer’s instructions. Long distance PCR for CD79A and CD79B was performed with the LA PCR kit (Takara Bio Inc.) using the following conditions: 94°C for 5 min followed by 30 cycles of denaturation: 30 sec. at 94°C, annealing: 30 sec. at 60°C, extension: 6–7 min. at 72°C and final extension for 10 min at 72°C. PCR primers used were:
| Gene | Primer name | Sequence |
|---|---|---|
| CD79A | CD79A_1_f CD79A_1_r |
5′-TCCACTCACAGCCTGAAGCATAC-3′ 5′-GGTTAGGAGGTGGGGCAGTTTAG-3′ |
| CD79B | CD79B_1_f CD79B_1_r |
5′-GGTGCAGTTACACGTTTTCCTCC-3′ 5′-TGGTTGCGGGAGAGGAATGATG-3′ |
The PCR products were visualized by electrophoresis on a 0.8% agarose gel and ethidium bromide staining. The templates were purified using the QIAquick PCR Purification Kit (Qiagen) and subsequently sequenced (BigDye sequencing system, Applied Biosystems). Mutations were confirmed on independent PCR products.
RT PCR and TA cloning
1 mg of total RNA from ABC DLBCL cell lines was transcribed using the GeneAmp RNA PCR Core Kit (Applied Biosystems) according to the manufacturer’s instructions. cDNA was amplified using the following conditions: 94°C for 10 min followed by 40 cycles of denaturation: 30 sec. at 94°C, annealing: 30 sec. at 58°C, extension: 1 min. at 72°C and final extension for 10 min at 72°C. The templates were purified using the QIAquick Gel Extraction Kit (Qiagen) and subsequently TA-cloned using the TOPO TA Cloning Kit (Invitrogen) according to the manufacturer’s instructions. Between 12 and 28 clones were picked, bacterial cultures were grown and plasmid DNA was isolated and subsequently sequenced.
PCR primers used were:
| CD79a | CD79a_2_f CD79a_2_r |
5′-GCAACTCAAACTAACCAACCCACTG-3′ 5′-CACTAACGAGGCTGCTACAATCAG-3′ |
| CD79b | CD79b_2_f CD79b_2-r |
5′-ATGGGATTCAGCACCTTGGC-3′ 5′-CCTCATAGGTGGCTGTCTGGTC-3′ |
Gene-expression profiling
Total RNA (Trizol reagent; Invitrogen, Carlsbad, CA) was prepared from HBL-1 cells following incubation with 25 μmol/L MLN120B (Millennium Pharmaceuticals, Cambridge, MA) for 2, 3, 4, 6, 8, 12, 16, and 24 hours. In addition, HBL-1 cells were infected with retroviral vectors expressing various shRNAs in a doxycycline-inducible fashion (Supplemental Fig. 4A, B; Supplemental Table 2), selected with puromycin, treated with doxycycline for 24 or 48 hr and then harvested for total RNA. Uninduced cultures were prepared in parallel.
Gene expression was measured using whole-genome Agilent 4×44K gene expression arrays (Agilent, Santa Clara, CA), following the manufacturer’s protocol. Signals from either untreated or uninduced HBL-1 cells (labelled with Cy3) were compared to signals from the respective MLN120B-treated or doxycycline-induced cells (labeled with Cy5). For each sample, 2 mg total RNA were used for the preparation of fluorescent probes.
A gene was selected as an NF-κB target gene in HBL-1 cells if MLN120B decreased expression of the gene by > 0.65 log2 (1.57-fold) at ≥ 4 time points. This NF-κB target gene signature was subsequently applied to the gene expression data following induction of shRNAs directed against BTK, CARD11, SYK, and CD79A.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and National Institute of Allergy and Infectious Disease. P.R. is an HHMI-NIH Research Scholar. We thank Lee Honigberg (Pharmacyclics) for PCI-32765, Lenny Dang (Millennium Pharmaceuticals) for IKKβ inhibitors, Sherry Ansher (CTEP, NCI) for dasatinib, and Shuji Tohda (Tokyo Medical and Dental University) for the TMD8 cell line.
Footnotes
Author contributions
RED, VNN, GL, PT, RY, HK, LL, and ALS designed and performed experiments. HZ, YY, and WX performed experiments. GW, WX, and JP analyzed data. JJ and CJT synthesized reagents. AR, GO, HKM-H, RDG, JMC, LMR, EC, ESJ, JD, EBS, RIF, RMB, RRT, JRC, DDW, and WCC supplied patient samples and reviewed pathological and clinical data. SKP supervised research. LMS designed and supervised research and wrote the manuscript.
References
- 1.Klein G, Klein E. Conditioned tumorigenicity of activated oncogenes. Cancer Res. 1986;46 (7):3211–3224. [PubMed] [Google Scholar]
- 2.Bahler DW, Levy R. Clonal evolution of a follicular lymphoma: evidence for antigen selection. Proc Natl Acad Sci U S A. 1992;89 (15):6770–6774. doi: 10.1073/pnas.89.15.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Refaeli Y, et al. The B cell antigen receptor and overexpression of MYC can cooperate in the genesis of B cell lymphomas. PLoS Biol. 2008;6 (6):e152. doi: 10.1371/journal.pbio.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441 (7089):106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 5.Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319 (5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 6.Reth M. Antigen receptor tail clue. Nature. 1989;338 (6214):383–384. doi: 10.1038/338383b0. [DOI] [PubMed] [Google Scholar]
- 7.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 8.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappa B activity is required for survival of activated B Cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194 (12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dal Porto JM, et al. B cell antigen receptor signaling 101. Mol Immunol. 2004;41 (6–7):599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, et al. SYK-dependent tonic B-cell receptor signaling is a rational treatment target in diffuse large B-cell lymphoma. Blood. 2008;111 (4):2230–2237. doi: 10.1182/blood-2007-07-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolar P, Hanna J, Krueger PD, Pierce SK. The Constant Region of the Membrane Immunoglobulin Mediates B Cell-Receptor Clustering and Signaling in Response to Membrane Antigens. Immunity. 2009 doi: 10.1016/j.immuni.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90 (6):1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 14.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117 (6):787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 15.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4 (5):348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 16.Gordon MS, Kanegai CM, Doerr JR, Wall R. Somatic hypermutation of the B cell receptor genes B29 (Igbeta, CD79b) and mb1 (Igalpha, CD79a) Proc Natl Acad Sci U S A. 2003;100 (7):4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gazumyan A, Reichlin A, Nussenzweig MC. Ig beta tyrosine residues contribute to the control of B cell receptor signaling by regulating receptor internalization. J Exp Med. 2006;203 (7):1785–1794. doi: 10.1084/jem.20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gauld SB, Cambier JC. Src-family kinases in B-cell development and signaling. Oncogene. 2004;23 (48):8001–8006. doi: 10.1038/sj.onc.1208075. [DOI] [PubMed] [Google Scholar]
- 19.Chan VW, Meng F, Soriano P, DeFranco AL, Lowell CA. Characterization of the B lymphocyte populations in Lyn-deficient mice and the role of Lyn in signal initiation and down-regulation. Immunity. 1997;7 (1):69–81. doi: 10.1016/s1074-7613(00)80511-7. [DOI] [PubMed] [Google Scholar]
- 20.Niiro H, et al. The B lymphocyte adaptor molecule of 32 kilodaltons (Bam32) regulates B cell antigen receptor internalization. J Immunol. 2004;173 (9):5601–5609. doi: 10.4049/jimmunol.173.9.5601. [DOI] [PubMed] [Google Scholar]
- 21.Ma H, et al. Visualization of Syk-antigen receptor interactions using green fluorescent protein: differential roles for Syk and Lyn in the regulation of receptor capping and internalization. J Immunol. 2001;166 (3):1507–1516. doi: 10.4049/jimmunol.166.3.1507. [DOI] [PubMed] [Google Scholar]
- 22.Kraus M, Saijo K, Torres RM, Rajewsky K. Ig-alpha cytoplasmic truncation renders immature B cells more sensitive to antigen contact. Immunity. 1999;11 (5):537–545. doi: 10.1016/s1074-7613(00)80129-6. [DOI] [PubMed] [Google Scholar]
- 23.Torres RM, Hafen K. A negative regulatory role for Ig-alpha during B cell development. Immunity. 1999;11 (5):527–536. doi: 10.1016/s1074-7613(00)80128-4. [DOI] [PubMed] [Google Scholar]
- 24.Hantschel O, et al. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc Natl Acad Sci U S A. 2007;104 (33):13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Z, et al. Discovery of selective irreversible inhibitors for Bruton’s tyrosine kinase. ChemMedChem. 2007;2 (1):58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 26.Jin S, DiPaola RS, Mathew R, White E. Metabolic catastrophe as a means to cancer cell death. J Cell Sci. 2007;120 (Pt 3):379–383. doi: 10.1242/jcs.03349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam LT, et al. Small Molecule Inhibitors of IkB-Kinase are Selectively Toxic for Subgroups of Diffuse Large B Cell Lymphoma Defined by Gene Expression Profiling. Clin Cancer Res. 2005;11:28–40. [PubMed] [Google Scholar]
- 28.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blethrow J, Zhang C, Shokat KM, Weiss EL. Design and use of analog-sensitive protein kinases. Curr Protoc Mol Biol. 2004;Chapter 18(Unit 18):11. doi: 10.1002/0471142727.mb1811s66. [DOI] [PubMed] [Google Scholar]
- 30.Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat Immunol. 2005;6 (11):1168–1176. doi: 10.1038/ni1262. [DOI] [PubMed] [Google Scholar]
- 31.Jiang S, et al. Chemical genetic transcriptional fingerprinting for selectivity profiling of kinase inhibitors. Assay Drug Dev Technol. 2007;5 (1):49–64. doi: 10.1089/adt.2006.032. [DOI] [PubMed] [Google Scholar]
- 32.Stewart DM, Kurman CC, Nelson DL. Production of monoclonal antibodies to Bruton’s tyrosine kinase. Hybridoma. 1995;14 (3):243–246. doi: 10.1089/hyb.1995.14.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.