Abstract
To understand how broad recognition of HIV-1 variants may be achieved we examined T-cell reactivity in newly-infected persons as well as vaccine recipients to a broad spectrum of potential T-cell epitope (PTE) variants containing conservative, semi-conservative and non-conservative amino acid substitutions. Among early-infected persons T-cells recognized epitope variants with one substitution at a significantly higher frequency versus those with two (P=0.0098) and three (P=0.0125) substitutions. Furthermore T cells recognized variants containing conservative substitutions at a higher frequency versus those containing semi-conservative (P=0.0029) and non-conservative (P<0.0001) substitutions. Similar effects were observed on recognition of variants by vaccine induced T-cells. Moreover even when variants were recognized, the IFN-γ and granzyme B responses as well as T-cell proliferation were of lower magnitude. Finally, we show that epitope distribution is strongly influenced by both processing preferences and amino acid entropy. We conclude that induction of broad immunity is likely to require immunogen sequences that encompass multiple variants. However, cost-effective design of peptide and sequence based vaccine immunogens that provide maximal coverage of circulating sequences may be achieved through emphasis on virus domains likely to be T-cell targets.
1. INTRODUCTION
The enormous genetic diversity in HIV-1 is a major challenge in the design of vaccines that would protect against the wide range of HIV-1 circulating strains. Group-M HIV-1 dominates the worldwide pandemic and consists of several distinct subtypes [1]. Proteins within subtypes vary by approximately 5–25% and between subtypes by 10–40%, the lower bound representing conserved proteins and the upper bound representing variable proteins. A globally efficacious vaccine will need to elicit broadly reactive immune responses [2,3]. Such a vaccine is likely to require immunogen sequences that induce T-cells recognizing epitopes that are similar to or cross-react with those in circulating viruses. Although, cross-clade T-cell responses have been reported [4–15] our understanding of the numbers and types of amino-acid differences that are tolerated and how these impact T-cell functional properties is limited. Such an analysis requires a definition of the immune responses at the epitope level comparing T-cell reactivity to variants containing conservative versus semi- conservative and non-conservative amino acid substitutions.
In this study we examined the ability of T-cells to recognize a broad spectrum of potential T cell epitope (PTE) variants in newly-infected persons within the context of autologous virus sequences and in uninfected persons who were recipients of HIV-1 vaccines. We chose subjects with early HIV-1 infection since the immune responses detected in these subjects are more likely to be relevant for immune control, compared to later in infection when the viruses may have escaped key CTL responses. Additionally, we chose to examine a group of vaccine recipients since it is not known if the spectrum of epitope variants recognized in the context of vaccination and infection are similar. We assessed responses to a standardized peptide panel that represented PTE contained in diverse circulating HIV-1 strains. Compared to the consensus (CON) peptides, which have at each site the most common amino acid residue across a sequence alignment, the peptide reagents based on the PTE approach offer improvements in the assessment of T-cell responses [16,17]. PTE sequences are naturally occurring in HIV-1 strains and encompass sequences among virus variants circulating at moderate to high frequencies [18].
Most T-cell epitopes identified by central-sequence peptides in infected persons cluster in domains with low sequence variability, with few epitopes being detected in regions of moderate and high sequence variability [19]. This could partially be attributed to sequence variability between infecting and testing antigen sequences and consequently poor sensitivity for detecting epitopes in more variable domains with such peptide sets. Indeed T-cell responses detected by stimulation with a PTE peptide set are significantly greater in breadth and magnitude compared to those detected with a CON peptide set [20]. Therefore in this study we examined epitope distribution characteristics in infected persons using the PTE peptide set with its expanded epitope coverage.
Our findings indicate that while Nef-specific T-cells capable of recognizing multiple variants are commonly induced during early infection, semi- and non-conservative substitutions and those affecting more than one residue are infrequently tolerated. Further investigations of immune responses in recipients of HIV-1 vaccines consisting of Ad5 encoding Gag/Pol (clade B) and Env (clades A, B and C) corroborated these findings. We provide further evidence that there are considerable differences in the quality of T-cell responses recognizing multiple epitope variants. The data have wider implications in relation to vaccine immunogenicity, suggesting that while vaccine-induced T-cells may be able to recognize epitopes both in and outside the subtype represented in the vaccine, the quality of the response to epitopes outside the subtype may not be adequate to provide protection against highly diverse circulating strains and rapidly changing virus populations. Thus, induction of broad immunity may require vaccines that encompass multiple sequence variants. Furthermore, we show that epitope distribution is strongly influenced by both processing preferences and amino acid entropy. This information provides critical insight into the cost-effective design of peptide and sequence based vaccine immunogens as well as peptide test reagents that provide maximal coverage of circulating sequences through emphasis on domains that are likely to be T-cell targets [21–24].
2. METHODS
Study population
Subjects with primary HIV-1 infection were recruited and enrolled at the University of Washington (UW) Primary Infection Clinic (PIC). The duration of infection was typically defined as the time from the onset of clinical signs and/or symptoms suggestive of acute retroviral syndrome [25–27]. All participants were men who have sex with men, and presumed to be infected with clade B viruses [28]. Some patients at various time points following diagnosis elected to receive combination antiretroviral therapy (ART) with nucleoside reverse transcriptase inhibitors, and either a protease inhibitor or a non-nucleoside reverse transcriptase inhibitor. Participants in the HVTN054 vaccine trial were recruited through the HIV Vaccine Trials Network (HVTN) and received one dose of Ad5 encoding Gag/Pol (clade B) and Env (clades A, B and C) [Producer: Vaccine Research Center (VRC), National Institutes of Health (NIH)]. Immunogenicity assessment was performed 28 days after vaccination. Uninfected volunteers at low risk for HIV-1 infection were recruited through the Fred Hutchinson Cancer Research Center HIV Vaccine Trials Unit to serve as donor controls. The Human Subjects Review Board at the institutions approved the studies, and all volunteers provided written consent prior to participation.
Plasma HIV-1 RNA levels, T-cell subset analyses, and HLA typing
Plasma HIV-1 RNA was determined by quantitative branched-chain DNA (bDNA, Chiron, Emeryville, CA, USA) and ultrasensitive reverse transcriptase-polymerase chain reaction (RT-PCR, Roche Molecular Systems, Branchburg, NJ, USA) assays. RNA levels were expressed as copies/ml, and the lower levels of sensitivity were 500 copies/ml (bDNA assay) and 50 copies/ml (RT-PCR assay) [29]. Absolute blood CD4+ T-cell counts were measured by consensus flow cytometry. HLA typing was performed at the Puget Sound Blood Center by sequence-specific primer PCR as previously described [30].
Analysis of PTEs from circulating strains and PTE coverage
The design of PTE peptides has been previously described [18]. In brief, given that MHC class I molecules typically present antigenic peptides that are 8–10 aa in length [31–33], we considered 9 aa as the minimal epitope length, and defined PTE as all possible unique 9-mers contained in the virus sequences. Within this mixture of unique PTEs, each was then classified by its frequency of occurrence relative to the other PTEs. To design longer peptides such as 15-mers, each unique 15 amino acid sequence embedded in the group of virus sequences is extracted. These naturally-occurring 15-mers are then ordered by their PTE coverage level, which is the summation of the PTE frequencies associated with each of the individual embedded 9-mers. Thus, the higher the PTE coverage, the greater the likelihood that the peptide represents the virus sequences. A peptide reagent set is then built in a step-wise fashion until the desired cumulative PTE coverage is reached.
The nef PTE peptide set used to assess immune responses in newly-infected subjects covered all PTE contained among 5% or more of the subtype B virus sequences [20]. The Env, Gag, and Pol PTE peptide sets used to assess immune responses in the vaccine recipients covered all PTE contained in 15% or more of the global viral sequences.
IFN-γ and granzyme B ELISpot assay
HIV-1 Nef PTE peptides (n=90) were synthesized at Mimotopes (Raleigh, NC, U.S.A.); and the Gag, Env, Pol, and truncated peptides to fine map epitopes were synthesized at BioSyn (Lewisville, TX, USA). All peptides were used at a final concentration of 2 µg/ml unless stated otherwise, and the final concentration of DMSO never exceeded 1% in the assays.
IFN-γ ELISpot assays were performed to detect HIV-1-specific IFN-γ secreting cells as previously described [34], with modifications as follows. Granzyme B ELISpot assays were performed using monoclonal antibodies from Mabtech (Sweden) and following the manufacturer’s directions. Phytohemagglutinin (Murex Biotech, Dartford, United Kingdom) stimulation at 2 µg/ml was used as a positive control and no peptide stimulation was used as a negative control. Spot-forming cells (SFC) were counted using the Immunospot (Cellular Technology Ltd., Cleveland, Ohio, USA) optical reader. The number of antigen-specific IFN-γ-and granzyme B-secreting cells was calculated by subtracting the SFC in the negative control wells from those in the antigen-stimulated wells. Peptides were first tested in pools and all responses were confirmed at the single peptide level [35,36]. Responses > 3-fold background and > 50 SFC/106 cells above background were scored positive.
To determine the functional avidity of the T-cell response, the standard IFN-γ ELISpot assay was performed with the optimal epitopic peptides at serial concentrations between 10,000 and 0.1 ng/ml. SFC frequencies were plotted against the log10 peptide concentration with ORIGIN 6.0 professional software (Microcal Software, Inc. Northampton, MA) and the effective peptide concentration that elicited 50% of the maximum T-cell response, defined as the EC50, was determined with the Sigmoidal Fit tool.
Measurement of T-cell proliferation by CFSE labeling [37,38]
Briefly, thawed, washed, and overnight-rested PBMC were incubated with 1.3µg/mL (1.25µM) Carboxyl Fluorescein Succinimidyl Ester (CFSE) (Molecular Probes, Eugene, OR) for 8 minutes; quenched with cold FBS; and washed with PBS. The cells were then resuspended in complete media (106 cells/ml), plated in 24-well plates at 1 ml per stimulus and cultured for 5 days. Positive and negative controls included cells incubated with anti-CD3 mAb (30 ng/mL) and anti-CD28 mAb (1 µg/mL), and with medium alone. The decrease in CFSE signal is used to monitor proliferation of each cell subset.
Sequencing of autologous viruses
DNA was purified from 2 × 106 cryopreserved PBMC using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA), according to the manufacture’s recommendations. HIV-1 Nef sequences were amplified by standard nested PCR as previously described [39]. Primers DS7 (F) 8169–8198 and TMMNEF6 (R) 9607–9631 were used in the first round PCR resulting in a 1.5 Kb product. Primers DS9 (F) 8678–8697 and DS8 (R) 9527–9550 were used in the second round PCR resulting in an 872 bp product. The PCR products were cloned into the TA vector using the TOPO TA Cloning Kit according to the manufacturer’s protocol (Invitrogen, San Diego, CA) and selected for sequencing as described previously [40] to avoid template resampling [41]. Plasmids from individual clones were isolated and sequenced using the M13 355–370 and T7 328–347 forward and M13 205–221 and T3 243–262 reverse primers with ABI Prism Big Dye Terminator Cycle sequence reagents (Applied Biosystems, Foster City, CA). Computational analysis of the sequences was performed using ExPASy Translate Tool (http://us.expasy.org/tools/dna.html), ClustalW (http://www.ebi.ac.uk/clustalw/) and BioEdit.
Scoring of changes in amino acid sequences
Amino acid substitutions between sequences were scored for similarity using the BLOSUM matrix [42]. The matrix is derived from blocks of aligned sequences from homologous proteins with a certain level of identity. Conservative changes have positive scores and non-conservative changes have negative scores with the scale extending from −4 to +3. Conservative amino acid substitutions are likely to conserve the physical and chemical properties necessary to maintain the structure and function of the protein, whereas non-conservative substitutions are likely to disrupt essential structural and functional features of the protein.
Calculation of amino acid entropy and proteasomal cleavage scores
Amino acid variability was calculated as the entropy. The average entropy score for each 9-mer or a defined optimal epitope was calculated from Shannon entropy for each residue of B clade protein alignments, using only complete Nef protein sequences and only a single protein from multi-sequence sets derived from a single subject. Proteasomal cleavage prediction scores were determined based on a cleavage prediction algorithm (NetChop 3.0, [43] that has been shown to be highly predictive of C-termini in epitopes.
Statistical analysis
Effects of numbers and relative conservation of amino acid substitutions on recognition of T-cell epitope variants in subjects with primary HIV-1 infection and in HIV-1 vaccine recipients, as well as entropy and C-terminus cleavage scores for targeted optimal epitopes and the group of all Nef subtype B 9-mers, were assessed using the Wilcoxon Signed Rank Test. Entropy vs. C-terminus cleavage scores for the group of all Nef subtype B 9-mers was examined using the Spearman correlation coefficient.
3. RESULTS
Recognition of epitope variants and autologous virus sequence analyses
T-cell recognition of a total of 150 Nef variants, encompassing 35 epitopes (mean 4.3 variants/epitope), was assessed in 17 newly HIV-1-infected subjects with demonstrated Nef responses. At the time of analysis of the immune response, the subjects were infected for a median duration of 45 (range 7 – 134) days post onset of symptoms of primary HIV-1 infection (DPS), and 2 subjects had received ART for > 1 week (duration 42 and 57 days). For each of the 35 epitopic domains, the optimal epitope was either confirmed using peptide truncations or inferred based on the subject’s HLA type and published MHC class I restricted CD8+ T-cell epitopes.
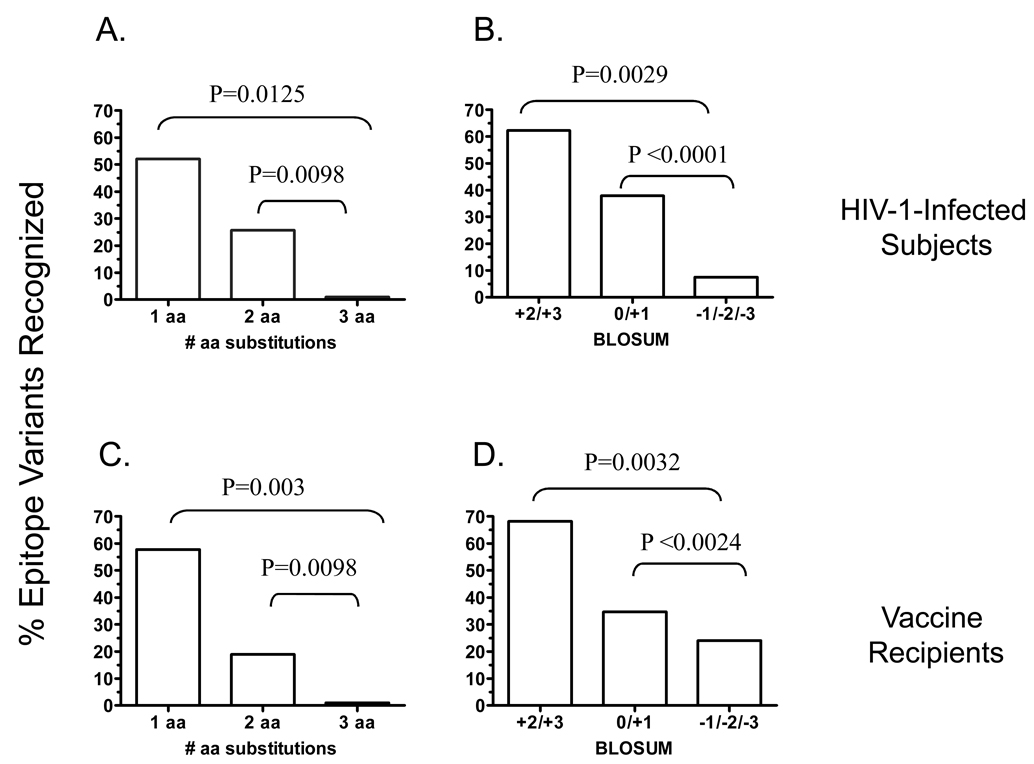
Autologous HIV-1 nef sequences were amplified by standard nested PCR in eight subjects and an average of 20 (range 10 – 33) Nef sequences were analyzed. Amino acid substitutions were scored for similarity using the BLOSUM matrix. Scores of +2 and +3 were imputed conservative, 0 and +1 semi-conservative and −1 and below non-conservative. Notably, of 19 epitopes sequenced in these subjects, the autologous consensus sequence either matched the subtype B consensus (n=14 epitopes) or differed by a single conservative amino acid (aa) substitution (n=4 epitopes) except for one epitope that contained two semi-conservative substitutions. With the referent sequence being either the autologous (when available, N=19), or the subtype B consensus (N=16), 73 of the 115 peptide variants examined across the 35 epitopes contained one aa substitution, 35 contained two, and seven contained three substitutions. T-cells recognized 52.1% (38 of 73) of the variant epitopes containing one substitution with a < 0.5 log difference in response magnitude in contrast to 25.7% (9 of 35, P=0.0098) that contained two substitutions and none (0 of 7, P=0.0125) that contained three substitutions (Figure 1A). Variants containing conservative substitutions were recognized at a significantly higher frequency [61.1% (22 of 36)] vs. those containing semi-conservative [37.9% (11 of 39), P=0.0029] and non-conservative substitutions [7.5% (3 of 40), P<0.0001] (Figure 1B). In no instance did we detect a response to a variant epitope in the absence of a response to the autologous consensus.
Figure 1. Effects of numbers and conservation of amino acid substitutions on recognition of T-cell epitope variants in subjects with primary HIV-1 infection (A, B) and in HIV-1 vaccine recipients (C, D).
PBMC were stimulated with HIV-1 PTE peptide sets in a matrix format and tested for IFN-γ production using an ELISpot assay. Responses were examined to the Nef protein in subjects with primary infection and to Gag, Env and Pol in the vaccinees. Responses > 3-fold background and > 50 SFC/106 cells above background were scored positive. All responses were confirmed at the single peptide level. Shown is the frequency of recognition of epitope variants containing one, two and three amino acid substitutions; and conservative, semi-conservative and non-conservative substitutions.
Next in HVTN 054 vaccine enrollees we characterized T-cell recognition of epitope variants 28 days after one dose of the Ad5 encoding Gag/Pol (clade B) and Env (clades A, B, C) using global PTE peptides encompassing Gag (n=330), Env (n=492) and Pol (n=509). Responses were analyzed in 7 subjects in whom T-cells targeted epitopes (n=16) with amino acid variability so that recognition or non-recognition of peptide variants could be assessed. With the referent sequence being the vaccine sequence, 45 of the 73 peptide variants examined across the 16 epitopes contained one aa substitution, 21 contained two, and seven contained three substitutions. T-cells recognized 57.8 % (26 of 45) of the variant epitopes containing one substitution with a < 0.5 log difference in response magnitude in contrast to 19.0% (4 of 21, P=0.003) that contained two substitutions and none (0 of 7, P=0.0098) that contained three substitutions (Figure 1C). Variants containing conservative substitutions were recognized at a significantly higher frequency [68.2% (15 of 22)] vs. those containing semi-conservative [34.6% (9 of 26), P=0.0032] and non-conservative substitutions [24% (6 of 25), P=0.0024] (Figure 1D).
In summary, HIV-1-specific T-cells capable of recognizing multiple variants are frequently induced during early infection as well as following vaccination. However, semi- and non-conservative substitutions and those affecting more than one residue are infrequently tolerated in the context of both infection and vaccination.
IFN-γ and granzyme B responses to epitope variants
We further characterized T-cell recognition of several optimal epitope variants in the Nef protein measuring the IFN-γ and granzyme B responses to serial dilutions of peptide variants in a sub-group of HIV-1-infected subjects where PBMC were available for these analyses. Due to lack of sufficient PBMC these studies could not be done in vaccine recipients.
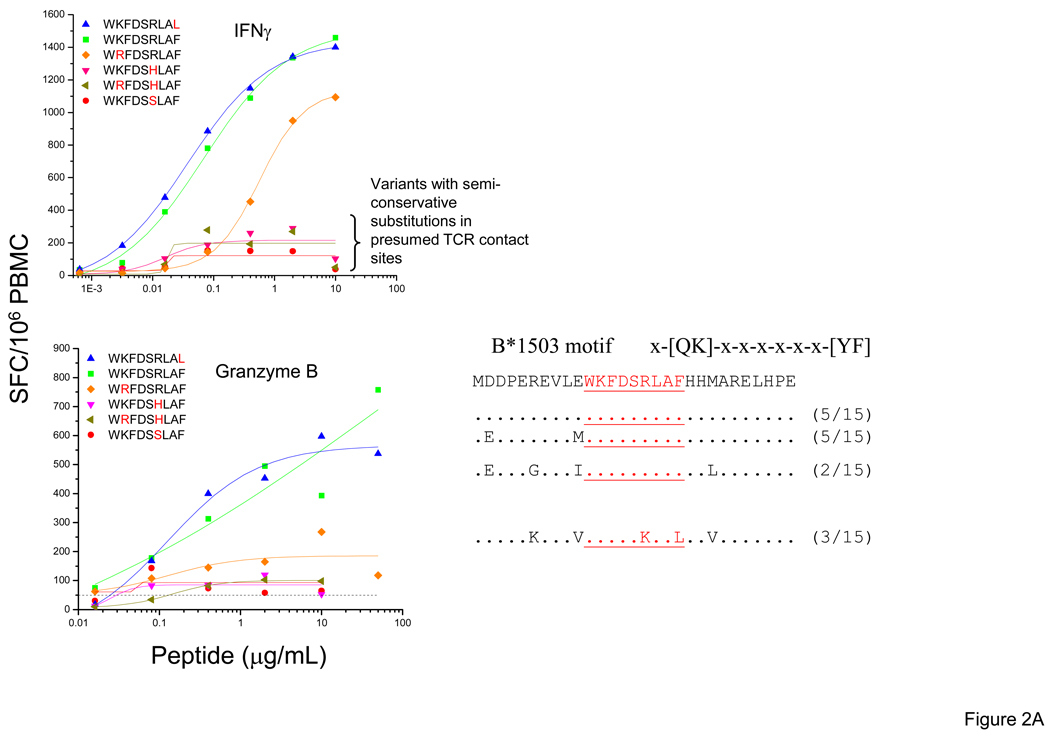
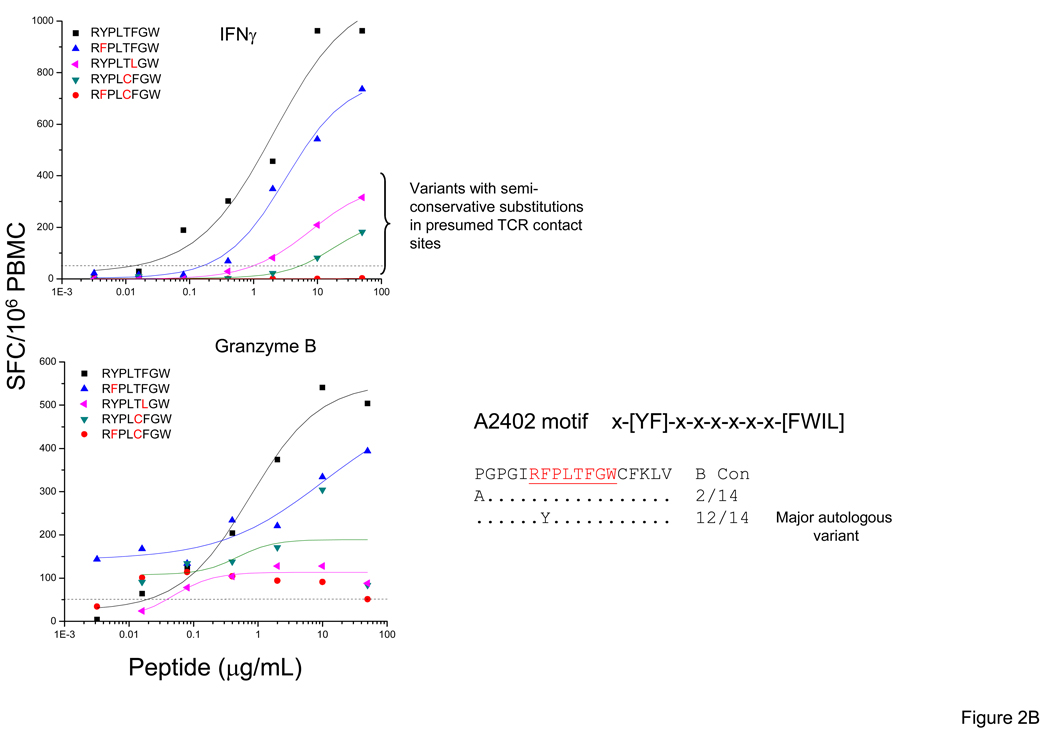
T-cells from subject 1188 recognized all five variants of the WF9/B1503 epitope tested producing IFN-γ [20], although only one of these variants was detected among the autologous sequences at 93 DPS. However, the magnitude and avidity of the responses to the variants showed significant differences and we found that the granzyme B response pattern mimicked the IFN-γ pattern. Amino acid substitutions in presumed T-cell receptor (TCR) contact sites e.g. WKFDSSLAF (R→S, position 6, BLOSUM score −1) and WKFDSHLAF (R→H, position 6, BLOSUM score 0) were associated with a 5–10-fold reduction in magnitude compared to responses to the WKFDSRLAF epitope (Figure 2A). Similarly in subject 1692 (at 50 DPS), the highest IFN-γ and granzyme B responses were elicited by the same two RW8 epitope variants (RYPLTFGW, RFPLTFGW). Semi-conservative substitutions at presumed TCR contact sites (RYPLTLGW, RYPLCFGW and RFPLCFGW) had large effects on IFN-γ and granzyme B responses (Figure 2B).
Figure 2. IFN-γ and Granzyme B responses to optimal epitope variants in the Nef protein.
Standard IFN-γ and granzyme B ELISpot assays were performed by stimulating with the optimal epitopic peptides and variants at serial concentrations between 10,000 and 0.1 ng/ml. Shown here are PBMC responses from subjects 1188 (WF9 epitope variants) (A) and 1692 (RW8 epitope variants) (B). The SFC frequencies are plotted against the log10 peptide concentration. CON B sequence for each epitope is indicated on the top. Identity of autologous sequence to amino acid in the CON sequence is indicated by a dot.
In summary, T-cells from newly-infected persons recognize multiple epitopic variants, although the magnitude and avidity of the responses can be highly variable. Multiple factors influence recognition, including the number, the type (i.e., conservative vs. non-conservative aa substitutions), and the position of the substitutions within the epitope. Even when epitope variants are recognized, the magnitude and/or avidity of IFN-γ and Granzyme B responses can be significantly lower.
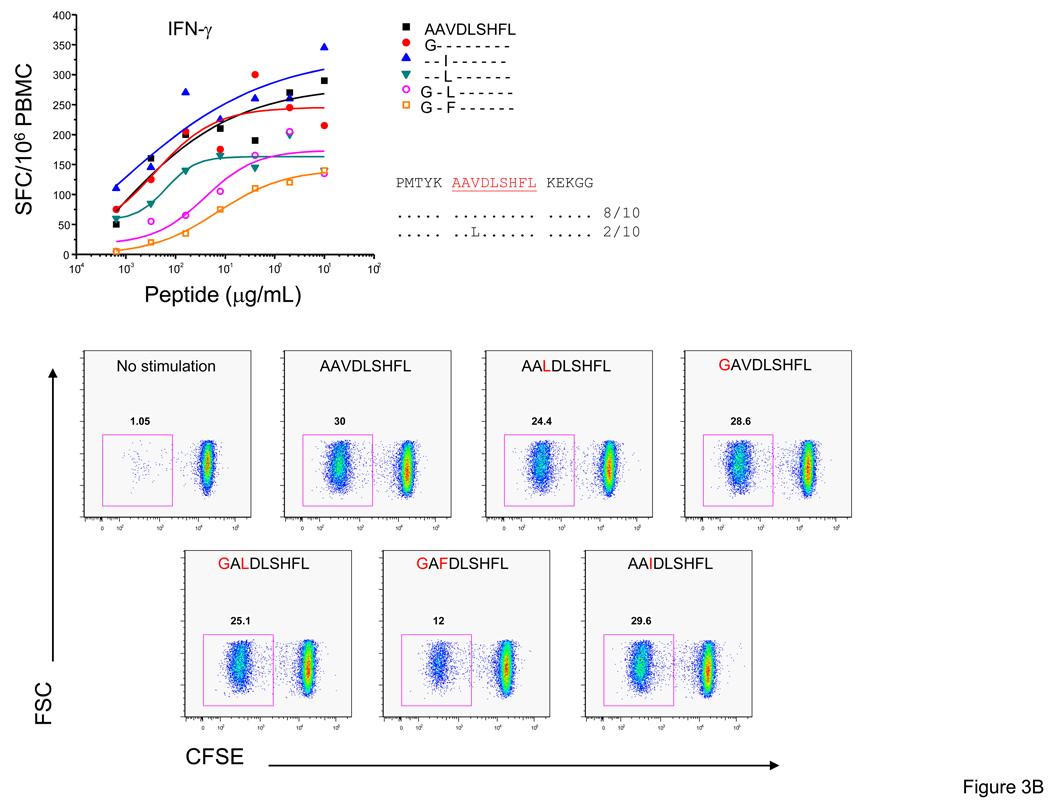
Functional heterogeneity among T-cell responses to epitope variants
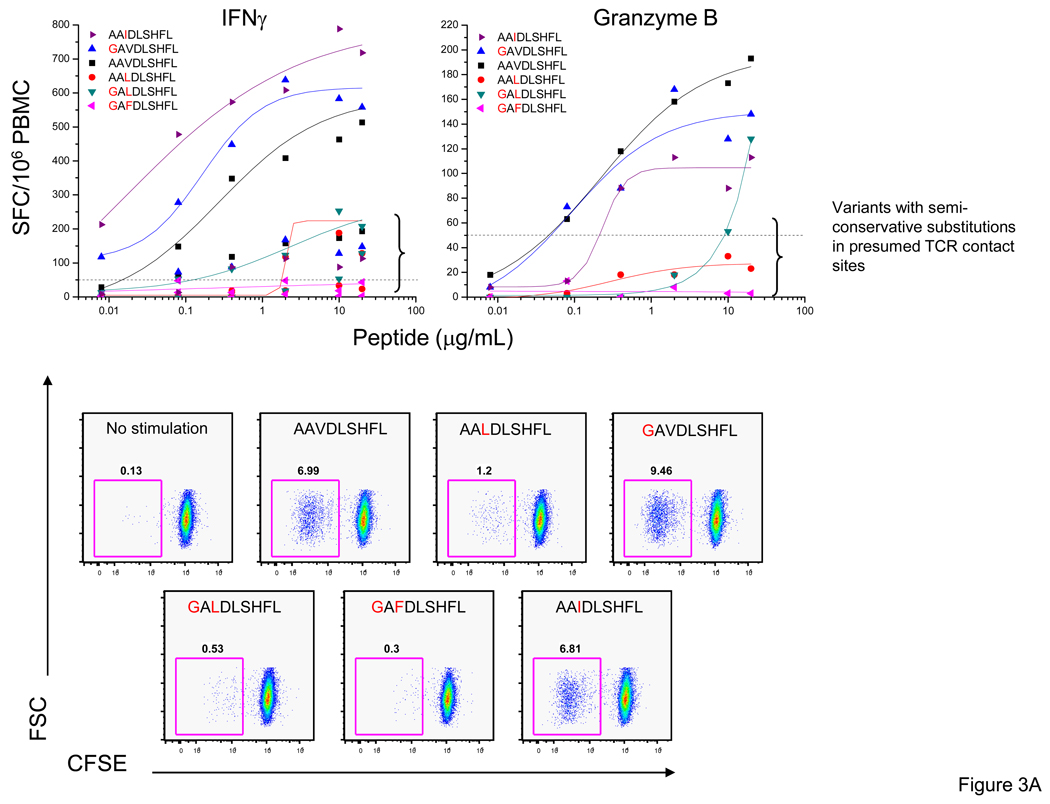
To further define the quality of the T-cell response to different variants, we examined T-cell proliferation by CFSE staining. In subject 1238 we characterized the AL9/Cw08-restricted response examining IFN-γ and granzyme B production as well as proliferation to six variants (AAVDLSHFL, AAV; GAVDLSHFL, GAV; AAIDLSHFL, AAI; GALDLSHFL, GAL; AALDLSHFL, AAL; and GAFDLSHFL, GAF). We observed high magnitude (542 – 726 SFC/106 PBMC, 1 µg/ml) and avidity (0.008 – 0.020) IFN-γ responses to the AAV, GAV and AAI variants and relatively lower magnitude (136 – 179 SFC/106 PBMC at 1 µg/ml) and avidity (0.202 – 0.909 µg/ml) responses to the GAL and AAL variants with no response to the GAF variant. To determine if there were differences in the granzyme B production and proliferative responses to the epitope variants we assessed granzyme B production and the CFSElo frequencies of cells. We detected the strongest granzyme B and T-cell proliferative responses to the same three variants, AAV, GAV and AAI, as the IFN-γ responses (Figure 3A).
Figure 3. T-cell proliferation in response to stimulation with Nef epitope variants relative to IFN-γ and granzyme B responses.
Standard IFN-γ and granzyme B ELISpot assays were performed by stimulating with the optimal epitopic peptides and variants at serial concentrations between 10,000 and 0.1 ng/ml. Shown here are PBMC responses from subjects 1238 (AL9 epitope variants) (A) and 1212 (AL9 epitope variants) (B). The SFC frequencies are plotted against the log10 peptide concentration. Autologous sequence where available is shown and identity of autologous sequence to amino acid in the CON sequence is indicated by a dot. The decrease in CFSE signal was used to monitor proliferation. Positive and negative controls included cells incubated with anti-CD3 mAb (30 ng/mL) and anti-CD28 mAb (1 µg/mL), and with medium alone.
In subject 1212 we characterized the AL9/B62 response examining IFN-γ production and T-cell proliferation to six variants (Figure 3B). The IFN-γ responses to all six variants were strongly avid (EC50 0.0004 – 0.07 µg/ml) and of high magnitude (120 – 270 SFC/106 PBMC, 1 µg/ml) and we observed robust T-cell proliferation to all AL9 epitope variants with CFSElo frequencies in the range of 12 – 30%. Overall, strongly avid, high magnitude IFN-γ responses were associated with granzyme B production and high frequencies of proliferating antigen-specific T-cells.
Distribution of reactive PTE peptides vs. frequency distribution in virus isolates
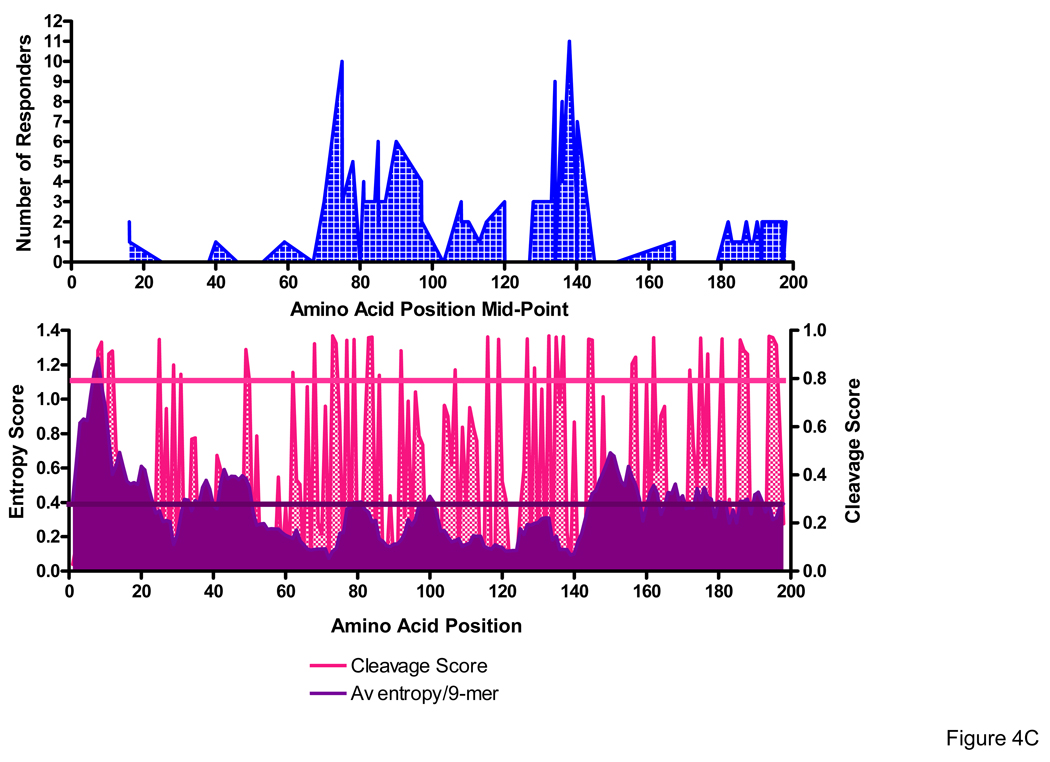
We previously showed that the reactive Nef domains detected with both the CON and PTE peptide sets were similar, with the greatest frequency of responses spanning amino acids 65–103 and 113–147, and minor clusters between amino acids 1–27 and 177–206 [20]. Although majority of T-cell responses were detected following stimulation with the most frequent PTE among subtype B virus isolates, there were several high frequency PTE (peptides # 5, 11, 12, and 13) that were not targeted at all by any of the 18 Nef responders [20]. (PTE peptides with lower numbers are those present at the highest frequency among the virus sequences in the database).
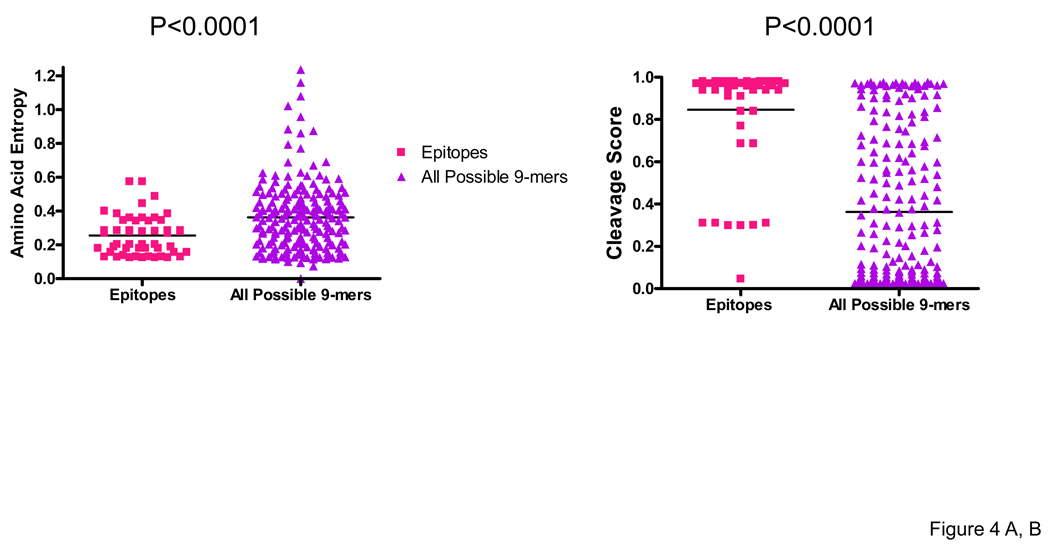
We next examined the extent to which processing likelihoods and amino acid sequence entropy influenced epitope distribution, comparing the average entropy and C-terminus cleavage scores for the defined optimal epitopes with that of the entire group of 9-mers (overlapping by one aa, N=206) spanning the entire Nef sequence. Sequence variability was addressed by averaging the Shannon entropy for each amino acid residue in the 9-mer using the subtype B Nef alignments in the Los Alamos database. We considered the epitope data set generated with the use of PTE peptides to be optimal to address these issues since the peptide set has been designed to encompass virus heterogeneity and thus to improve the detection of epitopes in domains with moderate heterogeneity.
The median entropy score for 50 optimally defined 8 to 10-mer epitopes targeted in 17 Nef responders, which included a total of 23 unique epitopes, was 0.236 (range 0.126 – 0.574) compared to 0.363 (range 0 – 1.237) for the entire group of 9-mers spanning the Nef protein sequence (P<0.0001) (Figure 4A). The median C-terminus cleavage scores for the targeted epitopes was 0.960 (0.046 – 0.980) compared to 0.202 (0.022 – 0.980) for the entire group of 9-mers (P<0.0001) (Figure 4B). Notably, 45 of the 50 (90%) targeted epitopes had entropy scores <0.3 and 43 of the 50 (86%) epitopes had cleavage scores >0.5, with scores for 38 of the 50 (76%) being > 0.9. No significant correlation was observed between the average entropy and the C-terminus cleavage scores for the 9-mers (P=0.453, r=−0.053). Epitope distribution relative to entropy (top panel) and cleavage scores (bottom panel) is shown (Figure 4C). Our findings provide evidence for a strong role for both processing preferences and amino acid entropy in determining epitope distribution.
Figure 4. Epitope distribution in the Nef protein relative to entropy and cleavage scores.
Amino acid entropy (A) and proteasomal cleavage scores (B) were compared between 50 optimal epitopes versus all possible 9-mers spanning the nef sequence. Epitope distribution relative to entropy and cleavage scores is shown from the N- to C-terminus (C). Entropy and C-terminus cleavage scores for targeted optimal epitopes and the group of all Nef subtype B 9-mers were compared by the use of Wilcoxon Signed Rank Test.
4. DISCUSSION
Here in a comprehensive analysis we defined the extent of amino acid diversity in epitope variants that is tolerated by HIV-1-specific T-cells in the context of infection and vaccination. We hypothesized a greater breadth of variant epitope recognition by T-cells among infected persons compared to vaccine recipients because of the generation of sequence variants through virus replication during infection. However, T-cell recognition of variant epitopes was similar in the two settings as also were the effects of numbers and conservation amino acid substitutions on the recognition of variant epitopes. It is plausible that the distinct antigen conditions during the priming of T-cell responses in infection versus vaccination and differences in antigen processing and T-helper function have counter effects on variant epitope recognition. Alternately, T-cell recognition of variant epitopes may be independent of these factors.
While Gag, Pol, and Env responses were analyzed in the vaccinees, Nef responses were examined in early infection. Gag, Pol, and Env responses could not be analyzed in early infection due to PBMC limitations. Instead we selected Nef as a prototype because, although a small protein, Nef contains many highly immunogenic regions, is frequently targeted in early infection, and has a broad range of sequence diversity with domains that are highly conserved and others that are highly variable [20,44]. Recognizing the caveat we have refrained from making direct comparisons in the response patterns between infected subjects and vaccinees. We have kept our primary focus on the analysis of the effects of numbers and conservation of substitutions in amino acid residues on variant epitope recognition. We speculate that our findings will be similar if we tested responses to Gag, Pol and Env epitope variants in early infection. We expect these data to be useful in developing algorithms to predict cross- and intra-clade recognition of variant epitopes by T-cells induced following vaccination and infection, accounting for the numbers and relative conservation of amino acid substitutions between the variants and the infecting/consensus/vaccine sequence.
Altfeld et al previously demonstrated that T-cell responses to 29% (12 of 42) of targeted peptides in Gag, Tat, and Vpr were only detected with peptides representing the autologous virus strain compared to the HIV-1 clade B consensus sequence [45]. Specifically, the use of autologous peptides allowed the detection of significantly stronger HIV-1-specific T-cell responses in the more variable regulatory and accessory HIV-1 proteins Tat and Vpr (P = 0.007). Overall our data are in agreement with these findings. While, we demonstrate that many epitope variants can be recognized even when not detected among the autologous virus populations, in further detailed analyses we show that cross-reactive responses are predominantly directed to variants containing single conservative or semi-conservative amino acid substitutions. It is difficult to say with certainty if T-cell responses to variant epitopes were primed without exposure to the variant sequences since the possibility that the variant sequences were present among the autologous virus population at a prior point in time cannot be excluded. Conversely, in no instance did we detect a response to a variant epitope without a response to the autologous consensus sequence epitope.
Furthermore even when T-cells showed an IFN-γ response upon reactivity with epitope variants, the magnitude and/or avidity of IFN-γ and Granzyme B responses and T-cell proliferation was significantly reduced. We postulate that amino acid differences in the peptide variants affect the binding of the peptide variants with the HLA molecules and that of the peptide-MHC (pMHC) complexes with the TCR which then contributes to the difference in responses. For example, epitope variants may deliver negative signals to T-cells specific for the agonist peptide-MHC complexes [46] and the relative avidity of the TCR-pMHC interaction may influence the cytokine response profiles [47,48].
Our findings lead us to hypothesize that vaccine-induced T-cells will proliferate and produce high magnitude IFN-γ and granzyme B responses when challenged with either homologous virus sequences or those that differ from the vaccine sequence by single conservative substitutions at the targeted specificities. By contrast, if the challenge sequences contain multiple or semi- and non-conservative substitutions in the targeted epitopes, then more likely than not, the T-cells will exhibit poor proliferation with low or no granzyme B production and low magnitude IFN-γ responses. We further hypothesize that broad T-cell recognition encompassing proliferation and high magnitude cytokine production directed at several epitope specificities may require vaccines that encompass multiple sequence variants.
Since PTE peptides, as well as similar [49] and combined central sequence [50] peptide sets made by others, are designed to maximally cover known circulating viral sequences, they are especially suited for analysis of the breadth of potentially protective immune responses in recipients of HIV-1 preventive vaccines and in infected individuals. While in this study we employed the PTE peptides to evaluate vaccine immunogenicity, this and similar approaches [21,23] lay the framework for the design of a vaccine immunogen with expanded T-cell epitope coverage for protection against highly diverse and rapidly changing virus populations.
Several prior studies have addressed factors that contribute to immunogenicity of highly targeted domains. These include processing preferences for proteasomal cleavage [22,51–53]; the frequency of C-terminus amino acids that inhibit HLA binding [53,54]; and constraints on the amino acids that can be accommodated in the peptide-binding groove of the HLA molecules [31,55,56]. Comparing published experimentally defined HIV-1 CTL epitope distributions to global protein sequence alignments, Yusim et al showed that epitopes are concentrated in relatively conserved regions and that highly variable regions that lack epitopes have a paucity of predicted proteasome processing sites and an enrichment for amino acids that do not serve as C-terminal anchor residues [19,57]. In a subsequent study of CTL responses using a clade B consensus sequence peptide set spanning all HIV proteins Frahm et al showed that the frequency of recognition for each peptide was highly correlated with the relative conservation of the peptide sequence, the presence of predicted proteasome cleavage sites within the C-terminal half, and a reduced frequency of amino acids that impair binding to the restricting class I molecules [19,57].
The unique aspect of our study was the use of PTE peptides to address the issue. Since, sequence variability between infecting and testing antigens, especially when the latter are based on central-sequences, could limit the ability to detect epitopes in relatively variable domains[45], we examined epitope distribution characteristics using the PTE peptide set with its expanded epitope coverage. T-cell responses detected by stimulation with these peptides are significantly greater in breadth and magnitude compared to those detected with CON peptides [20]. Our findings with the use of a peptide set with significantly expanded epitope coverage indicate that conservation of the peptide sequence remains a critical factor in driving epitope distribution in addition to the C-terminus cleavage scores. We show that the entropy scores for the targeted epitopes are significantly lower (P<0.0001) and the C-terminus cleavage scores significantly higher (P<0.0001) compared to the group of 9-mers spanning the protein sequence. We postulate that cost-effective design of peptide and sequence based vaccine immunogens as well as peptide test reagents should emphasize domains that are likely to be T-cell targets [21–24].
In summary, we have defined the extent of amino acid diversity in epitope variants that is tolerated by HIV-1-specific T-cells in the context of infection and vaccination. In more detailed analyses, although, in a small number of subjects we show significant differences in the magnitude of the IFN-γ response to the variants as well as in other functional properties such as proliferation and granzyme B production, particularly in the presence of multiple or semi- and non-conservative amino acid substitutions. Therefore, when assessing response to variant epitopes in studies of cross-clade T-cell responses, a qualitative assessment for the presence or absence of an IFN-γ response is not sufficient. Our data support the strategic design of vaccines that generate T-cell populations with optimal effector activities through inclusion of multiple sequence variants. Finally, we show that epitope distribution is strongly influenced by both processing preferences and amino acid entropy. Thus, cost-effective design of peptide and sequence based vaccine immunogens that provide maximal coverage of circulating sequences may be achieved through emphasis on virus domains with high likelihood of being T-cell targets.
Table 1.
Demographic, clinical, and virologic characteristics of the primary infection study population.
| Enrollment Time Point | Sampling Time Point | HLA Class I | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject IDA |
Age | EthnicityB | Days Post Infection |
HIV-1 RNA copies/ml |
CD4+ T- cells/mm3 |
Days Post Infection |
Treatment duration (days) |
HIV-1 RNA copies/mL |
CD4+ T- cells/mm3 |
A | B | Cw |
| 1188 | 23 | H | 79 | 73,074 | 409 | 93 | 5 | <500 | 375 | 23 | 35, 15 | 02, 07 |
| 1212 | 37 | C | 11 | 5,060,000 | 757 | 16 | No | 102,045 | 925 | 02, 23 | 44, 62 | 03, 04 |
| 1216 | 33 | C | 129 | 26,000 | 300 | 134 | 2 | 38,400 | 626 | 03,24 | 07,39 | 07 |
| 1238 | 42 | C | 45 | 99,600 | 367 | 45 | No | 99,600 | 367 | 01, 03 | 07, 14 | 07, 08 |
| 1242 | 30 | C | 42 | 96,900 | 874 | 105 | 57 | 343 | 787 | 03, 33 | 14, 35 | 04, 08 |
| 1362 | 24 | C | 8 | 8,240,000 | 878 | 8–680 | No | 117,077 | 871 | 02, 25 | 18, 51 | 01, 12 |
| 1484 | 27 | C | 12 | 4,340,700 | 300 | 19 | No | 4,340,700 | 830 | 02,03 | 07,35 | 04,07 |
| 1490 | 41 | C | 55 | 286,272 | 568 | 62 | No | 286,272 | 568 | 02, 11 | 15, 35 | 03, 04 |
| 1576 | 32 | C | 22 | 3,717,000 | 332 | 63 | 42 | 4,129 | 552 | 11, 33 | 49, 51 | 07, 15 |
| 1596 | 36 | C | 19 | 260,605 | 700 | 20 | No | 260,605 | 265 | 03,26 | 38,35 | 04,12 |
| 1599 | 30 | C | 46 | 404,463 | 265 | 46 | No | 404,463 | 265 | 01, 02 | 08, 18 | 7 |
| 1686 | 38 | H | 55 | 155,545 | 186 | 70 | No | 219,312 | 241 | 23, 25 | 35, 40 | 03, 04 |
| 1689 | 28 | H | 11 | 2,240,958 | 1206 | 11 | No | 94,708 | 745 | 02, 03 | 35, 40 | 03, 04 |
| 1690 | 42 | C | 21 | 1,031,477 | 396 | 21 | No | 400,163 | 528 | 01 | 08 | 07 |
| 1692 | 42 | B | 50 | 2,461 | 612 | 50 | No | 2,461 | 612 | 23, 30 | 08, 42 | 07, 17 |
| 1693 | 41 | H | 7 | 377,063 | 559 | 7 | No | 1,912,195 | 472 | 01, 03 | 08, 35 | 04, 07 |
| 1698 | 45 | H | 11 | 734,413 | 458 | 11 | No | 262,437 | 531 | NT | NT | NT |
All subjects were male.
Ethnicity: C, Caucasian; H, Hispanic; B, Black
ACKNOWLEDGMENTS
We thank Dr Janine Maenza and Claire Stevens for subject enrollment, Dr. Kurt Diem and his laboratory for specimen processing, T. Smith for data management, and the study participants for their time and effort. This work was supported by the National Institutes of Health grants AI64061, AI27757 and AI57005, and awards from the HIV Vaccine Trials Network (UM) and Puget Sound Partners in Global Health (UM).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Korber B, Muldoon M, Theiler J, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288(5472):1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 2.Nabel GJ. Challenges and opportunities for development of an AIDS vaccine. Nature. 2001;410(6831):1002–1007. doi: 10.1038/35073500. [DOI] [PubMed] [Google Scholar]
- 3.Mullins JI, Jensen MA. Evolutionary dynamics of HIV-1 and the control of AIDS. Curr Top Microbiol Immunol. 2006;299:171–192. doi: 10.1007/3-540-26397-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari G, Humphrey W, McElrath MJ, et al. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci U S A. 1997;94(4):1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buseyne F, Chaix ML, Fleury B, et al. Cross-clade-specific cytotoxic T lymphocytes in HIV-1-infected children. Virology. 1998;250(2):316–324. doi: 10.1006/viro.1998.9373. [DOI] [PubMed] [Google Scholar]
- 6.Lynch JA, deSouza M, Robb MD, et al. Cross-clade cytotoxic T cell response to human immunodeficiency virus type 1 proteins among HLA disparate North Americans and Thais. J Infect Dis. 1998;178(4):1040–1046. doi: 10.1086/515652. [DOI] [PubMed] [Google Scholar]
- 7.McAdam S, Kaleebu P, Krausa P, et al. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. Aids. 1998;12(6):571–579. doi: 10.1097/00002030-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Wilson SE, Pedersen SL, Kunich JC, et al. Cross-clade envelope glycoprotein 160-specific CD8+ cytotoxic T lymphocyte responses in early HIV type 1 clade B infection. AIDS Res Hum Retroviruses. 1998;14(11):925–937. doi: 10.1089/aid.1998.14.925. [DOI] [PubMed] [Google Scholar]
- 9.Dorrell L, Dong T, Ogg GS, et al. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J Virol. 1999;73(2):1708–1714. doi: 10.1128/jvi.73.2.1708-1714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao H, Mani I, Vincent R, et al. Cellular immunity to human immunodeficiency virus type 1 (HIV-1) clades: relevance to HIV-1 vaccine trials in Uganda. J Infect Dis. 2000;182(5):1350–1356. doi: 10.1086/315868. [DOI] [PubMed] [Google Scholar]
- 11.Ferrari G, Kostyu DD, Cox J, et al. Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines. AIDS Res Hum Retroviruses. 2000;16(14):1433–1443. doi: 10.1089/08892220050140982. [DOI] [PubMed] [Google Scholar]
- 12.Keating SM, Bollinger RC, Quinn TC, Jackson JB, Carruth LM. Cross-clade T lymphocyte-mediated immunity to HIV type 1: implications for vaccine design and immunodetection assays. AIDS Res Hum Retroviruses. 2002;18(14):1067–1079. doi: 10.1089/08892220260235425. [DOI] [PubMed] [Google Scholar]
- 13.De Groot AS, Jesdale B, Martin W, et al. Mapping cross-clade HIV-1 vaccine epitopes using a bioinformatics approach. Vaccine. 2003;21(27–30):4486–4504. doi: 10.1016/s0264-410x(03)00390-6. [DOI] [PubMed] [Google Scholar]
- 14.Coplan PM, Gupta SB, Dubey SA, et al. Cross-Reactivity of Anti-HIV-1 T Cell Immune Responses among the Major HIV-1 Clades in HIV-1-Positive Individuals from 4 Continents. J Infect Dis. 2005;191(9):1427–1434. doi: 10.1086/428450. [DOI] [PubMed] [Google Scholar]
- 15.Malhotra U, Nolin J, Mullins JI, McElrath MJ. Comprehensive epitope analysis of cross-clade Gag-specific T-cell responses in individuals with early HIV-1 infection in the US epidemic. Vaccine. 2007;25(2):381–390. doi: 10.1016/j.vaccine.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 16.Gaschen B, Taylor J, Yusim K, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 17.Nickle DC, Jensen MA, Gottlieb GS, et al. Consensus and ancestral state HIV vaccines. Science. 2003;299(5612):1515–1518. doi: 10.1126/science.299.5612.1515c. author reply 1515–1518. [DOI] [PubMed] [Google Scholar]
- 18.Li F, Malhotra U, Gilbert PB, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24(47–48):6893–6904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Yusim K, Kesmir C, Gaschen B, et al. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol. 2002;76(17):8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malhotra U, Li F, Nolin J, et al. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J Virol. 2007;81(10):5225–5237. doi: 10.1128/JVI.02564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nickle DC, Rolland M, Jensen MA, et al. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3(4):e75. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathog. 2007;3(11):e157. doi: 10.1371/journal.ppat.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer W, Perkins S, Theiler J, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13(1):100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- 24.Fischer W, Liao HX, Haynes BF, Letvin NL, Korber B. Coping with viral diversity in HIV vaccine design: a response to Nickle et al. PLoS Comput Biol. 2008;4(1):e15. doi: 10.1371/journal.pcbi.0040015. author reply e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berrey MM, Schacker T, Collier AC, et al. Treatment of primary human immunodeficiency virus type 1 infection with potent antiretroviral therapy reduces frequency of rapid progression to AIDS. J Infect Dis. 2001;183(10):1466–1475. doi: 10.1086/320189. [DOI] [PubMed] [Google Scholar]
- 26.Schacker TW, Hughes JP, Shea T, Coombs RW, Corey L. Biological and virologic characteristics of primary HIV infection. Ann Intern Med. 1998;128(8):613–620. doi: 10.7326/0003-4819-128-8-199804150-00001. [DOI] [PubMed] [Google Scholar]
- 27.Malhotra U, Holte S, Zhu T, Delpit E, Huntsberry C, Sette A, Shankarappa R, Maenza J, Corey L, McElrath MJ. Early induction and maintenance of env-specific T-helper cells following HIV-1 infection. J Virology. 2003;77(4) doi: 10.1128/JVI.77.4.2663-2674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbins KE, Lemey P, Pybus OG, et al. U.S. Human immunodeficiency virus type 1 epidemic: date of origin, population history, and characterization of early strains. J Virol. 2003;77(11):6359–6366. doi: 10.1128/JVI.77.11.6359-6366.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dewar RL, Highbarger HC, Sarmiento MD, et al. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J Infect Dis. 1994;170(5):1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 30.Bunce M, Fanning GC, Welsh KI. Comprehensive, serologically equivalent DNA typing for HLA-B by PCR using sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;45(2):81–90. doi: 10.1111/j.1399-0039.1995.tb02422.x. [DOI] [PubMed] [Google Scholar]
- 31.Pamer E, Cresswell P. Mechanisms of MHC class I--restricted antigen processing. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 32.Wenzel T, Eckerskorn C, Lottspeich F, Baumeister W. Existence of a molecular ruler in proteasomes suggested by analysis of degradation products. FEBS Lett. 1994;349(2):205–209. doi: 10.1016/0014-5793(94)00665-2. [DOI] [PubMed] [Google Scholar]
- 33.Momburg F, Roelse J, Hammerling GJ, Neefjes JJ. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. 1994;179(5):1613–1623. doi: 10.1084/jem.179.5.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of Human Immunodeficiency Virus type 1 (HIV-1)-specific Gamma Interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003;77(12):6867–6878. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kern F, Surel IP, Brock C, et al. T-cell epitope mapping by flow cytometry. Nat Med. 1998;4(8):975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 36.Betts MR, Casazza JP, Patterson BA, et al. Putative immunodominant human immunodeficiency virus-specific CD8(+) T- cell responses cannot be predicted by major histocompatibility complex class I haplotype. J Virol. 2000;74(19):9144–9151. doi: 10.1128/jvi.74.19.9144-9151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyons AB. Divided we stand: tracking cell proliferation with carboxyfluorescein diacetate succinimidyl ester. Immunol Cell Biol. 1999;77(6):509–515. doi: 10.1046/j.1440-1711.1999.00864.x. [DOI] [PubMed] [Google Scholar]
- 38.Lyons BA, Parish CR. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171(1):131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 39.Liu SL, Schacker T, Musey L, et al. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71(6):4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73(12):10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu S-L, Rodrigo AG, Shankarappa R, et al. HIV quasispecies and resampling. Science. 1996;273(5274):415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 42.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57(1–2):33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 44.Addo MM, Yu XG, Rathod A, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77(3):2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altfeld M, Addo MM, Shankarappa R, et al. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J Virol. 2003;77(13):7330–7340. doi: 10.1128/JVI.77.13.7330-7340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mirshahidi S, Ferris LC, Sadegh-Nasseri S. The magnitude of TCR engagement is a critical predictor of T cell anergy or activation. J Immunol. 2004;172(9):5346–5355. doi: 10.4049/jimmunol.172.9.5346. [DOI] [PubMed] [Google Scholar]
- 47.La Gruta NL, Turner SJ, Doherty PC. Hierarchies in Cytokine Expression Profiles for Acute and Resolving Influenza Virus-Specific CD8+ T Cell Responses: Correlation of Cytokine Profile and TCR Avidity. J Immunol. 2004;172:5553–5560. doi: 10.4049/jimmunol.172.9.5553. [DOI] [PubMed] [Google Scholar]
- 48.Price DA, O'Callaghan CA, Whelan JA, Easterbrook PJ, Phillips RE. Cytotoxic T lymphocytes and viral evolution in primary HIV-1 infection. Clin Sci (Lond) 1999;97(6):707–718. [PubMed] [Google Scholar]
- 49.Frahm N, Kaufmann DE, Yusim K, et al. Increased sequence diversity coverage improves detection of HIV-specific T cell responses. J Immunol. 2007;179(10):6638–6650. doi: 10.4049/jimmunol.179.10.6638. [DOI] [PubMed] [Google Scholar]
- 50.Frahm N, Nickle DC, Linde CH, et al. Increased detection of HIV-specific T cell responses by combination of central sequences with comparable immunogenicity. Aids. 2008;22(4):447–456. doi: 10.1097/QAD.0b013e3282f42412. [DOI] [PubMed] [Google Scholar]
- 51.Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16(1):76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Cardozo C, Kohanski RA. Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the "immunoproteasome". J Biol Chem. 1998;273(27):16764–16770. doi: 10.1074/jbc.273.27.16764. [DOI] [PubMed] [Google Scholar]
- 53.Bochtler M, Ditzel L, Groll M, Hartmann C, Huber R. The proteasome. Annu Rev Biophys Biomol Struct. 1999;28:295–317. doi: 10.1146/annurev.biophys.28.1.295. [DOI] [PubMed] [Google Scholar]
- 54.Beekman NJ, van Veelen PA, van Hall T, et al. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J Immunol. 2000;164(4):1898–1905. doi: 10.4049/jimmunol.164.4.1898. [DOI] [PubMed] [Google Scholar]
- 55.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257(5072):927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 56.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 57.Frahm N, Korber BT, Adams CM, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78(5):2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]