Abstract
Bile salts are the major end-metabolites of cholesterol and are also important in lipid and protein digestion, as well as shaping of the gut microflora. Previous studies had demonstrated variation of bile salt structures across vertebrate species. We greatly extend prior surveys of bile salt variation in fish and amphibians, particularly in analysis of the biliary bile salts of Agnatha and Chondrichthyes. While there is significant structural variation of bile salts across all fish orders, bile salt profiles are generally stable within orders of fish and do not correlate with differences in diet. This large data set allowed us to infer evolutionary changes in the bile salt synthetic pathway. The hypothesized ancestral bile salt synthetic pathway, likely exemplified in extant hagfish, is simpler and much shorter than the pathway of most teleost fish and terrestrial vertebrates. Thus, the bile salt synthetic pathway has become longer and more complex throughout vertebrate evolution. Analysis of the evolution of bile salt synthetic pathways provides a rich model system for the molecular evolution of a complex biochemical pathway in vertebrates.
Keywords: Cholesterol, enzymes, hagfishes, mass spectrometry, molecular evolution, steroids
Introduction
Cholesterol has played a major role in the evolution of vertebrates (Nes and Nes 1980), who utilize cholesterol to an extent not matched in any invertebrate species studied to date. Vertebrate animals take advantage of the versatile properties of cholesterol to regulate cell membrane fluidity, to insulate nerve fibers, and to serve as a precursor for the synthesis of steroid hormones and other endogenous compounds. The increased use of cholesterol by vertebrates compared to invertebrates is thought to have been a major evolutionary shift, requiring tightly regulated systems for controlling cholesterol synthesis and elimination from the body. The ability to remove cholesterol from the body in a regulated fashion was accomplished by the parallel development of a hepatobiliary tract and synthetic pathways for biosynthesis and conjugation of bile alcohols and bile acids, the major elimination products of cholesterol (Haslewood 1967; Nes and Nes 1980). Collectively, these conjugated amphipathic molecules are termed bile salts (Hofmann and Hagey 2008).
Bile salts are water-soluble, amphipathic end-metabolites of cholesterol that facilitate intestinal absorption of lipids, enhance proteolytic cleavage of dietary proteins, and exert potent antimicrobial activity in the small intestine (Hofmann and Hagey 2008). Bile salts are produced by every class of vertebrate animals and show remarkable structural diversity across species (Haslewood 1967; Moschetta et al. 2005; Une and Hoshita 1994). Bile salts have not been detected to date in invertebrate animals, although certain species such as sea squirts (e.g., Ciona intestinalis) synthesize bile salt-like compounds for physiological functions likely unrelated to digestion or cholesterol disposal (Yoshida et al. 2002). Bile salt derivatives are known to be pheromones in the sea lamprey (Petromyzon marinus) (Li et al. 2002). The olfactory systems of a number of teleost fish have been shown to be highly sensitive to detection of bile salts in the water, although the physiologic importance of this is as yet unclear (Hara 1994).
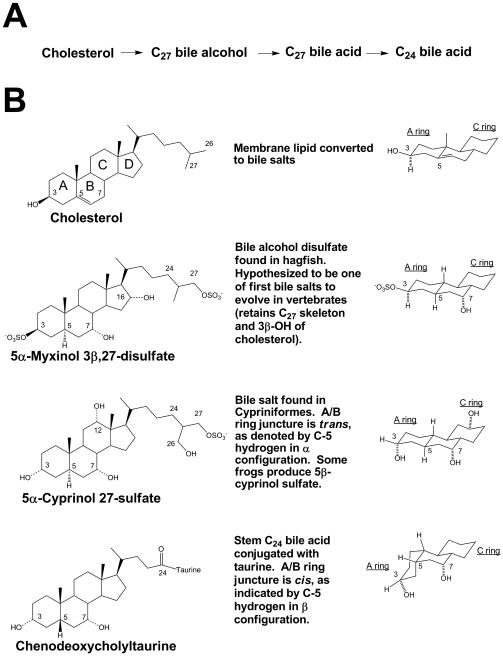
The two basic structural components of bile salts are the 19-carbon (C19) steroid nucleus and the side-chain (Figure 1). In all bile salts characterized to date, the cyclopentanophenanthrene nucleus (with four steroid rings labelled A, B, C, and D in Figure 1B) is fully saturated (unlike cholesterol which has a double bond at C5–C6). The A/B ring juncture is variable, being cis in most bile salts but trans in some species, and influences overall shape of the steroid nucleus. A/B cis bile salts have a ‘bent’ orientation of the A ring relative to the other three rings, while A/B trans bile salts have an extended, planar orientation of the steroid rings. Virtually all bile salts have hydroxyl groups at C-3 (from that of cholesterol) and at C-7. Additional common sites of hydroxylation are at C-12 and C-16, but other sites of hydroxylation have been found. Hydroxyl groups may also vary in their configuration, i.e. α or β, and in some cases have undergone dehydrogenation to an oxo group (Hofmann and Hagey 2008).
Figure 1.
Representative bile salts and their structures. (A) Simplified version of the bile salt synthetic pathway. (B) All bile salts are derived from cholesterol (topmost structure), illustrated with the four steroid rings labeled A, B, C, and D. Hagfish, sea lamprey, lobe-finned fish, and Cypriniform fish (e.g., carp, zebrafish) utilize bile salts that have an A/B ring juncture that is trans, resulting in an overall planar and extended structure of the steroid rings (see representation of A, B, and C rings on the right side). One of the two major primary bile salts in many ray-finned fish is chenodeoxycholyltaurine (taurochenodeoxycholic acid), which is the taurine N-acyl amidate of chenodeoxycholic acid (CDCA). CDCA has a stem (or root) hydroxylation pattern (3α,7α-hydroxy) and an A/B ring juncture that is cis, resulting in a bent conformation of the steroid rings. The most common primary bile acid in ray-finned fish is cholic acid, which has an additional hydroxyl group at C-12 (relative to CDCA).
Bile salts that retain all the carbon atoms of cholesterol have an 8-carbon (C8) side-chain and a total of 27 carbon atoms (C27). C27 Bile alcohols are likely the evolutionarily oldest bile salts (Hofmann et al. 2009; Reschly et al. 2008a). In many species, the bile salt side-chain is cleaved by three carbon atoms, resulting in a C5 side-chain and C24 bile acids. Further structural variation in the side-chain in addition to the side-chain length includes the presence and orientation of hydroxyl groups, the presence of unsaturation in the side-chain, and, above all, the substituents on the terminal carbon atom which is a hydroxyl (-OH) group in bile alcohols and a carboxyl (-COOH) group in bile acids. Primary bile salts are those synthesized by the liver. Secondary bile salts result from extra-hepatic modifications of bile salts, typically by host bacteria in the distal gut (Hofmann 2004; Hofmann and Hagey 2008).
Most bile salts can be assigned to one of three broad classes (Haslewood 1967; Moschetta et al. 2005; Une and Hoshita 1994): (1) C27 bile alcohols, (2) C27 bile acids, or (3) C24 bile acids. We use this classification throughout this manuscript. Bile alcohols are almost always present in bile as sulfate esters. Cholic acid (CA; 3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) are examples of more evolutionarily recent 24-carbon atom (C24) bile acids that have a ‘bent’ shape because of their cis A/B ring juncture. They are shown in Figure 1B conjugated (N-acylamidated) with taurine, which is the main form in which they are present in bile.
The details of bile salt biosynthetic pathways are known only in mammals such as humans and rodents, and have not yet been resolved in other species. The synthetic pathway in humans and rodents using C24 5β bile acids is long and complex, involving up to 16 enzymes and requiring the transport of intermediates between multiple organelles (Norlin and Wikvall 2007; Russell 2003). Although the details of the bile salt biosynthetic pathways used by non-mammalian species are unknown, animals using exclusively C27 bile alcohols likely have simpler and shorter synthetic pathways, as C27 bile alcohols do not require side-chain cleavage or oxidation of a terminal alcohol to a carboxylic acid (Krasowski et al. 2005a; Reschly et al. 2008a).
One of the main prerequisites for an effective bile salt is to maintain aqueous solubility in bile and small intestinal content. Cholesterol, with its single hydroxyl group at 3β, is essentially insoluble in water. Even the sulfated conjugate of cholesterol is insoluble, being a component of skin lipids (Gray and Yardley 1975). Although the physicochemical properties of C27 bile alcohols with two or three hydroxyl groups have not been studied, at least two nuclear hydroxyl groups are probably required to achieve adequate aqueous solubility. This opinion is based on the known insolubility of monohydroxy C24 bile acids at biological temperatures (Hofmann and Small 1967). We consider the C27 bile alcohol with hydroxyl groups at C-3, C-7, and C-27 to be the ‘minimal’ or ‘stem’ bile alcohol, i.e., the compound that requires the fewest structural changes from cholesterol that can function as a bile salt and maintain aqueous solubility in the biliary and intestinal tracts. CDCA can be considered as the ‘stem’ C24 bile acid as it possess the minimal hydroxylation pattern (3α,7α) common to the majority of C24 primary bile acids (Figures 1 and 2).
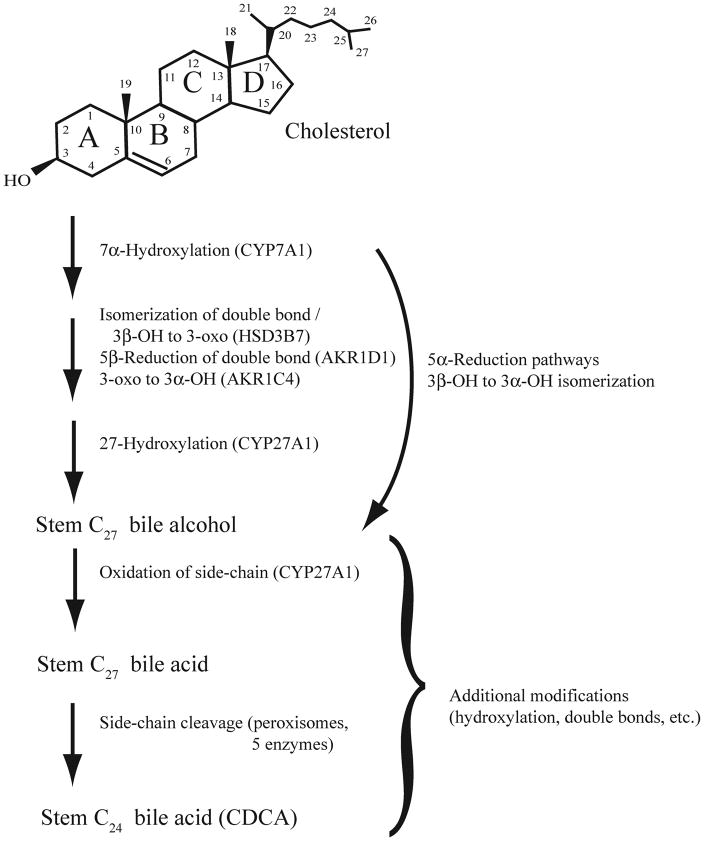
Figure 2.
The bile salt synthetic pathway. The enzymatic pathways involved in bile salt synthesis have been elucidated only in humans and rodents The pathway depicted is the so-called ‘neutral’ pathway, as opposed to an ‘acidic’ pathway that modifies the side-chain first (Russell 2003) and likely is a later evolutionary innovation. The ultimate production of C24 bile acids such as chenodeoxycholic acid (CDCA) require multiple changes to the cholesterol structure including 7α-hydroxylation, isomerization of the C-5–C-6 double bond to form a Δ4 compound coupled with oxidation of the 3β-hydroxy group to a 3-oxo group, stereospecific reduction of the 3-oxo group to a 3α-hydroxy group, hydroxylation of the terminal carbon of the side-chain, oxidation of the side-chain, and shortening of the side-chain. Animals using C27 bile alcohols or C27 bile acids as their primary bile salts presumably require fewer enzymatic steps than those synthesizing C24 bile acids. The enzymes involved in 5α-reduction leading to 5α-bile salts are unknown. The stem C27 bile alcohol would be 3α,7α,27-trihydroxy-5β-cholestane. The stem C27 bile acid would be 3α,7α-dihydroxy-5β-cholestan-27-oic acid, but this bile acid occurs very rarely in fish. Note that some steps in the pathway require more than one enzyme (e.g., isomerization of the 3-hydroxy group) whereas some enzymes (e.g., CYP27A1) can catalyze more than one reaction. Additional hydroxyl groups may be added to the nucleus or side-chain; hydroxylation may occur on an intermediate or on the completed stem bile acid.
A simplified version of the bile salt synthetic pathway is shown in Figure 2. There are five enzymes required to form the stem C27 bile alcohol. The first enzyme is cytochrome P450 (CYP) 7A1 which converts cholesterol to 7α-hydroxycholesterol. The second enzyme, 3β-hydroxysteroid-Δ5-C27 steroid oxidoreductase (HSD3B7), mediates simultaneous isomerization of the C-5–C-6 double bond to C-4–C-5 and oxidation of the 3β hydroxyl group to a 3-oxo group. The third enzyme, aldo-keto reductase 1D1 (AKR1D1), mediates stereospecific reduction of the Δ4 double bond to form a 5β (A/B cis) steroid. The fourth enzyme, AKR1C4, reduces the 3-oxo group to a 3α-hydroxy group (Jez and Penning 2001). The fifth and final enzyme is the mitochondrial C-27 hydroxylase (CYP27A1) that results in the formation of a C27 bile alcohol. In the Discussion, we speculate that variation in three of the five enzymes (AKR1C4, AKR1D1, and CYP27A1) required for C27 bile alcohol formation may account for some of the observed cross-species variation in bile salt profiles.
We greatly extend prior surveys of bile salt diversity in fish and amphibians. To provide insight into the ancestral bile salt type, we focused studies on Agnatha (jawless fish), a possibly paraphyletic superclass of the phylogenetically most basal vertebrates currently limited to extant hagfish and lampreys (Forey and Janvier 1993; Nelson 2006). We also surveyed a diverse array of cartilaginous and teleost fish species. The use of multiple analytical techniques enabled the identification of previously unreported bile salts and detailed resolution of biliary bile salt profiles. In this manuscript, we analyze and discuss the patterns of bile salt structural variation across fish and amphibians. Lastly, we also used bioinformatic techniques to identify putative orthologs of the human bile salt synthetic enzymes in fish and amphibians and used this information to form hypotheses for the evolution of the complex pathway for bile salt synthesis.
Material and Methods
Bile samples
Bile samples were obtained from a number of sources (see Acknowledgements) over the course of two decades of analysis. Many of the samples analyzed were from an extensive collection of bile samples of the late G.A.D. Haslewood donated to the University of California – San Diego. Many samples were taken from newly dead animals, but a few were obtained from preserved museum specimens. Some samples were obtained during necropsy of animals that died in captivity at the San Diego Zoo. Some bile samples were collected at the University of Pittsburgh from animals of the following sources: sea lamprey, Acme Lamprey Co., Harrison, ME, USA; Pacific hagfish (Eptatretus stouti), Marinus Scientific, Garden Grove, CA, USA; African clawed frog (Xenopus laevis) and Western clawed frog (Xenopus tropicalis), NASCO, Fort Atkinson, WI, USA; aquatic tiger salamander (Ambystoma tigrinum), Research Amphibians, Inc., Nashville, TN, USA. All animal studies were performed in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals, incorporated in the Institute for Laboratory Animal Research Guide for Care and use of Laboratory Animals. All vertebrate animal studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committees (approval number 0601348) or Committee on Animal Studies of the University of California, San Diego. Bile from the hagfish Eptatretus strickrotti (Møller and Jones 2007) was obtained from a preserved specimen from the California Academy of Science, museum number CAS 223480, and bile from several other deepwater fishes were obtained from the Zoological Museum, University of Copenhagen (ZMUC), Denmark.
Analysis of bile salts
We analyzed the bile salts in biliary bile from 13 jawless fish, 37 cartilaginous fish, 5 lobe-finned fish, 171 ray-finned fish, and 29 amphibian species. A number of different analytical methods were used to analyze the bile salts found in animal bile. Thin-layer chromatography (TLC) was helpful in identifying which classes of bile salts (bile alcohols, C27 bile acids, C24 bile acids) were present. Depending on the results of TLC analysis, the samples were subjected to further analyses optimized for the bile salt classes present. For C24 bile acids and some C27 bile acids, high-performance liquid chromatography (HPLC) analysis with suitable reference standards was sufficient to resolve the bile salt profiles. When possible, samples were further subjected to electrospray ionization mass spectrometry/mass spectrometry (ESI/MS/MS) analysis. Gas chromatography/mass spectrometry (GC/MS) was helpful when the orientations of hydroxyl or hydrogen groups were not certain from other analyses. However, GC/MS requires relatively large amounts of sample and could not be applied if limited bile was available. For previously unknown bile salts, nuclear magnetic resonance (NMR) was needed to provide definitive structural identification.
TLC
Whole bile was separated on silica gel G (E. Merck, Darmstadt, Germany) using two solvent systems: (1) isoamyl acetate: propionic acid: 1-propanol: water 4: 3: 2: 1 (v/v) (Hofmann 1962); and (2) a double development system for the resolution of conjugated bile acids, in which the plates were developed first in chloroform: methanol: water: acetone: propionic acid, 10: 2: 1: 4: 1 (v/v), and after drying overnight, followed by 1-butanol: propionic acid: water, 10: 1: 1 (v/v) (Elferink et al. 1989). Bile acids were visualized by spray reagents for hydroxyl groups (phosphomolybdic acid, 10% w/v in ethanol), oxo groups (2,4-dinitrophenyl-hydrazine), sugars (naphthoresorcinol), or vicinyl hydroxyl group (lead tetra-acetate).
HPLC
Conjugated bile acids were analyzed by HPLC using a modification of a previously reported technique (Rossi et al. 1987). An octadecylsilane column (RP C-18, Beckman Instruments, Fullerton, CA, USA) was used with isocratic elution at 0.75 mL/min. The eluting solution was composed of a mixture of methanol and 0.01 M KH2PO4 (67.4% v/v), adjusted to an apparent pH of 5.35 with H3PO4. Conjugated bile acids were quantified by measuring the absorbance of their amide bond at 205 nm. Unconjugated bile acids and bile alcohol sulfates are not detected by this method. Bile acids were tentatively identified by matching their relative retention times with those of known standards.
Mass spectrometry
ESI/MS/MS
Biliary contents were dissolved and diluted in methanol (Burdick & Jackson, Muskegon, MI, USA) and analyzed using ESI/MS/MS on a Hewlett-Packard HP 1100 MSD operated in the negative mode. The high-performance liquid chromatography (HPLC) column was removed, and the injector output coupled directly to the ESI inlet. Samples (2 μL) were injected in methanol: water, 90:10 (v/v) mobile phase running at a flow rate of 0.35 mL/min. The fragmenter was set to 200 V and the capillary voltage set to 5000 V. We have previously published ESI/MS/MS spectra of the biliary bile salts of Western clawed frog (Reschly et al. 2008b), sea lamprey, African clawed frog, and zebrafish (Reschly et al. 2008a).
GC/MS
Glycine and taurine conjugates of bile acids were deconjugated chemically using 1.0 N NaOH at 130°C for 4 hours. Bile alcohol sulfates were deconjugated enzymatically (Goto et al. 2003). Unconjugated bile acids were isolated by acidification and extraction into ethyl acetate. They were then analyzed by capillary GC/MS as methyl ester acetates (prepared using acetic anhydride in acetic acid with perchloric acid catalyst) or as methyl ester trimethylsilyl derivatives (prepared using Tri-Sil, Pierce Chemicals, Rockford, IL). GC was performed using a Hewlett-Packard 5890 Gas Chromatograph-5970 MSD, controlled by HP/UX Chem Station software. The column was a 30 m 0.25 mm ID intermediate polarity SPB-35 of 35% phenyl methyl silicone (Supelco Co., Bellefonte, PA) operated at 277°C (isothermal). A splitless injection was used with an injection temperature of 290°C. Helium was used as the carrier gas with a 7 psi column head pressure. Relative retention times and fragmentation spectra of peaks obtained by GC/MS were compared with those of known standards for identification.
NMR
Carbon-13 NMR (13C-NMR) spectra of deconjugated methyl ester per acetyl derivatives of bile acids were also recorded. Multiplicities were determined with the APT sequence. Chemical shifts were recorded in ppm relative to tetramethylsilan. The central peak of the signal of Cl3CD was used as a reference (δ 77.0 ppm).
Classification of bile salt profiles
We grouped bile salts into three broad types (C27 bile alcohols, C27 bile acids, and C24 bile acids). With only three exceptions, the fish and amphibian species analyzed had only one or two bile salt types present at 5% or greater of the total bile salt pool. Thus, we classified each species into one of six categories based on which one or two bile salt types are present at 5% or greater of the biliary bile salt pool (Reschly et al. 2008a): class I, C27 bile alcohols only; class II, C27 bile alcohols + C27 bile acids; class III, C27 bile alcohols + C24 bile acids; class IV, C27 bile acids only; class V, C27 bile acids + C24 bile acids; class VI, C24 bile acids only; and class VII (all three bile salt categories present at proportions greater than 10%) (see summary information on first pages of Supplemental Data Tables S1 and S2, Supporting Information). The three species with class VII bile salt profiles are black-bellied angler (Lophius budegassa), MacCarthy grassland frog (Ptychadena bibroni) and Western clawed frog (Xenopus tropicalis). The classification system used makes no assumptions about the physiological functions or effectiveness of bile salts.
Phylogeny of fishes in relation to bile salt variation
The phylogeny of fish is still actively studied and debated. With the rise of molecular-based phylogenetic studies (Inoue et al. 2003; Miya et al. 2003), the traditional position of many taxa have been either confirmed or challenged. Examples of recent significant changes are the splitting of the gasterosteiform families into three phylogenetically widely separated groups (Kawahara et al. 2008), and the discovery that three deep-water fish families with greatly differing morphologies, Mirapinnidae, Megalomycteridae, and Cetomimidae are actually larvae, males and females, of a single family Cetomimidae (Johnson et al. 2009). To provide an evolutionary framework, we overlay the bile salt profiles of fish orders onto the tree of Nelson (Nelson 2006) that represents a conservative summary of the current knowledge. An exception for the use of Nelson (Nelson 2006) is the recognition of Alepocephaliformes as a separate order and not a family in Argentiniformes (Lavoué et al. 2008).
Identification of bile salt synthetic enzyme orthologs
We utilized BLAST searches to try to identify putative orthologs of the human enzymes involved in bile salt biosynthesis (Table S1). While assignment of true orthology depends on both sequence comparisons and biochemical verification, tentative orthology can be assigned by reciprocal BLAST searching. Nonetheless, caution needs to be applied with comparisons of enzymes that are part of large superfamilies (e.g, CYPs, aldo-keto reductases). Indeed, we were not able to clearly identify putative orthologs in teleost fish to the gene for human AKR1C4 due to the presence of multiple apparent paralogs in fish in the AKR1C subfamily. Similarly, we had difficulty identifying putative orthologs in teleost fish to genes for two of the five peroxisomal enzymes known to be involved in bile acid side-chain shortening in humans and rodents. These enzymes were branched-chain acyl-CoA oxidase (ACOX2) and D-bifunctional protein (HSD17B4), both of which are part of complex superfamilies of enzymes (Russell 2003).
Results
Overview of bile salt variation in fish and amphibians
We analyzed fish from 38 different orders, with more than one fish species examined in 26 of these 38 orders (Tables S1 and S2). Many orders showed essentially no variation of bile salt profiles between species, even between species with different diets. In contrast for several orders (e.g., Beloniformes and Perciformes), there is variation in bile salt types among species (Table 1). However, in such cases, there was always overlap among species in the bile salt type.
Table 1.
Summary of bile salt variation in fish
| Order | Number of species analyzed | Bile salt classes present* | Main bile salts** |
|---|---|---|---|
| AGNATHA | |||
| Myxiniformes | 12 | I | 5α-Myxinol*** |
| Petromyzontiformes | 1 | I | C24 bile alcohols***, C27 bile alcohols |
| CHONDRICHTHYES | |||
| Carcharrhiniformes | 16 | I | 5β-Scymnol |
| Lamniformes | 3 | I | 5β-Scymnol |
| Orectolobiformes | 4 | I | 5β-Scymnol |
| Pristiformes | 1 | I | 5β-Scymnol |
| Rajiformes | 7 | I | 5β-Scymnol |
| Squaliformes | 3 | I | 5β-Scymnol |
| HOLOCEPHALI | |||
| Chimaeriformes | 3 | I | 5β-Chimaerol |
| SARCOPTERYGII | |||
| Ceratodontiformes | 1 | I | C26 to C30 bile alcohols |
| Coelacanthiformes | 1 | I | C27 bile alcohols |
| Lepidosireniformes | 4 | I | C26 to C30 bile alcohols |
| RAY-FINNED FISH | |||
| Acipenseriformes | 1 | III | CA, C27 bile alcohols |
| Alepocephaliformes | 1 | III | CA (95%), C27 bile alcohols (5%) |
| Amiiformes | 1 | III | CA, CDCA, C27 bile alcohols |
| Anguilliformes | 4 | III, VI | |
| Aulopiformes | 1 | VI | CA |
| Beloniformes | 5 | V, VI | CA, CDCA; medaka has 35% C27 bile acids |
| Beryciformes | 2 | VI | CA |
| Characiformes | 1 | V | CA, C27 bile acids |
| Clupeiformes | 3 | III, VI | CA; C27 bile alcohols (pilchard) |
| Cypriniformes | 10 | I | C27 bile alcohol (5α-cyprinol)*** |
| Esociformes | 1 | VI | CA |
| Gadiformes | 8 | VI | CA |
| Gonorynchiformes | 1 | VI | CA |
| Gymnotiformes | 1 | VI | CA |
| Lepisosteiformes | 2 | VI | CA |
| Lophiiformes | 3 | III | CA, C27 bile alcohols |
| Mugiliformes | 2 | VI | CA, CDCA |
| Osteoglossiformes | 5 | III | CA, C27 bile alcohols; 2β-OH bile acids (arapaima)*** |
| Perciformes | 77 | III, VI | CA, CDCA, C27 bile alcohols |
| Pleuronectiformes | 10 | VI | CA, CDCA |
| Polypteriformes | 4 | III | CA, C27 bile alcohols |
| Saccopharygiformes | 1 | VI | CA |
| Salmoniformes | 4 | VI | CA, CDCA |
| Scorpaeniformes | 9 | VI | CA, CDCA |
| Siluriformes | 4 | VI | CA, CDCA |
| Stomiiformes | 1 | VI | CA |
| Syngnathiformes | 2 | V | CA, CDCA, C27 bile acids |
| Tetraodontiformes | 6 | VI | CA, CDCA |
For description of bile salt classes, see Materials and Methods.
Abbreviations: CA, cholic acid; CDCA, chenodeoxycholic acid.
Bile salts unique to this order of fish.
The most common bile salts of ray-finned fish were C24 bile acids, particularly CA and the stem C24 bile acid CDCA. We only detected one C24 bile acid that had nuclear hydroxyl groups at positions other than 3α, 7α, or 12α. This was the unusual 2β-hydroxylated C24 bile acid of the arapaima (Arapaima gigas), a finding first reported by Haslewood and Tökés (Haslewood and Tökés 1972).
5α (A/B trans) bile alcohols were found in Agnatha, lobe-finned fish, Cypriniformes, and some amphibians (e.g., Caudata). 5β C27 bile acids were common in the frogs analyzed (Table S2) but relatively uncommon in the fish samples (Table S1). Forty of the fish species analyzed possessed type III bile salt profiles (C27 bile alcohols and C24 bile acids). In Perciformes, where 77 species were analyzed, 21 had type III profiles while the remainder were type VI (all C24 bile acids). None had appreciable amounts of C27 bile acids in their biliary bile. This suggests that this order of fish went through an evolutionary transition directly from C27 bile alcohols to C24 bile acids, without an ‘intermediate’ step of C27 bile acids (see Discussion for speculation on the mechanism of this transition). We did not detect any 5α C27 bile acids in the fish and amphibian species we analyzed.
The unusual bile salts of hagfish
All hagfish analyzed, including species from both Myxininae (Myxine, 7 spp.) and Eptratretinae (Eptatretus, 5 spp., including formerly recognized genera Paramyxine and Quadratus), used 5α-myxinol disulfate (3β,7α,16α,27-tetrahydroxy-5α-cholestan-3β,27-disulfate) (Haslewood 1966) as their major bile salt (Table S1; Supplemental Fig. S1, A-H). When we examined the bile of the Atlantic hagfish (Myxine glutinosa) in high detail with both ESI/MS/MS and GC/MS, we did detect a minor fraction of hagfish biliary bile salts that were 3α-hydroxylated. 5α-Myxinol disulfate is unusual relative to bile salts from other vertebrate species in retaining the 3β-hydroxy group of cholesterol and in being esterified with two sulfate groups (see Discussion).
Bile salts of the sea lamprey
We found that the sea lamprey produces unusual C24 bile alcohols, especially 5α-petromyzonal-24-sulfate (3α,7α,12α,24-tetrahydroxy-5α-cholan-24-sulfate), in addition to C27 bile alcohols. We focused our studies of lampreys on larval and juvenile (transformer or parasitic) life stages. Haslewood and colleagues four decades ago (Haslewood and Tökés 1969) reported C24 bile alcohol sulfates as the dominant bile salt of sea lampreys. In contrast, we found that the majority of the biliary bile salt pool in the lampreys was comprised of C27 bile alcohol sulfates. The differences may possibly relate to the lampreys we analyzed being more immature than those of Haslewood and colleagues. ESI/MS/MS also showed that approximately 15% of the C27 bile alcohols were found as disulfates (like the main bile salt of hagfish). Approximately half (52%) of the C27 bile alcohols had the 3α-hydroxy oxidized to 3-oxo. As in hagfish, no C24 or C27 bile acids were detected in lamprey bile.
Bile salts of chondrichthyan fish
In contrast to the unusual bile salts of agnathans, the biliary bile salt composition of all species of Elasmobranchii (rays, sharks, and skates) examined was dominated by a single primary C27 5β-bile alcohol, 5β-scymnol 27-sulfate (3α,7α,12α,24,26,27-hexahydroxy-5β-cholestan-27-sulfate) (Bridgwater et al. 1962). The default C27 bile alcohol has hydroxyl groups at C-3, C-7, and C-27 (Supplemental Figures S2 and S3). Therefore, the hexahydroxy bile alcohol scymnol has undergone three additional hydroxylations (one on the nucleus at C-12 and the other two on the side-chain).
C24 bile acids were not detected at more than 5% of the biliary bile salt pool in any of the cartilaginous fish examined. Small amounts (< 2%) of C24 bile acids detected in some elasmobranchs are likely due to absorption of these bile acids from ingested prey. The major bile salt of chimaerae was 5β-chimaerol (3α,7α,12α,24,27-pentahydroxy-5β-cholestane; Supplemental Fig. S1, I), a bile alcohol that differs from 5β-scymnol only by the lack of a hydroxyl group at the C-26 position.
Bile salts of ray-finned fish
The most common bile salts of ray-finned fish in our samples were C24 bile acids, especially CA and CDCA, but some species also have significant quantities of C27 bile alcohols or C27 bile acids (Table S1; Supplemental Fig. S1, J–U). Basal ray-finned fish orders (Polypteriformes, Acipenseriformes, Amiiformes) have species showing varying ratios of C24 bile acids and C27 bile alcohols. For a handful of ray-finned fish species analyzed (e.g., bowfin, Amia calva), C27 bile alcohols account for 20% or more of the biliary bile salt pool. Species analyzed for the remaining fish orders generally show either approximately 100% C24 bile acids (commonly CA) or mostly C24 bile acids with a minor (< 10%) fraction of C27 bile acids or alcohols. The arapaima was unusual in having 2β-hydroxylated C24 bile acids. This was the only ray-finned fish in our sample that had a dominant C24 bile acid other than CA or CDCA.
The bile salts of Cypriniformes were unusual in that all ten species analyzed had an evolutionarily ‘early’ bile salt profile, namely 5α C27 bile alcohols, confirming previous analyses of cypriniform species (Goto et al. 2003; Haslewood 1954). So far, no other ray-finned species analyzed by us or others has shown >5% 5α C27 bile alcohols in their bile salt pool. We discuss below some possible biochemical reasons for the unusual bile salt profile of Cypriniformes.
C27 bile acids appear to be uncommon in ray-finned fish. We found only 5 species with >5% C27 bile acids in bile, but 37 species with type III profiles (C27 bile alcohols + C24 bile acids). Many of the fish with type III profiles in our dataset were found in Perciformes, a group that otherwise had fish with type VI profiles (all C24 bile acids). In all 77 perciform species we examined, none had any appreciable amount (>1% of the total biliary bile salt pool) of C27 bile acids, suggesting that the bile salt transitions in this order of fish did not involve C27 bile acids as an intermediate step (see Discussion). However, some fish do synthesize C27 bile acids. The bile of the Japanese medaka (Oryzias latipes) contained both C24 and C27 bile acids, whereas four other species in Beloniformes had bile with predominantly C24 bile acids.
In two fish species where we analyzed animals of varying developmental stages, European eel (Anguilla anguilla) and Senegalese sole (Solea senegalensis), the bile salt profile of immature animals was ‘earlier’ than in mature animals. In the European eel and Senegalese sole, the bile of adult animals contains almost entirely tauro-CA. However, the bile of immature European eel contains C27 bile acids (dominant bile salt, ~50%) with lesser amounts of C24 bile acids (~30%), C27 bile alcohols (~10%), and C26 bile alcohols (~8%). The bile of immature Senegalese sole was found to contain C27 bile alcohols and mainly unconjugated CA.
Bile salts of lobe-finned fish
We analyzed the biliary bile salts of four lungfish and one coelacanth species. The bile salts of lobe-finned fish were unusual is being 25-hydroxylated (an uncommon site of hydroxylation) and in containing uncommon C26, C28, C29, and C30 bile alcohols (Amos et al. 1977; Anderson and Haslewood 1964). 25-Hydroxylated bile salts were not found in any of the other fish or amphibian species analyzed.
Bile salts of amphibians
The biliary bile salt profiles of species within Anura were often very complicated, in some cases containing all three major types of bile salts, each present at > 5% of the bile salt pool (Table S2). Many frogs synthesize bile salts with total carbon atoms other than 24 or 27. An example of a complicated bile salt profile is illustrated by the Efulen forest tree frog (Leptopelis calcaratus; Fig. S1, V–X). Our limited sample of 3 species from Caudata showed a dominance of 5α bile salts. We did not analyze the bile of any caecilian species.
Comparison of bile salt synthetic enzymes across species
We were able to identify putative orthologs to the genes for human CYP7A1, HSD3B7, and CYP27A1 in all fish analyzed (Table S1). Interestingly, we identified putative orthologs to the gene for human AKR1D1 in all teleost fish analyzed except for zebrafish, the only fish of the group that uses 5α and not 5β bile salts. In fact, our analyses did not reveal any gene in the zebrafish genome similar to human, mouse, chicken, frog, medaka, or pufferfish AKR1D1 or AKR1D1-like genes. The closest matches in the zebrafish genome were much more similar in sequence and substrate binding motifs to mammalian AKR1B enzymes which are not known to be involved in bile salt synthesis (Lindsay et al. 2008; Negishi et al. 2001). All teleost fish genomes analyzed, including that of zebrafish, had putative orthologs to genes for all four peroxisomal enzymes involved in shortening of the side-chain to produce C24 bile acids.
Bile salt variation in relation to fish phylogeny
Using the phylogeny of Nelson (Nelson 2006) as a conservative summation of fish phylogeny, we overlaid the bile salt variation patterns. Starting from the base of the tree, there is a progression from the 5α bile alcohols of Agnatha, coelacanths, and lungfishes to the 5β bile alcohols of Chondrichthyes to the C24 bile acids of ray-finned fish (Figure 3). The bile salt profile of Cypriniformes (C27 5α-bile alcohols) stands out as unusual relative to other related fish such as Clupeiformes and Alepocephaliformes (Lavoué et al. 2008). According to Lavoué et al., Alepocephaliformes is placed phylogenetically near Clupeiformes and Ostariophysi including Cypriniformes, Siluriformes, Gymnotiformes, Characiformes, and Gonorynchiformes. Cypriniformes is unique among these orders in bile salt composition.
Figure 3.
Bile salt variation overlaid on the fish tree proposed by Nelson (Nelson 2006). Fish orders in Nelson’s tree that were not analyzed in our study are not included in the figure. Alepocephaliformes is used instead of Argentiformes following recent results of Lavoué et al. (Lavoué et al. 2008), and Syngnathiformes is used instead of Gasterosteiformes following the results of Kawahara et al. (Kawahara et al. 2008). The major bile salts for each species are indicated by • • •; minor bile salts are indicated by •.
Discussion
We have greatly expanded previous surveys of bile salt structural diversity in fish and amphibians. In addition, our use of multiple analytical techniques (e.g., ESI/MS/MS, GC/MS, HPLC) allowed us to precisely resolve complicated bile salt profiles. We speculate on what these findings mean for the understanding of the evolution of the bile salt synthetic pathway, a key vertebrate innovation that permits cholesterol excretion to be regulated and at the same time generates amphipathic molecules with multiple physiological functions.
What was the structure of the first ‘ancestral’ bile salt? This question can likely never be answered directly. Bile salts are more stable than many endogenous molecules, having been recovered intact from coprolites up to 2000 years old (Lin et al. 1978). Nonetheless, they are unlikely to be able to survive intact hundreds of millions of years in fossils of the earliest vertebrates. It is tempting to hypothesize that the main bile salt of modern hagfish, 5α-myxinol, may closely resemble the ancestral bile salt structure, as do the general morphology of recent and 300 MYA old fossil hagfish (Bardack 1998). The presence of 5α-myxinol disulfate in geographically dispersed hagfish in multiple ecosystems suggests that bile salt composition can be as stable as the general morphology of hagfishes over the course of hundreds of millions of years of animal evolution (Bardack 1998).
Synthesis of 5α-myxinol requires only four alterations to the cholesterol structure: reduction of the cholesterol C5–C6 double bond, 7α hydroxylation, 16α hydroxylation, and 27 hydroxylation. These four changes could theoretically be accomplished by four (or possibly fewer) enzymes, compared to the sixteen enzymes known to be involved in human bile acid synthesis and the five enzymes (CYP7A1, HSD3B7, AKR1D1, AKR1C4, and CYP27A1) required for synthesis of a stem C27 5β-bile alcohol (Russell 2003). Hagfish are also the only known animals whose main primary bile salts retain the 3β-hydroxyl group found in cholesterol. It may also be of importance that the stereochemistry of reducing the cholesterol double bond in a manner that results in an α orientation of C-5 (as seen in the bile salts of agnathan species) is thermodynamically more favorable than reduction to a β orientation (Nes and Nes 1980). We speculate that the addition of a third nuclear hydroxyl group, in this case at C-16, may have been required for aqueous solubility of hagfish bile salts.
All other primary bile salts characterized in our dataset contained 3α-hydroxyl groups, meaning that there is an isomerization of the cholesterol 3β-hydroxyl group during bile salt synthesis. This seemingly simple switch from 3β to 3α in humans and rodents is accomplished by three enzymes (HSD3B7, AKR1D1, and AKR1C4) (Norlin and Wikvall 2007; Russell 2003). These reduce the double bond in cholesterol and use a 3-oxo intermediate step to flip the orientation of the lone hydroxyl group in cholesterol from a 3β to 3α orientation. It will be interesting to determine how hagfish reduce the cholesterol double bond during synthesis of 5α-myxinol, a pathway that does not need to be coupled with isomerization of the 3β-hydroxyl group. This could theoretically be catalyzed by an ortholog to mammalian AKR1D1 or 5α-reductases, or by another as yet uncharacterized enzyme.
The major bile salts of hagfish and lampreys are quite different from one another in both hydroxylation pattern (3β,7α,16α,27 for hagfish and 3α,7α,12α,24 for lamprey) and side-chain length (C8 for hagfish, C5 for lampreys). The phylogeny of Agnatha are in debate, with evidence from mitochondrial DNA that extant lampreys and hagfish are monophyletic (Delarbre et al. 2002) and morphological evidence that they are paraphyletic (Janvier 2007). The distinct differences between lamprey and hagfish bile salts are consistent with proposals that these two agnathans are either paraphyletic and not very closely related to one another, or have evolved independently for a long time. Estimates of diversification in cyclostome evolution based on nucleotide and amino acid sequences suggested that the split between hagfish and lampreys took place 470–390 Mya and that the two hagfish subfamilies Myxininae and Eptatretinae took place some 90–60 Mya in the Cretaceous-Tertiary periods (Kuraku and Kuratani 2006). We can then conclude that the first mentioned time-frame was enough for the evolution of multiple differences in bile salts, whereas the latter was not.
The unusual C24 bile alcohols found in the sea lamprey present an evolutionary challenge. The synthesis of C24 bile alcohols requires cleavage of the side-chain of cholesterol (reducing the side-chain from 8 to 5 carbon atoms and the total number of carbon atoms from 27 to 24), a process that in humans and rodents is accomplished by multiple peroxisomal enzymes that also catalyze β-oxidation of long-chain fatty acids (Norlin and Wikvall 2007; Russell 2003). All other non-bony fish species analyzed by us or others (Haslewood 1967; Haslewood 1966; Haslewood and Tökés 1969; Une and Hoshita 1994), including hagfish and many species within Chondrichthyes, only have C27 bile alcohols (Table S1). These data could be explained by independent evolution of the ability to cleave the cholesterol side-chain by lampreys or, less parsimoniously, by side-chain cleavage of bile salts being an ancestral phenotype that was lost during evolution of the ancestors of hagfish and Chondrichthyes. The sea lamprey is also unusual in having approximately 50% of their C27 bile alcohols with 3-oxo groups. In mammals, conversion of a 3-oxo group (as an intermediate step) to a 3α-hydroxy group is mediated by AKR1C4 (Norlin and Wikvall 2007; Russell 2003). It is possible that the lamprey has incomplete conversion to 3α-hydroxy by its AKR1C4 ortholog (if present) or, alternatively, uses a different enzymatic pathway.
The bile salt variation patterns in fish imply that 12α-hydroxylation and sulfation of bile salts evolved early in vertebrate evolution or even prior to the appearance of vertebrates. With the exception of hagfish bile salts, every fish in our dataset had bile salts with 12α-hydroxylation, either the common C24 bile acid CA or other bile salts (C27 bile alcohols, C27 bile acids, sea lamprey C24 bile alcohols) that have 12α-hydroxylation. The enzyme that carries out 12α-hydroxylation of bile acids in humans, CYP8B1, is unusual in being the only cytochrome P450 enzyme in mammals to lack introns (Gafvels et al. 1999). We also found that the putative orthologs of the CYP8B1 gene in Japanese medaka, green-spotted pufferfish, and zebrafish lack introns. As genome sequences of cartilaginous and jawless fish become available, CYP8B1 may be an interesting enzyme to compare across species. All bile alcohols analyzed in our study were esterified (conjugated) with sulfate. Although the enzymes catalyzing sulfation of bile alcohols have not been studied, mammalian sulfotransferases of the SULT2 family can sulfate bile acids (Alnouti 2009). Sulfation of steroidal compounds is common in invertebrates (Ivanchina et al. 2003; Kornprobst et al. 1998), including a bile salt-like compound that functions as a sperm chemoattractant in Ciona intestinalis (sea squirt) (Yoshida et al. 2002), suggesting that the sulfotransferases that esterify bile alcohols or similar compounds with sulfate may have an evolutionary history that predates the evolution of vertebrates.
In analyzing the bile salt variation patterns across fish, there appears to be at least two major pathways in the evolutionary transition from C27 bile alcohols to C24 bile acids: (a) a ‘direct’ pathway and (b) an ‘indirect’ pathway that uses C27 bile acids as an ‘intermediate’ step. Evidence to support pathway (a) comes from Perciformes and some other fish families where either type III (C27 bile alcohols and C24 bile acids) or type VI (C24 bile acids only) bile salt profiles are found, but not fish with appreciable amount of C27 bile acids. In our data set, seven fish orders had species with type III bile salt profiles; of these seven orders, four also had other species with type VI profiles but not any species with C27 bile acids. This suggests that the ancestors of these fish followed an evolutionary transition from C27 bile alcohols directly to C24 bile acids (see discussion of possible mechanism below). This differs from amphibians and reptiles, which use pathway (b), where C27 bile acids are common (Moschetta et al. 2005; Une and Hoshita 1994).
In contrast, C27 bile acids were common in the amphibians we analyzed (Table S2) (Moschetta et al. 2005; Une and Hoshita 1994). In fact, as mentioned above, there are examples of frogs with significant (>10%) amounts of C27 bile alcohols, C27 bile acids, and C24 bile acids all in the same bile specimen (Table S2). C27 bile acids are also common in reptiles other than snakes, with characteristic C27 bile acids found in each of the four extant reptilian orders (Crocodylia, Sphenodontia, Squamata, and Testudines), as well as ratite birds (Moschetta et al. 2005).
This raises the question of how the bile salt synthetic pathway can ‘stop’ at C27 bile alcohols, and not form C27 bile acids. If non-mammalian bile salt synthetic pathways are homologous to those in humans and rodents (an assumption currently lacking experimental verification), the multi-functional capabilities of CYP27A1 provide a possible explanation for the evolutionary transition from one bile salt class to another. Animals synthesizing only C27 bile alcohols (e.g., Cypriniformes such as zebrafish) would have CYP27A1 enzymes that can 27-hydroxylate but not oxidize the side-chain of synthetic intermediates. No high-resolution crystal structure of CYP27A1 is currently available, but a homology model of human CYP27A1 has been developed and used for docking studies of multiple substrates, including the bile salt synthetic intermediate 3α,7α,12α-trihydroxy-5β-cholestane (Prosser et al. 2006). Zebrafish CYP27A1 has substantial sequence divergence (more than other model teleost fish) from human CYP27A1, including multiple amino acid residues in or near the proposed substrate binding cavity (Prosser et al. 2006), possibly related to zebrafish synthesizing 5α and not 5β bile salts. It would be of interest to test the substrate specificities of zebrafish CYP27A1. Once CYP27A1 was capable of converting the C-27 methyl group to a carboxyl group, the intermediate could undergo adenylation and coenzyme A ester formation that in turn would permit N-acyl amidation with taurine (to form a taurine-conjugated C27 bile acid) or β-oxidation to form a C24 bile acid.
An alternative hypothesis would be that separate enzymes (e.g., alcohol or aldehyde dehydrogenases) introduce the hydroxyl group at C-27 and then oxidize this group to an aldehyde and then to a carboxylic acid. In mammalian cell culture systems, recombinant CYP27A1 can carry out both reactions, although the oxidation step is slow (Andersson et al. 1989; Dahlback and Holmberg 1990; Pikuleva et al. 1998); however, this has not been studied in any non-mammalian species. Further clarification will require study of bile salt synthetic enzymes in fish.
Genomic comparisons also provided some potential insight into the synthesis of 5α bile salts. Of all the model teleost fish whose genomes are being sequenced, zebrafish is the only species synthesizing mainly 5α bile salts and whose genome apparently lacks an ortholog to the gene for AKR1D1, the enzyme that in humans and rodents catalyzes the stereospecific 5β-reduction of the A-ring of the bile salt synthetic intermediate 4-cholesten-7α,12α-diol-3-one, thereby generating 5β-cholestan-7α,12α-diol-3-one. 5α-Bile acids have been detected in humans and other mammals as a minor component of circulating bile, but the origin of 5α-bile acids is not necessarily from biosynthesis, as 5α-bile acids can also be formed by the microbial flora (Kallner 1967). The hepatic enzyme(s) that perform 5α reduction of bile salt intermediates like 4-cholesten-7α,12α-diol-3-one in mammals are as yet uncharacterized (Russell 2003). There are ongoing sequencing projects for three animals other than zebrafish that produce 5α-bile salts: sea lamprey, coelacanth (Latimeria calumnae), and the green anole lizard (Anolis caroliensis). The genomes of these animals should provide insight into how they achieve a bile salt pool consisting of mainly 5α-bile salts.
The bile salt biosynthetic pathway has apparently evolved in vertebrates in a considerably different fashion from another set of cholesterol derivatives, namely the adrenocortical and sex steroid hormones. Estrogens are the terminal products of this pathway whereas other steroids such as glucocorticoids or mineralocorticoids are intermediates. The actions of the steroid hormones are mediated by nuclear hormone receptors. Thornton and colleagues have published multiple studies suggesting that estrogen receptors have a longer evolutionary history than receptors for intermediate products in the steroid hormone pathways (e.g., glucocorticoids or mineralocorticoides) (Bridgham et al. 2006; Thornton 2001; Thornton et al. 2003) (although their ancestral reconstruction in invertebrates have been challenged by other research (Paris et al. 2008)). In the ‘molecular exploitation’ model of a biochemical pathway proposed by Thornton, the terminal products of a synthetic pathway mediate the more ancestral activities. During evolution, functions (and receptors) develop for the intermediate products of the pathway, and structural diversity involving intermediates in the pathway can increase over time.
A closer parallel to the bile salt synthetic pathway may be found in the enzymatic pathways involved in the formation of triterpenoids, naturally occurring 30-carbon molecules derived from isoprene that serve as a building block for a diverse array of compounds. Studies of bacteria, animals, and plants have revealed two independent pathways for the formation of isoprene units, each involving multiple organelles (Rohmer 2008). Similar to bile salts, the synthetic intermediates in the triterpinoid pathways are low in diversity but the end-products have spectacular structural diversity with numerous physiological functions. Bile acid synthesis is now known to be catalyzed in humans and rodents by at least two independent pathways (acidic and neutral), with regulation of the pathways keeping intermediate products at very low concentrations in the hepatocytes and biliary bile (Russell 2003). Like triterpinoids, the end-products of bile salt synthesis have substantial structural diversity by variations in hydroxylation patterns, unsaturation, and side-chain length. The bile salt synthetic pathway has evolved from a relatively simple pathway that produces C27 bile alcohols (by saturation and polyhydroxylation of cholesterol) to more complex pathways that utilize multiple organelles (cytoplasm, mitochondria, and peroxisomes) to produce C24 bile acids. In the process, many ray-finned fish (and indeed most land animals and birds) have bile containing almost exclusively C24 bile acids and very low amounts of the synthetic precursors including C27 bile alcohols, a process achieved by the evolution of regulatory control and enzyme modifications.
There is still little understanding of what may have driven variation in bile salt chemical diversity. As our analysis of fish shows, there is essentially no apparent link between diet and bile salt structure. Typical 5β-C24 bile acids are found in fish with a wide variety of diets. It is unclear what benefit may be conferred to those fish and amphibians that use 5α bile salts. Studies in the rabbit have shown that 5α bile acids can be toxic (Hofmann et al. 1968). Our study also does not shed light on how animals regulate their bile salt profiles. It is logical to assume to that both bile acid biosynthesis and intestinal conservation are controlled by transcriptional regulators, although this is not well-understood even in mammals.
Although there is extensive data on the physicochemical and physiological properties of common bile acids such as CA and CDCA (and their conjugated derivatives), there has been much less study of other classes of bile salts. In part, this relates to the difficulty of synthesizing or isolating sufficient material to perform detailed studies. In cold marine environments, there would likely be selective pressure against bile salts that cannot form micelles to solubilize dietary lipid at cold temperatures. Along these lines, 5α-bile acids were not common in the fish we analyzed. Standard C24 5β-bile acids have critical micellar temperatures (also called Krafft points) well below the freezing point, meaning that these bile acids can still form micelles even in fish in extremely cold waters. In contrast, C24 5α-bile acids have Krafft points greater than 10°C and would thus be effective in warm-blooded animals but not animals with core body temperatures than can drop below 10°C (Hofmann and Mekhjian 1973). The only C27 bile alcohol to be analyzed in any detail for Krafft point was 5α-cyprinol 27-sulfate, the major bile salt of cypriniform fish (Goto et al. 2003). Its Krafft point was below 4°C and thus would be predicted to function well in lipid solubilization in fish living in cold waters. Based on the limited data available for 5α-cyprinol sulfate, one may hypothesize that the 5α-bile alcohols of hagfish, sea lamprey, and lobe-finned fish similarly can form micelles at cold temperatures, although this awaits experimental verification. The complicated variation of bile salts may also relate to other functions of bile salts that are not well understood, including communication, influencing of the microbial environment in the gut, and regulation of hepatobiliary development and regeneration. The interactions between bile salt profile and gut microbial colonization have been only studied in limited detail in mammals (Hofmann and Eckmann 2006; Hofmann and Hagey 2008), and not at all in non-mammalian species.
Lastly, the extensive variation of bile salt structures across vertebrate species predicts that protein receptors that bind bile salts will also show variation in structure and ligand-binding specificity. Indeed, preliminary research has already shown distinct patterns of ligand-receptor co-evolution for three nuclear hormone receptors (pregnane X receptor, PXR; farnesoid X receptor, FXR; and vitamin D receptor, VDR) that regulate various aspects of bile salt synthesis, transport, or metabolism (Ekins et al. 2008; Krasowski et al. 2005a, b; Reschly et al. 2008a). Putative orthologs to PXR, FXR, and VDR are found in invertebrates such as Ciona intestinalis, raising the evolutionary question of how these receptors shifted selectivity from as yet unknown ligand(s) in invertebrates to bile salts, a vertebrate-specific class of cholesterol end-metabolites. Ultimately, we aim to build up a complete multi-dimensional picture of ligand, protein, species, and commensal microbial evolution involving bile salts.
Supplementary Material
Figure S1. Bile salt profiles of fish and amphibians, with annotated ESI/MS/MS and HPLC traces.
Figure S2. Illustration of 5α bile salt pathways.
Figure S3. Illustration of 5β bile salt pathways.
Bile salts of fish, with bile salt profiles color-coded by bile salt class. The systematic organization of the fishes follows Nelson (Nelson 2006), except for the order status of Alepocephaliformes.
Bile salts of amphibians, with bile salt profiles color-coded by bile salt class.
Comparative genomics of enzymes involved in bile salt synthesis.
Acknowledgments
MDK is supported by K08-GM074238 from the National Institutes of Health. AFH and LRH are supported by NIH grant DDK 64891 (to AFH). The authors thank the Pathology Laboratory of the San Diego Zoo; Sea World in San Diego, Texas, and Coral Gables, FL. Samples were also provided by Dr. G.A.D. Haslewood (now deceased), Professor of Biochemistry at Guy’s Hospital (London, UK); Dr. Valentine Lance (Department of Biology, San Diego State University); Dr. Jan Raines, DVM (formerly of Dallas World Aquarium); Dr. Joseph Barrett and Michelle Becker (University of Memphis); Dr. Hubert. Johnston (former Veterinarian of the County of San Diego); Dr. Richard Norman (University of Melbourne); Dr. Bob Vrijenhoek (Monterey Bay Aquarium Research Institute), Dr. Cindy Van Dover (College of William and Mary) (funded by NSF grants OCE 0241613 and OCE 0350554), W. Joe Jones (University of South Carolina), and Steen W. Knudsen (Zoological Museum, University of Copenhagen, Denmark).
Literature cited
- Alnouti Y. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol Sci. 2009;108:225–246. doi: 10.1093/toxsci/kfn268. [DOI] [PubMed] [Google Scholar]
- Amos B, I, Anderson G, Haslewood GAD, Tökés L. Bile salts of the lungfishes Lepidosiren, Neoceratodus and Protopterus and those of the coelacanth Latimeria chalumnae Smith. Biochem J. 1977;161:201–204. doi: 10.1042/bj1610201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson IG, Haslewood GAD. Comparative studies of ‘bile salts’. 20. Bile salts of the coelacanth, Latimeria chalumnae Smith. Biochem J. 1964;93:34–39. doi: 10.1042/bj0930034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S, Davis DL, Dahlback H, Jornvall H, Russell DW. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- Bardack D. Relationships of living and fossil hagfishes. In: Jorgensen JM, Lomholt JP, Weber RE, Malte H, editors. The biology of hagfishes. Chapman & Hall; London: 1998. pp. 3–14. [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1681/01.asn.0000926836.46869.e5. [DOI] [PubMed] [Google Scholar]
- Bridgwater RJ, Briggs T, Haslewood GAD. Comparative studies of ‘bile salts’. 14. Isolation from shark bile and partial synthesis of scymnol. Biochem J. 1962;82:285–290. doi: 10.1042/bj0820285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlback H, Holmberg I. Oxidation of 5 beta-cholestane-3 alpha,7 alpha, 12 alpha-triol into 3 alpha,7 alpha,12 alpha-trihydroxy-5 beta-cholestanoic acid by cytochrome P-450(26) from rabbit liver mitochondria. Biochem Biophys Res Commun. 1990;167:391–395. doi: 10.1016/0006-291x(90)92034-w. [DOI] [PubMed] [Google Scholar]
- Delarbre C, Gallut C, Barriel V, Janvier P, Gachelin G. Complete mitochondrial DNA of the hagfish, Eptatretus burgeri: the comparative analysis of mitochondrial DNA sequences strongly supports the cyclostome monophyly. Mol Phylogenet Evol. 2002;22:184–192. doi: 10.1006/mpev.2001.1045. [DOI] [PubMed] [Google Scholar]
- Ekins S, Reschly EJ, Hagey LR, Krasowski MD. Evolution of pharmacologic specificity in the pregnane X receptor. BMC Evol Biol. 2008;8:103. doi: 10.1186/1471-2148-8-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink RO, Haan J, Lambert KJ, Hagey LR, Hofmann AF, Jansen PLM. Selective hepatobiliary transport of nordeoxycholate side chain conjugates in mutant rats with a canalicular transport defect. Hepatology. 1989;9:861–865. doi: 10.1002/hep.1840090612. [DOI] [PubMed] [Google Scholar]
- Forey P, Janvier P. Agnathans and the origin of jawed vertebrates. Nature. 1993;361:129–134. [Google Scholar]
- Gafvels M, Olin M, Chowdhary BP, Raudsepp T, Andersson U, Persson B, Jansson M, Bjorkhem I, Eggertsen G. Structure and chromosomal assignment of the sterol 12alpha-hydroxylase gene (CYP8B1) in human and mouse: eukaryotic cytochrome P-450 gene devoid of introns. Genomics. 1999;56:184–196. doi: 10.1006/geno.1998.5606. [DOI] [PubMed] [Google Scholar]
- Goto T, Holzinger F, Hagey LR, Cerre C, Ton-Nu HT, Schteingart CD, Steinbach JH, Shneider BL, Hofmann AF. Physicochemical and physiological properties of 5a-cyprinol sulfate, the toxic bile salt of cyprinid fish. J Lipid Res. 2003;44:1643–1651. doi: 10.1194/jlr.M300155-JLR200. [DOI] [PubMed] [Google Scholar]
- Gray GM, Yardley HJ. Lipid compositions of cells isolated from pig, human, and rat epidermis. J Lipid Res. 1975;16:434–440. [PubMed] [Google Scholar]
- Hara TJ. Olfaction and gustation in fish: an overview. Acta Physiol Scand. 1994;152:207–217. doi: 10.1111/j.1748-1716.1994.tb09800.x. [DOI] [PubMed] [Google Scholar]
- Haslewood GA. A bile alcohol from Cyprinidae. Biochem J. 1954;59:xi. [PubMed] [Google Scholar]
- Haslewood GA. Bile salt evolution. J Lipid Res. 1967;8:535–550. [PubMed] [Google Scholar]
- Haslewood GAD. Comparative studies of bile salts. Myxinol disulphate, the principal bile salt of hagfish (Myxinidae) Biochem J. 1966;100:233–237. doi: 10.1042/bj1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslewood GAD, Tökés L. Comparative studies of bile salts: bile salts of the lamprey Petromyzon marinus L. Biochem J. 1969;114:179–184. doi: 10.1042/bj1140179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslewood GAD, Tökés L. Comparative studies of bile salts. A new type of bile salt from Arapaima gigas (Cuvier) (family Osteoglossidae) Biochem J. 1972;126:1161–1170. doi: 10.1042/bj1261161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF. Thin layer absorption chromatography of free and conjugated bile acids on silicic acid. J Lipid Res. 1962;3:127–128. [Google Scholar]
- Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab Rev. 2004;36:703–722. doi: 10.1081/dmr-200033475. [DOI] [PubMed] [Google Scholar]
- Hofmann AF, Bokkenheuser V, Hirsch RL, Mosbach EH. Experimental cholelithiasis in the rabbit induced by cholestanol feeding: effect of neomycin treatment on bile composition and gallstone formation. J Lipid Res. 1968;9:244–253. [PubMed] [Google Scholar]
- Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci U S A. 2006;103:4333–4334. doi: 10.1073/pnas.0600780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. 2008;65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 2009 doi: 10.1194/jlr.R000042. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann AF, Mekhjian HS. Bile acids and the intestinal absorption of fat and electrolytes in health and disease. In: Nair PP, Kritchevsky D, editors. The bile acids: chemistry, physiology, and metabolism. Plenum Press; New York: 1973. pp. 103–152. [Google Scholar]
- Hofmann AF, Small DM. Detergent properties of bile salts: correlation with physiological function. Annu Rev Med. 1967;18:333–376. doi: 10.1146/annurev.me.18.020167.002001. [DOI] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Tsukamoto K, Nishida M. Basal actinopterygian relationships: a mitogenomic perspective on the phylogeny of the “ancient fish”. Mol Phylogenet Evol. 2003;26:110–120. doi: 10.1016/s1055-7903(02)00331-7. [DOI] [PubMed] [Google Scholar]
- Ivanchina NV, Kicha AA, Kalinovsky AI, Dmitrenok PS, Stonik VA. Hemolytic steroid disulfates from the Far Eastern starfish Pteraster pulvillus. J Nat Prod. 2003;66:298–301. doi: 10.1021/np010650w. [DOI] [PubMed] [Google Scholar]
- Janvier P. Evolutionary biology: born-again hagfishes. Nature. 2007;446:622–623. doi: 10.1038/nature05712. [DOI] [PubMed] [Google Scholar]
- Jez JM, Penning TM. The aldo-keto reductase (AKR) superfamily: an update. Chem Biol Interact. 2001;130–132:499–525. doi: 10.1016/s0009-2797(00)00295-7. [DOI] [PubMed] [Google Scholar]
- Johnson GD, Paxton JR, Sutton TT, Satoh TP, Sado T, Nishida M, Miya M. Deep-sea mystery solved: astonishing larval transformations and extreme sexual dimorphism unite three fish famlies. Biol Lett. 2009;5:235–239. doi: 10.1098/rsbl.2008.0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallner A. The transformation of deoxycholic acid into allodeoxycholic acid in the rat. Bile acids and steroids. 174. Acta Chem Scand. 1967;21:87–92. doi: 10.3891/acta.chem.scand.21-0087. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Lavoué S, Inoue JG, Satoh TP, Kawaguchi A, Nishida M. Interrelationships of the 11 gasterosteiform families (sticklebacks, pipefishes, and their relatives): a new perspective based on whole mitogenome sequences from 75 higher teleosts. Mol Phylogenet Evol. 2008;46:224–236. doi: 10.1016/j.ympev.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Kornprobst JM, Sallenave C, Barnathan G. Sulfated compounds from marine organisms. Comp Biochem Physiol B. 1998;119:1–51. doi: 10.1016/s0305-0491(97)00168-5. [DOI] [PubMed] [Google Scholar]
- Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol Endocrinol. 2005a;19:1720–1739. doi: 10.1210/me.2004-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Yasuda K, Hagey LR, Schuetz EG. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors) Nucl Recept. 2005b;3:2. doi: 10.1186/1478-1336-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Kuratani S. Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zool Sci. 2006;23:1053–1064. doi: 10.2108/zsj.23.1053. [DOI] [PubMed] [Google Scholar]
- Lavoué S, Miya M, Poulsen JY, Møller PR, Nishida M. Monophyly, phylogenetic position and inter-familial relationships of the Alepocephaliformes (Teleostei) based on whole mitogenome sequences. Mol Phylogenet Evol. 2008;47:1111–1121. doi: 10.1016/j.ympev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Li W, Scott AP, Siefkes MJ, Yan H, Liu Q, Yun SS, Gage DA. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 2002;296:138–141. doi: 10.1126/science.1067797. [DOI] [PubMed] [Google Scholar]
- Lin DS, Connor WE, Napton LK, Heizer RF. The steroids of 2000-year-old human coprolites. J Lipid Res. 1978;19:215–221. [PubMed] [Google Scholar]
- Lindsay J, Wang LL, Li Y, Zhou SF. Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab. 2008;9:99–105. doi: 10.2174/138920008783571819. [DOI] [PubMed] [Google Scholar]
- Miya M, Takeshima H, Endo H, Ishigura NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, Shirai SM, Nishida M. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 2003;26:121–138. doi: 10.1016/s1055-7903(02)00332-9. [DOI] [PubMed] [Google Scholar]
- Møller PR, Jones WJ. Eptatretus strickrotti n. sp. (Myxinidae): first hagfish captured from a hydrothermal vent. Biol Bull. 2007;212:55–66. doi: 10.2307/25066580. [DOI] [PubMed] [Google Scholar]
- Moschetta A, Xu F, Hagey LR, van Berge Henegouwen GP, van Erpecum KJ, Brouwers JF, Cohen JC, Bierman M, Hobbs HH, Steinbach JH, Hofmann AF. A phylogenetic survey of biliary lipids in vertebrates. J Lipid Res. 2005;46:2221–2232. doi: 10.1194/jlr.M500178-JLR200. [DOI] [PubMed] [Google Scholar]
- Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferase. Arch Biochem Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the World. John Wiley and Sons, Inc; Hoboken, NJ: 2006. [Google Scholar]
- Nes WR, Nes WD. Lipids in evolution. Plenum Press; New York: 1980. [Google Scholar]
- Norlin M, Wikvall K. Enzymes in the conversion of cholesterol into bile acids. Curr Mol Med. 2007;7:199–218. doi: 10.2174/156652407780059168. [DOI] [PubMed] [Google Scholar]
- Paris M, Pettersson K, Schubert M, Bertrand S, Pongratz I, Escriva H, Laudet V. An amphioxus orthologue of the estrogen receptor that does not bind estradiol: insights into estrogen receptor evolution. BMC Evol Biol. 2008;8:219. doi: 10.1186/1471-2148-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikuleva IA, Babiker A, Waterman MR, Bjorkhem I. Activities of recombinant human cytochrome P450c27 (CYP27) which produce intermediates of alternative bile acid biosynthetic pathways. J Biol Chem. 1998;273:18153–18160. doi: 10.1074/jbc.273.29.18153. [DOI] [PubMed] [Google Scholar]
- Prosser DE, Guo Y, Jia Z, Jones G. Structural motif-based homology modeling of CYP27A1 and site-directed mutational analysis affecting vitamin D hydroxylation. Biophys J. 2006;90:3389–3409. doi: 10.1529/biophysj.105.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res. 2008a;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Welsh WJ, Ekins S, Hagey LR, Krasowski MD. Ligand specificity and evolution of liver X receptors. J Steroid Biochem Mol Biol. 2008b;110:83–94. doi: 10.1016/j.jsbmb.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M. From molecular fossils of bacterial hopanoids to the formation of isoprene units: discovery and elucidation of the methylerythritol phosphate pathway. Lipids. 2008;43:1095–1107. doi: 10.1007/s11745-008-3261-7. [DOI] [PubMed] [Google Scholar]
- Rossi SS, Converse JL, Hofmann AF. High pressure liquid chromatography analysis of conjugated bile acids in human bile: simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugated bile acids. J Lipid Res. 1987;28:589–595. [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- Une M, Hoshita T. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima J Med Sci. 1994;43:37–67. [PubMed] [Google Scholar]
- Yoshida M, Murata M, Inaba K, Morisawa M. A chemoattractant for ascidian spermatozoa is a sulfated steroid. Proc Natl Acad Sci U S A. 2002;99:14831–14836. doi: 10.1073/pnas.242470599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Bile salt profiles of fish and amphibians, with annotated ESI/MS/MS and HPLC traces.
Figure S2. Illustration of 5α bile salt pathways.
Figure S3. Illustration of 5β bile salt pathways.
Bile salts of fish, with bile salt profiles color-coded by bile salt class. The systematic organization of the fishes follows Nelson (Nelson 2006), except for the order status of Alepocephaliformes.
Bile salts of amphibians, with bile salt profiles color-coded by bile salt class.
Comparative genomics of enzymes involved in bile salt synthesis.