Abstract
Targeted gene silencing by RNA interference (RNAi) requires loading of a short guide RNA (siRNA or miRNA) into an Argonaute protein to form the functional center of an RNA-induced silencing complex (RISC). In humans, Argonaute2 (Ago2) assembles with the guide RNA-generating enzyme Dicer and the RNA-binding protein TRBP to form a RISC-loading complex (RLC) necessary for efficient transfer of nascent siRNAs and miRNAs from Dicer to Ago2. Here we show, using single-particle electron microscopy analysis, that human Dicer exhibits an L-shaped structure. Withn the RLC Dicer's N-terminal DExH/D domain, located at the short base branch, interacts with TRBP, while its C-terminal catalytic domains in the main body are proximal to Ago2. A model generated by docking the available atomic structures of Dicer and Argonaute homologs into the RLC reconstruction suggests a mechanism for siRNA transfer from Dicer to Ago2.
In RNA interference (RNAi) and related pathways, ∼21 nt. guide RNAs assemble into large ribonucleoprotein complexes termed RNA-induced silencing complexes (RISCs) 1. Guide RNAs are generated from double-stranded RNA precursors by the enzyme Dicer, which binds directly to both Ago2 and TRBP to facilitate transfer of the guide RNA duplex 2,3. Once loaded onto the Ago2 protein, one strand of the duplex (termed the “passenger strand”) is cleaved by Ago2 and dissociates from the complex, leaving a short single-stranded “guide” RNA that base-pairs with complementary mRNAs and targets them for Ago2-catalyzed degradation or translational arrest 4,5.
Although crystal structures of Dicer and Argonaute homologs have provided insights into the guide RNA processing and targeting steps of this pathway 6-12, no structural information is available for any RNAi-related protein complexes for which in vivo functions have been established. Using reconstituted functional human RISC-loading complexes (RLCs) containing purified Dicer, Ago2 and TRBP proteins, we examined the architecture of both the complex and the Dicer component using electron microscopy (EM) and single particle image analysis. These results represent the first view of the overall molecular architecture of the RLC and, together with analysis of a Dicer deletion mutant and modeling based on available Dicer and Ago structures, provide insights into the mechanism of RNA strand-specific RISC loading.
Results
Single particle reconstruction of human Dicer
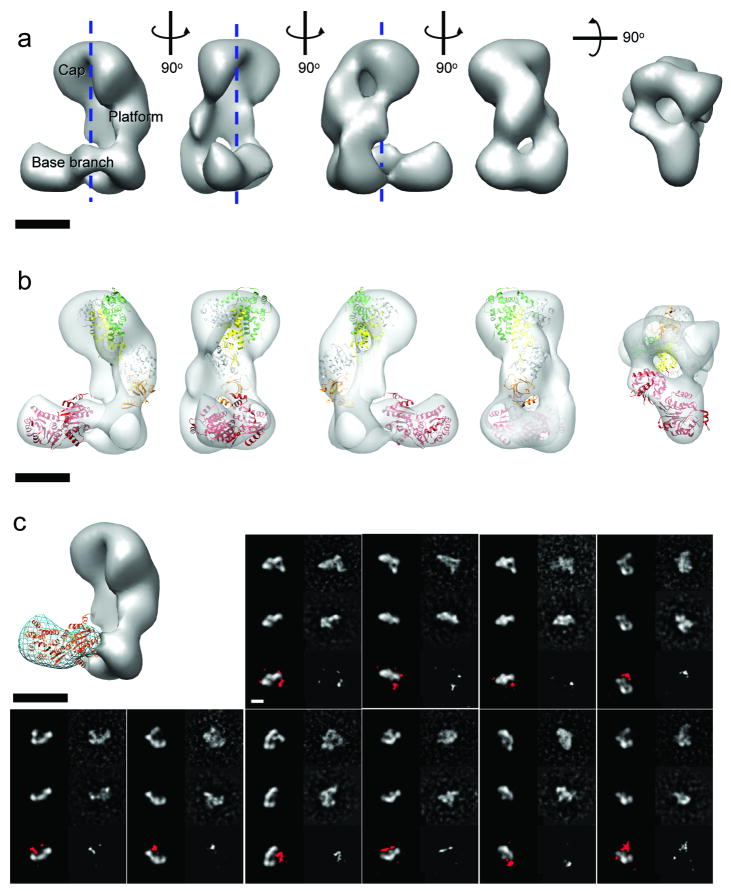
We began by examining the structure of the intact ∼220 kD human Dicer protein by negative-stain EM. The molecules appeared as well-dispersed, elongated particles with a length of ∼20 nm (Supplementary Fig. 1). Using Random Conical Tilt (RCT) methodology 13 followed by projection matching refinement 14, we obtained a three-dimensional (3D) reconstruction of human Dicer at a resolution of ∼34 Å (0.5 FSC criteria) from a final set of about 3400 particle images (Fig. 1a, Supplementary Fig. 1, see Supplementary Methods for image processing details). The final 3D density map appears as an “L” containing a main vertical platform, and an extended bi-lobed base branch extending perpendicularly at the bottom. The curved “cap” of the central platform protrudes in the same direction as the base branch. This arrangement defines an inner space or cavity delimited by an internal surface of the platform that is remarkably flat in the vertical direction and mostly concave in the horizontal direction (Fig. 1a).
Figure 1. Architecture of human Dicer.
(a) Three-dimensional (3D) reconstruction of human Dicer shown in different orientations. The main structural elements are labeled. The channel in the middle of the volume is indicated by dashed lines. (b) The most biologically relevant of the top docking results of the atomic model of Giardia Dicer (color coded as in Supplementary Fig. 2a) and the human DEAD-box RNA helicase DDX3X (PDB 2i4i) (red ribbon). (c) The synthetic 3D model (gray solid surface) generated by removing the density corresponding to the base branch (blue wire) in the human Dicer reconstruction is used to match the reference-free two-dimensional (2D) class averages of the ΔDExH/D Dicer mutant sample. In each of the 10 panels, the first rows are reprojections (left) and the corresponding reference-free class averages (right) of the intact human Dicer; the second rows are reprojections from the synthetic Δbase-branch model (left) and the corresponding reference-free class averages (right) of the ΔDExH/D mutant (second row); the third rows are the difference maps of the 2D class averages between intact human Dicer and the mutant at 3σ threshold (right) and the superimposition of them (in red) to the corresponding projections of the synthetic model (left). Scale bars in all figures are 5 nm.
With the whole protein's shape defined, we next tried to dissect human Dicer's architecture. Compared with the Giardia Dicer, whose crystal structure was recently solved 7, human Dicer is substantially larger in size and includes an extra domain at its N-terminal end, termed the DExH/D helicase and ATPase domain (Supplementary Fig. 2a). Despite lacking the DExH/D domain, the Giardia Dicer has full dicing activity as an intact molecule. Similarly, a human Dicer mutant that lacks the DExH/D domain also retains dicing activity 15. These data suggest that the DExH/D domain is likely to be a discrete structural element within the human Dicer. While the catalytic core of Dicer, as approximately defined by the Giardia crystal structure, cannot fit within the base branch, it could fit within the reconstruction's platform with an optimal cross-correlation coefficient of 0.55 in two possible orientations, the most relevant of which has the flat surface of the crystal structure, previously shown to be the site of RNA binding 16, aligned along the flat vertical surface facing the central cavity in our reconstruction (Fig. 1b) (see Methods for details of the docking and the discussion below about its biological relevance). On the other hand, the two-lobed volume of the base branch agrees well with the size of the DExH/D domain (68 kDa compared to the calculated 70 kDa from the volume at 3σ threshold). The crystal structure of its homolog, the human DEAD-box RNA helicase DDX3X (PDB 2i4i), can be docked within the base branch volume with a cross-correlation coefficient of 0.40 (Fig. 1b; see Supplementary Fig. 2b for sequence alignment of human DDX3X and the DExH/D domain). These observations lead us to propose a working model in which the DExH/D domain of Dicer corresponds to the base branch region in our reconstruction.
To test this model we examined a DExH/D-domain truncated version of human Dicer (ΔDExH/D) by EM. The truncated protein showed a strong tendency to dimerize, limiting the number of useful particles and precluding us from obtaining a reliable 3D reconstruction of this smaller protein (about 140 kDa). However, we were able to use reference-free two-dimensional (2D) alignment and classification to sort the ΔDExH/D particles and to obtain good quality class averages. The class averages are more rod-like than seen for the full length and appear to lack the base branch, in agreement with our proposed assignment of the base branch to the DExH/D domain. To further test our hypothesis on the location of the DExH/D domain, we generated a “synthetic” 3D model of the ΔDExH/D protein by erasing the base branch from the full-length Dicer 3D reconstruction (Fig. 1b). Reprojections of this 3D model were then compared to the experimental, reference-free 2D class averages of the ΔDExH/D mutant. For each class average, we selected the best match out of the reprojections based on the highest cross-correlation coefficient. This analysis retrieved more than 20 good matches out of 100 experimental class averages to different views of the hypothesized 3D model, covering most of the views. When compared with the same views of the full Dicer model and the experimental class averages, the density corresponding to the base branch is obviously missing in the ΔDExH/D mutant (Fig. 1c, Supplementary Fig. 3). This result, together with our docking of the DDX3X helicase, supports our initial proposal that the DExH/D domain of human Dicer is located at the base arm in the 3D reconstruction.
Architecture of human RISC-loading complex
Previous work has shown that the human RISC-loading complex can be reconstituted from individually purified Dicer, Ago2 and TRBP in a 1:1:1 stoichiometric ratio 17. While the complex appears stable at the concentrations of around 1 μM used during biochemical reconstitution, it dissociates at concentrations around 50 nM in the stain used for EM (see Methods). In addition to the L-shaped particles corresponding to Dicer, a substantial number of globular particles with the shape and size of the dissociated Ago2 could be easily recognized in the micrographs of this sample (Supplementary Fig. 4a).
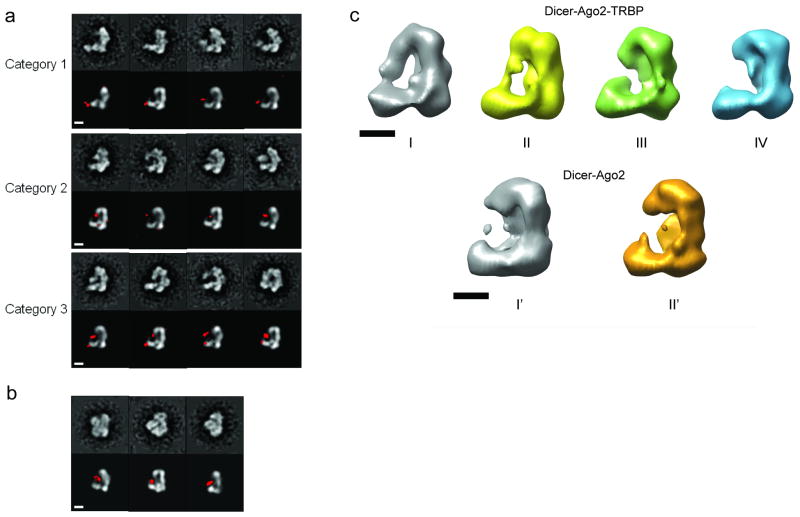
In order to overcome the instability of the complex at low concentrations (or potentially in the negative stain solution) and achieve better definition of the RLC's architecture, we sought to obtain intact assemblies by cross-linking them with glutaraldehyde immediately after elution from a gel-filtration column and before dilution for negative staining. Titration experiments showed that 0.02% (v/v) glutaraldehyde cross-links a significant amount of the complex in its native 1:1:1 ratio, but does not generate larger species (Supplementary Fig. 5a). Under the electron microscope, the cross-linked specimen showed fewer disassociated components in the background than the non-cross-linked sample, without the presence of large aggregates (Supplementary Fig. 5b). We performed iterative 2D reference-free alignment and multivariate statistical analysis (MSA) classification 18 of about 4000 negatively stained particles and sorted them into 200 classes. Some class averages emerged immediately with apparent additional densities when compared to the Dicer-only sample. We matched the 200 class averages with different projection views of Dicer's 3D density using multi-reference alignment and found that more than two-thirds of the class averages have significant additional densities (Supplementary Fig. 5c). This indicated that although one-third of the particles still correspond to Dicer alone, the rest are likely to retain all or part of the RLC components.
The class averages with additional densities were further sorted manually into three categories based on location and size of the extra densities. The first category includes particles with a small additional density attached to Dicer's base branch, the region of Dicer assigned to the DExH/D domain (Fig 2a, top row). The location of this density is variable around the tip of the DExH/D domain. The second category includes particles with a larger additional density located on the inner side of the L between the base arm and the cap at the top of the platform (Fig 2a, middle row). This density also shows a range of positions and shapes, but consistently remains close to the central cavity. The particles in the third category appear to have both the additional density connected to the DExH/D domain and the density facing the inner surface of the platform (Fig. 2a, bottom row). We propose that these additional densities correspond to TRBP and Ago2, respectively. It should be noted that the above categorization is rather subjective. To assign more firmly and objectively the extra densities, we examined the cross-linked complex comprising only Ago2 and Dicer, which was reconstituted in the same manner as the RLC (Supplementary Fig. 6a). Similar image analysis revealed class averages clearly falling in the second category that show a large density occupying the inner space between Dicer's platform and the base branch (Fig. 2b). The larger size of the Ago2 in these class averages compared to those in RLC suggests a more rigid configuration of Ago2 in the complex or a different orientation within the complex. These results further suggest that particles in category 1 likely correspond to the TRBP-Dicer complex (however, an alternative is that it may represent Ago2-TRBP-Dicer, see below); particles in category 2 likely correspond to the Ago2-Dicer complex; and that category 3 is formed by particles corresponding to the full RLC. Consistent with these assignments, our efforts to reconstitute Ago2 and TRBP with the ΔDExH/D Dicer mutant showed that Ago2 can still bind to Dicer but TRBP cannot (Supplementary Fig. 6b, 6c). This finding further supports a model in which Ago2 interacts with the C-terminal region of Dicer and TRBP interacts directly with the DExH/D domain.
Figure 2. Architecture of the human RISC-loading complex (RLC).
(a) Comparison of the cross-linked RLC reference-free class averages (odd rows) with corresponding projection views of the 3D map of human Dicer (even rows). The difference maps of the 2D class averages between the cross-linked RLC and the apo-Dicer at 3σ level (in red) are superimposed onto the projection views. The class averages are classified into three distinct categories as discussed in the text. For categorization purposes, only the front L-shape views are shown here. (b) Comparison of the cross-linked Dicer-Ago2 complex reference-free class averages (top row) with corresponding projection views of the 3D map of human Dicer. The difference maps between the cross-linked Dicer-Ago2 complex and apo-Dicer class averages at 3σ threshold (red) are superimposed onto the projection views. Shown here are the three clearest class averages out of a total of 100. (c) Maximum-likelihood heterogeneity analysis of the uncross-linked RLC and Dicer-Ago2 samples. From about 4000 particles of uncross-linked RLC, we generated four subclasses and calculated their 3D reconstructions (upper panel). Among the four models, Model I has an obvious additional density between the platform and the base branch, while Model IV doesn't appear to have additional densities besides the apo-Dicer. About 2000 particle images of uncross-linked Dicer-Ago2 complex underwent similar analysis to generate two subclasses, each having its 3D model reconstructed (lower panel). Both models lack the additional density that exists in the RLC. The scale bars in all panels are 5 nm.
TRBP may stabilize the Ago2-Dicer interaction
Because we matched the 200 class averages of the cross-linked RLC sample to the corresponding projection views of the 3D density of Dicer, we were able to reconstruct a 3D map by back-projecting all the class averages with their assigned Eulerian angles. This “all-in” reconstruction looked basically like the Dicer reconstruction, but had some additional density in the proposed location of Ago2 (Supplementary Fig. 7a, golden structure as a low-pass filtered model at 40 Å resolution). We then performed supervised classification 19 on all the particles by matching them against two initial reference models, one being the low-pass filtered Dicer reconstruction (Supplementary Fig. 7a, grey structure) and the other the low-pass filtered “all-in” RLC reconstruction (Supplementary Fig. 7a, golden structure). The histogram of cross-correlation values roughly agrees with our 2D analysis in that about one-third of particles are likely Dicer only (Supplementary Fig. 7b, model I). The one-third of particles that correlated better to the “all-in” model produced a final 3D reconstruction with a prominent globular density narrowly attached to the cap and located in the inner space of the “L”, between the platform and the base branch (Supplementary Fig. 7b, model III). The remaining third of the particles in the center of the histogram produced a 3D reconstruction with intermediate features, suggesting that they are a mixture of the two states (Supplementary Fig. 7b, model II). In none of the three models was there a clear density extending from the base branch corresponding to the putative TRBP protein, likely due to the flexibility of the TRBP molecule and/or of its attachment to the DExH/D domain (based on sequence analysis, TRBP is composed of three small RNA-binding domains connected by flexible linkers). On a similar note, the density corresponding to the putative Ago2 in the RLC reconstruction III could not account for the entire Ago2 atomic model (at the 3σ threshold, this density corresponds roughly to 50 kDa, about half of Ago2's molecular weight). We propose, based on this result and on the initial reference-free 2D analysis in Fig. 2a, that even when Ago2 is present in the complex, it is able to take on different positions within the RLC. Therefore, it is likely that flexibility results in the reduced density seen in reconstruction III. The low occupancy of Ago2 and TRBP even in the cross-linked samples reflects a relatively low affinity among the RLC components (our estimations indicate that the Kd is around 0.2∼1 μM). In addition to the data just described, we carried out an analysis of the heterogeneity present in the uncross-linked Ago2-Dicer complex, both in the presence and absence of TRBP, using a maximum-likelihood strategy 20. This study showed that in the absence of cross-linking, a prominent Ago2 density is observed in some particles of the sample that includes TRBP, in agreement with the 3D reconstruction results (Fig. 2c, upper panel). In the absence of TRBP, however, the Ago2-Dicer complex disassociates entirely under the dilute conditions used for EM grid preparation, such that only the apo-Dicer structure is present in the particle data set (Fig. 2c, lower panel). These results suggest that TRBP increases the affinity of Ago2 for Dicer and generally stabilizes the complex during dilution.
Ago2 may bind to Dicer at its two termini
In order to further verify RLC's architecture and to better characterize the components' interaction, we used the GraFix technique to prepare RLC complexes for single particle analysis. This recently developed methodology has been proven to be valuable for structural analysis of low-stability molecular assemblies 21,22 by cross-linking the complexes gradually as they are separated based on molecular weight and shape in a double gradient of glutaraldehyde and glycerol. The homogeneity of the RLC samples improved using this method, with the fraction containing the 360 kDa species appearing monodisperse by EM observation (Supplementary Fig. 8a, b).
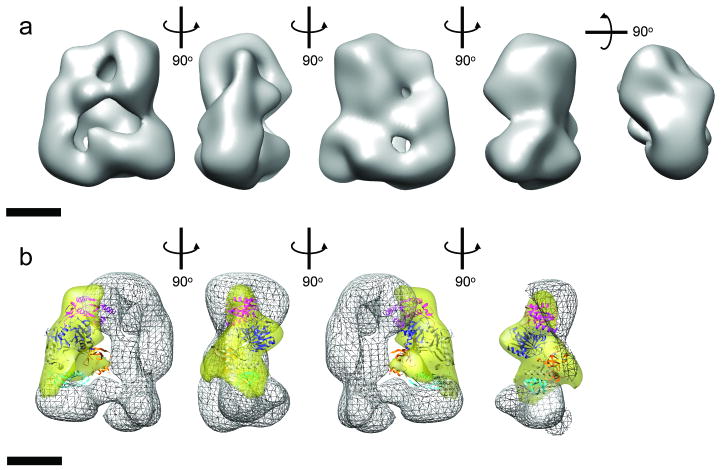
We performed de novo single particle reconstruction of the dataset collected from this sample using the RCT method, maximum-likelihood analysis and projection matching refinement, and obtained a reconstruction at a resolution of ∼33 Å (Fig. 3a, Supplementary Fig. 8c, d). When compared with the reconstruction of Dicer only, the reconstruction of this GraFix-prepared RLC has a prominent density in the middle of the molecule that connects the top of the platform and the tip of the base-branch. The location of the density agrees well with the category 2 2D class averages and the maximum-likelihood reconstructed model I and II of the uncross-linked RLC, suggesting the density to be Ago2. In agreement with this observation, the atomic model of Argonaute can be docked unambiguously in the difference map between RLC and Dicer reconstructions (Fig. 3b, Supplementary Video 1). The docking suggests that Ago2 connects the top of the Dicer platform and the tip of Dicer base-branch, giving rise to a tri-angular architecture for the complex. On one end, Ago2's PIWI domain and Mid domain interact with Dicer's platform; on the other end, Ago2's N-terminal domain interacts with Dicer's base-branch density. Although there are additional densities in this reconstruction, beyond those corresponding to Ago2 and Dicer, that could represent TRBP, especially at the base-branch (see Supplementary Video 1), it is difficult to interpret the location and shape of TRBP because of its possible elongated shape and flexibility. Nonetheless, these data together with those derived from the conventionally-cross-linked sample suggest that Ago2 may transiently interact with TRBP to form a closed complex with Dicer. We also note the significant difference density observed in the region assigned to the DExH/D domain of Dicer in the GraFix sample when compared to Dicer alone (Supplementary Video 1). This implies a possible structural rearrangement or change in relative conformational dynamics of the helicase region upon association with Ago2 and/or TRBP.
Figure 3. Reconstruction of GraFix-prepared human RLC.
(a) 3D reconstruction of RLC shown in different orientations. (b) Docking of the atomic model of Thermus thermophilus Argonaute in the major part of the difference map (yellow transparent map) calculated between the 3D reconstructions of RLC shown in (a) and that of Dicer alone (shown as wire map). Argonaute domain code: cyan, N-terminal domain; orange, PAZ domain; pink, Mid domain; blue, PIWI domain. The docking is shown in four different views along the vertical axis. The front part of the RLC in the fourth view was removed to show the Ago docking more clearly. The scale bars represent 5 nm.
Discussion
The importance of RNAi to the biomedical field necessitates a detailed structural understanding of the precise mechanism of this process in a human system. The model presented here represents a first view of the molecular architecture of the human RLC and provides a structural framework for testing how siRNA duplexes may be passed from Dicer to Ago2 during RISC loading (Fig. 4). Ago2 appears to bind Dicer at the opposite end from the DExH/D domain, where it also exhibits substantial conformational flexibility in its attachment to Dicer. Previous biochemical studies suggest that this interface corresponds to the Ago2 PIWI domain in contact with the Dicer RNase IIIa domain 23. This information suggested the overall orientation of Dicer's catalytic core structure in the reconstruction map, with the C-terminal RNase III domains located near the top of the platform density. Beginning with this approximate orientation, quantitative docking of the Giardia Dicer crystal structure into the platform region of the 3D volume of human Dicer was conducted using an automatic software algorithm (Fit-Model-In-Map in UCSF-Chimera followed by COLACOR in Situs 2.0)7 (Fig. 1b). The most biologically relevant of the two top docking results positions the flat surface of the crystal structure, demonstrated biochemically to accommodate dsRNA 16, facing the flat surface of the reconstruction (see Supplementary Fig. 9b for the other docking). Such a model implies that this is the initial binding site of substrate dsRNA. Our RLC reconstruction shows considerable extra density for the human Dicer located at the proposed interface with Ago2, which may account for the extra 127-amino acid region of human Dicer that is not present in Giardia Dicer 23. The RLC reconstruction presented here can accommodate the atomic model of the Thermus thermophilus Argonaute containing a guide-strand RNA adjacent to the long axis of the Dicer density 11. Modeling a 22 base-pair siRNA onto Dicer shows that the position of Ago2 leaves ample room for dsRNA binding to Dicer (Fig. 4a). Intriguingly, this docking positions the Ago2 PAZ domain above the Dicer DExH/D domain in such a manner that the distance between the PAZ domains of Dicer and Ago2 can also accommodate a 22nt siRNA. Thus, it would in theory be possible for these domains to simultaneously bind opposite ends of the siRNA product (Fig. 4b). The class averages in Fig. 2a indicate that Ago2 may be able to orient itself close enough to the Dicer active site to engage the 3′ overhang of the newly cleaved siRNA, achieving this siRNA-bound intermediate.
Figure 4. Proposed working model of the human RLC.
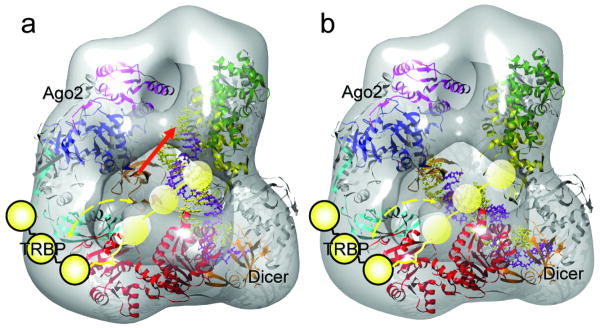
The 3D density map of RLC is shown as a semi-transparent iso-surface. The atomic model of the DExH/D domain (red ribbon), the Giardia Dicer atomic model (gray-yellow-green-orange ribbon, color coding the same as in Figure 1b), and the Thermus thermophilus Argonaute (gray-cyan-orange-pink-blue, color coding the same as in Figure 3b) are docked in the density map. TRBP is illustrated as a string of three yellow spheres with a flexible linker connecting it to the DExH/D domain. Its motion range, based on our experimental results, is marked by the dashed yellow arrows. In (a), an atomic model of the siRNA (paired spirals with the guide strand in purple color and the passenger strand in yellow color) is aligned vertically between the Dicer's RNase III domain flat surface and Ago2's PAZ domain. This panel illustrates the state proposed in our model for the dicing of dsRNA by Dicer. In this state the PAZ domain of Ago2 could engage the newly diced end of the siRNA, as illustrated by the red arrow. In (b), the distance between the PAZ domains of Ago2 and Dicer allows a perfect accommodation of the 22 nt siRNA between them. This state of Ago2 could be stabilized by interactions with TRBP. Thus, this panel illustrates the hypothetical state after transfer of the newly diced siRNA's onto Ago2's PAZ domain while the other end remains bound to Dicer's PAZ domain. The flexible TRBP could help the transfer efficiency and correctness.
TRBP is known to be important both for Dicer binding to its siRNA product and for recruiting siRNAs to Ago2 3. Our analysis indicates that TRBP is flexibly bound to the Dicer DExH/D domain, in agreement with existing biochemical data 24,25 (Supplementary Fig. 6b, 6c). The position of TRBP near the Dicer DExH/D domain, and its flexibility, allowing access to the PAZ domain of Ago2 in our model, makes biochemical sense, and suggests a role for TRBP in bridging the steps between release of the siRNA by Dicer and loading of the duplex into Ago2. Binding by TRBP may allow the siRNA intermediate to stay associated with the RLC after release from Dicer and may also help optimize the orientation of the siRNA for Ago2-loading. Additionally, TRBP may be important for proper strand selection during RISC loading, acting as a proofreading mechanism for strand-selectivity that ensures the proper orientation of the siRNA once it has been handed off to Ago2. In general, the strand of an asymmetric siRNA duplex whose 5′-end is less thermodynamically stable will become the guide strand when loaded into Ago2 26. In flies, asymmetric siRNAs are bound in a directional manner by Dicer-2 and R2D2, a TRBP homolog in Drosophila. R2D2 binds the more thermodynamically stable end, while Dicer-2 binds the less stable end. The Dicer-2 PAZ domain thus specifically interacts with the 3′-end of the siRNA passenger strand 27. Although it is currently unknown whether TRBP binds the more stable siRNA end in humans, it is tempting to propose this specificity, as it would allow TRBP to act as a sensor to detect proper loading of the 3′-end of the guide strand onto the Ago2 PAZ domain. In the case of an exogenous siRNA, the complex architecture supports the notion that TRBP could also function to orient the guide strand for direct handoff to Ago2. Likewise, if an siRNA produced by dicing a long dsRNA is positioned in the less-favored direction with respect to RISC loading, then the siRNA could be reoriented by release to the bulk solvent followed by rebinding in the opposite orientation, as has been observed in Drosophila extracts 28,29. PACT, another human dsRBP involved in RNAi that binds to the Dicer DExH/D domain 30, may also serve this role. Future biochemical and structural studies of this basic RLC associated with its other binding partners may shed more light on the mechanism of the final handoff to Ago2.
Methods
Protein purification and characterization
A detailed cloning strategy and primers used for amplification of target genes were described elsewhere 17. Briefly, PCR-amplified DNA fragments were cloned into SfoI/XhoI pre-digested pFastBac plasmids to obtain the constructs to produce N-terminally His6-tagged proteins. In order to obtain corresponding recombinant Bacmid DNAs, we transformed the pFastBac plasmids into competent DH10Bac E. coli cells (Invitrogen), and then used these Bacmid DNAs to produce baculoviruses by transfecting them into Sf9 cells with FuGene Transfection Reagent (Roche Applied Science). To generate the recombinant proteins, we infected Sf9 cells with each baculovirus for 72 hours before harvesting for protein purification. We purified the N-terminally His6-tagged proteins by Ni2+-affinity chromatography followed by TEV protease cleavage to remove the His6 tag, and finally by gel filtration chromatography using HiLoad 16/60 Superdex 200 (GE Healthcare).
Reconstitution and cross-linking of RLC and Ago-Dicer complexes
To reconstitute the RLC in vitro, we mixed recombinant human Dicer, Ago2, and TRBP in a 500 μL reaction volume with a molar ratio of 1:2:4 (Dicer:Ago2:TRBP) in a buffer containing 20 mM HEPES, pH 7.5, 150 mM KCl, 10% (v/v) glycerol, and 1 mM TCEP. In the case of the Dicer-Ago2 complex, the molar ratio used was 1:2 (Dicer:Ago2). We incubated this reconstitution reaction for 30 minutes on ice. We then spun the reaction for 5 minutes at 4°C at 16,000 g and applied the supernatant to a pre-equilibrated Superose 6 10/300 column. We collected the fraction with peak RLC content, as assessed by SDS-PAGE, and immediately stained or cross-linked the fraction for EM analysis. Cross-linking reactions contained 0.5-1 μM complex and 0.02% (v/v) glutaraldehyde and were brought to a final volume of 14 μl with a buffer containing 20 mM HEPES, pH 7.5, 150 mM KCl, 5 mM EDTA, and 1 mM TCEP. We incubated the reactions at room temperature for 10 minutes, quenched them with 100 mM Tris, pH 7.5, and incubated them an additional 5 minutes at room temperature before placing on ice. The extent of cross-linking was analyzed by SDS-PAGE.
We used the recently described GraFix sample preparation technique to obtain intact RLC 21. Briefly, the RLC was reconstituted by incubation of a 1:2:4 molar ratio mixture of Dicer (∼0.2 mg ml-1), Ago2, and TRBP in a buffer containing 20 mM Hepes, 150 mM KCl, pH 7.5 at 0°C for 1 hr. We applied 100 μl of the in-vitro reconstituted complex to a 2 ml 5-25% (v/v) glycerol/0.02-0.1% (v/v) glutaraldehyde gradient in a 2.2 ml ultracentrifuge tube. After centrifugation at 45,000 g in a TLS-44 rotor (Beckman) for 6 hrs at 4°C, 200 μl fractions of the gradient were collected using a glass capillary with a peristaltic pump from the bottom of the tube. We then analyzed the fractions by SDS-PAGE and used the fraction corresponding to the ∼350 kDa cross-linked complex for single particle analysis.
Electron microscopy
We diluted human Dicer, RLC or RLC-subcomplex samples, either with or without cross-linking, to a concentration of about 30∼40 nM in 20 mM Tris-HCl pH 7.5, 100 mM KCl, 3 mM MgCl2, 1 mM DTT immediately before applying them to glow-discharged holey carbon grids with a thin layer of carbon over the holes. After adsorption for 1 minute, the sample was stained consecutively with three droplets of 2% (w/v) uranyl formate solution, blotted gently of the residual stain, and air-dried in a hood. We examined the specimens under an FEI Tecnai-12 Bio-Twin electron microscope equipped with a LaB6 filament and operated at 120 kV acceleration voltage, using a nominal magnification of 49,000. For tilt-pair images, the same area of a specimen was recorded first at 55° and then at 0° tilt angles. We recorded images either on SO-163 film or on a Gatan 1k×1k CCD camera using low-dose mode with an exposure dose of 20-30 e-/Å2. The defoci range used to collect the 0°-tilt image was -0.8 to -1.1 μm and that used for the 55°-tilt image was -1.5 μm. We developed the films using full-strength D19 for 12 minutes and digitized the micrographs without apparent drift and astigmatism using a Nikon Coolscan 8000 with a scanning step of 12.7 μm, resulting in a pixel size of 2.59 Å. The scanned images were first converted from their transmission values to optical densities using the PROC2D program of the EMAN image processing package 31. The CCD-recorded micrographs had a pixel size of 3.61 Å and were directly used in the image processing.
Image processing
We used random conical tilt (RCT) method to obtain the initial model of human Dicer and of the GraFix prepared RLC 13, and then used projection matching refinement 14 combined with a maximum-likelihood algorithm analysis 20 to obtained the refined structures of human Dicer and the GraFix prepared RLC. We also used reference-free 2D alignment and classification 18 through several iterations of multi-variant statistical analysis and multi-reference alignment to analyze different samples. We performed supervised heterogeneity analysis on cross-linked RLC samples. For more detail of the image processing, please see Supplementary Methods.
Docking of atomic models in the EM 3D density map
The docking of the atomic models was done either using rigid-body docking programs within the Situs2.0 software package 32, or by visual inspection and manual docking using Chimera 33. More specifically, the atomic model of human DEAD-box RNA helicase DDX3X (PDB 2i4i) was docked in the DExH/D domain using Colores within Situs2.0; the Giardia intestinalis Dicer crystal structure (PDB 2qvw) was docked in the human Dicer platform density using Fit-Model-In-Map function in Chimera guided by the biochemical evidence for the orientation, and then adjusted locally using Colacor of Situs2.0; the guide strand-containing Thermus thermophilus Argonaute crystal structure (PDB 3dlh), the Giardia intestinalis Dicer crystal structure (PDB 2qvw), and the atomic model of human DEAD-box RNA helicase DDX3X (PDB 2i4i) were docked in the GraFix prepared RLC 3D map by Fit-Model-in-Map of Chimera. All the figures with 3D models were generated using UCSF-Chimera.
Supplementary Material
Acknowledgments
We thank Ian MacRae at the Scripps Institute for the purified Dicer and un-crosslinked RLC samples used in the initial stages of this work, P. Gabriel for help with particle picking, M. Jinek and S. Chakravarthy for help with sample preparation, members of the Nogales and Doudna labs for valuable insights and technical support, A. Fischer for tissue culture assistance, and the Keck MacroLab for the use of their resources. This work was supported in part by grants from the NIH. J.A. Doudna and E. Nogales are HHMI investigators.
Footnotes
Author contributions: H.-W.W. planned the experiments, performed electron microscopy, analyzed and interpreted the data, and wrote the paper; C.N. purified the proteins, prepared the specimen of cross-linked RLC, performed electron microscopy, and analyzed the data; B.S. prepared the specimen of uncross-linked RLC, performed electron microscopy, and analyzed the data; D.W.T. prepared the specimen of GraFix prepared RLC, performed electron microscopy, and analyzed the data; E.M. purified the proteins and reconstituted the RLC; K.F. prepared the specimen of human Dicer, performed electron microscopy, and analyzed the data; J.A.D. planned the experiments, interpreted the data, and wrote the paper; E.N. planned the experiments, analyzed and interpreted the data, and wrote the paper.
References
- 1.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 3.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rand TA, Petersen S, Du FH, Wang XD. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 5.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 6.Rivas FV, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–9. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 7.MacRae IJ, et al. Structural basis for double-stranded RNA processing by dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 8.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–7. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 9.Yuan YR, Pei Y, Chen HY, Tuschl T, Patel DJ. A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure. 2006;14:1557–65. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–19. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang YL, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–U34. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–6. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–36. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 14.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–70. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 15.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. Journal of Molecular Biology. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–40. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 17.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 19.Gao H, Valle M, Ehrenberg M, Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol. 2004;147:283–90. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Scheres SH, et al. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–9. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 21.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–5. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 22.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki T, Shimizu N. Evolutionary conservation of a unique amino acid sequence in human DICER protein essential for binding to Argonaute family proteins. Gene. 2007;396:312–320. doi: 10.1016/j.gene.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Haase AD, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. Embo Reports. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kok KH, Ng MHJ, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. Journal of Biological Chemistry. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 27.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 28.Preall JB, He Z, Gorra JM, Sontheimer EJ. Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila. Curr Biol. 2006;16:530–5. doi: 10.1016/j.cub.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 29.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y, et al. The role of PACT in the RNA silencing pathway. Embo Journal. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 32.Wriggers W, Birmanns S. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J Struct Biol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.